Abstract
A variety of systems have been developed to study biofilm formation. However, most systems are based on the surface-attached growth of microbes under shear stress. In this study, we designed a microfluidic channel device, called a microfluidic agarose channel (MAC), and found that microbial cells in the MAC system formed an embedded cell aggregative structure (ECAS). ECASs were generated from the embedded growth of bacterial cells in an agarose matrix and better mimicked the clinical environment of biofilms formed within mucus or host tissue under shear-free conditions. ECASs were developed with the production of extracellular polymeric substances (EPS), the most important feature of biofilms, and eventually burst to release planktonic cells, which resembles the full developmental cycle of biofilms. Chemical and genetic effects have also confirmed that ECASs are a type of biofilm. Unlike the conventional biofilms formed in the flow cell model system, this embedded-type biofilm completes the developmental cycle in only 9 to 12 h and can easily be observed with ordinary microscopes. We suggest that ECASs are a type of biofilm and that the MAC is a system for observing biofilm formation.
INTRODUCTION
In natural habitats, bacterial cells often form biofilms to structurally organize and protect their communities. A biofilm is an aggregate of microbial cells that adhere to a live/nonliving surface, and the adherent cells in biofilms are enclosed in a self-produced matrix of extracellular polymeric substances (EPS), including nucleic acids, proteins, polysaccharides, and lipids (1). Bacterial biofilm formation is a complex developmental process involving several stages, including migration and initial attachment, EPS production and irreversible attachment, maturation, destruction, and cell dispersion (2). The significance of biofilms is witnessed in the environment, industry, and human health (3). Importantly, pathogenic bacteria build biofilms on various medical implants and human tissues, and they have a strong resistance to antimicrobial agents, as well as persistently colonizing patients with chronic diseases. In the United States, the fouling of microorganisms on industrial equipment and architecture costs billions of dollars annually (4).
To study biofilms, many researchers have developed a variety of in vitro model systems that mimic the environment of biofilm habitats (5), and several models are commonly used for biofilm assays, such as the agar plate system (6), multiwell plate system (7), biofilm reactor system (8, 9), and flow cell system (10). Flow and biofilm reactor systems are based on the surface-attached growth of microbes under shear stress. While these conventional models may mimic some aspects of natural environments, biofilms sometimes form under very different conditions that conventional systems cannot mimic entirely. The clinically relevant biofilms in host tissues are formed without shear stress within the mucosal layer on epithelial cells or inside host cells. Pseudomonas aeruginosa, which infects cystic fibrosis (CF) patients, does not attach to the pulmonary epithelial surfaces of CF patients, but it forms persistent biofilms within the thick mucosal layer on the epithelial cells of these patients (11). In urinary tract infections, intracellular bacteria mature into biofilms, forming pod-like bulges in the mouse bladder lumen, showing a different structure from biofilms formed on surfaces with shear stress (12). Additionally, bacteria reside on soil matrices or plant surfaces as aggregates of cells in terrestrial habitats (non-shear stress conditions), and they are embedded in exopolymeric substances that they generate and that can be considered a biofilm. These biofilms are commonly unsaturated, and the size of biofilms varies depending on the environmental conditions (13–15). Therefore, it is necessary to develop a new biofilm modeling system that more closely resembles the clinical and terrestrial relevance of biofilms.
To address this need, we developed a new biofilm-forming system based on the embedded growth of bacteria, using a microfluidic channel device called a microfluidic agarose channel (MAC) (Fig. 1A and B), instead of based on conventional biofilm generation using shear stress, such as the multiwell plate system (7), biofilm reactor system (8, 9), or flow cell system (10). The developed device generated a novel type of biofilm-like structure under shear stress-free conditions. In the MAC system, bacterial cells multiplied while embedded in an agarose matrix, and they generated embedded cell aggregative structures (ECASs) that may represent the clinical biofilms formed in mucosal layers or intracellular space (11, 12), while conventional systems can model attached biofilms only under shear stress conditions. The ECASs formed in our system demonstrated a sufficient number of biofilm characteristics to be considered a type of biofilm. We suggest that the MAC can be a good in vitro system that reenacts biofilm formation in the mucosal or viscous environment and that ECAS-derived biofilms provide a valuable model for studying the clinical relevance of biofilms that are generated from the embedded growth of bacteria in host tissues.
FIG 1.
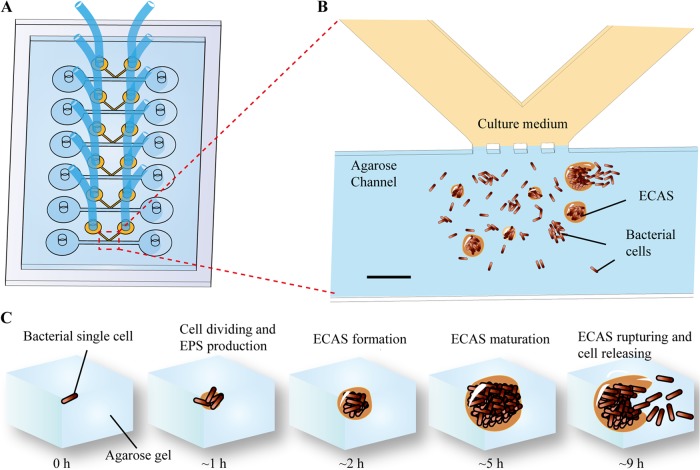
Schematic diagram of the MAC for developing an ECAS. (A) The MAC chip was fabricated with PDMS and assembled with a glass slide. One chip contained six parallel channel sets. The tube connected to the PDMS chip was used to supply the culture medium. (B) Expanded version of panel A. The bacteria were mixed with agarose and then injected into the left hole of the straight channel, and liquid medium was applied from a V-shaped channel. The interface of two channels was connected by open capillary valves. Bacterial cells embedded in the agarose grew in two forms: individually dividing cells and ECASs. (C) The full developmental cycle of ECASs in the MAC system. Diagrams show, from left to right, a single cell, cell division and EPS production, ECAS formation, ECAS maturation, and ECAS rupture and cell release. Bar, 100 μm.
