Abstract
Multiple anthropogenic disturbances to bacterial diversity have been investigated in coastal ecosystems, in which temporal variability in the bacterioplankton community has been considered a ubiquitous process. However, far less is known about the temporal dynamics of a bacterioplankton community responding to pollution disturbances such as toxic metals. We used coastal water microcosms perturbed with 0, 10, 100, and 1,000 μg liter−1 of cadmium (Cd) for 2 weeks to investigate temporal variability, Cd-induced patterns, and their interaction in the coastal bacterioplankton community and to reveal whether the bacterial community structure would reflect the Cd gradient in a temporally varying system. Our results showed that the bacterioplankton community structure shifted along the Cd gradient consistently after a 4-day incubation, although it exhibited some resistance to Cd at low concentration (10 μg liter−1). A process akin to an arms race between temporal variability and Cd exposure was observed, and the temporal variability overwhelmed Cd-induced patterns in the bacterial community. The temporal succession of the bacterial community was correlated with pH, dissolved oxygen, NO3−-N, NO2−-N, PO43−-P, dissolved organic carbon, and chlorophyll a, and each of these parameters contributed more to community variance than Cd did. However, elevated Cd levels did decrease the temporal turnover rate of community. Furthermore, key taxa, affiliated to the families Flavobacteriaceae, Rhodobacteraceae, Erythrobacteraceae, Piscirickettsiaceae, and Alteromonadaceae, showed a high frequency of being associated with Cd levels during 2 weeks. This study provides direct evidence that specific Cd-induced patterns in bacterioplankton communities exist in highly varying manipulated coastal systems. Future investigations on an ecosystem scale across longer temporal scales are needed to validate the observed pattern.
INTRODUCTION
In the last decades, heavy metal concentrations have increased significantly, reaching up to several mg liter−1, in seriously polluted estuaries or harbors (1, 2) as a consequence of industrial discharge and river outflows. Cadmium (Cd) is one of the most toxic metals frequently detected in the coastal areas of China, and its presence exceeds the permissible environmental limit in several bays, including Bohai Bay and Jiaozhou Bay (3, 4), which can harm human health via seafood consumption because of its accumulative and carcinogenic properties (3, 5, 6). One of the major scientific challenges is to evaluate threats of pollution disturbances to the biodiversity. Bacterioplankton plays crucial roles in the mineralization of organic matter and nutrient cycling in marine ecosystems (7, 8), and bacterioplankton community composition (BCC) quickly responds to anthropogenic disturbances such as chemical pollution, nutrient enrichment, and the introduction of antibiotics and pathogens (9). Therefore, bacterioplankton communities might be used to evaluate the ecological effect of pollution in a coastal environment.
The responses of marine macrobenthic communities to Cd have been extensively revealed (10–12). Also, heavy metals such as lead (Pb), copper (Cu), and Cd exhibited strong selective pressures on microbial communities, shifting community composition to be dominated by metal-tolerant or -resistant taxa in sediments (13). But how the marine bacterioplankton community responds to toxic metal exposure was poorly understood. A previous study indicated that Pb inhibited the growth of heterotrophic picoplankton (14); however, no further information about the phylogenetic diversity and structure of bacterioplankton communities affected by toxic metals has been revealed.
The temporal variability of bacterioplankton communities is one of the fundamental issues in marine microbial ecology (15). Multiple environmental factors involved in the temporal dynamics in BCC in experimental and natural coastal environments have been reported, such as temperature (16), nutrient conditions (9), phytoplankton succession (17), and predation (18). Bacterial communities in surface seawater were shown to be highly diverse and dynamic on a temporal scale (19). However, several recent studies demonstrate repeatable and predictable patterns rather than random distributions in BCCs across different temporal scales (19–22). In other words, variation in BCC could reflect multiple underlying environmental changes. The whole bacterioplankton community may change rapidly to maintain the completed ecological function in highly variable environments (23). For example, high turnover and predictability of bacterioplankton community succession were observed in seawater ponds during a 77-day shrimp cultivation using the similarity-time decay model (21).
In relatively stable systems like sediments, microbial assemblages showed a low temporal turnover rate (15), indicating that specific patterns in the microbial community induced by pollution disturbances can be identified more easily. In ecosystems with high temporal variability such as coastal water, in contrast, the influence of toxic metals such as Cd on the diversity patterns of the bacterial community may be difficult to separate from temporal variation. Current knowledge on how and to what extent the coastal bacterioplankton community responds to the competition and interaction between toxic metals and temporal succession is scarce. In addition, it is unclear whether bacterial assemblages with robust temporal variability can be applied to reflect Cd pollution in seawater, although several recent studies have found significant correlations between the bacterial community and specific environment changes such as spatial and seasonal gradients, nitrogen pollution, and outbreak of shrimp disease (19, 24, 25).
In this study, we assessed the effects of four levels of Cd exposure (from no perturbation to strong perturbation) on bacterial community phylogenetic composition, richness, and structure over 2 weeks, using coastal water microcosms as a manipulated system. Moreover, we expanded beyond beta-diversity patterns to focus on the key taxa associated with Cd pollution level over time and gleaned insights into their ecological roles in bacterial community responses to Cd pollution in coastal water. We hypothesized that temporal succession of BCC fits the similarity-time decay model and could be strongly correlated with the dynamic changes of seawater variables. Therefore, we used amplicon pyrosequencing of 16S rRNA genes of seawater samples from microcosms to achieve the following goals: (i) to investigate Cd-induced patterns, temporal variability, and their interaction in the bacterial community; (ii) to estimate the influence of Cd exposure on the temporal turnover rate of the bacterial community; (iii) to determine the factors shaping the temporal pattern in the bacterial community and their relationships with Cd; (iv) to identify key bacterial taxa associated with Cd pollution levels across the experimental duration.
MATERIALS AND METHODS
Seawater collection and microcosm setup.
Surface seawater samples at a 0.5-m depth were collected at a coastal site (29°32′26.43″N, 121°45′41.53″E) located in Xiangshan Bay, East China Sea, on 2 May 2013 and then transported within 2 h to the laboratory to construct the experimental microcosms. Acid (10% HCl)-washed 5-liter polyethylene terephthalate bottles were filled with 4 liters unfiltered seawater and immediately incubated at a constant room temperature of 20 ± 2°C. Sixty-eight microcosms were established in an illumination shelf under 16 treatments (four replicates for each treatment) with four Cd levels (control, 10, 100, and 1,000 μg liter−1) combined with five incubation times (0 [original status], 1, 4, 7, and 14 days). The Cd treatments were spiked with a stock solution of CdCl2, which was prepared in 0.2-μm-filtered preautoclaved original seawater. The microcosms were exposed to a 12 h/12 h light/dark cycle with a light intensity of 650 lx and manually shaken twice per day.
Seawater sampling and environmental parameter analyses.
On the sampling days, pH and dissolved oxygen (DO) in seawater were quantified using a probe (YSI 550A; Instrumart, USA), and then the whole water column in the microcosms was gently mixed. Dissolved organic carbon (DOC) was quantified using an automated C/N analyzer (multi N/C 3100; Analytik Jena, Germany). Total phosphorus (TP), PO43−-P, NO3−-N, NH4+-N, and NO2−-N were analyzed following the standard methods (26). Chlorophyll a (Chl-a) was measured as described previously (21).
