Abstract
The diversity and genetic differentiation of populations of Fusarium oxysporum associated with tomato fields, both endophytes obtained from tomato plants and isolates obtained from soil surrounding the sampled plants, were investigated. A total of 609 isolates of F. oxysporum were obtained, 295 isolates from a total of 32 asymptomatic tomato plants in two fields and 314 isolates from eight soil cores sampled from the area surrounding the plants. Included in this total were 112 isolates from the stems of all 32 plants, a niche that has not been previously included in F. oxysporum population genetics studies. Isolates were characterized using the DNA sequence of the translation elongation factor 1α gene. A diverse population of 26 sequence types was found, although two sequence types represented nearly two-thirds of the isolates studied. The sequence types were placed in different phylogenetic clades within F. oxysporum, and endophytic isolates were not monophyletic. Multiple sequence types were found in all plants, with an average of 4.2 per plant. The population compositions differed between the two fields but not between soil samples within each field. A certain degree of differentiation was observed between populations associated with different tomato cultivars, suggesting that the host genotype may affect the composition of plant-associated F. oxysporum populations. No clear patterns of genetic differentiation were observed between endophyte populations and soil populations, suggesting a lack of specialization of endophytic isolates.
INTRODUCTION
Fusarium oxysporum is a well-known phytopathogenic fungus that affects hundreds of crops worldwide. Most studies of the fungus focus on the ability of F. oxysporum to cause vascular wilt on a particular host. However, environmental surveys of a wide range of habitats frequently find F. oxysporum in the absence of plant disease (1). These F. oxysporum populations, often referred to as nonpathogenic, although lack of pathogenicity is usually not confirmed, are a cosmopolitan component of soil communities. F. oxysporum can also infect and colonize plants asymptomatically as an endophyte. This endophytic lifestyle has not been well researched, despite reports of endophytic colonization by F. oxysporum on nearly 100 plant species (2).
The interaction of endophytic F. oxysporum and plants may give insight into how pathogen recognition and plant resistance operate. In susceptible plants, pathogenic F. oxysporum penetrates the root, colonizes the root cortex, and then spreads through the xylem to the rest of the plant, causing occlusion of the xylem and ultimately wilting and death (3, 4). In host cultivars that are resistant to the pathogen, the spread of the fungus is blocked, and only limited colonization of host tissue occurs (5, 6, 7). Nonpathogenic F. oxysporum inoculated onto plants can colonize the root cortex similarly to pathogens, but host responses, such as thickening of cell walls and protuberances into intercellular spaces, seem to limit the growth of endophytes, similar to defenses limiting the growth of pathogens in resistant plants (3, 4, 8). Nonpathogenic F. oxysporum can also have a differential ability to colonize and multiply in the root cortex of different plant species. For example, research done by Leoni et al. (9) showed how F. oxysporum f. sp. cepae (which causes Fusarium basal rot of onions) can reach high population levels in the roots of plants, such as black bean or Sudan grass, in the absence of disease. Although the above observations suggest that endophytic F. oxysporum is not systemic, F. oxysporum endophytes can be recovered from naturally colonized plant stems (10). However, most population genetics studies have focused on nonpathogenic F. oxysporum isolated from roots (11, 12, 13, 14, 15).
Despite the cosmopolitan nature of F. oxysporum and its ecological diversity, the structure and diversity of soil and endophytic F. oxysporum populations are not well understood. A recent typing scheme proposed by O'Donnell et al. (16) used DNA sequences of the ribosomal DNA intergenic spacer region (IGS) and the translation elongation factor 1α (TEF) gene to define sequence types (STs) and found 256 sequence types among 850 isolates, mainly plant pathogens. Subsequent studies using the same typing scheme found 26 new sequence types among isolates obtained from soil in Sardinia (17) and 46 new sequence types from noncultivated soils across Australia (18). These novel sequence types suggest that focusing only on plant-pathogenic F. oxysporum may underestimate the total species diversity. Other studies indicate that F. oxysporum populations from soil are geographically heterogeneous and greatly influenced by resident plant populations (19, 20, 21), suggesting that some plants may exert a selective effect on soil populations of F. oxysporum (22). Studies comparing pathogenic and nonpathogenic F. oxysporum populations associated with the same host have found much higher levels of diversity in the nonpathogenic populations than in the pathogens (11, 14, 23, 24).
An additional unaddressed topic is the diversity of F. oxysporum within an individual plant. Endophytic F. oxysporum is commonly isolated from plants infected with pathogenic F. oxysporum (for examples, see references 23 and 25), suggesting that colonization of plants by multiple genotypes of F. oxysporum may be common. Also, different genotypes may interact with their host plants in different ways; for example, studies focusing on developing nonpathogenic F. oxysporum fungi as biocontrol agents have found particular isolates to be better biocontrol agents than others (26, 27, 28). A few studies have reported overrepresentation of a few genotypes within endophytic populations (12, 22), suggesting a possible selective effect by the host on soil populations.
This study investigated the population structure of F. oxysporum associated with asymptomatic tomato plants grown under field conditions. We hypothesized that endophyte populations would be genetically differentiated from soil populations. Additionally, if only specific genotypes of F. oxysporum could colonize plants endophytically, we expected that endophyte populations would be less diverse than soil populations. The objective of the study was to compare the compositions and diversities of F. oxysporum populations to look for evidence of specialization of F. oxysporum to different environmental niches, such as soil or plants. A further goal was to assess the diversity of F. oxysporum fungi found within individual plants and associated with different plant parts, namely, roots, crowns, and stems.
MATERIALS AND METHODS
Sampling and fungal isolation.
F. oxysporum populations were sampled from two tomato fields located in the Russell E. Larson Research Center, The Pennsylvania State University, Pennsylvania Furnace, PA. These are experimental fields used for agricultural research and are located approximately 1.5 km from each other; they were chosen for sampling of F. oxysporum populations because they had no known history of Fusarium wilt of tomato. Within each field, four plots were randomly chosen for sampling of tomato plants and soil. Each plot consisted of four plants in a row with 1-foot spacing. Within a plot, six 4-inch soil cores were collected, two cores from between each pair of plants, and bulked together to make a composite soil sample per plot, for a total of 16 plant samples and 4 soil samples per field. Plants in field 1 were all of the processing tomato cultivar Heinz 9907. In field 2, each set of four plants consisted of a different cultivar, the processing tomato cultivars Gem 611 and Heinz 3402 and the fresh market cultivars FL 47 and Mountain Fresh. These cultivars are all resistant to F. oxysporum f. sp. lycopersici races 1 and 2. No fungicides had been applied to the plants. Plants in field 1 were sampled at the green fruit stage, and plants in field 2 were sampled after the fruits were mature.
