Abstract
Scant attention has been paid to Lyme disease, Borrelia burgdorferi, Ixodes scapularis, or reservoirs in eastern North Dakota despite the fact that it borders high-risk counties in Minnesota. Recent reports of B. burgdorferi and I. scapularis in North Dakota, however, prompted a more detailed examination. Spirochetes cultured from the hearts of five rodents trapped in Grand Forks County, ND, were identified as B. burgdorferi sensu lato through sequence analyses of the 16S rRNA gene, the 16S rRNA gene-ileT intergenic spacer region, flaB, ospA, ospC, and p66. OspC typing revealed the presence of groups A, B, E, F, L, and I. Two rodents were concurrently carrying multiple OspC types. Multilocus sequence typing suggested the eastern North Dakota strains are most closely related to those found in neighboring regions of the upper Midwest and Canada. BALB/c mice were infected with B. burgdorferi isolate M3 (OspC group B) by needle inoculation or tick bite. Tibiotarsal joints and ear pinnae were culture positive, and B. burgdorferi M3 was detected by quantitative PCR (qPCR) in the tibiotarsal joints, hearts, and ear pinnae of infected mice. Uninfected larval I. scapularis ticks were able to acquire B. burgdorferi M3 from infected mice; M3 was maintained in I. scapularis during the molt from larva to nymph; and further, M3 was transmitted from infected I. scapularis nymphs to naive mice, as evidenced by cultures and qPCR analyses. These results demonstrate that isolate M3 is capable of disseminated infection by both artificial and natural routes of infection. This study confirms the presence of unique (nonclonal) and infectious B. burgdorferi populations in eastern North Dakota.
INTRODUCTION
Eastern North Dakota borders some Minnesota counties (i.e., Kittson, Marshall, Polk, and Norman) where the risk of contracting the tick-borne diseases Lyme disease and human granulocytic anaplasmosis is moderate to high based on confirmed human cases (1, 2), the abundance of Borrelia burgdorferi-positive Ixodes scapularis ticks (3), and the density of nymphal I. scapularis (1). Despite this close geographical proximity, there has been a paucity of studies on the migration of I. scapularis or B. burgdorferi into North Dakota (4). Eastern North Dakota is classified as a transition zone for Lyme disease based on studies investigating the expansion of I. scapularis and B. burgdorferi in the Midwest (3). However, in 2011, the North Dakota Department of Health reported the results of a 2010 survey showing established I. scapularis populations in six eastern North Dakota counties, including Grand Forks County (5). In addition, the pathogens Anaplasma phagocytophilum, Babesia sp., and B. burgdorferi were detected in I. scapularis via PCR (5).
Surveillance of I. scapularis has shown an increase in its geographic distribution (6–8), which has been accompanied by a concomitant increase in the distribution of confirmed Lyme disease cases (9). The primary causative agents of Lyme disease differ between the United States and Europe, with B. burgdorferi sensu stricto being the primary agent in the United States and Borrelia afzelii and Borrelia garinii being the primary agents in Europe (10, 11). While it is generally accepted that B. burgdorferi sensu stricto is the sole cause of Lyme disease in the United States, there is increasing evidence that other members of the B. burgdorferi sensu lato complex, a group consisting of approximately 18 closely related species, could also cause Lyme disease in the United States (12). For example, Borrelia bissettii has been identified in DNA isolated from human serum samples in California residents by sequence analysis of p66; however, it has not been clearly associated with Lyme disease in the United States because many of the samples that were positive for B. bissettii were also positive for B. burgdorferi (13). It has been demonstrated, however, to be associated with Lyme disease in Europe (10, 14) and has been shown to be infectious and pathogenic in a mouse model (15). Borrelia americana, found in Ixodes pacificus and Ixodes minor, is predominantly found in California and South Carolina and has not yet been associated with Lyme disease in humans (10), but antibodies to B. americana have reportedly been detected in blood from Lyme disease patients (16). These data underscore the need to correctly identify newly isolated Borrelia species in order to assess their potential contributions to human disease.
Several schemes have emerged to classify presumptive Lyme disease Borrelia. One is based on outer surface protein C (OspC), a protein expressed only during transmission from vector to host (17, 18) that is required for B. burgdorferi to infect mammals (19, 20). While ospC is highly polymorphic, many groups or types have been described (21, 22). OspC types are commonly used to determine a strain's ability to cause disseminated infections in humans (21, 23, 24). OspC groups A to U have been identified (21, 22), with A to O, T, and U found in North America (21, 23, 25). Groups A, B, I, K, and N have been found most commonly associated with disseminated infections in humans (21, 26, 27). Groups C, D, E, F, G, H, and M have also been found to be capable of causing disseminated infection in humans, although the occurrence of infection with these types is much lower (27, 28). B. burgdorferi small-mammal reservoirs have been found to carry specific OspC types (29). For example, it has previously been shown that Peromyscus leucopus (the white-footed mouse) tested positive for groups A, B, D, F, G, I, and K, while Tamias striatus (the eastern chipmunk) tested positive for groups A, D, F, G, I, K, T, and U (29).
Antigenic outer surface proteins (e.g., OspC) are highly variable, and there are indications they are subject to horizontal gene transfer (30–32), which makes them less than ideal candidates for evaluating the evolutionary history and geographical relationships of B. burgdorferi strains. Multilocus sequence typing (MLST) schemes have been developed that have proven to be the most reliable method for determining the histories of and relationships within many bacterial genera and species, including B. burgdorferi sensu lato strains (33, 34). MLST is based on the analysis of housekeeping genes, which are under strong pressure to minimize large-scale mutation events, such as the events seen in outer surface proteins. The B. burgdorferi MLST scheme utilizes eight housekeeping genes: clpA, clpX, nifS, pepX, pyrG, rplB, recG, and uvrA (34). Each gene is assigned an allele number based on the sequence identity to previously submitted alleles. The profiles produced from all eight loci correspond to a sequence type (ST), which can be used to compare strains to determine the evolutionary history and relationships.
With the discovery of I. scapularis and B. burgdorferi in eastern North Dakota, we sought to characterize previously unidentified spirochete cultures obtained from five hearts of Peromyscus spp. (deer mice) and Myodes gapperi (Southern red-backed vole) trapped in the Turtle River and Forest River areas of eastern North Dakota. All five small mammals harbored unique or nonclonal populations of spirochetes determined to be B. burgdorferi sensu lato. The presence of nonclonal populations is significant, as it indicates the B. burgdorferi populations present in eastern North Dakota are not the result of a recent or single migration event. Two of the five small mammals were carrying at least two different OspC types. OspC typing showed that the infectious B. burgdorferi types A, B, E, F, and I are disproportionately represented in our samples. However, one isolate typed to the noninfectious group L and one typed to none of the previously described groups. B. burgdorferi M3 (ospC group B) was obtained from M. gapperi at Forest River and was predicted to be a highly infectious isolate. Using this isolate, we determined that B. burgdorferi M3 is infectious in laboratory mice via both artificial and natural routes of exposure, is culturable from mouse tissues, and survives I. scapularis molting. These data confirm B. burgdorferi is present in eastern North Dakota and is infectious and transmissible in a laboratory model.
MATERIALS AND METHODS
Animal care and use.
Infection experiments were performed according to a University of North Dakota Institutional Animal Care and Use Committee (UND IACUC)-approved protocol (1101-2) at the University of North Dakota Center for Biomedical Research. Four- to 6-week-old BALB/c mice (Harlan, Madison, WI) were cared for in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care guidelines (Animal Welfare Assurance A3917-01) and the National Research Council of the National Academies Guide for the Care and Use of Laboratory Animals, 8th ed. Wild rodents were collected, euthanized, and necropsied in the field as described in the UND IACUC-approved protocol 1304-3. All efforts were made to minimize animal suffering.
Sample collection and culturing of spirochetes.