DNA extraction.
Approximately 500 ml of seawater from each of 68 microcosms was filtered onto a 0.2-μm polycarbonate membrane (Type GTTP; Millipore, USA). Community DNA was extracted using a Power Soil DNA isolation kit (MO BIO Laboratories, USA) according to the manufacturer's instruction. The DNA extracts were quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, USA) and stored at −80°C prior to amplification.
Bacterial 16S rRNA gene amplification and pyrosequencing.
An aliquot of 50 ng purified DNA from each sample was used as the template for amplification. The V4-V5 regions of bacterial 16S rRNA gene were amplified using the primer sets F515 (GTGCCAGCMGCCGCGG), with the Roche 454 “A” pyrosequencing adapter and a unique 11-bp barcode sequence, and R907 (CCGTCAATTCMTTTRAGTTT), with the Roche 454 “B” sequencing adapter. Each sample was amplified in triplicate with a 50-μl reaction system under the following conditions: 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 10 min. PCR products were pooled, purified with a PCR fragment purification kit (TaKaRa, Japan), and then quantified using a Quant-It Pico Green kit (Invitrogen, USA). Equimolar amounts of PCR products for each sample were combined in a single tube and run on a Roche FLX 454 pyrosequencing platform (Roche, USA), producing reads from the forward direction (F515 with bar code). The pyrosequencing service was provided by Majorbio Co., Shanghai, China.
Processing of pyrosequencing data.
All sequences were run through the QIIME v. 1.7.0 pipeline (27). The sequences were grouped into each sample according to the bar codes and quality controlled using the “split_libraries.py” script with default settings (minimum length, 200; maximum length, 1,000; minimum mean quality score, 25; maximum ambiguous bases, 0; maximum homopolymer length, 6; maximum primer mismatch, 0). The remaining sequences were chimera detected and removed using Usearch (28). Bacterial phylotypes were assigned to operational taxonomic units (OTUs; 97% sequence similarity) using the “pick_de_novo_otus.py” script with the Uclust method (29). The most abundant sequence of each phylotype was selected as the representative sequence and then was aligned in the Greengenes database (30) using PyNAST (31) to identify the taxonomy of the phylotype. A phylogenic tree was generated from the filtered alignment using FastTree (32). After this, Archaea and chloroplast sequences were removed. The remaining clean sequences of each parallel sample (as biological replicate) were used for the subsequent analysis. To normalize the sequencing depth of each sample, we used a randomly selected subset of 3,380 sequences per sample for downstream analysis, based on the sample with the least number of sequences.
Statistical analysis.
Two-way analysis of variance (ANOVA) was applied to investigate the influences of Cd level, incubation time, and their interaction on alpha-diversity metrics and the relative abundance of dominant phyla/classes of the bacterial community using SPSS 16.0. Nonmetric multidimensional scaling (NMDS) based on unweighted UniFrac distance was applied to evaluate the overall differences in bacterial community structure. Two-way crossed analysis of similarity (ANOSIM) was applied to investigate the influence of Cd level and incubation time on the bacterial community using the Primer-E v5 software package (33). Permutational multivariate analysis of variance (with ADONIS function) based on unweighted UniFrac distance was applied for partitioning the community variance constrained by incubation time, Cd level, and their interaction. Constrained analysis of principal coordinates (CAP) was applied to identify the composition of seawater variables correlated with the community variance based on unweighted UniFrac distance. Both ADONIS and CAP were performed in the R environment (http://www.r-project.org) using the “vegan” package (34). The bacterial community similarity-time decay model (15) and temporal turnover rate of bacterial community based on the Sørensen index were determined with a contiguous sampling approach with the power law model (S = cTw) as described in our previous work (21). Here, the exponent w was considered an index of the temporal turnover rate of the bacterial community and estimated directly with a linear regression based on the function expressed in log-log scale: log10 S = log10 c + w log10 T. The Sørensen index was calculated using PAST (palaeontological statistics program, V1.21).
To find representative phylotypes associated with the Cd levels, a similarity percentage (SIMPER) analysis was first applied to screen OTUs primarily responsible for the overall dissimilarity (contributing more than 0.7% dissimilarity each) in the bacterial communities along the Cd gradient at each time point using PAST. Then, the driving OTUs with abundance significantly correlated with Cd level (P < 0.05) were screened. Pearson's correlation coefficients (r) between the Cd level and the relative abundances of the OTUs were analyzed using SPSS 16.0. The representative sequences of these OTUs were aligned in the EzTaxon database (35) to find the closest-matched type strains as a supplement for taxonomic identification. The representative OTUs of each time point were extensively validated at the other time points to reveal their frequency of correlation with Cd level across the whole duration. Heat maps of average relative abundances of the key phylotypes, with high frequency (≥3 time points) of correlation with Cd level, were made in the R environment using the “pheatmap” package.
Nucleotide sequence accession number.
The sequence data generated in this study were deposited in the DDBJ Sequence Read Archive and are available under the project number DRA001855.
RESULTS
Dynamic changes of seawater chemical characteristics.
The chemical variables were monitored at each sampling day to investigate the dynamic changes of environmental conditions in the microcosms (see Table S1 in the supplemental material). All the measured parameters were temporally changed (P < 0.05). The concentrations of DOC, Chl-a, PO43−-P, NO3−-N, NH4+-N, and NO2−-N were also significantly influenced by Cd level. Furthermore, the interactions of time and Cd level significantly influenced DO, DOC, Chl-a, PO43−-P, and NH4+-N.
Taxon distribution and alpha diversity of bacterial community.
The full data set (n = 68) after quality control procedures and removing Archaea and chloroplast sequences yielded 12,329 OTUs comprising 396,452 clean reads (mean reads per sample, 5,830). About 99.2% of those sequences can be classified at the phylum level. The abundances of primary dominant phyla/classes (relative abundance, >5%), including Actinobacteria, Bacteroidetes, Alphaproteobacteria, Gammaproteobacteria, and Planctomycetes, showed obvious temporal variability across the experimental duration (see Fig. S1 in the supplemental material). At day 1, BCC was stable across all the Cd levels compared with the original status. However, BCC shifted a lot later, during days 4 to 14, with a great decrease in the abundance of Actinobacteria. Linear increase and decrease were observed in the abundances of Bacteroidetes and Alphaproteobacteria, respectively, along the Cd gradient at both day 4 and day 7. The increase in the abundance of Planctomycetes at day 14 reflected the specific pattern of BCC, except for the Cd3 level. Two-way ANOVA showed the abundances of all the dominant phyla and classes (relative abundance, >1%), except for Deltaproteobacteria, which were significantly changed over time (Table 1). On the other hand, the Cd level significantly influenced the abundances of Bacteroidetes, Planctomycetes, Proteobacteria, and Verrucomicrobia. Most bacterial classes affected by Cd addition were affiliated to these phyla.
TABLE 1.