MATERIALS AND METHODS
Design and manufacture of the MAC.
For the microfluidic channel-based biofilm assay, MACs were designed and manufactured by QuantaMatrix Inc. at Seoul National University, as shown in Fig. 1 and Fig. S1 in the supplemental material, with a straight channel for filling agarose-embedded cells that merges into a V-shaped channel for flowing medium. The junction region of two channels has an open barrier to allow for the diffusion of nutrients, and it was manufactured to be optically transparent for direct microscopic observation. The embedded biofilms form and grow in this area (indicated by dotted lines in Fig. 1A and B). The MAC chip was fabricated with polydimethylsiloxane (PDMS) (Sylgard 184; Dow Corning) and then assembled with a PDMS-coated glass slide. The size of the chip was 50 mm by 25 mm (Fig. 1).
Bacterial strains and biofilm formation.
Four standard bacteria from the Clinical and Laboratory Standards Institute (Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, and Staphylococcus aureus ATCC 29213) and laboratory bacteria (Pseudomonas aeruginosa PA14 and its mutant strains, as well as Bacillus subtilis ATCC 6633) were studied for biofilm formation using the MAC system. Bacterial strains were grown overnight and subcultured in 5 ml of fresh medium to an optical density at 600 nm (OD600) of ≈1.0. The cells were mixed with agarose liquid gel at a 1:3 ratio (final concentrations of agarose, 0.5%, 1%, 1.5%, and 2%) and injected into the straight channel by use of a syringe pump (model 781230; KD Scientific). After solidification of the agarose at room temperature, nutrients were continuously supplied with flowing Luria-Bertani (LB) broth (BD Science Ltd.) or pseudominimal medium 7 (PMM7) (Kisan Biotech Ltd., South Korea) through the V-shaped channel, at a flow rate of 10 μl/h, for 5 h with a syringe pump. The entire MAC system was incubated at 37°C. As the ECASs formed in the microfluidic channel, as illustrated in Fig. 1, they were monitored by bright-field microscopy.
EPS staining.
The ECASs were allowed to form for 5 h in the MAC system. For SYTO 9 and propidium iodide (PI) staining (LIVE/DEAD BacLight bacterial viability kit L7012; Molecular Probes, Inc.) and for FilmTracer calcein (Invitrogen) staining, the staining solutions were supplied with LB broth through a V-shaped channel for 30 min; the ECASs were then observed by fluorescence microscopy. The concentrations of the staining dyes were prepared as recommended in the suppliers' manuals. For concanavalin A (ConA) (conjugated with Alexa Fluor 350; Invitrogen) staining, the tiny agarose block containing ECASs was carefully removed from the MAC chip and placed on a glass slide, which was stained overnight in ConA solution (200 μg/ml) and then rinsed with phosphate-buffered saline (PBS). The stained ECASs were then observed using fluorescence microscopy. For Congo red staining, the agarose block was carefully removed and stained on a glass slide for 20 min in a solution containing Congo red (40 mg/ml) and 10% Tween 80 (3:1). After rinsing of the slides with water, the samples were observed using bright-field microscopy.
Microscopic analysis.
ECASs were monitored using an S Plan Fluor ELWD 40× lens (numerical aperture [NA] = 0.6; Nikon Instruments, Tokyo, Japan) of an inverted optical microscope (Eclipse Ti-Nikon IX71; Olympus), and micrographs were obtained by employing an electron-multiplying charge-coupled device (CCD) camera (QuantEM:512SC; Photometrics for Eclipse Ti) and a true-color CCD camera (DP71 for IX71). ConA staining was examined by epifluorescence with a Nikon inverted microscope equipped with a filter set for UV radiation (for UV-1A, DM400; for excitation [Ex] at 365/10 nm, BA 400). SYTO 9, FilmTracer, and green fluorescent protein (GFP) staining was observed under a filter set for blue light (for fluorescein isothiocyanate [FITC], DM 505; for Ex at 465 to 495 nm, BA 515 to 555). For PI staining, a filter set for green light (for tetramethyl rhodamine isocyanate [TRITC], DM 565; for Ex at 540/25 nm, BA 605/55) was used.
Image analysis.
For comparison of the sizes of ECASs, ImageJ (V. 1.48) was used (16). After adjusting the threshold values, the ECASs were detected by binarization of the raw images. The areas of the ECASs were calculated and plotted accordingly.
SEM.