Live trapping of rodents was conducted during June and July 2012 in the two largest forested areas within an otherwise agricultural landscape in Grand Forks County, i.e., Turtle River State Park (lat 47.94, long −97.50; ca. 254 ha) and Forest River Biological Station and Wildlife Management Area (lat 48.17, long −97.66; ca 349 ha). Sherman live traps (H. B. Sherman Traps, Tallahassee, FL) were baited with peanut butter and oatmeal, supplied with cotton bedding, set in the evening, and recovered in the morning. Captured mammals were identified as M. gapperi and Peromyscus spp. based on morphological characteristics. This method makes it difficult to identify Peromyscus spp. in the field; thus, Peromyscus mice were identified only to the genus level. However, all of the Peromyscus mice in this study were trapped deep in deciduous forests, a preferred habitat for Peromyscus leucopus, while Peromyscus maniculatus prefers more open terrain and/or coniferous forest, strongly suggesting the captured Peromyscus mice were P. leucopus (35; Robert Seabloom, personal communication). Peromyscus spp. and M. gapperi were euthanized with isoflurane and necropsied, and the hearts were immediately inoculated into modified Barbour-Stoenner-Kelly II (BSK-II) medium containing 6% rabbit serum and 50 μg/ml rifampin (36). Surgical tools were sterilized with 95 to 99% ethanol prior to each necropsy to prevent possible cross-contamination between animals. Three days later, uncontaminated cultures were blind passed into modified BSK-II with 6% rabbit serum without rifampin, incubated for three additional days, and then examined for spirochetes via dark-field microscopy.
Amplification and sequencing of ospA, ospC, flaB, the 16S rRNA gene, the 16S rRNA gene-ile tRNA IGS, and p66.
ospA, ospC, p66, and the 16S rRNA gene-ile tRNA intergenic spacer region (IGS) were obtained under amplification conditions previously described (37), except that the annealing temperature was adjusted to 48°C (Table 1 lists the primers used). The 16S rRNA gene and flaB were amplified under the following conditions: 1 cycle of 94°C for 5 min; 40 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 1 min; and 1 cycle of 72°C for 5 min. Sequencing was performed at Davis Sequencing (Davis, CA). The chromatograms were visually inspected, and then the forward (coding) and reverse (template) strand sequences were aligned to obtain a double-stranded consensus sequence. 16S rRNA gene sequences were queried using the Ribosomal Database Project's Sequence Match (Seqmatch) program (38) and the Nucleotide Collection (nr/nt) BLAST database (blast.ncbi.nlm.nih.gov) to obtain genus and species identifications. flaB, ospA, ospC, p66, and the IGS nucleotide sequences were queried using BLAST.
TABLE 1.
Primer sequences used in this study and predicted amplicon sizes
| Primer name | Sequence (5′–3′) | Predicted amplicon size (nt)a |
|---|---|---|
| 16S For | GGT CAA GAC TGA CGC TGA GTC A | 136 |
| 16S Rev | GGC GGC ACA CTT AAC ACG TTA G | |
| flaB For | GGG TCT CAA GCG TCT TGG | 139 |
| flaB Rev | GAA CCG GTG CAG CCT GAG | |
| ospA For | ATG AAA AAA TAT TTA TTG GGA ATA GG | 829 |
| ospA Rev | ATT CTC CTT ATT TTA AAG CG | |
| ospC Forb | ATG AAA AAG AAT ACA TTA AGT GC | 638 |
| ospC Rev | CTT AAT TAA GGT TTT TTT GG | |
| p66 For | GAT TTT TCT ATA TTT GGA CAC AT | 755 |
| p66 Rev | TGT AAA TCT TAT TAG TTT TTC AAG | |
| 16S IGS For | AGT GCG GCT GGA TCA CCT CC | 950 |
| ileT IGS Revb | GTC TGA TAA ACC TGA GGT CGG A | |
| nid1 For | CCA GCC ACA GAA TAC CAT CC | 153 |
| nid1 Rev | GGA CAT ACT CTG CTG CCA TC | |
| recA For | GTG GAT CTA TTG TAT TAG ATG AGG CT | 171 |
| recA Rev | GCC AAA GTT CTG CAA CAT TAA CAC CT | |
| I. scap 16S For | CGG TCT GAA CTC AGA TCA AG | 300 |
| I. scap 16S Rev | GGG ACA AGA AGA CCC TAT C | |
| MLST primers for amplification and sequencingc | ||
| clpA For | AAA GAT AGA TTT CTT CCA GAC | 982 |
| clpA Rev | GAA TTT CAT CTA TTA AAA GCT TTC | |
| clpX For | GCT GCA GAG ATG AAT GTG CC | 884 |
| clpX Rev | GAT TGA TTT CAT ATA ACT CTT TTG | |
| nifS For | ATG GAT TTC AAA CAA ATA AAA AG | 1,049 |
| nifS Rev | GAT ATT ATT GAA TTT CTT TTA AG | |
| pepX For | ACA GAG ACT TAA GCT TAG CAG | 811 |
| pepX Rev | GTT CCA ATG TCA ATA GTT TC | |
| pyrG For | GAT TGC AAG TTC TGA GAA TA | 801 |
| pyrG Rec | CAA ACA TTA CGA GCA AAT TC | |
| recG For | CCC TTG TTG CCT TGC TTT C | 805 |
| recG Rev | GAA AGT CCA AAA CGC TCA G | |
| rplB For | TGG GTA TTA AGA CTT ATA AGC | 760 |
| rplB Rev | GCT GTC CCC AAG GAG ACA | |
| uvrA For | GAA ATT TTA AAG GAA ATT AAA AGT AG | 911 |
| uvrA Rev | CAA GGA ACA AAA ACA TCT GG |
The predicted amplicon sizes were determined using the following accession versions: B. burgdorferi B31, AE000783.1 (chromosome), AE000790.2 (lp54), and AE000792.1 (cp26); mouse, NC_000079.6 (nid1). nt, nucleotides.
Primers ospC For and ileT IGS Rev were obtained from reference 36.
NCBI Nuccore and BLAST database searches.
Sequences were obtained by searching the NCBI Nuccore database (http://www.ncbi.nlm.nih.gov/nuccore) using the following terms: Borrelia plus p66, Borrelia plus ospC, Borrelia plus “outer surface protein C,” Borrelia plus ospA, Borrelia plus “outer surface protein A,” Borrelia plus “intergenic spacer region,” Borrelia plus IGS, Borrelia plus 16S plus 23S plus IGS, and Borrelia plus 16S plus 23S plus “intergenic spacer region.” A search was performed in the nonredundant protein sequence (nr) BLAST database using the complete B. burgdorferi B31 protein sequences for p66 (chromosome accession no., NC_001318.1), OspC (cp26 accession no., NC_001903.1), and OspA (lp54 accession no., NC_001857.2).
Alignments and phylogeny.
Sequences for ospA, ospC, p66, and the IGSs from eastern North Dakota isolates were aligned, along with BLAST and NCBI database sequences, in ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The shaded alignment was generated using BoxShade (ExPASy [http://www.ch.embnet.org/software/BOX_form.html]). Duplicate sequences (identified as the same species and found to be 100% identical) were represented in the analyses by a single sequence. Sequences obtained from BLAST were included in analyses if the query coverage was greater than 90%. The obtained sequences were manually trimmed to conserved regions aligning with the shortest sequence obtained for the eastern North Dakota isolates. Trimmed sequences meeting the above criteria were then used in phylogenetic analyses. For OspA and OspC, a subset of sequences from each clade was chosen to create representative phylogenetic trees. Final phylogenetic analyses for OspA, OspC, and p66 were performed using the PHYLIP programs SeqBoot, Proml, and Consense (version 3.695 [http://evolution.genetics.washington.edu/phylip/]). Briefly, sequence files were put into SeqBoot and analyzed with 1,000 bootstrap replicates. The SeqBoot output file was analyzed in Proml using the Jones-Taylor-Thornton model (48) with multiple data sets, slower analysis, a random number seed of 9, data sets jumbled 5 times, and an outgroup root when appropriate. The resulting file was input into Consense to obtain a single consensus tree using the majority rule (extended) consensus type, and the tree was treated as rooted when appropriate. DNA trees were created using Dnaml in PHYLIP. The trees were visualized using FigTree (version 1.4 [http://tree.bio.ed.ac.uk/software/figtree]) and labeled using Adobe (San Jose, CA) Illustrator CS3.