Two-way analysis of variance (ANOVA) for relative abundances of the dominant bacterial phyla and classes (>1%) in the microcosms across different sampling days and Cd levelsa
| Taxon | Time |
Cd |
Time × Cd |
|||
|---|---|---|---|---|---|---|
| F | P | F | P | F | P | |
| Phyla | ||||||
| Unclassified | 20.3 | <0.001 | 1.30 | 0.287 | 2.73 | 0.012 |
| Actinobacteria | 417.3 | <0.001 | 1.87 | 0.148 | 4.96 | <0.001 |
| Bacteroidetes | 39.1 | <0.001 | 11.9 | <0.001 | 3.09 | 0.005 |
| Cyanobacteria | 4.20 | 0.010 | 1.69 | 0.181 | 1.49 | 0.180 |
| Lentisphaerae | 13.4 | <0.001 | 1.20 | 0.320 | 1.20 | 0.315 |
| Planctomycetes | 20.7 | <0.001 | 9.13 | <0.001 | 8.39 | <0.001 |
| Proteobacteria | 108.5 | <0.001 | 7.76 | <0.001 | 4.29 | <0.001 |
| Verrucomicrobia | 53.8 | <0.001 | 35.2 | <0.001 | 39.5 | <0.001 |
| Classes | ||||||
| Acidimicrobiia | 316.4 | <0.001 | 2.34 | 0.085 | 3.77 | 0.001 |
| Actinobacteria | 222.0 | <0.001 | 5.24 | 0.003 | 4.90 | <0.001 |
| Unclassified Bacteroidetes | 21.2 | <0.001 | 3.26 | 0.030 | 1.63 | 0.134 |
| Flavobacteria | 26.6 | <0.001 | 20.9 | <0.001 | 3.08 | 0.005 |
| Sphingobacteria | 32.2 | <0.001 | 15.2 | <0.001 | 3.22 | 0.004 |
| Oscillatoriophycideae | 3.75 | 0.017 | 2.32 | 0.087 | 1.96 | 0.066 |
| Synechococcophycideae | 4.44 | 0.008 | 1.27 | 0.294 | 1.14 | 0.354 |
| Lentisphaeria | 13.4 | <0.001 | 1.20 | 0.320 | 1.20 | 0.315 |
| Phycisphaerae | 7.43 | <0.001 | 27.8 | <0.001 | 7.41 | <0.001 |
| Planctomycetia | 17.0 | <0.001 | 7.88 | <0.001 | 8.08 | <0.001 |
| Alphaproteobacteria | 36.0 | <0.001 | 5.44 | 0.003 | 1.82 | 0.090 |
| Betaproteobacteria | 11.3 | <0.001 | 1.85 | 0.151 | 1.04 | 0.425 |
| Deltaproteobacteria | 1.57 | 0.210 | 2.65 | 0.059 | 1.37 | 0.231 |
| Gammaproteobacteria | 13.3 | <0.001 | 0.45 | 0.717 | 0.43 | 0.913 |
| Opitutae | 28.0 | <0.001 | 18.8 | <0.001 | 24.2 | <0.001 |
Bold values represent significant influence of factors on metrics (P < 0.05).
Overall, the alpha diversity of the bacterial community was significantly influenced by time, Cd level, and their interaction (P < 0.05; see Table S2 in the supplemental material). All the alpha-diversity metrics exhibited similar trends across different sampling days along the Cd gradient. In general, they remained similar across different Cd levels at days 1, 7, and 14; however, an acute increase was found in Cd2 and Cd3 microcosms compared with the control and Cd1 ones at day 4.
Bacterial community structure.
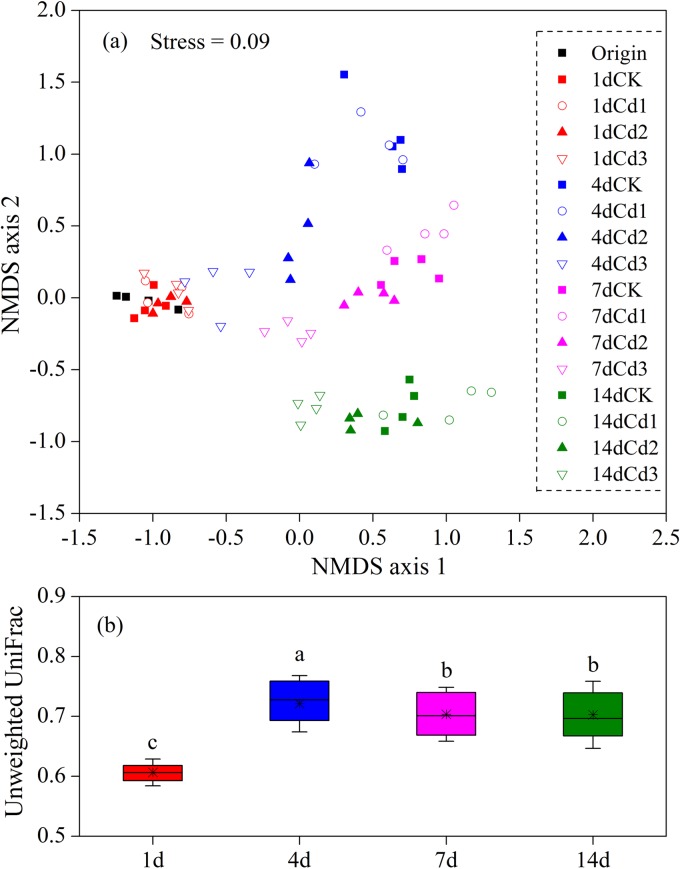
Nonmetric multidimensional scaling (NMDS) plots showed that the samples were phylogenetically segregated by incubation time (Fig. 1a). At the initial stage of incubation (day 1), the bacterial community structure seemed relatively stable along the Cd gradient compared with original samples. With the growth of incubation time, a gradual variation in the community along the Cd gradient at each sampling day was clearly observed. The repeatability of parallel samples in each Cd level is visualized in Fig. 1a. In general, significantly more similarity between communities from parallel samples than between those with different Cd levels was observed at each time point (see Fig. S2 in the supplemental material), thus indicating repeatability of parallel samples to a certain extent. The mean unweighted UniFrac distance between communities at each time point reached its peak at day 4 and showed a decreasing trend later (Fig. 1b), indicating that the bacterial communities under elevated Cd levels tended to converge at days 7 and 14. Two-way crossed ANOSIM indicated that time and Cd level were both significant factors in shaping the BCC (Table 2), and time (global R = 0.914, P = 0.001) showed greater influence on community variation than Cd did (global R = 0.618, P = 0.001). Furthermore, pairwise ANOSIM comparisons showed significant differences in communities between any two time points. Similar results were found for each pair of Cd levels except CK versus Cd1. Accordingly, ADONIS analysis showed that incubation time constrained 56.5% of community variance, which is much more than Cd did (13.6%), and their interaction constrained an additional 11.5% of community variance (Table 3).
FIG 1.
(a) Nonmetric multidimensional scaling (NMDS) plots of community dissimilarities based on unweighted UniFrac distance (OTU 0.03 level) across different sampling days and Cd levels. CK represents control microcosms; Cd1, Cd2, and Cd3 represent 10, 100, and 1,000 μg liter−1 Cd-spiked microcosms, respectively. (b) Boxplots of unweighted UniFrac distance between communities within each sampling day. Lines at the top, bottom, and middle of the box correspond to the 75th, 25th, and 50th percentiles (median), respectively. Whiskers at the top and bottom of the box represent ± standard deviations (SD). The asterisk in the box represents the mean value, and means with different letters are significantly different based on least significant difference (P < 0.05).