Sampling from plants was done by carefully digging up the plants, washing off the soil under tap water, and surface disinfesting entire plants with a solution of 2% commercial bleach (6.15% NaOCl) for 1.5 min. Small (approximately 5-mm-long) pieces of the root and stem were placed on Nash-Snyder Fusarium-selective growth medium for fungal isolation (29). Soil samples were air dried for at least 10 days and sieved to remove large particles. Five grams of soil was suspended in 50 ml sterile water by mixing for 20 min, and then 10-fold serial dilutions of 10−1 to 10−4 were plated on Nash-Snyder medium (24). A maximum of 10 isolates were collected per plant: 4 from the roots and stem, respectively, and 2 from the crown, for a maximum total of 40 isolates for each set of four plants (one plot), and a maximum of 40 isolates were collected per composite soil sample (one plot), selected randomly from dilution plates. F. oxysporum isolates were identified based on colony morphology, and monoconidial cultures were grown on potato dextrose agar. Isolates were stored long-term at 4°C in 1.5-ml microcentrifuge tubes in sterile sand mixed with 250 μl of potato dextrose broth.
DNA extraction and sequencing.
DNA was extracted following the protocol of Cenis (30). Briefly, mycelium was grown in potato dextrose broth in 1.5-ml microcentrifuge tubes for at least 3 days at room temperature, centrifuged to remove the medium, and ground in an SDS-based extraction buffer. Cellular debris was precipitated with sodium acetate, and finally, DNA was precipitated with isopropanol. The TEF gene, a region commonly used for Fusarium identification, was amplified using the primers EF1 and EF2 under conditions described previously (16). Amplification was checked on agarose gels stained with ethidium bromide, and the PCR products were cleaned with Exo-Sap (Affymetrix, Santa Clara, CA) before sequencing. Sequencing was done using the primers EF1 and EF2 at the Pennsylvania State University Genomics Core Facility (University Park, PA).
Data analysis.
Sequences were compared to reference sequences in GenBank and the Fusarium-ID database (31) to confirm the isolates' identities as F. oxysporum and to identify if the sequences were present in the Fusarium-ID database or if they represented new sequence types. Isolates that were not F. oxysporum (5% of the sequenced samples) were excluded from further analysis. Sequence types were defined and randomly numbered for each allele, including gaps as informative characters. Population size was estimated using the nonparametric estimator Chao 2, calculated in EstimateS 8.2.0 (R. K. Colwell, 2006 [http://www.purl.oclc.org/estimates]). Population genetic diversity was measured with Shannon's diversity index (SHA) based on the frequency of sequence types, using the program GENALEX 6 (32). Diversities were compared between subpopulations based on Shannon's mutual-information index (SHUA), and significance was assessed using the log-likelihood test statistic, G (33). Genetic differentiation among subpopulations was measured using two analyses: the nearest-neighbor statistic (Snn) (34), which is based on the nucleotide sequence and determines how often isolates with the most similar DNA sequences (the nearest neighbors) are found in the same population, and the haplotype statistic (HST), which is based on allele frequency, specifically the following equation: HST = 1 – (average haplotype diversity in the subpopulations/average haplotype diversity in the total population) (35). Analyses were performed using DnaSP 5.10 (36), and statistical significance was assessed with 1,000 permutations using Monte Carlo simulations (35). Gaps were considered a fifth character for HST analysis but not for Snn. Sequences from different locations (i.e., plants or soil from each of the eight plots) were compared using principal-coordinate analysis to look for clustering of sequences from particular environments. The principal-coordinate analysis was performed using UniFrac (37), with sequences weighted for abundance and nonnormalized. A phylogenetic network of the sequence types was estimated with statistical parsimony analysis and performed using TCS v. 1.21 (38). Maximum-parsimony analysis was inferred from the DNA sequences using the program PAUP* v.4.0b10 (D. L. Swofford, 2002; Sinauer Associates, Sunderland, MA). Parsimony analysis was performed using the heuristic-search option with the MulTrees option on, 1,000 random addition sequences, the tree-bisection-reconnection branch-swapping algorithm, and gaps not considered characters. Support for each branch was assessed with 1,000 bootstrap replicates. Included in the phylogenetic analysis were sequences representative of the known diversity in F. oxysporum based on previous phylogenetic analyses (16, 39, 40) downloaded from GenBank or the Fusarium-ID database (31).
Nucleotide sequence accession numbers.
The sequences identified in this work have been deposited in GenBank under accession numbers KJ920404 to KJ920429.
RESULTS
Fungal isolation.
F. oxysporum was successfully isolated from all eight soil samples and all 32 plants from both fields, with a total of 609 isolates collected and analyzed: 314 isolates from soil and 295 from plants. A total of 16 populations were analyzed, 8 from each field, with four plots per field. From each plot, we established one population from plants and one from the corresponding soil. For this research, we used a very broad definition of population as a group of individuals of the same species occupying a particular space or niche (in our case, a plant or a soil) at a particular time (41). Isolates from plants were considered to be endophytes, because the plants were asymptomatic and surface disinfested. A maximum of 10 isolates were collected for each plant, and the majority of the plants yielded 9 or 10 isolates. The plant that yielded the fewest isolates (six) was heavily colonized by Geotrichum spp. Soil samples contained approximately 1,000 CFU of F. oxysporum per gram soil, based on estimations from soil dilution plating.
Within each plant, F. oxysporum was isolated from the roots, crown, and stem, although some plants yielded only one isolate per plant part. F. oxysporum was isolated from the top third of the stem (approximately 60 cm above the soil) for 19 of the 32 plants, suggesting that colonization of the entire stem is not uncommon. Whether F. oxysporum could also be found in the leaves and fruit was not assessed.
F. oxysporum was the most common Fusarium sp. observed. Other Fusarium species observed but excluded from further study (5% of the sequenced samples) were identified, using the Fusarium-ID database (31), as Fusarium commune, Fusarium proliferatum, and members of the Fusarium solani species complex, the Gibberella fujikuroi species complex, and the Fusarium incarnatum-Fusarium equiseti species complex. F. commune was frequently isolated, but this was due to difficulties in distinguishing it morphologically from F. oxysporum and not to its relative abundance in the samples.
Sequence types and population diversity.