For scanning electron microscopy (SEM) observation of the biofilm, the tiny agarose block containing the ECASs was carefully removed from the MAC chip and placed on a glass slide. For ECAS fixation, the agarose block was exposed to osmium tetroxide vapor (2% in H2O; Electron Microscopy Sciences) for 9 h in a petri dish. The fixed agarose block was observed under an SEM (Hitachi S-48000; acceleration voltage, 0.5 kV; current, 10 μA).
RESULTS
ECAS formation in the MAC system.
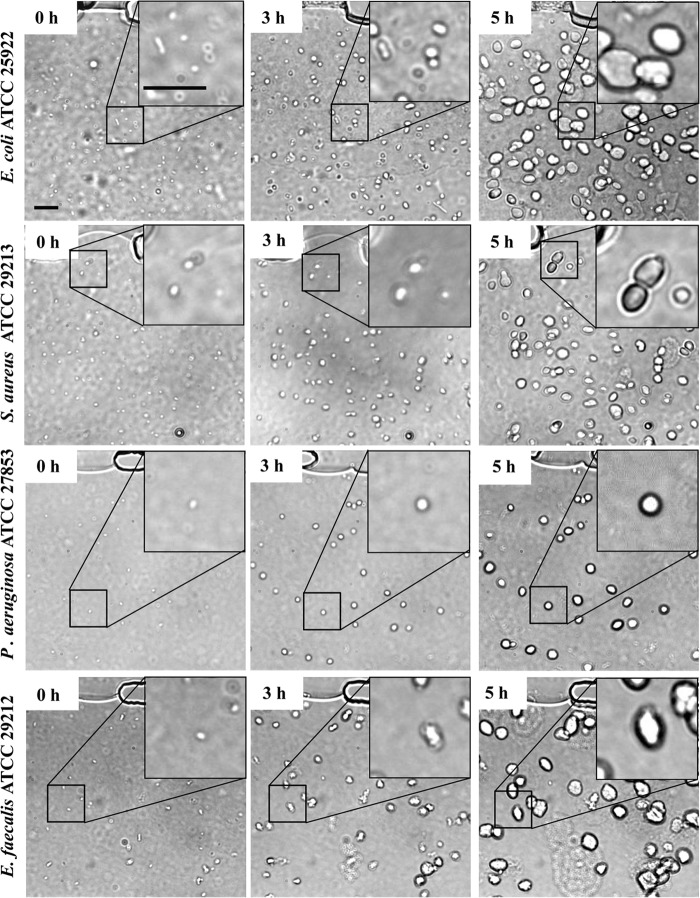
When injected into the MAC system and incubated at 37°C, all bacteria tested in this study, such as E. coli, E. faecalis, S. aureus, and P. aeruginosa, formed spherical ECASs within a few hours (Fig. 2). When the growth of the ECASs was further monitored by time-lapse microscopy, the ECASs grew larger and matured into a large spherical shape, and they eventually burst to release individual cells (see Fig. S2 and Movies S1 and S2 in the supplemental material). This growth pattern resembles the developmental cycle of canonical biofilms. Although this embedded-type biofilm structure, derived from the embedded growth of bacteria in a gelling agent, has not previously been suggested to be a biofilm, the appearance was similar to that of clinical biofilms found in mucosal tissues or inside host cells (11, 12). Additionally, while the mature ECASs were derived from the aggregates of cells, they were apparently distinct from the simple aggregation of individual cells or premature ECASs in microscopic images. While the individual cells retained their cellular shapes and Brownian movement even in the aggregation, the ECASs lacked cell shape or movement (Fig. 2, 3A and B, 4, and 5; see Movie S3). Both individual cells and ECASs were often detected together in the MAC system, providing a clear contrast (Fig. 3A and B; see Movie S3). The ECASs generated by a GFP-expressing P. aeruginosa strain showed a much stronger GFP signal than that of the simple aggregation of individual cells in the MAC system (see Fig. S3), demonstrating that the bacterial cells in the biofilms of the MAC system were highly compact and also alive. We also confirmed ECAS formation with other bacterial strains, including the well-known lab strain E. coli DH5α (data not shown) and Bacillus subtilis ATCC 6633 (see Fig. S4A, B, and C).
FIG 2.
ECAS formation by various bacteria in a microfluidic channel. Four bacterial strains, namely, P. aeruginosa ATCC 27853, E. coli ATCC 25922, E. faecalis ATCC 29212, and S. aureus ATCC 29213, were applied to the MAC system for biofilm formation. The subcultured bacterial cells were mixed with agarose liquid gel (final concentration, 1.5%) and injected into a straight channel by use of a syringe injector. After solidification of the agarose at room temperature, nutrients were supplied by flowing LB broth through the V-shaped channel at a flow rate of 10 μl/h for 5 h. The entire system was incubated at 37°C. ECAS formation in the microfluidic channel was monitored by bright-field microscopy for 5 h. The large, white, pebble-like objects are ECASs. Some images of the biofilms were magnified for clearer visualization. Bars, 50 μm.
FIG 3.