TABLE 2.
Two-way crossed analysis of similarity (ANOSIM) of bacterioplankton community structure (based on the occurrences of bacterial OTUs) from four treatments with different Cd levels across 14 days
| Factor | R | Pa |
|---|---|---|
| Time | ||
| Global ANOSIM | 0.914 | 0.001 |
| 1 day versus 4 days | 0.958 | 0.001 |
| 1 day versus 7 days | 1.000 | 0.001 |
| 1 day versus 14 days | 1.000 | 0.001 |
| 4 days versus 7 days | 0.503 | 0.001 |
| 4 days versus 14 days | 1.000 | 0.001 |
| 7 days versus 14 days | 0.934 | 0.001 |
| Cdb | ||
| Global ANOSIM | 0.618 | 0.001 |
| CKc versus Cd1 | 0.031 | 0.352 |
| CK versus Cd2 | 0.375 | 0.001 |
| CK versus Cd3 | 0.941 | 0.001 |
| Cd1 versus Cd2 | 0.393 | 0.001 |
| Cd1 versus Cd3 | 0.917 | 0.001 |
| Cd2 versus Cd3 | 0.906 | 0.001 |
Significant P values are given in bold.
Cd1, Cd2, and Cd3 represent 10, 100, and 1,000 μg liter−1 Cd-spiked microcosms, respectively.
CK, control microcosms.
TABLE 3.
Quantitative effects of incubation time, Cd level, and their interaction on the bacterial community variance by permutational multivariate analysis of variance (with ADONIS function) based on unweighted UniFrac distance
| Time |
Cd |
Time × Cd |
|||
|---|---|---|---|---|---|
| R2a | P | R2 | P | R2 | P |
| 0.563 | <0.001 | 0.136 | <0.001 | 0.115 | <0.001 |
R2 values represent the proportion of variance constrained by factors.
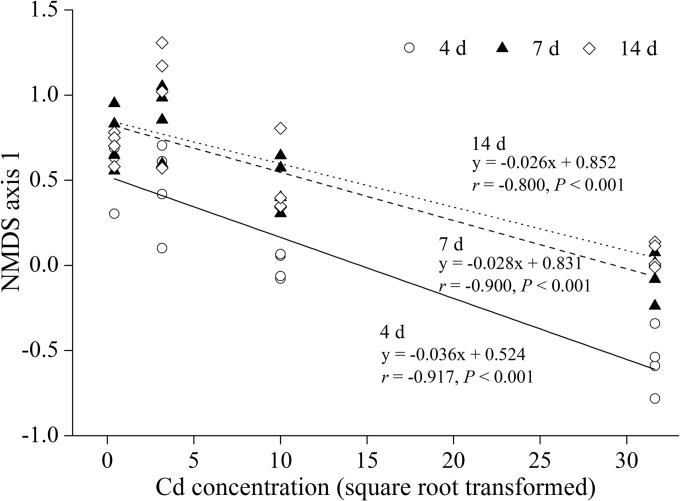
We applied the linear function to build the linkage between NMDS axis 1 (as a proxy for the bacterial community dissimilarity) and Cd levels in each time point after the initial stage to answer the following question: is there any specific Cd-induced pattern in the beta diversity of bacterial community under robust temporal variation? A significant Pearson's correlation (P < 0.01) between NMDS axis 1 and Cd level was observed at each time point (Fig. 2). A similar trend of the linear model in three time points indicated a consistent Cd-induced pattern in bacterial community variation. Moreover, the absolute value of slope decreased over time, corresponding to the converging trend of communities among different Cd levels during days 4 to 14.
FIG 2.
Relationships between the first axis of NMDS (as a proxy for the bacterial community dissimilarity) with Cd level for different sampling days. The regression slopes of the linear relationships are shown. Refer to Fig. 1 for treatment notation. d, days.
Similarity-time relationships along the Cd gradient.
A significant time-similarity decay was observed across all the Cd levels (P < 0.01) (Fig. 3), with the turnover rate ranging from −0.220 to −0.343. In general, there was an obvious decreasing trend in bacterial community turnover under elevated Cd levels, especially at the higher Cd levels (Cd2 and Cd3). However, well-overlapped linear regression curves of control and Cd1 treatments indicated that a slight level of Cd exposure did not influence the temporal succession of bacterial community.
FIG 3.
Similarity-time decay models of bacterial communities. The power-law exponent w was estimated directly with a linear regression (log-log space approach) fit between the average Sørensen similarity values and days between observations. Refer to Fig. 1 for treatment notation.
Linking bacterial community succession to seawater variables.
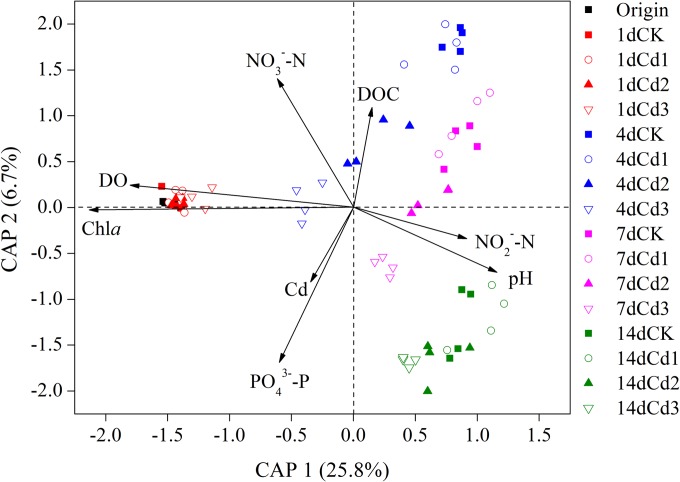
Constrained analysis of principal coordinates (CAP), a useful method to constrain environmental factors to community variation based on any distance/dissimilarity metrics (36), was applied to link the phylogenetic structure of the bacterial community with Cd level and seawater variables, listed in Table S1 in the supplemental material. Eight parameters showing significant correlation (P < 0.05) were selected to make the biplot, with the correlation coefficient (represented by the relative length of the arrow) changed in the following order: Chl-a > DO > PO43−-P > pH > NO3−-N > DOC > NO2−-N > Cd (Fig. 4). The combination of these parameters constrained 44.3% of the community variance.
FIG 4.
Constrained analysis of principal coordinates (CAP) of detected OTUs and the selected seawater parameters. The percentage of variation explained by each axis is shown. Refer to Fig. 1 for treatment notation.
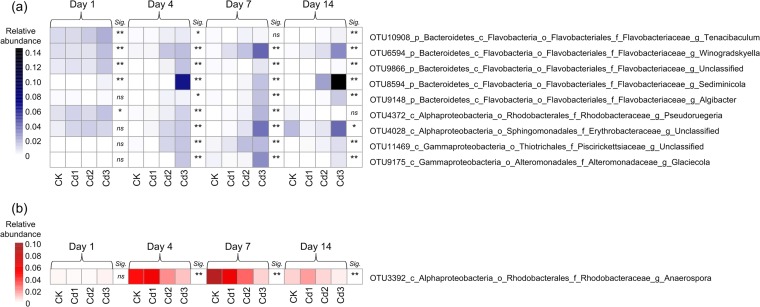
Representative phylotypes associated with Cd pollution level.