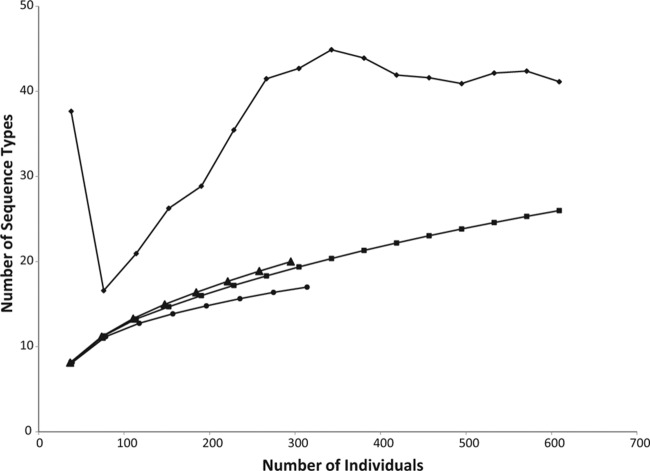
A fragment of approximately 630 bp of the TEF gene was amplified for all isolates. Fifty-seven sites were polymorphic, with 45 single-nucleotide polymorphisms and 12 insertions/deletions (indels). Including indels as informative characters, 26 different STs were observed (GenBank accession numbers KJ920404 to KJ920429). The species accumulation curves of populations from plants, soil, and plants and soil combined show that sampling was approaching but did not reach saturation (Fig. 1). The total population richness was estimated to be 41 STs based on the Chao 2 estimator, with a 95% confidence interval of 29 to 88 STs. The Chao 2 richness estimate for soil was 21 STs, compared to 17 observed STs, and it was 40 STs in plants, compared to 20 observed STs. Twenty-one of the 26 STs were found in field 1, and 18 of the 26 STs were found in field 2. Two STs, 5 and 13, were especially common and composed 35% and 30% of all isolates, respectively. These two STs were equally common in soil and in plant tissues and were found in both fields, although ST 5 was twice as common in field 2 as in field 1 and ST 13 was three times more common in field 1 than in field 2. The most abundant STs were found in both locations and in both soil and endophyte populations, but some of the rare STs, including 10 that were observed only once, were unique to field 1 (8 STs), field 2 (5 STs), plant tissues (8 STs only in roots or crowns and 1 ST only in stems), or soil (6 STs). Of the average of 9.2 isolates per plant, an average of 4.2 STs were found per plant, ranging from 2 to 7. One to 4 STs were found per stem and per root system, with an average of 2.2 STs per stem (out of an average of 3.5 isolates per stem) and 2.6 STs per root system (out of an average of 3.8 isolates per root system).
FIG 1.
Species accumulation curves showing the expected number of sequence types when sampling a given number of individuals. The curves show the observed numbers of sequence types for samples from plants and soil combined (squares), plants only (triangles), and soil only (circles). The Chao 2 nonparametric richness estimator, calculated in EstimateS 8.2.0, is also shown for samples from plants and soil combined (diamonds).
The diversities of populations from plants and from soil were compared using Shannon's diversity index (Table 1). Based on this index, endophytic populations were significantly more diverse than soil populations for field 2 and for both fields combined. The diversities were not significantly different between plant and soil populations for individual plots, except for plot 6 in field 2, in which plant populations were significantly more diverse than soil populations.
TABLE 1.
Pairwise comparisons of Shannon's diversity indices between populations from soil and plants
| Sampling location |
SHA |
SHUA | G | P valuea | |
|---|---|---|---|---|---|
| Plant population | Soil population | ||||
| All locations | 2.729 | 2.672 | 0.081 | 68.437 | <0.001 |
| Field 1 | 2.607 | 2.440 | 0.077 | 33.354 | 0.031 |
| Field 2 | 2.529 | 2.364 | 0.147 | 60.822 | <0.001 |
| Plot 1 (Heinz 9907) | 2.318 | 2.478 | 0.150 | 16.044 | 0.098 |
| Plot 2 (Heinz 9907) | 1.653 | 2.084 | 0.117 | 13.001 | 0.224 |
| Plot 3 (Heinz 9907) | 2.709 | 1.703 | 0.225 | 24.041 | 0.020 |
| Plot 4 (Heinz 9907) | 2.436 | 2.512 | 0.185 | 19.702 | 0.073 |
| Plot 5 (FL 47) | 2.542 | 2.190 | 0.180 | 18.484 | 0.047 |
| Plot 6 (Gem 611) | 2.710 | 1.619 | 0.351 | 37.426 | <0.001 |
| Plot 7 (Heinz 3402) | 1.386 | 2.052 | 0.158 | 16.405 | 0.022 |
| Plot 8 (Mountain Fresh) | 2.277 | 2.748 | 0.218 | 21.746 | 0.026 |
Values are P values of the log-likelihood test of differences between soil and endophyte populations, with statistically significant values in boldface (Bonferroni-adjusted P < 0.0045 [0.05/11]).
Sequences were compared to the Fusarium-ID database to determine if the sequence types had been found in previous samplings. Only 12 of the 26 sequence types had a 100% match to sequences in the database. The two most common sequence types, ST 5 and ST 13, have been previously found associated with many different plant species. Most of the 14 novel sequence types were rare, with only one or two isolates of each, although two of the novel sequence types, ST 4 and ST 8, were found 44 and 32 times, respectively.
Genetic differentiation among populations.
The genetic differentiation between various subpopulations was calculated based on both haplotypes (HST) and the nucleotide sequence (Snn) (Table 2). Gaps in the sequence alignment were considered in calculations based on haplotypes, but the results were similar when gaps were excluded (results not shown). Populations were compared between the two fields to determine if they were differentiated based on physical separation. Total populations were significantly different between the two fields, which were approximately 1.5 km apart (P < 0.001 for both Snn and HST), as were the soil and endophyte populations in each field (P < 0.001 for both Snn and HST). Populations from plots in field 1, all planted with the same tomato cultivar, were compared to determine if populations are also differentiated among locations several meters apart. Based on Snn, endophyte populations for plot 1 were significantly different than for plots 2 and 3, but not in any other pairwise comparison. Soil populations were not significantly differentiated among the plots based on Snn or HST. Physical separation, therefore, does not appear to be a significant cause of population differentiation on a small scale (meters) but is significant on a larger scale (kilometers).
TABLE 2.