OspC typing.
To sequence ospC from mixed populations, PCR products for ospC from samples M6, M7, and M9 were gel purified and cloned into Escherichia coli using the pCR2.1 TOPO TA cloning kit according to the manufacturer's instructions (Life Technologies, Carlsbad, CA). Plasmids were purified using the Qiagen (Valencia, CA) Plasmid Mini Prep kit and sequenced at Davis Sequencing. OspC amino acid sequences for previously typed isolates (21, 25) were obtained from NCBI. Nucleotide sequences from the eastern North Dakota isolates were translated (ExPASy [http://web.expasy.org/translate/]) and aligned with the previously typed isolates in ClustalW2. A percent identity matrix (PIM) was obtained, and an OspC type group was assigned to each eastern North Dakota isolate if the sequence did not diverge more than 2% from a particular group (25).
MLST.
MLST was performed for each eastern North Dakota sample (34). Primer sequences for the eight housekeeping genes used (clpA, clpX, nifS, pepX, pyrG, recG, rplB, and uvrA) were obtained from the Imperial College London's B. burgdorferi MLST website (http://borrelia.mlst.net). For amplification and sequencing, the outer forward and outer reverse primers for each gene were used. For each gene, a 50-μl reaction was set up using the HotStarTaq Plus master mix (Qiagen) according to the manufacturer's instructions. Primers were added to a final concentration of 1 μM, and 1 μl of purified genomic DNA was added. Previously described amplification conditions were used (34) with the following modifications: (i) the initial denaturing step was decreased to 5 min according to the HotStarTaq Plus master mix instructions, and (ii) the annealing temperature for recG was decreased to 48°C. Sequencing was performed at Eton BioSciences, Inc. (San Diego, CA). Chromatograms were inspected for double peaks, which indicated a mixed population. Chromatograms indicating mixed populations were omitted from further analyses. Single-locus queries were performed for each sequence to obtain an allele number. An allelic-profile query was performed with the available loci for each eastern North Dakota sample using the B. burgdorferi MLST website. When data for eight loci were available, the query type chosen was “exact or nearest match.” When fewer loci were available, the query type chosen was n − 1, where n is the number of available loci.
Infectivity of B. burgdorferi M3.
Six female 4- to 6-week-old BALB/c mice were each subcutaneously injected with 106 spirochetes/ml. Two weeks postinjection, infection was preliminarily determined by assaying pre- and postinfection sera by enzyme-linked immunosorbent assay (ELISA). Larval I. scapularis ticks were allowed to feed on infected mice as previously described (39). Briefly, approximately 200 to 300 uninfected larval I. scapularis ticks (Oklahoma State University—Stillwater) were placed on infected BALB/c mice and allowed to attach and feed. Four to 7 days after attachment, engorged larval I. scapularis ticks dropped off and were collected and placed in a humidified chamber until they molted to nymphs. The mice were euthanized 24 h after all I. scapularis ticks detached (i.e., day 8). One tibiotarsal joint and ear pinna and the heart were collected for quantitative PCR (qPCR) analysis. The second tibiotarsal joint and ear pinna were cultured in BSK-II medium with 6% rabbit serum and 50 μg/ml rifampin, blind passed, and examined by dark-field microscopy as described above. After molting, approximately 15 infected nymphal I. scapularis ticks were placed on 6 naive female 4- to 6-week-old BALB/c mice. Engorged I. scapularis ticks, mouse tissues, and cultures were treated as described above.
ELISA.
Anti-B. burgdorferi IgG in sera from inoculated mice was detected as previously described (39). Briefly, 96-well plates were coated with 10 μg/ml B. burgdorferi lysate in a carbonate coating buffer and incubated overnight at 4°C. All washes were performed with phosphate-buffered saline (PBS)-Tween. Serum samples were diluted 1:100 in PBS. Anti-mouse IgG was diluted 1:5,000 in PBS. Each serum sample was analyzed in triplicate.
DNA extraction.
DNA from bacterial cultures was extracted with a 25:24:1 phenol-chloroform-isoamyl alcohol extraction. The DNA was further purified with two consecutive ethanol precipitations. Total (mouse and spirochete) DNA for use in qPCR was extracted from tibiotarsal joints using a phenol-chloroform-isoamyl alcohol protocol and further purified with the Qiagen DNeasy blood and tissue kit according to the manufacturer's specifications. Total DNA from hearts and ear pinnae was extracted using a modified DNeasy blood and tissue kit protocol. Briefly, minced tissues were suspended in buffer ATL (Qiagen) with proteinase K and incubated overnight at 56°C. Samples were further purified according to the manufacturer's specifications and as previously described (39).
qPCR.
The primers used in qPCR are listed in Table 1. Reactions were performed using Bio-Rad (Hercules, CA) iQ SYBR green Supermix. Mouse DNA was detected using primers for nidogen (nid1) and quantified against 500-, 50-, 5-, 0.5-, 0.05-, and 0.005-ng mouse DNA standards. The amplification conditions were as follows: 95°C for 3 min; 40 cycles of 95°C for 30 s, 49°C for 1 min, 1 cycle of 95°C for 1 min, and 50°C for 1 min; and 1 cycle of 49°C for 1 min and 49 to 95°C at 0.5°C for 10 s for each step. B. burgdorferi DNA was detected using primers for recA and quantified against six B. burgdorferi DNA standards ranging in concentration from 10−6 to 10−1 copy number. The amplification conditions were as follows: 95°C for 3 min; 40 cycles of 95°C for 30 s and 50°C for 1 min; 1 cycle of 95°C for 1 min and 50°C for 1 min; 1 cycle of 50°C for 1 min and 50 to 95°C at 0.5°C for 10 s for each step. Each sample and a no-template control were run in triplicate.
Detection of B. burgdorferi DNA in nymphal I. scapularis.
Total DNA was extracted from I. scapularis larvae allowed to molt to nymphs after feeding as larvae using a modified Qiagen DNeasy blood and tissue kit protocol. Ten molted I. scapularis nymphs per mouse were homogenized in buffer ATL (600 μl) with proteinase K (20 μl) and incubated overnight at 56°C. Buffer AL (200 μl; Qiagen) was added, and the tubes were vortexed and incubated at 70°C for 10 min. Wheat germ tRNA, type V (1 μl; 10-mg/ml; R-7876; Sigma-Aldrich, St. Louis, MO) was added, and the tubes were vortexed. Ethanol (230 μl; 95%) was added, and the tubes were vortexed, transferred to DNeasy spin columns, and centrifuged for 1 min at 8,000 rpm. Buffer AW1 (500 μl; Qiagen) was added, and the tubes were centrifuged for 1 min at 8,000 rpm. Buffer AW2 (500 μl; Qiagen) was then added, and the tubes were centrifuged at 14,000 rpm for 3 min. DNA was eluted from the spin column with 100 μl nuclease-free water twice. PCR was performed using primers for the I. scapularis 16S rRNA gene, the B. burgdorferi 16S rRNA gene, and B. burgdorferi flaB (Table 1) under the following amplification conditions: initial denaturation at 94°C for 3 min; 40 cycles of 94°C for 30 s, 50°C for 30 s, and 65°C for 30 s; and final elongation at 65°C for 5 min. The reactions were run on a 2.5% NuSieve gel.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in GenBank with the following accession numbers: KM676013, KM676014, KM676015, KM676016, KM676017, KM676018, KM676019, KM676020, KM676021, KM676022, KM676023, KM676024, KM676025, KM676026, KM676027, KM676028, KM676029, KM676030, KM676031, KM676032, KM676033, KM676034, KM676035, KM676036, KM676037, KM676038, KM676039, KM676040, KM676041, KM676042, KM676043, KM676044, KM676045, KM676046, KM676047, KM676048, KM676049, KM676050, KM676051, KM676052, KM676053, KM676054, KM676055, KM676056, KM676057, KM676058, KM676059, KM676060, KM676061, KM676062, KM676063, KM676064, KM676065, KM676066, KM676067, KM676068, KM676069, and KM676070.