Twenty-five representative phylotypes (OTUs) correlated with Cd level were separated into positive and negative groups according to the correlation coefficients across the whole duration (see Table S3 in the supplemental material). The closely matched type strains provide additional information on taxonomic identification of some OTUs (see Table S4 in the supplemental material). Nine key phylotypes in the positive group, affiliated to the families Flavobacteriaceae (members of genera Tenacibaculum, Winogradskyella, Sediminicola, and Algibacter and an unclassified one), Rhodobacteraceae (a member of genus Pseudoruegeria), Erythrobacteraceae (a member of the genus Erythrobacter), Piscirickettsiaceae, and Alteromonadaceae (a member of the genus Glaciecola), showed high frequency (3 to 4 times) of positive correlation with the Cd level across the whole duration (Fig. 5a). Generally, the abundances of these phylotypes were greater at higher Cd levels (Cd2, Cd3). Most phylotypes in the negative group were from the families Flavobacteriaceae, Saprospiraceae, and Rhodobacteraceae. However, only one phylotype from the genus Anaerospora (OTU3392), affiliated to Rhodobacteraceae, showed a negative correlation with the Cd level at three time points (Fig. 5b).
FIG 5.
Heat maps of average relative abundances of key taxa positively (a) and negatively (b) associated with Cd level across the whole duration. At each time point, significance (Sig.) is noted as follows: **, significant Pearson's correlation (P < 0.01); *, significant Pearson's correlation (P < 0.05); ns, not significant (n = 16). Taxonomy of the OTUs was identified in the Greengenes database. Refer to Fig. 1 for treatment notation.
DISCUSSION
Temporal variability overwhelms Cd-induced patterns in the bacterial community.
In this study, high temporal variation in BCC was observed regardless of Cd level (see Fig. S1 in the supplemental material), and incubation time exhibited a more-extensive influence on dominant taxa than did the Cd level (Table 1). Accordingly, a similar trend in the beta diversity of the bacterial community was observed (Fig. 1a). Cd-induced patterns were obscured by temporal variability among time points and could be detected only within individual days. This is confirmed by the variation partitioning analysis indicating that temporal variability contributed more to bacterial community variance than Cd did (Table 3). Although the bacterial community structure shifted along the Cd gradient consistently from day 4, a slight Cd disturbance (10 μg liter−1) did not shift the community structure, suggesting that the response of the bacterioplankton community to Cd is dose dependent. This is in line with results from a previous study focusing on the response of bacterial community to pentachlorophenol (PCP) in tree hole microcosms (37).
Incubation time showed more-significant influences (P ≤ 0.01) on observed richness, Chao1 richness index, and phylogenetic diversity than Cd level did (P < 0.05; see Table S2 in the supplemental material). Cadmium pollution did not cause a decrease in the overall alpha diversity; on the contrary, we found an increase in all the metrics of bacterial communities in Cd2 and Cd3 microcosms at day 4. Actually, high diversity and evenness seemed to be common features in marine microbial communities of many chemically polluted environments (9), probably because pollution disturbance generated conditions for the proliferation of rare taxa with stress resistance. According to the widely reported metal resistance of bacteria (38–40), the acute response of the bacterial community to the higher Cd levels at the early stage might be due to the proliferation rate of resistant taxa being higher than the extinction rate of sensitive taxa. However, this trend disappeared later, indicating that the temporal succession overwhelms Cd-induced patterns in the alpha diversity of bacterial community.
Specific Cd-induced patterns of bacterial community in systems with high temporal variability.
Since marine bacterioplankton communities generally show high temporal and spatial variability (19, 41, 42) and less is known about the temporal dynamics of BCC under heavy metal pollution, we used here the manipulated microcosm experiment as a pilot study to explore the temporal trend of coastal BCC responding to Cd pollution level. Temporal succession of bacterial communities fit well with the similarity-time decay model (Fig. 3), which is consistent with our hypothesis. Here, the bacterial community turnover was much higher than those in marine environments in situ with gradual variability (15). This can be explained partly by the robust dynamics of environmental variables in a small water body (see Table S1 in the supplemental material) and further by the short duration of this study (15). Particularly, we found that elevated Cd levels decreased the temporal turnover rate (significant in Cd2 and Cd3 microcosms), meaning that Cd addition made the succession of bacterial community come about slowly. Relatively low temporal dynamics of microbial communities were observed in environments with high selective pressure such as wastewater treatment systems (43). Here, it seems that Cd showed a certain selective pressure to the temporal variability of BCC. Since the temporal turnover rate of microbial community was likely not correlated with taxon richness (15) and the Cd dose (at 0.01 mM) seemed not to decrease the abundance of bacterioplankton as previously reported (44), the Cd-induced trend in temporal turnover of BCC could result from the relatively high and stable evenness (presented by the greater mean value and lesser coefficient of variation [CV])of Pielou's J index over time at higher Cd levels [Cd2 and Cd3; data not shown]), leading to a more-stable bacterial community structure (15). Moreover, the Cd-induced inhibition of protozoan growth (45) leading to the decrease in predation pressure to bacterioplankton might contribute to this pattern. These points should be addressed in depth to reveal the mechanisms behind this pattern in future work.
A comprehensive study concluded that determining the temporal variability of microbial communities could provide a baseline for community changes to separate these processes from deterministic responses (15). Although a high temporal variability in the bacterial community compared with those previously reported in coastal ecosystems in situ (15) was observed, we can find similar Cd-induced patterns of communities at each time point from day 4 (Fig. 2), meaning that temporal variability did not mask the selective pressure of Cd on BCC. Particularly, the Cd-induced pattern of community structure showed some extent of predictability. The resilience of microbial communities to disturbances has been frequently reported (46, 47) and can be defined by the fact that the community presents a certain degree of recovery when the disturbance disappears or the disturbance intensity tends to decrease. For example, Ager et al. found that both the alpha diversity and the beta diversity of a bacterial community recovered due to the great dissipation rate of PCP after 4 days (37). Metals cannot be degraded and are supposed to influence the bacterial community more persistently. In this study, the alpha diversity tended to be uniform across Cd levels in the later stage of incubation (7 to 14 days; see Table S2 in the supplemental material). Also, phylogenetic beta diversity of bacterial communities among different Cd levels tended to converge during days 4 to 14 (Fig. 1b). Moreover, the slope of the NMDS axis 1-Cd linear model became less negative over time (Fig. 2). These results reflected the fact that the differences in community diversity along the Cd gradient became smaller in the later stage of the experiment than they were in the early stage, thus indicating some adaption of the bacterial community to Cd exposure. Another possible explanation could be the decrease in the concentrations of soluble Cd in the later duration due to the sorption of Cd to small particles (48).
Environmental variables involved in the interaction of temporal variability and Cd-induced patterns in bacterial community.