Genetic differentiation among F. oxysporum subpopulations from different fields, plots, and substrates
| Sampling location and comparisona | Snn | P | HST | P |
|---|---|---|---|---|
| All soil isolates (314) vs. all plants isolates (295) | 0.52244b | <0.001 | 0.00292 | 0.039 |
| Field 1 (311) vs. field 2 (298) | 0.59486 | <0.001 | 0.05196 | <0.001 |
| Soil in field 1 (158) vs. soil in field 2 (156) | 0.63467 | <0.001 | 0.07442 | <0.001 |
| Plants in field 1 (153) vs. plants in field 2 (142) | 0.55456 | <0.001 | 0.03237 | <0.001 |
| Field 1 | ||||
| Soil (158) vs. plants (153) | 0.51963 | 0.011 | −0.00064 | 0.5140 |
| Soil (158) vs. roots (61) | 0.61855 | 0.016 | 0.00134 | 0.1930 |
| Soil (158) vs. stems (61) | 0.60957 | 0.1050 | −0.00169 | 0.6130 |
| Roots (61) vs. stems (61) | 0.52225 | 0.0690 | 0.00364 | 0.2000 |
| Plot 1, soil (39) vs. plants (38) | 0.53453 | 0.0820 | 0.00000 | 0.3740 |
| Plot 2, soil (40) vs. plants (40) | 0.48901 | 0.5250 | 0.00354 | 0.2630 |
| Plot 3, soil (39) vs. plants (38) | 0.52804 | 0.0930 | 0.03640 | 0.005 |
| Plot 4, soil (40) vs. plants (37) | 0.54687 | 0.037 | 0.00360 | 0.2490 |
| By plot (plants only) | 0.29965 | <0.001 | 0.04364 | 0.001 |
| Plot 1 (38) vs. plot 2 (40) | 0.61930 | <0.001 | 0.08801 | <0.001 |
| Plot 1 (38) vs. plot 3 (38) | 0.59271 | 0.001 | 0.01693 | 0.044 |
| Plot 1 (38) vs. plot 4 (37) | 0.57874 | 0.004 | 0.03091 | 0.014 |
| Plot 2 (40) vs. plot 3 (38) | 0.51912 | 0.1230 | 0.03503 | 0.008 |
| Plot 2 (40) vs. plot 4 (37) | 0.49443 | 0.4640 | 0.01174 | 0.1020 |
| Plot 3 (38) vs. plot 4 (37) | 0.48607 | 0.6080 | −0.00411 | 0.6420 |
| By plot (soil only) | 0.25322 | 0.2640 | 0.02106 | 0.029 |
| Plot 1 (39) vs. plot 2 (40) | 0.51622 | 0.1570 | 0.01159 | 0.1170 |
| Plot 1 (39) vs. plot 3 (39) | 0.53279 | 0.0860 | 0.05149 | 0.003 |
| Plot 1 (39) vs. plot 4 (40) | 0.47066 | 0.8000 | −0.00825 | 0.9010 |
| Plot 2 (40) vs. plot 3 (39) | 0.48180 | 0.6470 | 0.00149 | 0.2910 |
| Plot 2 (40) vs. plot 4 (40) | 0.51827 | 0.1790 | 0.00086 | 0.3270 |
| Plot 3 (39) vs. plot 4 (40) | 0.52057 | 0.1310 | 0.03139 | 0.020 |
| Field 2 | ||||
| Soil (156) vs. plants (142) | 0.52643 | <0.001 | 0.01069 | 0.007 |
| Plot 5 (Gem 611), soil (40) vs. plants (37) | 0.62040 | <0.001 | 0.08284 | <0.001 |
| Plot 6 (FL 47), soil (38) vs. plants (36) | 0.50618 | 0.2780 | −0.00176 | 0.4900 |
| Plot 7 (Heinz 3402), soil (39) vs. plants (36) | 0.48576 | 0.5740 | 0.01350 | 0.0920 |
| Plot 8 (Mountain Fresh), soil (39) vs. plants (33) | 0.51499 | 0.2470 | 0.01975 | 0.044 |
| By cultivar (plants only) | 0.28595 | 0.006 | 0.06590 | <0.001 |
| Gem 611 (37) vs. FL 47 (36) | 0.50582 | 0.3120 | 0.00977 | 0.1190 |
| Gem 611 (37) vs. Heinz 3402 (36) | 0.57121 | 0.003 | 0.07524 | <0.001 |
| Gem 611 (37) vs. Mountain Fresh (32) | 0.47132 | 0.7830 | 0.00284 | 0.3060 |
| FL 47 (36) vs. Heinz 3402 (36) | 0.51714 | 0.1860 | 0.01999 | 0.048 |
| FL 47 (36) vs. Mountain Fresh (32) | 0.52684 | 0.1540 | 0.03992 | 0.008 |
| Heinz 3402 (36) vs. Mountain Fresh (32) | 0.61245 | <0.001 | 0.13219 | <0.001 |
| By plot (soil only) | 0.26894 | 0.025 | 0.01992 | 0.020 |
| Plot 5 (Gem 611) (40) vs. plot 6 (FL 47) (38) | 0.56327 | 0.008 | 0.02852 | 0.020 |
| Plot 5 (Gem 611) (40) vs. plot 7 (Heinz 3402) (39) | 0.50211 | 0.3490 | 0.00425 | 0.2150 |
| Plot 5 (Gem 611) (40) vs. plot 8 (Mountain Fresh) (39) | 0.55374 | 0.022 | 0.04594 | 0.002 |
| Plot 6 (FL 47) (38) vs. plot 7 (Heinz 3402) (39) | 0.49057 | 0.4670 | −0.00354 | 0.5510 |
| Plot 6 (FL 47) (38) vs. plot 8 (Mountain Fresh) (39) | 0.51342 | 0.2180 | −0.00240 | 0.5500 |
| Plot 7 (Heinz 3402) (39) vs. plot 8 (Mountain Fresh) (39) | 0.51798 | 0.1570 | 0.00912 | 0.1120 |
The number of samples from each sampling location is given in parentheses.
Statistically significant values are in boldface (Bonferroni-adjusted P < 0.001 [0.05/45]).
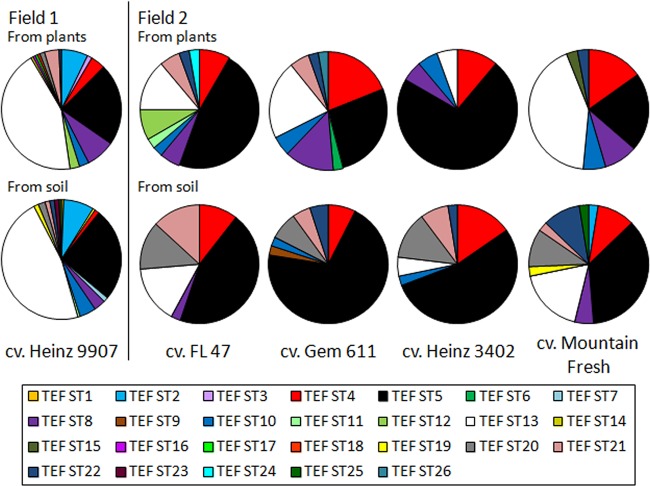
Populations were compared among plots in field 2, planted with different tomato cultivars, to determine if the host genotype affects F. oxysporum populations. Different sequence types predominated in each cultivar, most notably ST 5, composing 72% of the endophyte population from Heinz 3402 compared to 47% in Gem 611, 27% in FL 47, and 21% in Mountain Fresh (Fig. 2). These differences were reflected in the genetic differentiation analyses using HST, which is based on allele frequency. Endophyte populations from Heinz 3402 were significantly different from populations in Gem 611 and Mountain Fresh (Table 2). However, based on Snn, which incorporates the genetic distance between alleles, Heinz 3402 was significantly differentiated only from Mountain Fresh. Differences in allele frequency between Heinz 3402 and Gem 611, therefore, may be based on closely related clones. Soil populations were also compared to determine if differences between cultivars are biased by the physical separation among plots. In pairwise comparisons, no soil populations were significantly differentiated based on either Snn or HST, suggesting that the genetic differentiation observed among endophyte populations was due to the host genotype rather than different F. oxysporum populations in different locations in the field.
FIG 2.