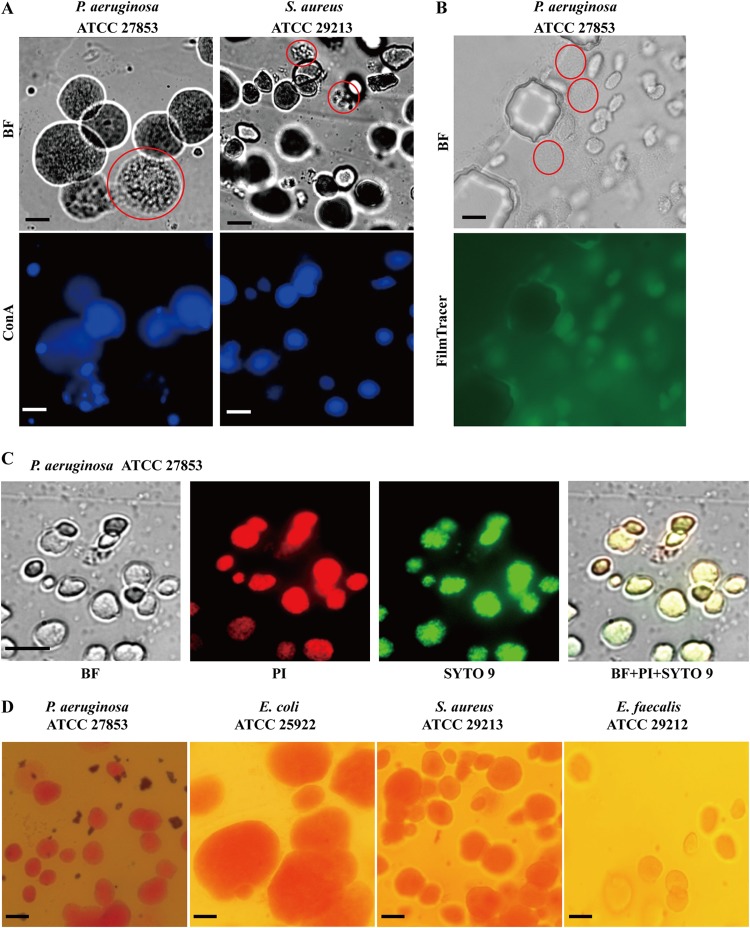
EPS production in ECASs. (A) ConA staining. The agarose patches containing the ECASs were carefully separated from the MAC chip and directly observed by use of a bright-field (BF) microscope (top row) or stained overnight in ConA solution. After rinsing in PBS, the samples were observed using a fluorescence microscope (bottom row). The bright-field image of P. aeruginosa was also recorded in a movie file, which is provided in Movie S3 in the supplemental material. (B and C) FilmTracer calcein (B) and SYTO 9 and PI (C) staining. The dyes were supplied through a V-shaped channel for 30 min, and ECASs and individual cells were observed with bright-field and fluorescence microscopes. Premature ECASs or simple aggregations of cells, which are poorly stained, are indicated by dotted red circles for comparison with mature ECASs. (D) Congo red staining. The agarose block was removed from the MAC system, stained with a Congo red solution, and observed with a microscope. Microscopes with 40× objective lenses and bright-field (BF), green (FilmTracer and SYTO 9), red (PI), and blue (ConA) filters were used in this study. The exposure time was 400 ms for ConA, 300 ms for SYTO 9 and PI, and 3 s for FilmTracer. Bars, 50 μm.
FIG 4.
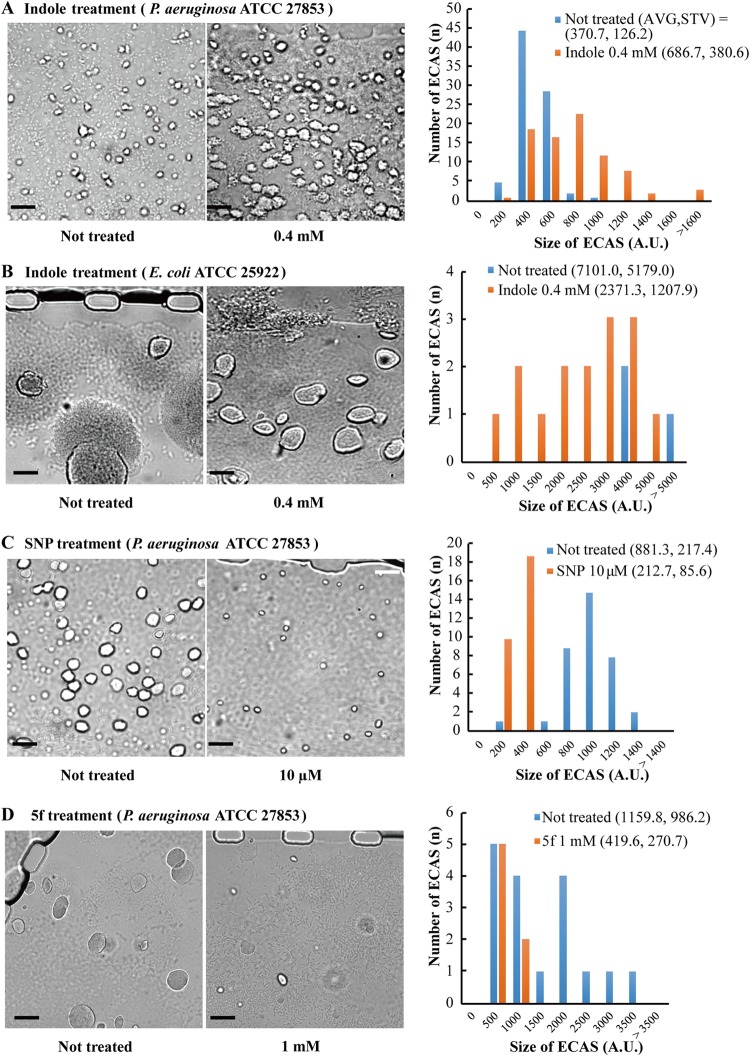
Effects of indole, SNP, and a quorum sensing inhibitor (5f) on ECAS formation. Bacterial cells were incubated for ∼5 h in the MAC system, and during ECAS formation, the chemicals were continually provided with LB medium through the side V-shaped channel. ECAS formation was monitored by bright-field microscopy. (A) P. aeruginosa ATCC 27853 was treated with 0.4 mM indole and compared with the untreated strain. (B) With E. coli ATCC 25922, the cells were incubated longer, until ECAS rupture. Without indole treatment, most ECASs ruptured. (C and D) P. aeruginosa ATCC 27853 was treated with 10 μM SNP (C) and 1 mM 5f (D) and compared with the untreated strain. The images were processed to measure the sizes of the biofilms by using ImageJ software (V. 1.48). The aggregative structures in each strain were changed to a binary code, and the pixel numbers were measured to represent the sizes of biofilms (see Fig. S7 in the supplemental material). The averages (AVG) and standard deviations (STV) of the biofilm sizes were calculated for comparison among different conditions. A.U., arbitrary units. Bars, 50 μm.