Robust temporal dynamics in seawater variables indicated that the system is highly unstable. Similar results have been documented in some ex situ microcosm/mesocosm studies (16, 23), possibly due to the higher metabolic rate in the smaller water body (23), in which the available nutrients could almost be transformed from local fractions rather than from the source at catchment. Moreover, the lack of sediment might also alter the chemical equilibrium and buffering capacity of the system (49, 50). We monitored the dynamic changes of the chemical variables to find key factors correlated with temporal variation of BCC. A number of studies have demonstrated that BCC can reflect biotic and abiotic processes over time (17, 19, 21, 47). In this study, Chl-a and DO showed the strongest correlations with community variation and shared a similar shaping direction (Fig. 4). Overall, a gradual vanishing of phytoplankton biomass over time was observed, while there was a slight decrease in DO in the later stage of incubation (see Table S1 in the supplemental material). The importance of phytoplankton and DO in shaping the marine bacterioplankton community has been confirmed before (51–53), as were other driving factors here (7, 16, 24). Phosphate and Cd share a similar direction in shaping the bacterial community (Fig. 4). The pattern might be due to the enhanced microbial transformation rate of PO43− resulting from the higher requirement of bioavailable polyphosphates by bacterioplankton cells for Cd detoxification (54), as indicated by generally higher concentrations of PO43−-P at higher Cd levels. Remarkably, Cd showed weaker correlation with community variation than any of the other driving variables did. Moreover, all these variables changed temporally (see Table S1 in the supplemental material), thus partly explaining why temporal variability overwhelms Cd-induced patterns in the BCC variation.
A combination of the key factors can only explain 44.3% of community variation, and thus some unmeasured factors could contribute to the unexplained variation. For example, the predation pressure of flagellates in shaping BCC through the size selection in bacterial communities has been frequently reported (23, 55). In addition, bacterial adherence to the bottle's inner surface could be considered a formation process of biofilm on an artificial surface, where a unique bacterial community composition was often observed compared with that in the surrounding water column (56, 57). But the influence of this process on the BCC in the water column was rarely revealed, which might also be a shaping force in bacterioplankton. The Cd-induced pattern referred to here was the community variation caused by the Cd addition, which could be directly induced by Cd stress (toxicity) and also could be due to its influence on the growth of phytoplankton (58) and protozoa (45). Furthermore, the interaction of Cd level and incubation time in seawater variables (see Table S1 in the supplemental material) could explain their interaction in the variation of BCC (Table 3). In conclusion, the variation of BCC could be an integrated reflection of the complicated interactions among time, Cd, and environmental factors (including both biotic and abiotic ones).
The purpose of this pilot study is to provide insight into the existence of specific Cd-induced patterns in BCC temporal variation in coastal water. However, the results from small-scale studies like microcosm experiments usually cannot be directly extrapolated to an in situ ecosystem because of inadequate or absent consideration of whole communities, sediment/air-water interactions, physical phenomena like wind and water renewal, and temporal events like seasonality (59). The 4-liter manipulated system here did not include sediment, which could influence the Cd-induced pattern of BCC variation through bacterial dispersal (60), release of nutrient (61), and interactions of Cd and sediment particles (62) during sediment-water exchange. Moreover, the weaker physical mixing, limited source of nutrient, and lack of activities of wide-ranging higher organisms in the bottles could also contribute to a reduced complexity compared to that of a coastal ecosystem (59). Therefore, the differentiation in the variation of BCC in coastal ecosystems compared with the microcosms here could be not only the lower temporal turnover but also the shaping forces in temporal patterns of bacterioplankton (19, 41, 42) and their interactions with Cd, suggesting the nondeterminacy and complexity in the Cd-induced pattern of BCC variation on an ecosystem scale. However, using 16S pyrosequencing technique, recent studies reported repeatable and predictable patterns in BCC across different temporal scales (15, 19, 21), as observed in this study. Therefore, the temporal dynamics of BCC responses to Cd exposure in coastal water in situ might show a trend similar to the one that we observed here. Future investigations on larger scales with more-complete components of whole ecosystems like mesocosm studies in situ or even ecosystem-scale experiments across longer temporal scales are needed to validate this observed pattern.
Key taxa associated with Cd pollution level on a temporal scale.
Recent studies have focused on screening key taxa associated to pollutants (63) and the health status of humans/animals (21, 64). A comprehensive study suggested that if a certain taxon could be consistently and repeatedly detected at great abundance in a location, there would be a high possibility that this taxon is environment specific rather than a transient passer-by (19). We extend this conception to propose that if a taxon shows frequent and stable correlation with Cd level, it is possible that this taxon plays an important ecological role in response to Cd exposure. Metal resistance of bacteria affiliated to various species has been widely reported in soils (65), sediments (66), and aquatic environments (39). Five of nine key taxa positively associated with Cd level were from Flavobacteriaceae, including a member of the genus Tenacibaculum (Fig. 5). A recent study reported the mercury resistance of a member of Tenacibaculum from marine Flavobacteriaceae (67). Moreover, flavobacteria made up 70% of a Cd-resistant bacterial community in soil amended with sewage sludge (68) and have not been extensively studied but could be major components of the bacterial community in metal-polluted marine environments (67). We also found a key taxon (OTU4028) from the genus Erythrobacter (Fig. 5; see also Table S4 in the supplemental material), and some members of this genus have genes encoding metal efflux proteins associated with metal resistance (69). However, the members of Rhodobacteraceae, Piscirickettsiaceae, and Alteromonadaceae were not associated with metal resistance in previous reports. In conclusion, these nine taxa significantly contributed to the Cd-induced pattern in the BCC variation over time, and high temporal variability of BCC did not suppress the characteristics of their responses to Cd exposure. On the other hand, most taxa in the negative group showed a one-time negative correlation with Cd level, except for a member of the genus Anaerospora (OTU3392), which showed a high frequency of correlation with the Cd level. The underlying functional roles that these taxa play in Cd-polluted coastal water should be considered in further works.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the anonymous reviewers for their valuable comments and suggestions to improve the manuscript.
This work was financially supported by the National High Technology Research and Development Program of China (863 Program, 2012AA092000), Zhejiang Provincial Natural Science Foundation of China (LQ14D060003), the Natural Science Foundation of Ningbo (2014A610095), the Scientific Research Project of Education Department of Zhejiang Province (Y201430360), Experimental Technology Research and Development Project (SYJS201402) and Natural Science Foundation (XYL14009) of Ningbo University, the Research Fund from 2011 Center of Modern Marine Aquaculture of East China Sea, and the KC Wong Magna Fund of Ningbo University.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02562-14.