Comparison of sequence type frequencies among populations. The frequency of each sequence type is shown for all plots in field 1 (153 isolates from plants and 158 isolates from soil) and for each cultivar in field 2: FL 47 (36 isolates from plants and 38 isolates from soil), Gem 611 (37 isolates from plants and 40 isolates from soil), Heinz 3402 (36 isolates from plants and 39 isolates from soil), and Mountain Fresh (33 isolates from plants and 39 isolates from soil). Sequence type frequencies were calculated using GENALEX 6 (32).
Populations were also compared between endophyte and soil populations to determine if populations were differentiated based on the environmental niche. The total soil population was differentiated from the total endophyte population for both fields combined and for field 2 based on Snn (P < 0.001), but they were not differentiated in field 1. The differentiation observed between the total soil population and the total endophyte population could be biased by the combination of populations from different locations and different tomato cultivars, so soil and endophyte populations were compared for each plot. Endophyte and soil populations were significantly differentiated for plot 5, planted with Gem 611 (P < 0.001 for both Snn and HST), but not for any of the other plots.
A principal-coordinate analysis was performed on the sequence types to look for clustering of isolations from different substrates. The first coordinate explained 56.79% of the variation, the second coordinate explained 22.40% of the variation, and the third coordinate explained 12.49% of the variation. In the plot of the first and second coordinates (Fig. 3), the samples were clustered based on whether they were from field 1 or field 2, but there was no clear pattern of clustering based on plot or substrate, for either soil or plants, although samples from soil tended to be less dispersed in the plot than samples from plants.
FIG 3.
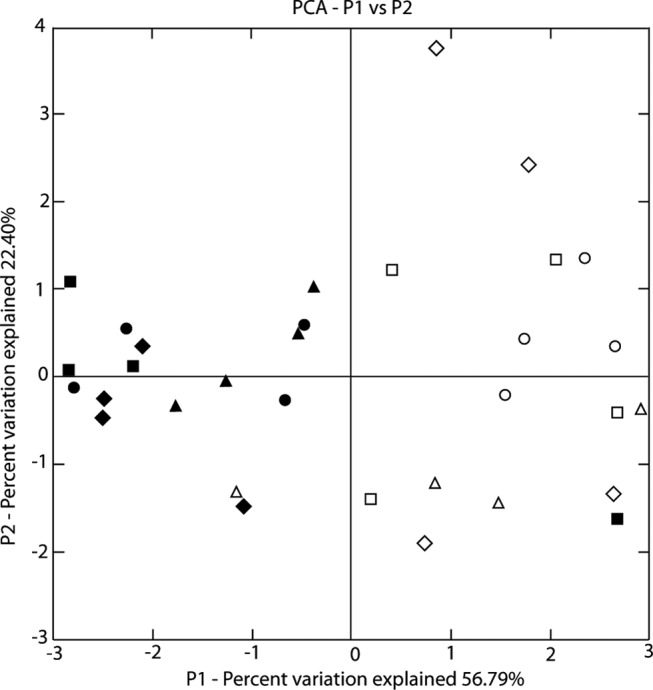
Principal-coordinate analysis (PCA) of sequences from different plots (P) and substrates. The symbols represent samples from different substrates (circles, soil; squares, root; diamonds, crown; triangles, stem). The black symbols represent samples from field 1, and the white symbols represent samples from field 2. Sequences were weighted for abundance, and the analysis was not normalized.
Phylogenetic analysis.
A statistical parsimony network of the sequence types was inferred, and the sequence types grouped into two networks (Fig. 4). Three loops were present in the larger network, indicating homoplastic relationships or possibly recombination between isolates. Sequence types mainly from soil were frequently closely related to sequence types mainly from plants, although one branch of the tree was composed entirely of endophytic isolates (STs 17, 14, and 15).
FIG 4.
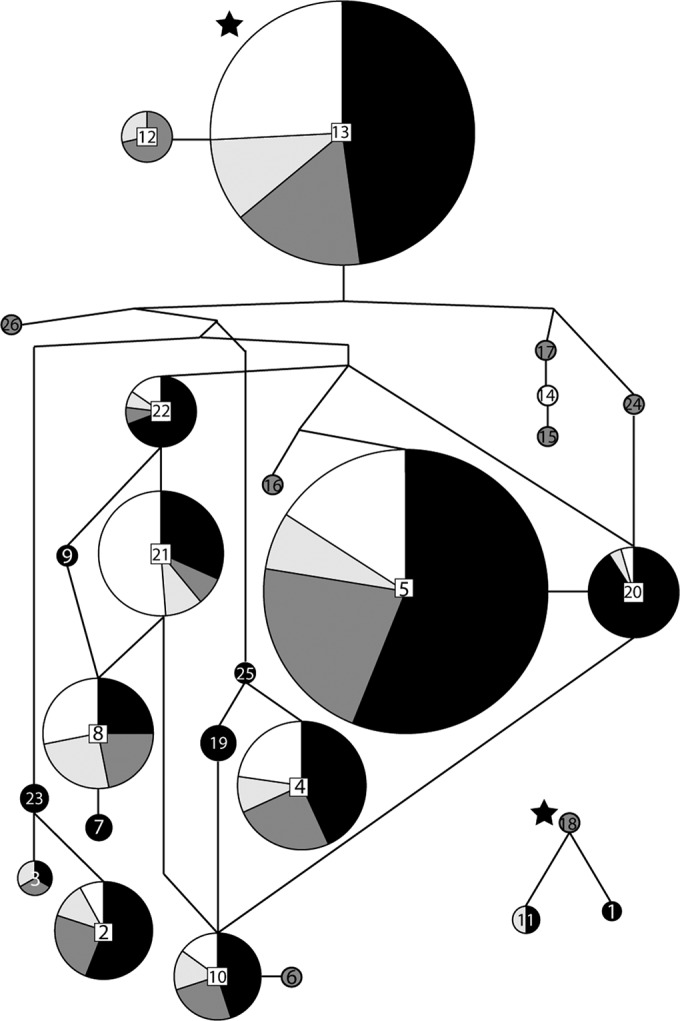
Statistical parsimony network of the translation elongation factor 1α sequences types, constructed in TCS v. 1.21 (38). Sequence types are represented by circles, with the number given to each sequence type in the center of the circle, and the size of the circle is proportional to the number of isolates with that sequence type. The shading represents the percentage of isolates in each sequence type from different substrates (black, soil; dark gray, roots; light gray, crowns; white, stems). The inferred ancestral sequence types are indicated with stars. Branch lengths are not to scale.
A phylogenetic analysis of the tomato-associated sequence types compared to other F. oxysporum isolates showed that the isolates were highly diverse and belonged to many different clades within F. oxysporum (Fig. 5). While some STs were closely related, no clear pattern was observed based on whether isolates were from plant tissues or soil, and isolates from soil and plants often shared the same sequence type. The two most common STs, 5 and 13, were not closely related. Most STs, including ST 5 and ST 13, were not closely related to isolates known to be pathogenic to tomato, F. oxysporum f. sp. lycopersici races 1, 2, and 3 and F. oxysporum f. sp. radicis-lycopersici.