FIG 5.
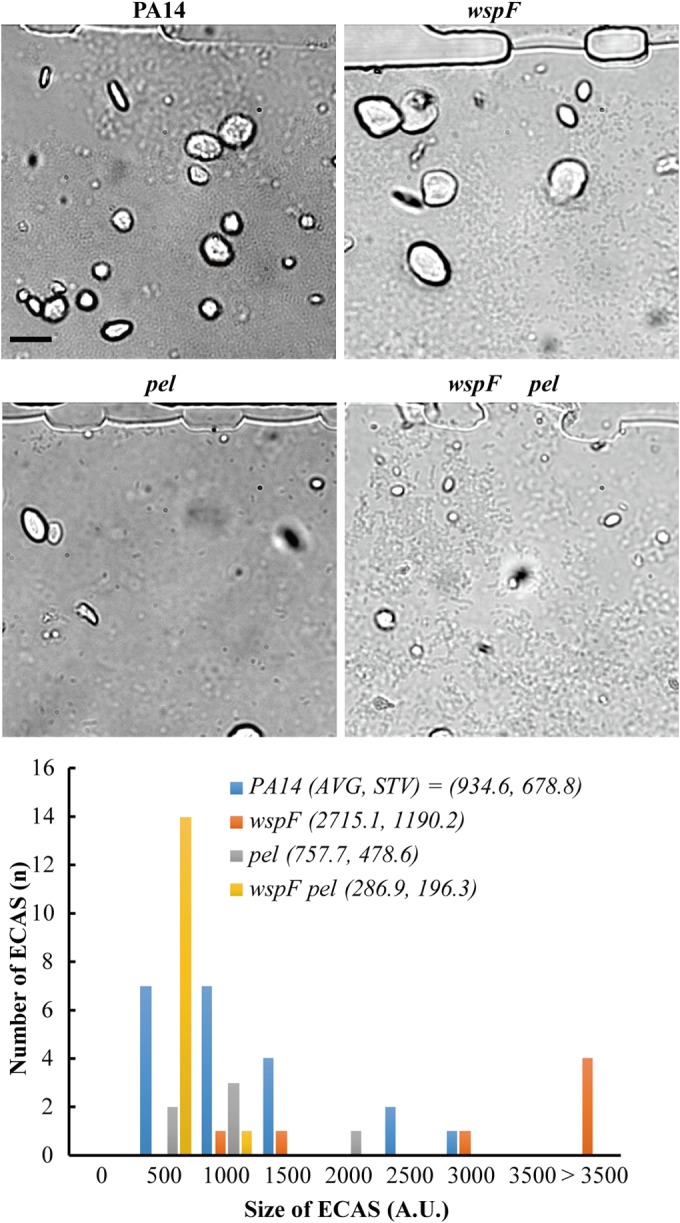
ECAS formation of biofilm mutants. The strains were incubated for ∼5 h, and ECAS growth was observed under a bright-field microscope. The images were processed to measure the sizes of the biofilms by using ImageJ software (V. 1.48). The aggregative structures in each strain were changed to a binary code, and the pixel numbers were measured to represent the sizes of the biofilms (see Fig. S8 in the supplemental material). The average (AVG) and standard deviation (STV) values for the biofilms from the biofilm images of the strains (P14, wspF mutant, pel mutant, and wspF pel mutant) were compared with each other. Bars, 50 μm.
The maturation and integrity of the ECASs were affected by the hardness of the agarose matrix. In 0.5% agarose, ECAS formation in the MAC system seemed to be delayed and less rigid than ECAS formation in 1% and 2% agarose, implying that fixing stress may influence ECAS formation (see Fig. S5 in the supplemental material). ECAS formation was also affected by nutrients; formation was delayed in minimum medium and was faster in complex medium (see Fig. S6). The results imply that ECAS formation may be influenced by environmental conditions, such as fixing stress, viscosity, and nutrition.
ECASs are a type of biofilm containing EPS.