RESULTS
Sequence and phylogeny confirm spirochetes are B. burgdorferi and represent nonclonal populations.
To confirm that the eastern North Dakota samples were B. burgdorferi sensu lato, we sequenced 136 and 139 nucleotides of the 16S rRNA gene and flaB, respectively. The 16S rRNA gene sequences were queried against the Ribosomal Database Project database. The sequences for each of the five samples returned hits for various Borrelia species (data not shown). A BLAST search of the flaB sequences obtained from all five samples showed 100% sequence identity matches to B. burgdorferi (data not shown). These data confirmed that the spirochetes were members of the B. burgdorferi sensu lato group.
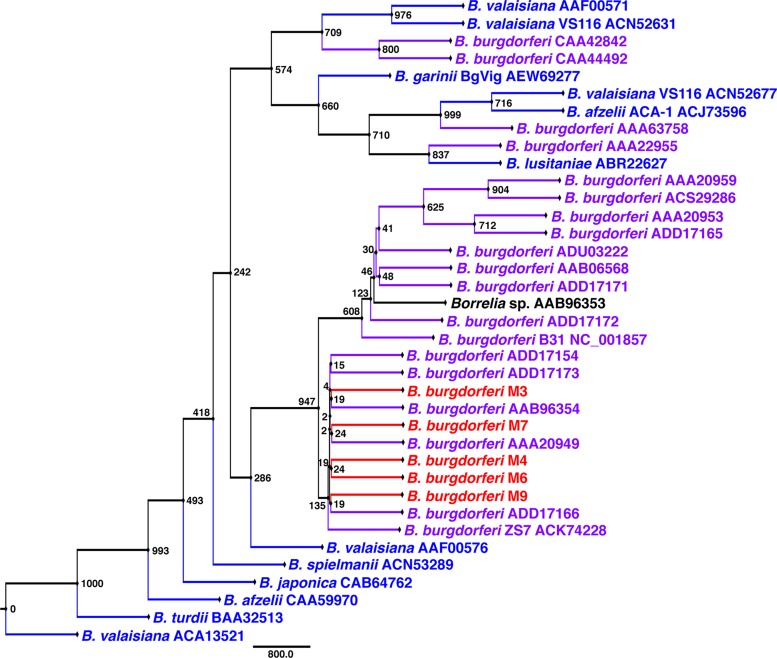
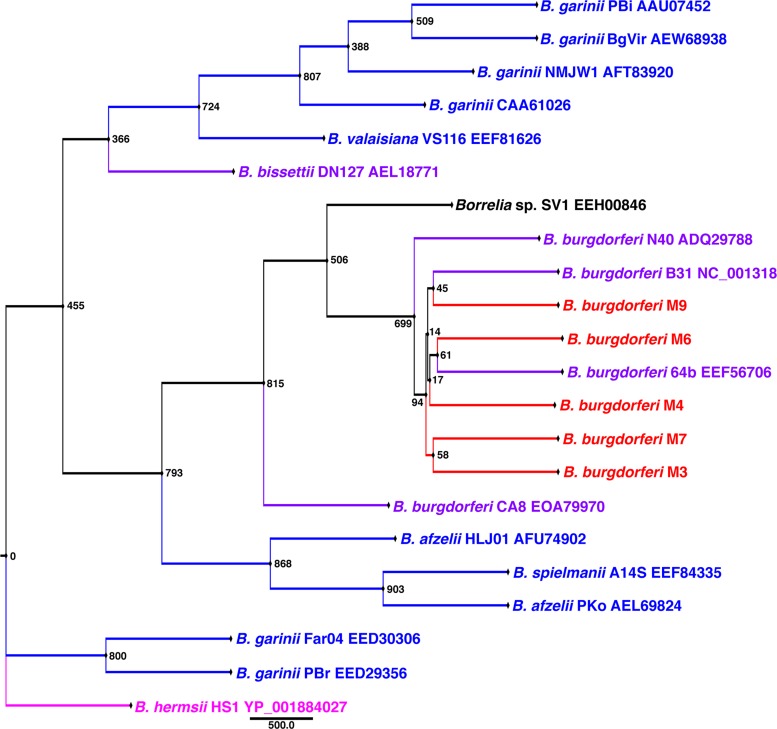
To determine whether the samples represented multiple B. burgdorferi populations or a single population, sequencing and phylogenetic analyses were performed for ospA, ospC, p66, and the 16S rRNA gene-ile tRNA IGS. For comparison, various B. burgdorferi sensu lato and Borrelia hermsii sequences were obtained from the NCBI and BLAST databases. A BLAST search using the 16S rRNA gene-ile tRNA DNA sequence from B. burgdorferi B31 returned results for B. burgdorferi sensu lato (data not shown). There were no differences in the sequences of ospA and p66 for the five samples. Protein maximum-likelihood analyses of OspA (Fig. 1) and p66 (Fig. 2) grouped the eastern North Dakota samples with B. burgdorferi sensu stricto.
FIG 1.
Unrooted protein maximum-likelihood analysis of OspA showing that the eastern North Dakota isolates group with North American B. burgdorferi. The sequences included (approximately) residues 41 to 236. The node values represent bootstrap values from 1,000 replicates. Red, eastern North Dakota isolates; purple, North American Lyme disease-associated Borrelia; blue, Eurasian Lyme disease-associated Borrelia; black, unknown species.
FIG 2.
Rooted protein maximum-likelihood analysis of p66 showing that the eastern North Dakota isolates group with North American B. burgdorferi. The sequences included (approximately) residues 364 to 548. The node values represent bootstrap values from 1,000 replicates. Red, eastern North Dakota isolates; purple, North American Lyme disease-associated Borrelia; blue, Eurasian Lyme disease-associated Borrelia; black, unknown species; pink, relapsing fever outgroup.