FIG 5.
One of two most parsimonious trees based on the translation elongation factor 1α gene using maximum-parsimony analysis (81 steps; consistency index = 0.926; retention index = 0.968; rescaled consistency index = 0.897). The tree is rooted with F. commune NRRL 38348, and bootstrap support of >50% based on 1,000 replicates is indicated for each branch. Sequence types generated in this study from tomato fields are in boldface, with the number of isolates per sequence type and the original substrates (plants or soil) given in parentheses. The arrows indicate the two most abundant sequences types, ST 5 and ST 13.
DISCUSSION
The main objectives of this study were to investigate the genetic diversity of F. oxysporum populations associated with soil and asymptomatic tomato plants and to look for evidence of specialization to particular environmental niches among nonpathogenic F. oxysporum. Over 600 isolates were collected from plants and soil samples from two fields, and endophytic F. oxysporum was successfully isolated from all plants sampled.
Populations of F. oxysporum living saprophytically in soil or nonpathogenically in asymptomatic plant roots have been previously reported to be highly diverse. Most studies of F. oxysporum populations used vegetative compatibility groups (VCGs) as a genetic diversity estimator and usually found nonpathogenic populations to comprise large numbers of VCGs even within one field, with the majority of isolates belonging to single-isolate VCGs (13, 15, 23, 24, 42). Studies using various molecular markers have reported 25 mitochondrial DNA (mtDNA) haplotypes among 197 isolates from soil and asymptomatic melon roots (11), 23 ribosomal IGS-restriction fragment length polymorphism (RFLP) types among 129 isolates from soil and asymptomatic carnation roots (24), and 46 TEF sequence types among 214 F. oxysporum isolates from soils across Australia (18). Twenty-six TEF sequence types were found in the two tomato fields, suggesting a level of diversity comparable to those in previous studies, given the differences in sampling strategies and markers used.
F. oxysporum populations in the two tomato fields for both soil and plant samples were significantly different (P < 0.001 for both Snn and HST) (Table 2), although the two fields were only approximately 1.5 km apart. The populations in the two fields were not entirely isolated, as half of the 26 total STs were found in both fields, but the distribution of sequence types was heterogeneous between the two fields. The populations of F. oxysporum in each field might differ due to factors such as management practices, previous planting history, or soil type (21).
F. oxysporum populations were also surprisingly diverse within each plant, with an average of 4.2 STs found per plant, including roots and stems. F. oxysporum was isolated from the stems of all plants, a niche that has not been studied for F. oxysporum, as previous population genetics studies of nonpathogenic F. oxysporum have focused on the roots or crown of plants (11, 12, 13, 15, 23). These isolations from tomato show that endophytic F. oxysporum also commonly colonizes plant stems. Within an individual stem, the presence of multiple STs was common, with an average of 2.2 STs per plant. The ability of multiple inoculated F. oxysporum genotypes to infect the same plant has been previously reported, such as for two pathogenic isolates in muskmelon (43), two endophytic isolates in carnation (15), and a pathogen and an endophyte in tomato (44) and in carnation (28). In addition, isolation of nonpathogenic F. oxysporum from plants also infected with pathogenic F. oxysporum is common (for example, see references 25 and 45). Endophytic F. oxysporum fungi, therefore, do not necessarily exclude other F. oxysporum isolates from colonizing the same plant, and wild plants might commonly harbor a population of F. oxysporum endophytes rather than just a single individual.
One unexpected result of this study was that endophytic populations were equally or sometimes more genetically diverse than populations from the surrounding soil (Table 1 and Fig. 1). Our initial hypothesis was that endophytic populations would be less diverse than soil populations if the interaction between the plant and the fungal endophyte is somewhat specific, leading to selection by the plant acting as a filter on the resident fungal soil population. Instead, Shannon's diversity index indicated that overall, populations isolated from plants were significantly more diverse than populations from soil (P < 0.001). In individual plots, the levels of diversity were not significantly differentiated, except for plot 6, in which levels of diversity were much higher for the endophyte population (P < 0.001). In the principal-coordinate analysis (Fig. 3), isolates from the crown and root are the most widely spread out, while isolates from the soil tend to cluster together. These results might be an artifact of the sampling scheme, as a greater volume of plant material than of soil was collected, but calculations of the population sizes based on the Chao 2 nonparametric richness estimator suggest that the populations were only slightly undersampled. One possible explanation is that STs found only in the plants could have colonized the plants prior to transplantation, either by seed transmission or during the seedling stage before planting. This hypothesis is supported by the higher degree of genetic differentiation among plots for endophyte populations than for soil populations (Table 2), suggesting that soil populations are relatively uniform across the field and not the source of the unique endophytic genotypes. Given that some F. oxysporum isolates obtained from soil have been found to suppress disease caused by pathogenic F. oxysporum (46) while other endophytic F. oxysporum isolates may be pathogenic to plant species besides the endophyte host (10, 47), the introduction of new genotypes of endophytic F. oxysporum conceivably could increase or decrease the incidence of Fusarium wilt and other diseases and may have important implications for agriculture.
No definitive evidence was found to support the hypothesis that certain genotypes of F. oxysporum are specialized to colonize tomato plants as endophytes. If only specific genotypes can form an endophytic association with tomato, we hypothesized that populations within the plants would be genetically differentiated from populations in the soil. This was true across both fields combined and for field 2 (based on Snn analysis; P < 0.001) (Table 2). However, closer analysis did not show consistent differentiation between populations from different parts of the plants (roots and stems) and the soil or between soil and plant populations for each plot within the fields (Table 2). Moreover, diversity was generally not statistically different for populations from plants and populations from soil (Table 1). If only specialized genotypes could colonize the plant, diversity within the plant should be lower than diversity in the soil, as is seen for pathogenic isolates of F. oxysporum compared to soil populations (14).
One result that may suggest selection of specialized genotypes within the population is the predominance of two STs, 5 and 13, in all samples. The majority of isolates belonged to ST 5 and ST 13, including isolates from both fields and from both plants and soil. ST 5 and ST 13 may be better adapted to colonizing tomato plants, or perhaps plants in general, than other isolates, leading these STs to prevail in endophytic populations, as was suggested for a population of F. oxysporum isolated from asymptomatic celery roots in which most isolates belonged to one of two VCGs (12). The high prevalence of these sequence types in the soil could be due to a selective effect of the plant roots on soil populations, or larger populations in the plant may lead to larger populations in the surrounding soil. Alternatively, the frequent isolation of ST 5 and ST 13 may reflect their abundance in the local F. oxysporum soil population and not a selective effect by tomato. Soil was not sampled from fields not cultivated with tomato, so it cannot be determined if STs 5 and 13 are equally common in other local fields in the absence of tomato plants.