Biofilms have some peculiar features. The most important feature of a biofilm is the production of an extracellular polymeric substance (EPS) enclosing the bacterial cells. To verify that ECASs are biofilms, the production of EPS was investigated by staining. Several different EPS staining agents were used, including ConA (to detect extracellular polysaccharides), Congo red (to detect cellulose), SYTO 9 (to detect nucleic acids), propidium iodide (to detect nucleic acids in dead cells), and FilmTracer (to detect both the cellular and matrix esterase activities within biofilms). While ConA, which binds to α-mannopyranosyl and α-glucopyranosyl residues of polysaccharides, stained the ECASs well, simple aggregations of individual cells or premature ECASs were poorly stained, demonstrating that mature ECASs contain a significant level of EPS (Fig. 3A; see Fig. S4A in the supplemental material). FilmTracer also specifically stained ECASs (Fig. 3B; see Fig. S4B). As for ConA, FilmTracer poorly stained individual cells even when they were densely aggregated (Fig. 3B; see Fig. S4B). The staining dyes for nucleic acids, i.e., SYTO 9 and PI, stained the ECASs at the same locations (Fig. 3C; see Fig. S4C). PI is known to stain dead cells, because the dye cannot pass through intact cell membranes. However, because extracellular DNA is also an important EPS component, PI can stain biofilms. Many bacteria, including P. aeruginosa, S. aureus, and E. faecalis, generate extracellular DNA by the lysis of subpopulations (17, 18). Congo red, a dye that is often used for biofilm staining due to its strong affinity for polysaccharides, such as cellulose (19), also stained the ECASs well (Fig. 3D). All of these data indicated that the ECASs contained abundant EPS and that cells were enclosed in this self-producing EPS.
ECASs reflect the previously reported features of biofilms.
Several agents have been studied for their effects on bacterial biofilm formation. Indole has been reported to repress the biofilm formation of E. coli but to enhance the biofilm formation of P. aeruginosa (20). When we applied indole to our MAC system, the ECAS formation of P. aeruginosa was significantly enhanced, whereas the ECAS formation of E. coli was reduced (Fig. 4A and B; see Fig. S7A and B in the supplemental material). With E. coli, cells were incubated longer, until most ECASs were ruptured without indole treatment. However, indole slowed the development of ECASs, leaving most ECASs intact (Fig. 4B). Another biofilm-modulating agent, sodium nitroprusside (SNP), which is a nitric oxide generator, significantly reduced ECAS formation (Fig. 4C; see Fig. S7C). Nitric oxide has been reported to repress biofilm formation by inducing biofilm dispersion (21, 22). A previously reported antibiofilm agent, 5f (23), also inhibited ECAS formation in the MAC system (Fig. 4D; see Fig. S7D). These results demonstrate that ECASs are a type of biofilm.
In P. aeruginosa, the wspF mutant is known to overproduce biofilm, and the pel mutant is known to form less biofilm when generated using conventional systems (24, 25). When these mutants and their parental wild-type strain, PA14, were applied to the MAC system and their growth was monitored, the wspF mutant formed ECASs more vigorously, but the pel mutant did not (Fig. 5; see Fig. S8 in the supplemental material). The effect of the wspF mutation was not observed in the presence of the pel mutation (Fig. 5). These results obtained using the MAC system are consistent with previous results obtained using conventional systems.
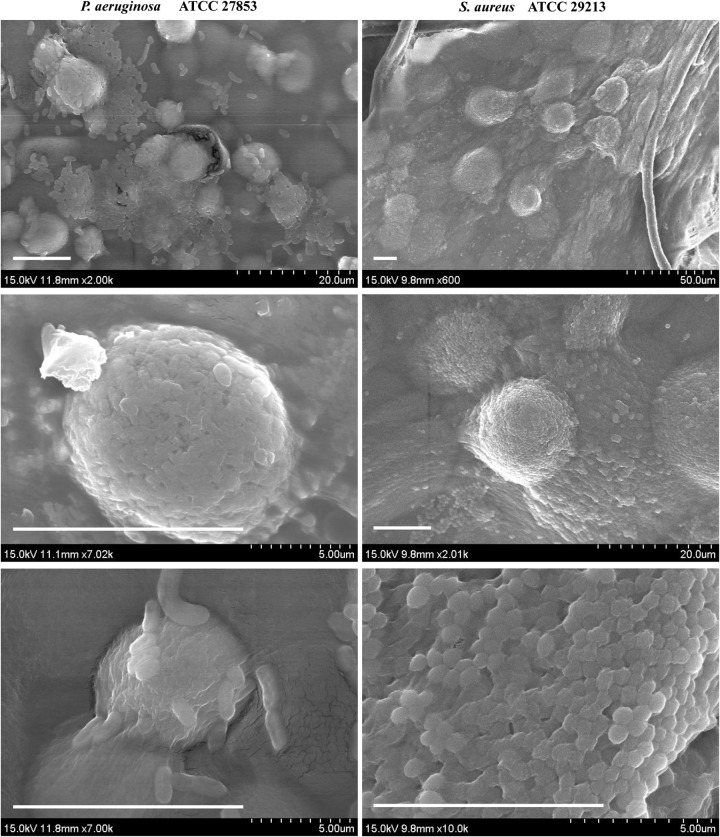
We investigated direct images of the ECASs by SEM. The SEM images show that ECASs exhibit a biofilm-like structure in which cells are densely aggregated, showing a slimy appearance that is distinct from that of individual cells (Fig. 6). These images also demonstrate that ECASs have morphological characteristics consistent with a biofilm. Considering the combined results, because the ECASs formed in the MAC system show most of the previously reported features of a biofilm, we define an ECAS as an embedded-type biofilm and suggest that this type of biofilm is derived from the embedded growth of bacteria within a matrix.