REFERENCES
- 1.Süren E, Yilmaz S, Türkoglu M, Kaya S. 2007. Concentrations of cadmium and lead heavy metals in Dardanelles seawater. Environ Monit Assess 125:91–98. doi: 10.1007/s10661-006-9242-5. [DOI] [PubMed] [Google Scholar]
- 2.Yilmaz S, Sadikoglu M. 2011. Study of heavy metal pollution in seawater of Kepez harbor of Canakkale (Turkey). Environ Monit Assess 173:899–904. doi: 10.1007/s10661-010-1432-5. [DOI] [PubMed] [Google Scholar]
- 3.Ma HQ, Song Q, Wang XC. 2009. Accumulation of petroleum hydrocarbons and heavy metals in clams (Ruditapes philippinarum) in Jiaozhou Bay, China. Chin J Ocean Limnol 27:887–897. doi: 10.1007/s00343-009-9223-y. [DOI] [Google Scholar]
- 4.Pan K, Wang WX. 2012. Trace metal contamination in estuarine and coastal environments in China. Sci Total Environ 421-422:3–16. doi: 10.1016/j.scitotenv.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Herve-Fernandez P, Houlbreque F, Boisson F, Mulsow S, Teyssie JL, Oberhaensli F, Azemard S, Jeffree R. 2010. Cadmium bioaccumulation and retention kinetics in the Chilean blue mussel Mytilus chilensis: seawater and food exposure pathways. Aquat Toxicol 99:448–456. doi: 10.1016/j.aquatox.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Waisberg M, Joseph P, Hale B, Beyersmann D. 2003. Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology 192:95–117. doi: 10.1016/S0300-483X(03)00305-6. [DOI] [PubMed] [Google Scholar]
- 7.del Giorgio PA, Newell REI. 2012. Phosphorus and DOC availability influence the partitioning between bacterioplankton production and respiration in tidal marsh ecosystems. Environ Microbiol 14:1296–1307. doi: 10.1111/j.1462-2920.2012.02713.x. [DOI] [PubMed] [Google Scholar]
- 8.Sebastian M, Pitta P, Gonzalez JM, Thingstad TF, Gasol JM. 2012. Bacterioplankton groups involved in the uptake of phosphate and dissolved organic phosphorus in a mesocosm experiment with P-starved Mediterranean waters. Environ Microbiol 14:2334–2347. doi: 10.1111/j.1462-2920.2012.02772.x. [DOI] [PubMed] [Google Scholar]
- 9.Nogales B, Lanfranconi MP, Pina-Villalonga JM, Bosch R. 2011. Anthropogenic perturbations in marine microbial communities. FEMS Microbiol Rev 35:275–298. doi: 10.1111/j.1574-6976.2010.00248.x. [DOI] [PubMed] [Google Scholar]
- 10.Lu L, Wu RSS. 2003. Recolonization and succession of subtidal macrobenthic infauna in sediments contaminated with cadmium. Environ Pollut 121:27–38. doi: 10.1016/S0269-7491(02)00210-5. [DOI] [PubMed] [Google Scholar]
- 11.Mucha AP, Vasconcelos MTSD, Bordalo AA. 2004. Vertical distribution of the macrobenthic community and its relationships to trace metals and natural sediment characteristics in the lower Douro estuary, Portugal. Coast Shelf Sci 59:663–673. doi: 10.1016/j.ecss.2003.11.010. [DOI] [Google Scholar]
- 12.Trannum HC, Olsgard F, Skei JM, Indrehus J, Øverås S, Eriksen J. 2004. Effects of copper, cadmium and contaminated harbour sediments on recolonisation of soft-bottom communities. J Exp Mar Biol Ecol 310:87–114. doi: 10.1016/j.jembe.2004.04.003. [DOI] [Google Scholar]
- 13.Kang S, Van Nostrand JD, Gough HL, He ZL, Hazen TC, Stahl DA, Zhou JZ. 2013. Functional gene array-based analysis of microbial communities in heavy metals-contaminated lake sediments. FEMS Microbiol Ecol 86:200–214. doi: 10.1111/1574-6941.12152. [DOI] [PubMed] [Google Scholar]
- 14.Caroppo C, Stabili L, Aresta M, Corinaldesi C, Danovaro R. 2006. Impact of heavy metals and PCBs on marine picoplankton. Environ Toxicol 21:541–551. doi: 10.1002/tox.20215. [DOI] [PubMed] [Google Scholar]
- 15.Shade A, Caporaso JG, Handelsman J, Knight R, Fierer N. 2013. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J 7:1493–1506. doi: 10.1038/ismej.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindh MV, Riemann L, Baltar F, Romero-Oliva C, Salomon PS, Granéli E, Pinhassi J. 2013. Consequences of increased temperature and acidification on bacterioplankton community composition during a mesocosm spring bloom in the Baltic Sea. Environ Microbiol Rep 5:252–262. doi: 10.1111/1758-2229.12009. [DOI] [PubMed] [Google Scholar]
- 17.Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A, Marrase C. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol 70:6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langenheder S, Jürgens K. 2001. Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol Oceanogr 46:121–134. doi: 10.4319/lo.2001.46.1.0121. [DOI] [Google Scholar]
- 19.Fortunato CS, Eiler A, Herfort L, Needoba JA, Peterson TD, Crump BC. 2013. Determining indicator taxa across spatial and seasonal gradients in the Columbia River coastal margin. ISME J 7:1899–1911. doi: 10.1038/ismej.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris R, Vergin K, Cho J, Rappe M, Carlson C, Giovannoni S. 2005. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50:1687–1696. doi: 10.4319/lo.2005.50.5.1687. [DOI] [Google Scholar]
- 21.Xiong JB, Zhu JL, Wang K, Wang X, Ye XS, Liu L, Zhao QF, Hou MH, Qiuqian LL, Zhang DM. 2014. The temporal scaling of bacterioplankton composition: high turnover and predictability during shrimp cultivation. Microb Ecol 67:256–264. doi: 10.1007/s00248-013-0336-7. [DOI] [PubMed] [Google Scholar]
- 22.Wu QL, Hahn MW. 2006. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ Microbiol 8:1660–1666. doi: 10.1111/j.1462-2920.2006.01049.x. [DOI] [PubMed] [Google Scholar]
- 23.Ren LJ, He D, Zeng J, Wu QL. 2013. Bacterioplankton communities turn unstable and become small under increased temperature and nutrient-enriched conditions. FEMS Microbiol Ecol 84:614–624. doi: 10.1111/1574-6941.12089. [DOI] [PubMed] [Google Scholar]
- 24.Xiong JB, Ye XS, Wang K, Chen HP, Hu CJ, Zhu JL, Zhang DM. 2014. The biogeography of the sediment bacterial community responds to a nitrogen pollution gradient in the East China Sea. Appl Environ Microbiol 80:1919–1925. doi: 10.1128/AEM.03731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang DM, Wang X, Xiong JB, Zhu JL, Wang YN, Zhao QF, Chen HP, Guo AN, Wu JF, Dai HP. 2014. Bacterioplankton assemblages as biological indicators of shrimp health status. Ecol Indic 38:218–224. doi: 10.1016/j.ecolind.2013.11.002. [DOI] [Google Scholar]
- 26.APHA. 1976. Standard methods for the examination of water and wastewater, 14th ed. American Public Health Association, Washington, DC. [Google Scholar]
- 27.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez-Peña A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows integration and analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 30.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price M, Dehal P, Arkin A. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke KR, Gorley RN. 2001. PRIMER v5: user manual/tutorial. PRIMER-E, Plymouth Marine Laboratory, Lutton, Ivybridge, Devon, United Kingdom. [Google Scholar]
- 34.R Development Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://cran.r-project.org. [Google Scholar]
- 35.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 36.Anderson MJ, Willis TJ. 2003. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84:511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2. [DOI] [Google Scholar]