Based on haplotype network results, isolates were not necessarily more closely related to other isolates from the same niche than to isolates from other substrates, whether soil or the roots, crown, or stem of the plants (Fig. 4). Likewise, in a phylogenetic analysis of the isolates from tomato compared to F. oxysporum isolated from other hosts, genotypes associated with asymptomatic tomato, either from plants or from soil, were not necessarily closely related (Fig. 5). Some sequence types differed by only one single-nucleotide polymorphism and therefore clustered together in the phylogeny, while other sequence types were quite different. These results agree with the findings of a large phylogenetic analysis of F. oxysporum (16), in which the included putative nonpathogens from several host plants did not belong to any specific lineage within the complex.
The potential effect of the host genotype on the population composition was studied using the samples from field 2, where four different tomato cultivars were sampled. Populations of endophytic isolates were significantly different among cultivars based on HST (P < 0.0001) (Table 2). Isolates from Heinz 3402 in particular were differentiated from those of Gem 611 and Mountain Fresh based on HST. No clear patterns were observed based on whether the cultivars were bred for fresh market (FL 47 and Mountain Fresh) or for processing (Gem 611 and Heinz 3402). The differentiation of endophyte populations among cultivars suggests that particular host genotypes interact differently with F. oxysporum genotypes, leading to different F. oxysporum populations within each cultivar. This interpretation is tenuous, however, as differentiation among endophyte populations was also observed among the plots in field 1, which contained only one cultivar, Heinz 9907, and an uneven distribution of STs in the field could influence the results. However, Gem 611 and Heinz 3402 were planted next to each other and harbored distinct populations, and no significant differentiation was seen among soil populations in field 2, which suggests that differences in endophyte populations are not due to uneven spatial distribution of STs. While the patterns observed are suggestive of the host genotype influencing endophyte populations, firm conclusions cannot be drawn from these results.
Overall, the large amount of diversity observed among these populations highlights the importance of nonpathogenic lifestyles to the biology of F. oxysporum. Both soil and endophyte populations were composed of many sequence types, even associated with just one plant. Some results were suggestive of specialization of endophytic populations, but generally, they support the hypothesis that most, if not all, F. oxysporum isolates are capable of living as endophytes. These endophytic and soil populations may represent a source of unstudied diversity within F. oxysporum. F. oxysporum is well known for its variety of agriculturally important characteristics, such as pathogenicity on many plant hosts; production of secondary metabolites, including mycotoxins; and biocontrol activity, and better exploration of endophytic and soil environments may discover even more economically or scientifically relevant traits.
ACKNOWLEDGMENTS
This study was partially funded by a grant to J.E.D. through the Pennsylvania State University College of Agricultural Sciences graduate student competitive-grants program.
We thank Majid Foolad for graciously allowing us to sample in his tomato field and Tiana Montgomery-Noel for technical assistance.
REFERENCES
- 1.Stoner MF. 1981. Ecology of Fusarium in noncultivated soils, p 276–286. In Nelson PE, Toussoun TA, Cook RJ (ed), Fusarium: diseases, biology, and taxonomy. The Pennsylvania State University, University Park, PA. [Google Scholar]
- 2.Kuldau GA, Yates IE. 2000. Evidence for Fusarium endophytes in cultivated and wild plants, p 85–117. In Bacon CW, White JF Jr (ed), Microbial endophytes. Marcel Dekker, New York, NY. [Google Scholar]
- 3.Olivain C, Alabouvette C. 1999. Process of tomato root colonization by a pathogenic strain of Fusarium oxysporum f. sp. lycopersici in comparison with a non-pathogenic strain. New Phytol 141:497–510. doi: 10.1046/j.1469-8137.1999.00365.x. [DOI] [PubMed] [Google Scholar]
- 4.Olivain C, Trouvelot S, Binet MN, Cordier C, Pugin A, Alabouvette C. 2003. Colonization of flax roots and early physiological responses of flax cells inoculated with pathogenic and nonpathogenic strains of Fusarium oxysporum. Appl Environ Microbiol 69:5453–5462. doi: 10.1128/AEM.69.9.5453-5462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baayen RP. 1988. Responses related to lignification and intravascular periderm formation in carnations resistant to Fusarium wilt. Can J Bot 66:784–792. doi: 10.1139/b88-115. [DOI] [Google Scholar]
- 6.Beckman CH, Mueller WC, Tessier BJ, Harrison NA. 1982. Recognition and callose deposition in response to vascular infection in Fusarium wilt-resistant or susceptible tomato plants. Physiol Plant Pathol 20:1–10. doi: 10.1016/0048-4059(82)90018-2. [DOI] [Google Scholar]
- 7.Harling R, Taylor GS. 1985. A light microscope study of resistant and susceptible carnations infected with Fusarium oxysporum f. sp. dianthi. Can J Bot 63:638–646. doi: 10.1139/b85-080. [DOI] [Google Scholar]
- 8.Benhamou N, Garand C. 2001. Cytological analysis of defense-related mechanisms induced in pea root tissues in response to colonization by nonpathogenic Fusarium oxysporum Fo47. Phytopathology 91:730–740. doi: 10.1094/PHYTO.2001.91.8.730. [DOI] [PubMed] [Google Scholar]
- 9.Leoni C, de Vries M, ter Braak CJF, van Bruggen AHC, Rossing WAH. 2013. Fusarium oxysporum f. sp. cepae dynamics: in-plant multiplication and crop sequence simulations. Eur J Plant Pathol 137:545–561. doi: 10.1007/s10658-013-0268-6. [DOI] [Google Scholar]
- 10.Katan J. 1971. Symptomless carriers of the tomato Fusarium wilt pathogen. Phytopathology 61:1213–1217. doi: 10.1094/Phyto-61-1213. [DOI] [Google Scholar]
- 11.Appel DJ, Gordon TR. 1994. Local and regional variation in populations of Fusarium oxysporum from agricultural field soils. Phytopathology 84:786–791. doi: 10.1094/Phyto-84-786. [DOI] [Google Scholar]
- 12.Correll JC, Puhalla JE, Schneider RW. 1986. Vegetative compatibility groups among nonpathogenic root-colonizing strains of Fusarium oxysporum. Can J Bot 64:2358–2361. doi: 10.1139/b86-310. [DOI] [Google Scholar]
- 13.Elias KS, Schneider RW, Lear MM. 1991. Analysis of vegetative compatibility groups in nonpathogenic populations of Fusarium oxysporum isolated from symptomless tomato roots. Can J Bot 69:2089–2094. doi: 10.1139/b91-263. [DOI] [Google Scholar]
- 14.Katan T, Katan J. 1988. Vegetative compatibility grouping of Fusarium oxysporum f. sp. vasinfectum from tissue and the rhizosphere of cotton plants. Phytopathology 78:852–855. doi: 10.1094/Phyto-78-852. [DOI] [Google Scholar]