FIG 6.
SEM images of ECASs. Agarose patches containing ECASs were segregated from the channel. The agarose block was fixed in osmium tetroxide vapor for 9 h before SEM observation. Bars, 10 μm.
DISCUSSION
The MAC system was originally designed for rapid antibiotic susceptibility testing (26). In this system, bacteria were immobilized using agarose in a microfluidic culture chamber in which different concentrations of antibiotics were applied, and single-cell growth was tracked by microscopy. This system reduced the time for the antimicrobial susceptibility test to 3 to 4 h. Interestingly, there were two types of bacterial growth in the MAC system: a dividing type and an aggregative type (26). We characterized the aggregative type as a new type of biofilm.
Biofilms are often considered “an aggregate of microbial cells adherent to a surface, enclosed in EPS they produce.” However, it does not seem necessary to include “adherence to a surface” in the biofilm definition. Flemming and Wingender included other cell aggregate types as biofilms, such as floating biofilm and sludge, which are not attached to a surface but show the characteristics of a biofilm (27). The aggregated forms of floating S. aureus and P. aeruginosa cells exhibit higher tolerances to antibiotics (28, 29). In the MAC system, bacterial cells are not attached to a surface but are fixed within an agarose matrix and form aggregates which exhibit most biofilm characteristics. Bacterial cells in the MAC system initially grew and were embedded in the matrix, produced EPS that formed ECASs, and then dispersed.
The ECASs showed previously reported features of biofilms, such as indole-mediated biofilm enhancement with P. aeruginosa, biofilm repression by antibiofilm agents, and some mutational effects. Therefore, we suggest that ECASs in an agarose matrix are a new type of biofilm, defined as an embedded-type biofilm. The advantage of this embedded-type biofilm as a new biofilm model is that it mimics infection-related clinical biofilms formed in host mucosal tissues and inside host cells, as clinical biofilms do not attach to hard surfaces in many cases but are embedded in a matrix of viscous mucus or in the cytoplasm of host cells, like the case for the ECASs in the MAC system. This type of system is clearly distinct from conventional flow cell systems based on surface-attached biofilms. The flow cell system is a more appropriate model system for studying environmental or industrial biofilms formed on hard surfaces with shear stress under liquid flow, such as water-supplying pipes, membranes of water-purifying systems, industrial reactors, and medical implants. In the chronic lung infection associated with cystic fibrosis (CF), the aggregates of P. aeruginosa are not attached to epithelial surfaces; instead, they are located within the thickened viscous mucosal layer associated with airways (30, 31). Bjarnsholt et al. (31) observed a compact biofilm that formed microcolonies in chronic P. aeruginosa-infected CF patient sputum. In urinary tract infections, E. coli forms biofilms, shown as pod-like bulges in the bladder lumen, and bacteria in pods are embedded in a matrix in the host cell cytoplasm (12). Anderson et al. (12) discovered that the pods contained bacterial cells encased in a polysaccharide-rich matrix surrounded by a protective shell of uroplakin. Pod-shaped biofilms resemble the gel microcolony biofilms from the MAC system. There are other biofilms on soil matrices or plant surfaces where bacterial cells aggregate by producing EPS to form a biofilm (13–15). However, the type of biofilm that forms on a surface might have rare shear stress, and its morphology may be similar to that of the embedded type of biofilm in the MAC system. Therefore, the MAC system, using embedded growth in a gelling agent such as agarose, may be a more suitable model system for studying clinical biofilms, providing researchers with an alternative choice of biofilm.
The MAC system provides a much faster, easier, and cheaper way to study biofilms. Conventional flow cell systems take a few days to 1 week to observe the full developmental cycle of biofilms. In the MAC system, biofilms form a few hours (3 to 4 h) after the inoculation of a bacterial cell with agarose (Fig. 2), and the biofilms mature in 5 to 7 h. The full developmental cycle, including dispersion, mostly terminates in 9 to 12 h (see Fig. S2 in the supplemental material). Moreover, conventional biofilm model systems require large and expensive equipment, including complex tube and pump connections, bulky media for long incubation times, and pricy fluorescence microscopy. The MAC system requires small microfluidic channels (using a slide glass-sized chip that includes multiple channels), simple tiny tubing, and several milliliters of medium. Biofilm formation can be observed under a normal microscope without additional accessories, and the MAC system makes it easy to observe single cells because they are immobilized in a gel.
Notably, biofilm formation in the MAC system depended on the concentration of agarose. Cell-fixing stress in the agarose matrix might be a significant factor for biofilm formation and therefore also influence clinical biofilm formation. In flow cell systems, the flow rate of fluid generating a shear force influences biofilm formation, EPS production, and the induction of biofilm deformation and detachment (32, 33). These are other important features discriminating the MAC system from conventional systems.