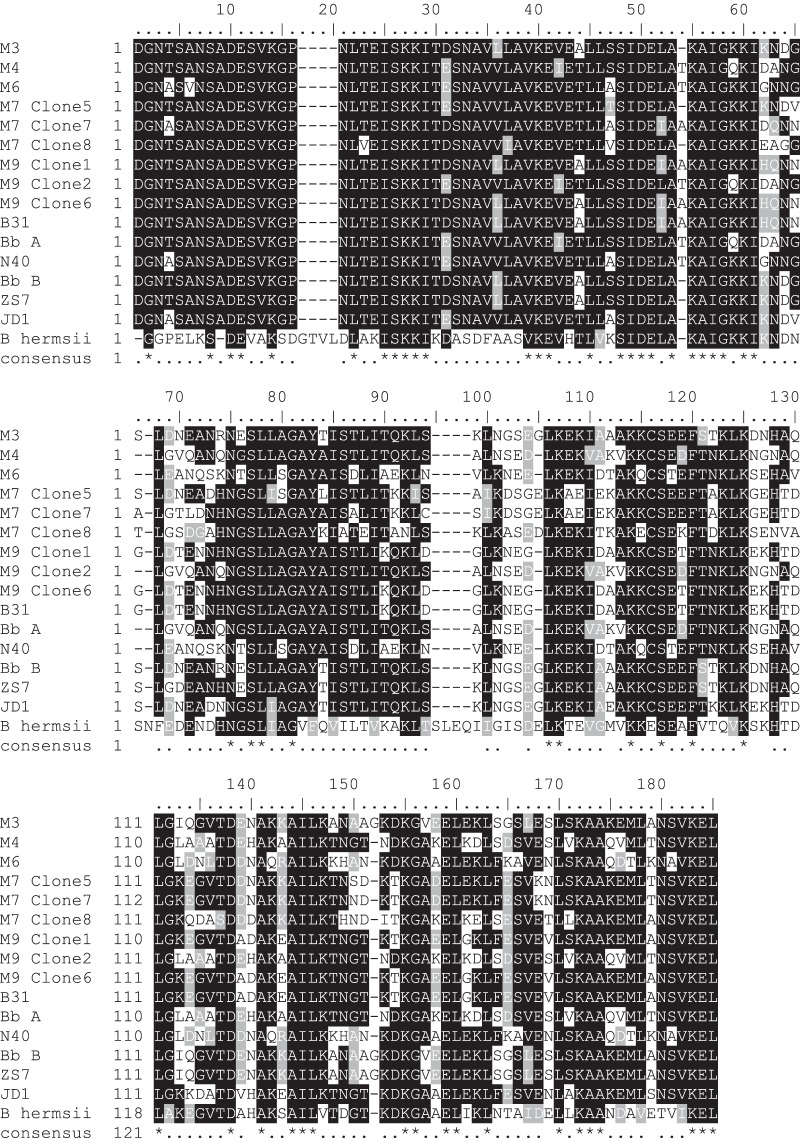
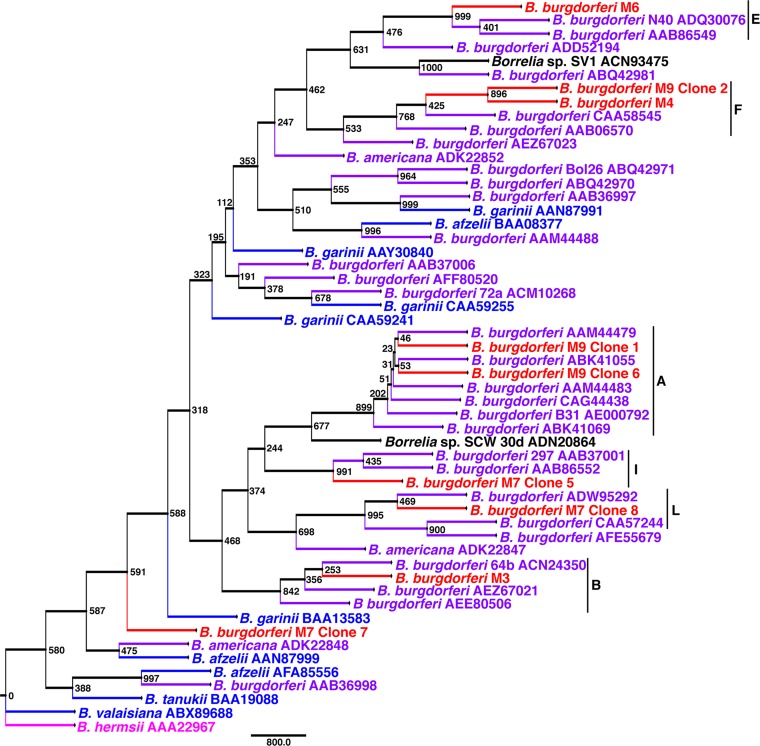
Unlike OspA and p66, OspC showed variation among the eastern North Dakota samples, sharing between 66 and 100% identity (Fig. 3 and 4). In two of the small mammals sampled (M7 and M9), multiple B. burgdorferi strains were detected. With the exception of B. burgdorferi M7 clone 7, which grouped with a clade consisting of both North American and Eurasian Borrelia spp., the eastern North Dakota populations grouped most closely with B. burgdorferi sensu lato (Fig. 4). B. burgdorferi M9 clones 1 and 6 were 100% identical across the region used for OspC analyses but showed variation outside the region, particularly at 4 residues immediately downstream. Further sequencing would be required to determine whether clones 1 and 6 are indeed different strains. OspC typing revealed a diverse group of spirochetes (Table 2). Interestingly, one of the isolates identified, B. burgdorferi M7 clone 8, belongs to one of the rarer groups (L) to be identified in reservoir animals (40). One isolate, B. burgdorferi M7 clone 7, did not group with OspC groups A to U. This is not surprising, given that it did not clearly group with North American B. burgdorferi sensu lato. Taken together, these data confirm that nonclonal, invasive populations of B. burgdorferi are present in eastern North Dakota.
FIG 3.
Alignment of OspC suggesting the eastern North Dakota isolates are genetically distinct strains of B. burgdorferi. Samples M7 and M9 contain a mixture of clones. The sequences included (approximately) residues 25 to 198. B. burgdorferi B31 (AE000792.1), N40 (DQ437463.1), A (a non-type strain, ABQ42987.1), B (a non-type strain, ABK41066.1), ZS7 (AF500204.1), and JD1 (DQ437462.1), as well as Vsp3, an OspC ortholog found in B. hermsii (relapsing fever; AAA22967.1), were included for comparison. Alignment was performed in ClustalW2, and shading was performed using ExPASy's BoxShade (black indicates identical residues, gray indicates residues with biochemical properties similar to those of the majority of the residues in the same position, and white indicates unrelated residues). Symbols: dashes indicate gaps, dots in the consensus line indicate moderate to high conservation, gaps indicate no conservation, and asterisks indicate fully conserved residues.
FIG 4.
Rooted protein maximum-likelihood analysis of OspC showing that the eastern North Dakota isolates group with North American B. burgdorferi. OspC groups are indicated by a vertical black line and a single-letter code. The sequences included (approximately) residues 25 to 198. The node values represent bootstrap values from 1,000 replicates. Red, eastern North Dakota isolates; purple, North American Lyme disease-associated Borrelia; blue, Eurasian Lyme disease-associated Borrelia; black, unknown species; pink, relapsing fever outgroup.
TABLE 2.
OspC groups for eastern North Dakota isolatesa
| Eastern ND B. burgdorferi isolate(s) | OspC group |
|---|---|
| M9 clones 1 and 6 | A |
| M3 | B |
| M6 | E |
| M4 and M9 clone 2 | F |
| M7 clone 5 | I |
| M7 clone 8 | L |
| M7 clone 7 | None |
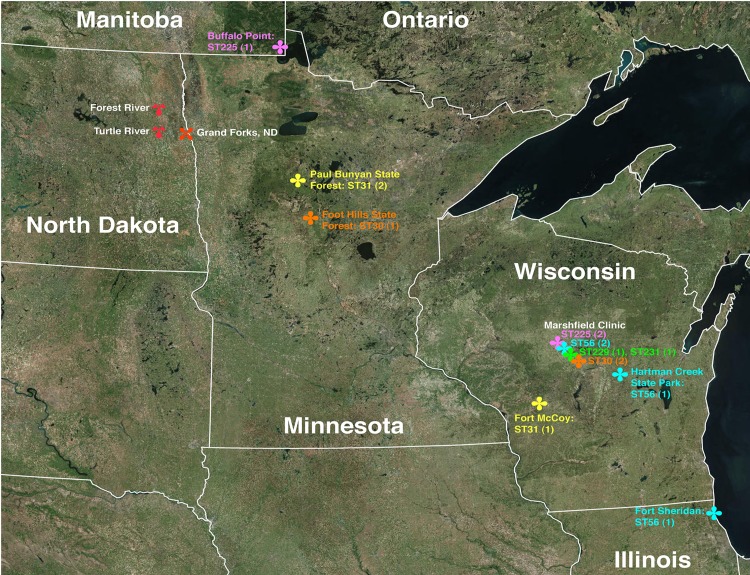
The eastern North Dakota populations are most closely related to B. burgdorferi found in the upper Midwest.
MLST analysis was performed to determine the regional source of the eastern North Dakota populations. A locus was omitted from further analysis if the chromatograms suggested a mixed population for that locus (i.e., a double peak at a single nucleotide location) (Tables 3 and 4). Sequences for all eight loci could be obtained only for one sample (B. burgdorferi M9), and thus, single-locus analyses were performed for the remaining samples. The database profiles that most closely matched the incomplete eastern North Dakota sample profiles were obtained (Tables 3 and 4). B. burgdorferi M3 most closely matched ST30 strains for all loci except nifS, which was unavailable. The available locus information for B. burgdorferi M4 matched ST56 strains. B. burgdorferi M6 matched ST31 and ST229 at five loci but had a different recG locus than ST31 or ST229. B. burgdorferi M7 differed at clpA from ST225 strains. B. burgdorferi M9 matched ST56 strains at all eight loci. Each database strain that was the closest match to the eastern North Dakota samples was initially identified in the upper Midwest (Illinois, Wisconsin, and Minnesota and Manitoba, Canada) (Fig. 5 and Table 4). These data suggest the eastern North Dakota samples are most closely related to upper Midwest strains, and thus, the upper Midwest is the most probable source of the eastern North Dakota samples.