- 15.Katan T, Berliner R, Katan J. 1994. Vegetative compatibility in populations of Fusarium oxysporum from wild carnation. Mycol Res 98:1415–1418. doi: 10.1016/S0953-7562(09)81072-1. [DOI] [Google Scholar]
- 16.O'Donnell K, Gueidan C, Sink S, Johnston PR, Crous PW, Glenn A, Riley R, Zitomer NC, Colyer P, Waalwijk C, Lee TVD, Moretti A, Kang S, Kim H-S, Geiser DM, Juba JH, Baayen RP, Cromey MG, Bithell S, Sutton DA, Skovgaard K, Ploetz R, Kistler HC, Elliott M, Davis M, Sarver BAJ. 2009. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet Biol 46:936–948. doi: 10.1016/j.fgb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Balmas V, Migheli Q, Scherm B, Garau P, O'Donnell K, Ceccherelli G, Kang S, Geiser DM. 2010. Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia 102:803–812. doi: 10.3852/09-201. [DOI] [PubMed] [Google Scholar]
- 18.Laurence MH, Burgess LW, Summerell BA, Liew ECY. 2012. High levels of diversity in Fusarium oxysporum from non-cultivated ecosystems in Australia. Fungal Biol 116:289–297. doi: 10.1016/j.funbio.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Edel V, Steinberg C, Gautheron N, Recorbet G, Alabouvette C. 2001. Genetic diversity of Fusarium oxysporum populations isolated from different soils in France. FEMS Microbiol Ecol 36:61–71. doi: 10.1111/j.1574-6941.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 20.Gordon TR, Okamoto D. 1992. Variation within and between populations of Fusarium oxysporum based on vegetative compatibility and mitochondrial DNA. Can J Bot 70:1211–1217. doi: 10.1139/b92-152. [DOI] [Google Scholar]
- 21.Gordon TR, Okamoto D, Milgroom MG. 1992. The structure and interrelationship of fungal populations in native and cultivated soils. Mol Ecol 1:241–249. doi: 10.1111/j.1365-294X.1992.tb00183.x. [DOI] [Google Scholar]
- 22.Edel V, Steinberg C, Gautheron N, Alabouvette C. 1997. Populations of nonpathogenic Fusarium oxysporum associated with roots of four plant species compared to soilborne populations. Phytopathology 87:693–697. doi: 10.1094/PHYTO.1997.87.7.693. [DOI] [PubMed] [Google Scholar]
- 23.Alves-Santos FM, Benito EP, Eslava AP, Díaz-Mínguez JM. 1999. Genetic diversity of Fusarium oxysporum strains from common bean fields in Spain. Appl Environ Microbiol 65:3335–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lori G, Edel-Hermann V, Gautheron N, Alabouvette C. 2004. Genetic diversity of pathogenic and nonpathogenic populations of Fusarium oxysporum isolated from carnation fields in Argentina. Phytopathology 94:661–668. doi: 10.1094/PHYTO.2004.94.6.661. [DOI] [PubMed] [Google Scholar]
- 25.Fiely MB, Correll JC, Morelock TE. 1995. Vegetative compatibility, pathogenicity, and virulence diversity of Fusarium oxysporum recovered from spinach. Plant Dis 79:990–993. doi: 10.1094/PD-79-0990. [DOI] [Google Scholar]
- 26.Bao J, Fravel D, Lazarovits G, Chellemi D, van Berkum P, O'Neill N. 2004. Biocontrol genotypes of Fusarium oxysporum from tomato fields in Florida. Phytoparasitica 32:9–20. [Google Scholar]
- 27.Larkin RP, Fravel DR. 1998. Efficacy of various fungal and bacterial biocontrol organisms for control of Fusarium wilt of tomato. Plant Dis 82:1022–1028. doi: 10.1094/PDIS.1998.82.9.1022. [DOI] [PubMed] [Google Scholar]
- 28.Postma J, Luttikholt AJG. 1996. Colonization of carnation stems by a nonpathogenic isolate of Fusarium oxysporum and its effect on Fusarium oxysporum f. sp. dianthi. Can J Bot 74:1841–1851. doi: 10.1139/b96-221. [DOI] [Google Scholar]
- 29.Nash SM, Snyder WC. 1962. Quantitative estimations by plate counts of propagules of bean root rot Fusarium in field soils. Phytopathology 52:567–572. [Google Scholar]
- 30.Cenis JL. 1992. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res 20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiser DM, Jiménez-Gasco MM, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O'Donnell K. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol 110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 32.Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherwin WB, Jabot F, Rush R, Rossetto M. 2006. Measurement of biological information with applications from genes to landscapes. Mol Ecol 15:2857–2869. doi: 10.1111/j.1365-294X.2006.02992.x. [DOI] [PubMed] [Google Scholar]
- 34.Hudson RR. 2000. A new statistic for detecting genetic differentiation. Genetics 155:2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson RR, Boos DB, Kaplan NL. 1992. A statistical test for detecting geographic subdivision. Mol Biol Evol 9:138–151. [DOI] [PubMed] [Google Scholar]
- 36.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 37.Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clement M, Posada D, Crandall K. 2000. TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 39.Baayen RP, O'Donnell K, Bonants PJM, Cigelnik E, Kroon LPNM, Roebroeck EJA, Waalwijk C. 2000. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 90:891–900. doi: 10.1094/PHYTO.2000.90.8.891. [DOI] [PubMed] [Google Scholar]
- 40.O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci U S A 95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krebs CJ. 1972. Ecology: the experimental analysis of distribution and analysis. Harper and Row, New York, NY. [Google Scholar]
- 42.Appel DJ, Gordon TR. 1996. Relationships among pathogenic and nonpathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA. Mol Plant Microbe Interact 9:125–138. doi: 10.1094/MPMI-9-0125. [DOI] [PubMed] [Google Scholar]
- 43.Meyer JA, Maraite H. 1971. Multiple infection and symptom mitigation in vascular wilt diseases. Trans Br Mycol Soc 57:371–377. doi: 10.1016/S0007-1536(71)80051-7. [DOI] [Google Scholar]
- 44.Olivain C, Humbert C, Nahalkova J, Fatehi J, L'Haridon F, Alabouvette C. 2006. Colonization of tomato root by pathogenic and nonpathogenic Fusarium oxysporum strains inoculated together and separately into the soil. Appl Environ Microbiol 72:1523–1531. doi: 10.1128/AEM.72.2.1523-1531.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skovgaard K, Bødker L, Rosendahl S. 2002. Population structure and pathogenicity of members of the Fusarium oxysporum complex isolated from soil and root necrosis of pea (Pisum sativum L.). FEMS Microbiol Ecol 42:367–374. doi: 10.1111/j.1574-6941.2002.tb01026.x. [DOI] [PubMed] [Google Scholar]
- 46.Alabouvette C. 1990. Biological control of Fusarium wilt pathogens in suppressive soils, p 27–43. In Hornby D. (ed), Biological control of soilborne plant pathogens. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 47.Helbig JB, Carroll RB. 1984. Dicotyledonous weeds as a source of Fusarium oxysporum pathogenic on soybean. Plant Dis 68:694–696. doi: 10.1094/PD-68-694. [DOI] [Google Scholar]