The MAC system can also provide some clues about single-cell behavior during biofilm formation. In the 1 to 1.5% agarose matrix of the MAC system, there were two cell types simultaneously: individual and ECAS-included cells. This observation reveals that responses to the same fixing stress are different among the members of the bacterial population, although the cells originated from the same inoculum and were cultured under the same conditions. It will be interesting to identify the factors that predestine each cell to form a biofilm or remain as an individual cell. The MAC system can help researchers to perform single-cell-level studies of biofilm formation and inhibition.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (grants HI13C1468 and HI13C0866); the Pioneer Research Center Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (grant NRF-2012-0009555); an NRF grant funded by the Korean Government (grant 2012M3A7A9671610); and Basic Science Research Programs through the NRF, funded by the Ministry of Education (formerly the Ministry of Education, Science and Technology) (grants 2010-0006622 and 2013R1A1A2012220).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02311-14.
REFERENCES
- 1.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Monroe D. 2007. Looking for chinks in the armor of bacterial biofilms. PLoS Biol 5:e307. doi: 10.1371/journal.pbio.0050307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 5.McBain AJ. 2009. Chapter 4: in vitro biofilm models: an overview. Adv Appl Microbiol 69:99–132. doi: 10.1016/S0065-2164(09)69004-3. [DOI] [PubMed] [Google Scholar]
- 6.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 7.Ali L, Khambaty F, Diachenko G. 2006. Investigating the suitability of the Calgary biofilm device for assessing the antimicrobial efficacy of new agents. Bioresour Technol 97:1887–1893. doi: 10.1016/j.biortech.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Goeres DM, Loetterle LR, Hamilton MA, Murga R, Kirby DW, Donlan RM. 2005. Statistical assessment of a laboratory method for growing biofilms. Microbiology 151:757–762. doi: 10.1099/mic.0.27709-0. [DOI] [PubMed] [Google Scholar]
- 9.Crabbe A, De Boever P, Van Houdt R, Moors H, Mergeay M, Cornelis P. 2008. Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01. Environ Microbiol 10:2098–2110. doi: 10.1111/j.1462-2920.2008.01631.x. [DOI] [PubMed] [Google Scholar]
- 10.Palmer RJ., Jr 1999. Microscopy flowcells: perfusion chambers for real-time study of biofilms. Methods Enzymol 310:160–166. doi: 10.1016/S0076-6879(99)10014-4. [DOI] [PubMed] [Google Scholar]
- 11.Hall-Stoodley L, Stoodley P, Kathju S, Høiby N, Moser C, William Costerton J, Moter A, Bjarnsholt T. 2012. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol Med Microbiol 65:127–145. doi: 10.1111/j.1574-695X.2012.00968.x. [DOI] [PubMed] [Google Scholar]
- 12.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 13.Monier J-M, Lindow S. 2005. Aggregates of resident bacteria facilitate survival of immigrant bacteria on leaf surfaces. Microb Ecol 49:343–352. doi: 10.1007/s00248-004-0007-9. [DOI] [PubMed] [Google Scholar]
- 14.Chang W-S, Halverson LJ. 2003. Reduced water availability influences the dynamics, development, and ultrastructural properties of Pseudomonas putida biofilms. J Bacteriol 185:6199–6204. doi: 10.1128/JB.185.20.6199-6204.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danhorn T, Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 16.Rasband W. 1997. ImageJ. US National Institutes of Health, Bethesda, MD. [Google Scholar]
- 17.Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011. Extracellular DNA in biofilms. Int J Artif Organs 34:824–831. doi: 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 18.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 19.McKinney RE. 1953. Staining bacterial polysaccharides. J Bacteriol 66:453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Jayaraman A, Wood T. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C, Kim J, Park H-Y, Park H-J, Lee J, Kim C, Yoon J. 2008. Furanone derivatives as quorum-sensing antagonists of Pseudomonas aeruginosa. Appl Microbiol Biotechnol 80:37–47. doi: 10.1007/s00253-008-1474-6. [DOI] [PubMed] [Google Scholar]
- 24.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung IY, Choi KB, Heo YJ, Cho YH. 2008. Effect of PEL exopolysaccharide on the wspF mutant phenotypes in Pseudomonas aeruginosa PA14. J Microbiol Biotechnol 18:1227–1234. [PubMed] [Google Scholar]
- 26.Choi J, Jung YG, Kim J, Kim S, Jung Y, Na H, Kwon S. 2013. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 13:280–287. doi: 10.1039/c2lc41055a. [DOI] [PubMed] [Google Scholar]
- 27.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 28.Fux CA, Wilson S, Stoodley P. 2004. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol 186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, Jensen PO, Nielsen AK, Parsek M, Wozniak D, Molin S, Tolker-Nielsen T, Hoiby N, Givskov M, Bjarnsholt T. 2011. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One 6:e27943. doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol 44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 32.Stoodley P, Cargo R, Rupp CJ, Wilson S, Klapper I. 2002. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J Ind Microbiol Biotechnol 29:361–367. doi: 10.1038/sj.jim.7000282. [DOI] [PubMed] [Google Scholar]
- 33.Menniti A, Kang S, Elimelech M, Morgenroth E. 2009. Influence of shear on the production of extracellular polymeric substances in membrane bioreactors. Water Res 43:4305–4315. doi: 10.1016/j.watres.2009.06.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.