TABLE 3.
Allele scores for clpA, clpX, nifS, pepX, pyrG, recG, rplB, and uvrA and closest matching database STs
| Eastern ND B. burgdorferi isolate | Allele score |
ST | |||||||
|---|---|---|---|---|---|---|---|---|---|
| clpA | clpX | nifS | pepX | pyrG | recG | rplB | uvrA | ||
| M3 | 19 | 1 | −a | 1 | 2 | 1 | 1 | 10 | 30 |
| M4 | 24 | − | − | 18 | − | 19 | 1 | − | 56, 231 |
| M6 | 20 | 4 | − | 3 | 3 | 1 | 3 | − | 31, 229 |
| M7 | 18 | 2 | − | − | 2 | 8 | 1 | − | 225 |
| M9 | 24 | 14 | 4 | 18 | 11 | 19 | 1 | 12 | 56, 231 |
−, chromatogram indicated a mixed population.
TABLE 4.
Allelic profiles and STs for the eastern North Dakota samples and the most closely matching MLST database strains
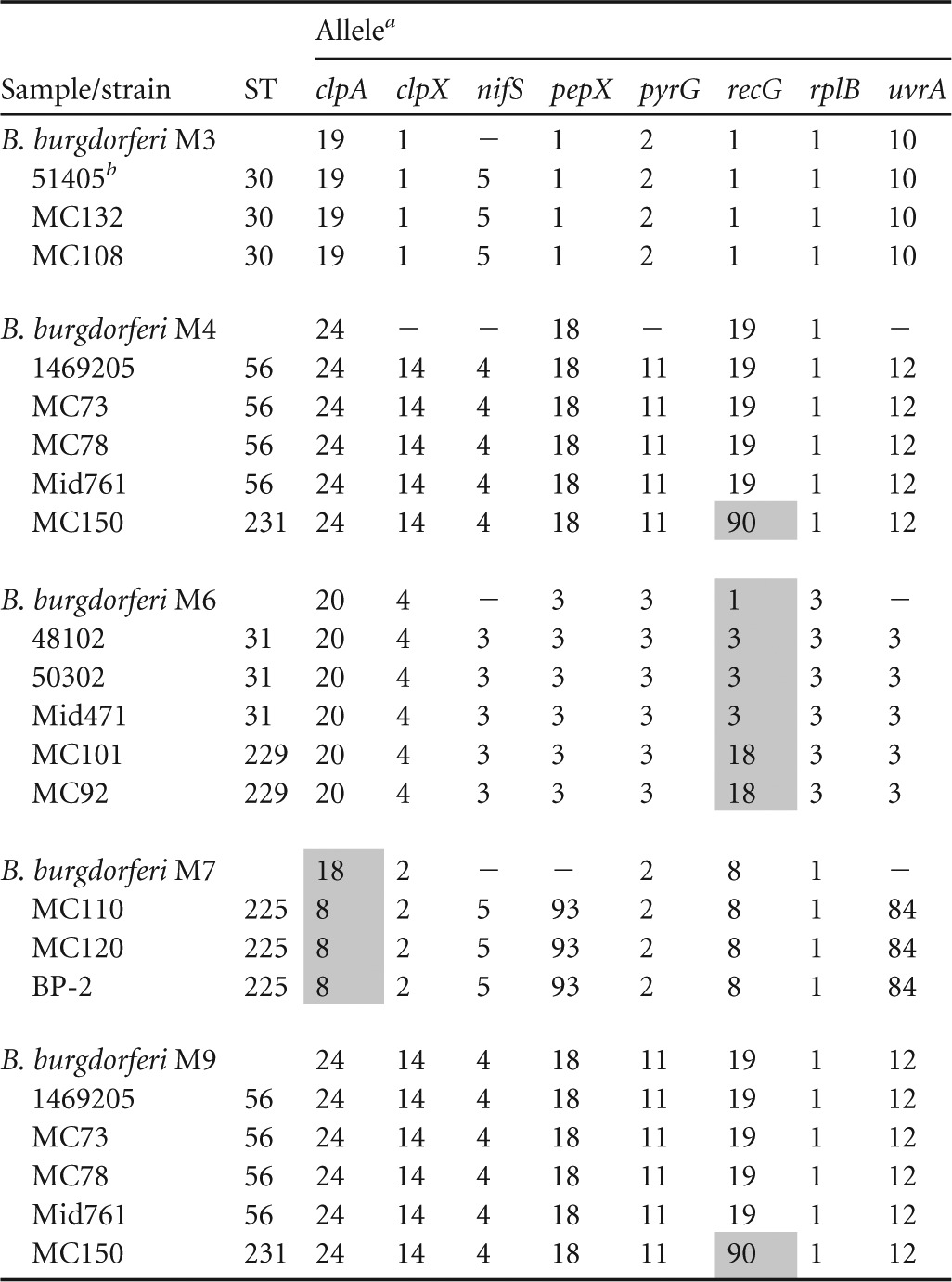
Alleles that differ from the respective eastern North Dakota allele are shaded. −, chromatogram indicated a mixed populations.
Database strain identifications.
FIG 5.
Locations of database STs from Table 4. Strains from Marshfield Clinic (Wisconsin) were obtained from unknown locations in Wisconsin. All were identified from human erythema migrans samples. The remaining strains were reported to have been isolated from the indicated locations. Pink, ST225; yellow, ST31; orange, ST30; green, ST229 and ST231; cyan, ST56. The numbers in parentheses indicate the number of unique strains comprising each ST. Forest River and Turtle River were the sites of sample collection in this study. The satellite images were obtained from NOAA via Google Earth.
Needle-injected B. burgdorferi M3 infects and survives in BALB/c mice and is acquired by larval I. scapularis during feeding.
To determine whether infectious and transmissible populations were present in eastern North Dakota, an infection-and-transmission study was performed using B. burgdorferi M3. M3 was the only sample obtained from M. gapperi, whose reservoir status is unknown. Further, the OspC data suggested M3 was a clonal population belonging to the infectious B group. An ELISA of pre- and postinfection sera from mice subcutaneously injected with 106 B. burgdorferi M3 spirochetes/ml showed increased anti-Borrelia antibodies 2 weeks postinjection. Tibiotarsal joints and ear pinnae were culture positive, except for one mouse for which culture data were unavailable due to contamination. B. burgdorferi flaB was detected in molted nymphs fed on five of the six needle-injected mice. Neither the B. burgdorferi 16S rRNA gene nor flaB could be positively identified in molted nymphs fed on mouse 6. B. burgdorferi recA was detected in one of three hearts, six of six tibiotarsal joints, and three of six ear pinnae. The mouse nidogen gene, nid1, was detected in all tissues. These data demonstrate B. burgdorferi M3 is infectious to mice, able to disseminate to multiple tissues, and capable of being acquired by I. scapularis during a blood meal.
B. burgdorferi M3 survives the I. scapularis larval molt and is subsequently transmitted to naive BALB/c mice during a blood meal.
To determine whether B. burgdorferi M3 was transtadially maintained during the I. scapularis larval molt and capable of transmission to naive mice, I. scapularis nymphs were fed on naive BALB/c mice. Infection was confirmed by ELISA, which showed an increase in absorbance postfeeding. Tibiotarsal joints and ear pinnae were all culture positive. nid1 and recA were detected in all six hearts, tibiotarsal joints, and ear pinnae. These data show B. burgdorferi M3 is transtadially maintained in I. scapularis and capable of dissemination in naive hosts.
DISCUSSION
Studies on the prevalence and spread of B. burgdorferi, I. scapularis, and Lyme disease in the Upper Midwest typically focus on Minnesota and Wisconsin (8, 41, 42). The vast majority of Lyme disease cases, and thus B. burgdorferi and I. scapularis, reported in the upper Midwest are found in Minnesota and Wisconsin. However, there were numerous factors that demanded a detailed investigation of B. burgdorferi in eastern North Dakota (e.g., the close proximity to high-risk Minnesota counties with a history of Lyme disease, B. burgdorferi, and I. scapularis; the presence of known small-mammal reservoirs; and recent studies [5, 43] identifying stable I. scapularis populations and B. burgdorferi in North Dakota, further confirming the expansion of I. scapularis described in other U.S. regions and Canada [3, 6, 7, 44, 45]).