- 37.Ager D, Evans S, Li H, Lilley AK, Van Der Gast CJ. 2010. Anthropogenic disturbance affects the structure of bacterial communities. Environ Microbiol 12:670–678. doi: 10.1111/j.1462-2920.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 38.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Jose J, Giridhar R, Anas A, Loka-Bharathi PA, Nair S. 2011. Heavy metal pollution exerts reduction/adaptation in the diversity and enzyme expression profile of heterotrophic bacteria in Cochin estuary, India. Environ Pollut 159:2775–2780. doi: 10.1016/j.envpol.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Trevors JT, Oddie KM, Belliveau BH. 1985. Metal resistance in bacteria. FEMS Microbiol Lett 32:39–54. doi: 10.1111/j.1574-6968.1985.tb01181.x. [DOI] [Google Scholar]
- 41.Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. 2012. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J 6:554–563. doi: 10.1038/ismej.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, Field D. 2012. Defining seasonal marine microbial community dynamics. ISME J 6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, Cummings TA, Beers AR, Knight R, Angenent LT. 2011. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci U S A 108:4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stepanauskas R, Glenn TC, Jagoe CH, Tuckfield RC, Lindell AH, King CJ, McArthur JV. 2006. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ Microbiol 8:1510–1514. doi: 10.1111/j.1462-2920.2006.01091.x. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez-Leborans G, Olalla-Herrero Y. 2000. Toxicity and bioaccumulation of lead and cadmium in marine protozoan communities. Ecotoxicol Environ Saf 47:266–276. doi: 10.1006/eesa.2000.1944. [DOI] [PubMed] [Google Scholar]
- 46.Allison SD, Martiny JB. 2008. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shade A, Read JS, Youngblut ND, Fierer N, Knight R, Kratz TK, Lottig NR, Roden EE, Stanley EH, Stombaugh J, Whitaker RJ, Wu CH, McMahon KD. 2012. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J 6:2153–2167. doi: 10.1038/ismej.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi N, Jamet JL. 2008. In situ heavy metals (copper, lead and cadmium) in different plankton compartments and suspended particulate matter in two coupled Mediterranean coastal ecosystems (Toulon Bay, France). Mar Pollut Bull 56:1862–1870. doi: 10.1016/j.marpolbul.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 49.Farías L, Graco M, Ulloa O. 2004. Temporal variability of nitrogen cycling in continental-shelf sediments of the upwelling ecosystem off central Chile. Deep-Sea Res Part II 51:2491–2505. doi: 10.1016/j.dsr2.2004.07.029. [DOI] [Google Scholar]
- 50.Burt WJ, Thomas H, Fennel K, Horne E. 2013. Sediment-water column fluxes of carbon, oxygen and nutrients in Bedford Basin, Nova Scotia, inferred from 224Ra measurements. Biogeosciences 10:53–66. doi: 10.5194/bg-10-53-2013. [DOI] [Google Scholar]
- 51.Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J 1:660–662. doi: 10.1038/ismej.2007.79. [DOI] [PubMed] [Google Scholar]
- 52.Niu Y, Shen H, Chen J, Xie P, Yang X, Tao M, Ma ZM, Qi M. 2011. Phytoplankton community succession shaping bacterioplankton community composition in Lake Taihu, China. Water Res 45:4169–4182. [DOI] [PubMed] [Google Scholar]
- 53.Zeng J, Bian YQ, Xing P, Wu QL. 2012. Macrophyte species drive the variation of bacterioplankton community composition in a shallow freshwater lake. Appl Environ Microbiol 78:177–184. doi: 10.1128/AEM.05117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seufferheld MJ, Alvarez HM, Farias ME. 2008. Role of polyphosphates in microbial adaptation to extreme environments. Appl Environ Microbiol 74:5867–5874. doi: 10.1128/AEM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jürgens K, Matz C. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie van Leeuwenhoek 81:413–434. doi: 10.1023/A:1020505204959. [DOI] [PubMed] [Google Scholar]
- 56.Elifantz H, Horn G, Ayon M, Cohen Y, Minz D. 2013. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol Ecol 85:348–357. doi: 10.1111/1574-6941.12122. [DOI] [PubMed] [Google Scholar]
- 57.Jones PR, Cottrell MT, Kirchman DL, Dexter SC. 2007. Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb Ecol 53:153–162. doi: 10.1007/s00248-006-9154-5. [DOI] [PubMed] [Google Scholar]
- 58.Echeveste P, Agustí S, Tovar-Sánchez A. 2012. Toxic thresholds of cadmium and lead to oceanic phytoplankton: cell size and ocean basin-dependent effects. Environ Toxicol Chem 31:1887–1894. doi: 10.1002/etc.1893. [DOI] [PubMed] [Google Scholar]
- 59.Schindler DW. 1998. Replication versus realism: the need for ecosystem-scale experiments. Ecosystems 1:323–334. doi: 10.1007/s100219900026. [DOI] [Google Scholar]
- 60.Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA. 2013. Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci U S A 110:4651–4655. doi: 10.1073/pnas.1217767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalnejais LH, Martin WR, Bothner MH. 2010. The release of dissolved nutrients and metals from coastal sediments due to resuspension. Mar Chem 121:224–235. doi: 10.1016/j.marchem.2010.05.002. [DOI] [Google Scholar]
- 62.Schlekat CE, Decho AW, Chandler GT. 1998. Sorption of cadmium to bacterial extracellular polymeric sediment coatings under estuarine conditions. Environ Toxicol Chem 17:1867–1874. doi: 10.1002/etc.5620170930. [DOI] [Google Scholar]
- 63.Sun MY, Dafforn KA, Johnston EL, Brown MV. 2013. Core sediment bacteria drive community response to anthropogenic contamination over multiple environmental gradients. Environ Microbiol 15:2517–2531. doi: 10.1111/1462-2920.12133. [DOI] [PubMed] [Google Scholar]
- 64.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ansari MI, Malik A. 2007. Biosorption of nickel and cadmium by metal resistant bacterial isolates from agricultural soil irrigated with industrial wastewater. Bioresour Technol 98:3149–3153. doi: 10.1016/j.biortech.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Li KF, Ramakrishna W. 2011. Effect of multiple metal resistant bacteria from contaminated lake sediments on metal accumulation and plant growth. J Hazard Mater 189:531–539. doi: 10.1016/j.jhazmat.2011.02.075. [DOI] [PubMed] [Google Scholar]
- 67.Allen RC, Tu YK, Nevarez MJ, Bobbs AS, Friesen JW, Lorsch JR, McCauley JA, Voet JG, Hamlett NV. 2013. The mercury resistance (mer) operon in a marine gliding flavobacterium, Tenacibaculum discolor 9A5. FEMS Microbiol Ecol 83:135–148. doi: 10.1111/j.1574-6941.2012.01460.x. [DOI] [PubMed] [Google Scholar]
- 68.Barkay T, Tripp SC, Olson BH. 1985. Effect of metal-rich sewage sludge application on the bacterial communities of grasslands. Appl Environ Microbiol 49:333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouzat JL, Hoostal MJ. 2013. Evolutionary analysis and lateral gene transfer of two-component regulatory systems associated with heavy-metal tolerance in bacteria. J Mol Evol 76:267–279. doi: 10.1007/s00239-013-9558-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.