The data presented here demonstrate that the spirochetes isolated in eastern North Dakota from Peromyscus spp. and M. gapperi hearts are members of the B. burgdorferi sensu lato complex. While OspA and p66 are identical among all of the eastern North Dakota populations, OspC typing shows they are distinctly nonclonal populations. The M9 population consisted of at least two OspC types, A and F; M7 consisted of at least two types, I and L (Table 2 and Fig. 4). B. burgdorferi M7 clone 7 does not appear to belong to any of the previously defined OspC groups. In phylogenetic analyses, clone 7 also does not appear to group clearly with either North American B. burgdorferi sensu lato or Eurasian Lyme disease Borrelia (Fig. 4). A BLAST analysis with the sequence from clone 7 returns results for B. burgdorferi sensu lato OspC but with a maximum identity score of 87% (data not shown). It is clear clone 7 is a member of the B. burgdorferi sensu lato complex, but its OspC type and infectivity remain unknown. Despite obtaining sequence data for a single clone, the M6 population may consist of multiple OspC types due to difficulties in obtaining ospC sequence prior to cloning ospC. Since less than five ospC clones were obtained and sequenced from three of the five eastern North Dakota samples, it is not possible to determine the proportion of OspC types in each sample. It is clear at least three of the most common OspC types known to cause disseminated infection in humans (A, B, and I) (21, 25) are present in eastern North Dakota (Table 2 and Fig. 4). A more comprehensive survey is required to determine the presence and distribution of OspC types.
The MLST data, though limited, suggest the eastern North Dakota populations are derived from the upper Midwest populations and are not a recent transplant from another region. Specifically, the eastern North Dakota populations appear to be most closely related to strains found in Minnesota, Wisconsin, Illinois, and southeastern Manitoba on the Minnesota-Canada border. A number of the housekeeping genes appeared to be identical in the populations that the OspC data showed were mixed populations. Likewise, a number of housekeeping genes appeared to indicate mixed populations in a single sample when the OspC data suggested that the populations in each sample were clonal. This was surprising, because the ospC gene is highly polymorphic, while the housekeeping genes are generally more conserved. Viewing the OspC and MLST data together suggests that, in addition to small-scale random mutation events, large-scale mutation events have also occurred with OspC. The sequence analyses, taken as a whole, suggest a regional population structure larger and more complex than was captured by the five samples partially characterized here.
The results of the infection study show B. burgdorferi M3 is infectious through both an artificial and a natural route of infection. B. burgdorferi M3 is capable of disseminating from the site of inoculation to the heart, tibiotarsal joints, and ear pinnae, indicating it is highly infectious (46). The ability to disseminate is not surprising, since B. burgdorferi M3 belongs to the ospC group B, a group associated with disseminated disease in humans (21, 26, 27).
In the United States, Lyme disease remains a significant public health issue. From 2001 to 2011, the number of confirmed cases reported to the CDC averaged 24,000, making it the most reported tick-borne disease in the United States. In 2013, the CDC released revised yearly estimates based on continuing studies, including analysis of data from tests conducted by seven participating commercial laboratories in 2008 (47). Based on these analyses, the estimated number of individuals infected with B. burgdorferi in the United States was revised to approximately 288,000 per year, about 10 times more than the average yearly number of reported and confirmed Lyme disease cases. North Dakota is not the Lyme disease hot spot Minnesota is, but Lyme disease is poised to be a significant public health issue in North Dakota. The number of Lyme disease cases reported yearly in North Dakota is low (126 reported cases between 1996 and 2012 [North Dakota Department of Health]) compared to the number of cases reported in neighboring Minnesota (nearly 13,000 confirmed cases between 1996 and 2012 [Minnesota Department of Health]). However, just as the national cases are underestimated, there are a number of factors that make a reasonable argument for cases in North Dakota being underestimated: the classification of eastern North Dakota as a transition zone (3), the conventional opinion that B. burgdorferi and I. scapularis are not found in North Dakota, the evolving criteria for reporting and confirming Lyme disease, the increasing number of Lyme disease cases in North Dakota, and the rural nature of North Dakota.
To develop comprehensive, informed public health policies in both the United States and Canada, it is imperative to understand whether I. scapularis, and subsequently B. burgdorferi, are expanding outside the previously identified geographical regions. While changes in habitat, and the reasons for those changes, are outside the scope of this study, it is clear that B. burgdorferi and I. scapularis have migrated westward in the upper Midwest. This information is relevant to North Dakota residents, visitors, and medical professionals, who should be aware of the risk of contracting Lyme disease in eastern North Dakota. This information is also important beyond the borders of North Dakota, as it provides additional data on the ever-evolving state of Lyme disease.
ACKNOWLEDGMENTS
We acknowledge Brian Stevenson, Brandon Jutras, Jean Tsao, Patty Rosa, and Paul Mead for helpful discussions and suggestions and Megan Quinlan for obtaining a high-resolution map.
This study was funded by ND EPSCoR and UND SMHS startup funds to C.A.B.
REFERENCES
- 1.Pepin KM, Eisen RJ, Mead PS, Piesman J, Fish D, Hoen AG, Barbour AG, Hamer S, Diuk-Wasser MA. 2012. Geographic variation in the relationship between human Lyme disease incidence and density of infected host-seeking Ixodes scapularis nymphs in the eastern United States. Am J Trop Med Hyg 86:1062–1071. doi: 10.4269/ajtmh.2012.11-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minnesota Department of Health. 2013. Lyme disease in Minnesota: Lyme disease areas of highest risk. Minnesota Department of Health, St. Paul, MN. [Google Scholar]
- 3.Diuk-Wasser MA, Hoen AG, Cislo P, Brinkerhoff R, Hamer SA, Rowland M, Cortinas R, Vourc'h G, Melton F, Hickling GJ, Tsao JI, Bunikis J, Barbour AG, Kitron U, Piesman J, Fish D. 2012. Human risk of infection with Borrelia burgdorferi, the Lyme disease agent, in eastern United States. Am J Trop Med Hyg 86:320–327. doi: 10.4269/ajtmh.2012.11-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russart NM, Dougherty M, Vaughan JA. 2014. Survey of ticks (Acari: Ixodidae) and tick-borne pathogens in North Dakota. J Med Entomol. 51:1087–1090. doi: 10.1603/ME14053. [DOI] [PubMed] [Google Scholar]
- 5.North Dakota Department of Health, Division of Disease Control. 2011. Tick surveillance in North Dakota—the pump handle. North Dakota Department of Health, Division of Disease Control, Bismarck, ND. [Google Scholar]
- 6.Koffi JK, Leighton PA, Pelcat Y, Trudel L, Lindsay LR, Milord F, Ogden NH. 2012. Passive surveillance for I. scapularis ticks: enhanced analysis for early detection of emerging Lyme disease risk. J Med Entomol 49:400–409. doi: 10.1603/ME11210. [DOI] [PubMed] [Google Scholar]
- 7.Rand PW, Lacombe EH, Dearborn R, Cahill B, Elias S, Lubelczyk CB, Beckett GA, Smith RP Jr. 2007. Passive surveillance in Maine, an area emergent for tick-borne diseases. J Med Entomol 44:1118–1129. doi: 10.1603/0022-2585(2007)44[1118:PSIMAA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Lee X, Hardy K, Johnson DH, Paskewitz SM. 2013. Hunter-killed deer surveillance to assess changes in the prevalence and distribution of Ixodes scapularis (Acari: Ixodidae) in Wisconsin. J Med Entomol 50:632–639. doi: 10.1603/ME12234. [DOI] [PubMed] [Google Scholar]
- 9.Bacon RM, Kugeler KJ, Mead PS. 2008. Surveillance for Lyme disease—United States, 1992–2006. MMWR Surveill Summ 57:1–9. [PubMed] [Google Scholar]
- 10.Rudenko N, Golovchenko M, Grubhoffer L, Oliver JH Jr. 2011. Updates on Borrelia burgdorferi sensu lato complex with respect to public health. Ticks Tick-Borne Dis 2:123–128. doi: 10.1016/j.ttbdis.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strle F, Stanek G. 2009. Clinical manifestations and diagnosis of Lyme borreliosis. Curr Probl Dermatol 37:51–110. doi: 10.1159/000213070. [DOI] [PubMed] [Google Scholar]
- 12.Clark KL, Leydet BF, Threlkeld C. 2014. Geographical and genospecies distribution of Borrelia burgdorferi sensu lato DNA detected in humans in the USA. J Med Microbiol 63:674–684. doi: 10.1099/jmm.0.073122-0. [DOI] [PubMed] [Google Scholar]
- 13.Girard YA, Fedorova N, Lane RS. 2011. Genetic diversity of Borrelia burgdorferi and detection of B. bissettii-like DNA in serum of north-coastal California residents. J Clin Microbiol 49:945–954. doi: 10.1128/JCM.01689-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudenko N, Golovchenko M, Rùzõek D, Piskunova N, Mallátová N, Grubhoffer L. 2009. Molecular detection of Borrelia bissettii DNA in serum samples from patients in the Czech Republic with suspected borreliosis. FEMS Microbiol Lett 292:274–281. doi: 10.1111/j.1574-6968.2009.01498.x. [DOI] [PubMed] [Google Scholar]
- 15.Schneider BS, Schriefer ME, Dietrich G, Dolan MC, Morshed MG, Zeidner NS. 2008. Borrelia bissettii isolates induce pathology in a murine model of disease. Vector Borne Zoonotic Dis 8:623–633. doi: 10.1089/vbz.2007.0251. [DOI] [PubMed] [Google Scholar]
- 16.Clark KL. 2013. Lyme borreliosis in human patients in Florida and Georgia, USA. Int J Med Sci 10:915–931. doi: 10.7150/ijms.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest 113:220–230. doi: 10.1172/JCI200419894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci U S A 92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, VanRaden MJ, Stewart P, Rosa P. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun 74:3554–3564. doi: 10.1128/IAI.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, Schwan TG, Policastro PF, Elias AF, Rosa PA. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci U S A 101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang I-N, Wormser GP, Schriefer ME, Luft BJ. 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun 67:3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theisen M, Borre M, Mathiesen MJ, Mikkelsen B, Lebech A-M, Hansen K. 1995. Evolution of the Borrelia burgdorferi outer surface protein OspC. J Bacteriol 177:3036–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagal V, Postic D, Ruzic-Sabljic E, Baranton G. 2003. Genetic diversity among Borrelia strains determined by single-strand conformation polymorphism analysis of the ospC gene and its association with invasiveness. J Clin Microbiol 41:5059–5065. doi: 10.1128/JCM.41.11.5059-5065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brisson D, Drecktrah D, Eggers CH, Samuels DS. 2012. Genetics of Borrelia burgdorferi. Annu Rev Genet 46:515–536. doi: 10.1146/annurev-genet-011112-112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang I-N, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wormser GP, Brisson D, Liveris D, Hanincová K, Sandigursky S, Nowakowski J, Nadelman RB, Ludin S, Schwartz I. 2008. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis 198:1358–1364. doi: 10.1086/592279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dykhuizen DE, Brisson D, Sandigursky S, Wormser GP, Nowakowski J, Nadelman RB, Schwartz I. 2008. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am J Trop Med Hyg 78:806–810. [PMC free article] [PubMed] [Google Scholar]
- 28.Earnhart CG, Buckles EL, Dumler JS, Marconi RT. 2005. Demonstration of OspC type diversity in invasive human Lyme disease isolates and identification of previously uncharacterized epitopes that define the specificity of the OspC murine antibody response. Infect Immun 73:7869–7877. doi: 10.1128/IAI.73.12.7869-7877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brisson D, Dykhuizen DE. 2004. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics 168:713–722. doi: 10.1534/genetics.104.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livey I, Gibbs CP, Schuster R, Dorner F. 1995. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol Microbiol 18:257–269. doi: 10.1111/j.1365-2958.1995.mmi_18020257.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, van Dam AP, Dankert J. 1999. Evidence for frequent OspC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol Lett 177:289–296. doi: 10.1111/j.1574-6968.1999.tb13745.x. [DOI] [PubMed] [Google Scholar]
- 32.Barbour AG, Travinsky B. 2010. Evolution and distribution of the ospC gene, a transferable serotype determinant of Borrelia burgdorferi. mBio 1:e00153-10. doi: 10.1128/mBio.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aanensen DM, Spratt BG. 2005. The multilocus sequence typing network: mlst.net. Nucleic Acids Res 33:W728–W733. doi: 10.1093/nar/gki415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margos G, Gatewood AG, Aanensen DM, Hanincová K, Terekhova D, Vollmer SA, Cornet M, Piesman J, Donaghy M, Bormane A. 2008. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc Natl Acad Sci U S A 105:8730–8735. doi: 10.1073/pnas.0800323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seabloom R. 2011. White-footed mouse; deer mouse, p 305–313. In The mammals of North Dakota. North Dakota Institute for Regional Studies, Fargo, ND. [Google Scholar]
- 36.Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 37.Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D, Barbour AG. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741–1755. doi: 10.1099/mic.0.26944-0. [DOI] [PubMed] [Google Scholar]
- 38.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Floden AM, Gonzalez T, Gaultney RA, Brissette CA. 2013. Evaluation of RevA, a fibronectin-binding protein of Borrelia burgdorferi, as a potential vaccine candidate for Lyme disease. Clin Vaccine Immunol 20:892–899. doi: 10.1128/CVI.00758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alghaferi MY, Anderson JM, Park J, Auwaerter PG, Aucott JN, Norris DE, Dumler JS. 2005. Borrelia burgdorferi ospC heterogeneity among human and murine isolates from a defined region of northern Maryland and southern Pennsylvania: lack of correlation with invasive and noninvasive genotypes. J Clin Microbiol 43:1879–1884. doi: 10.1128/JCM.43.4.1879-1884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steiner FE, Pinger RR, Vann CN, Grindle N, Civitello D, Clay K, Fuqua C. 2008. Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp., Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J Med Entomol 45:289–297. doi: 10.1603/0022-2585(2008)45[289:IACROA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 42.Caporale DA, Johnson CM, Millard BJ. 2005. Presence of Borrelia burgdorferi (Spirochaetales: Spirochaetaceae) in southern Kettle Moraine State Forest, Wisconsin, and characterization of strain W97F51. J Med Entomol 42:457–472. doi: 10.1603/0022-2585(2005)042[0457:POBBSS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 43.Russart NM. 2013. Ticks and tick-borne pathogens in North Dakota. M.S. thesis University of North Dakota, Grand Forks, ND. [Google Scholar]
- 44.Ogden NH, Lindsay LR, Hanincová K, Barker IK, Bigras-Poulin M, Charron DF, Heagy A, Francis CM, O'Callaghan CJ, Schwartz I, Thompson RA. 2008. Role of migratory birds in introduction and range expansion of Ixodes scapularis ticks and of Borrelia burgdorferi and Anaplasma phagocytophilum in Canada. Appl Environ Microbiol 74:1780–1790. doi: 10.1128/AEM.01982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogden NH, St-Onge L, Barker IK, Brazeau S, Bigras-Poulin M, Charron DF, Francis CM, Heagy A, Lindsay LR, Maarouf A, Michel P, Milord F, O'Callaghan CJ, Trudel L, Thompson RA. 2008. Risk maps for range expansion of the Lyme disease vector, Ixodes scapularis, in Canada now and with climate change. Int J Health Geogr 7:24. doi: 10.1186/1476-072X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinckley AF, Connally NP, Meek JI, Johnson BJ, Kemperman MM, Feldman KA, White JL, Mead PS. 2014. Lyme disease testing by large commercial laboratories in the United States. Clin Infect Dis 59:676–681. doi: 10.1093/cid/ciu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]