Abstract
Biocatalysts are essential for the development of bioprocesses efficient for plant biomass degradation. Previously, a metagenomic clone containing DNA from termite gut microbiota was pinpointed in a functional screening that revealed the presence of arabinofuranosidase activity. Subsequent genetic and bioinformatic analysis revealed that the DNA fragment belonged to a member of the genus Bacteroides and encoded 19 open reading frames (ORFs), and annotation suggested the presence of hypothetical transporter and regulator proteins and others involved in the catabolism of pentose sugar. In this respect and considering the phenotype of the metagenomic clone, it was noted that among the ORFs, there are four putative arabinose-specific glycoside hydrolases, two from family GH43 and two from GH51. In this study, a thorough bioinformatics analysis of the metagenomic clone gene cluster has been performed and the four aforementioned glycoside hydrolases have been characterized. Together, the results provide evidence that the gene cluster is a polysaccharide utilization locus dedicated to the breakdown of the arabinan component in pectin and related substrates. Characterization of the two GH43 and the two GH51 glycoside hydrolases has revealed that each of these enzymes displays specific catalytic capabilities and that when these are combined the enzymes act synergistically, increasing the efficiency of arabinan degradation.
INTRODUCTION
It is a widely held opinion that biocatalysts will play an essential role in the development of the bio-based economy, basically because these are exquisitely adapted to the modification of complex biomolecules and can perform cascade reactions. In this respect, in nature microorganisms generally deploy incredibly complex arsenals of enzymes, notably glycoside hydrolases (GHs), to break down complex plant biomass in sophisticated processes that are not yet completely elucidated but which are very often characterized by complex interplay between different GHs and between GHs and other enzymes and abiotic reagents (1, 2). Harnessing the power of enzymes for the industrial conversion of plant biomass is thus not an easy task, since microbial strategies are not easily imitated, especially when process economics has to be taken into account. However, in-depth understanding of the processes involved and the interplay between individual enzymes is a prerequisite for the formulation of cocktails that are used to hydrolyze specific plant cell wall structures.
The plant cell wall contains a large number of polysaccharides (3), whose complexity can be exemplified by that of pectins, a heterogeneous group of polysaccharides that can comprise as much as 30% of dicot cell walls and gymnosperms (4, 5). Among pectins, rhamnogalacturonan I (RGI) is composed of a backbone made up of repeating units of α-d-galacturonic acid and α-l-rhamnose, which is highly decorated with side chain structures, arabinan being the most abundant of these (6). Arabinan consists of a backbone containing α-1,5-linked l-arabinofuranosyl residues that can be substituted with α-1,2 and α-1,3-linked l-arabinofuranosyl side chains (6). Previously, in vitro studies have provided evidence of interactions between neutral pectins, such as arabinan, and cellulose (7), an observation that provides a rationale for the fact that the inclusion of pectinases in cellulase cocktails can be beneficial for the hydrolysis of certain biomass pulps (8, 9).
The CAZy classification groups GHs into families according to sequence similarities (10) (www.cazy.org). Regarding arabinan degradation, at least three distinct enzymatic activities are required and the associated CAZymes are found in different CAZy families. Exo-α-l-1,2- and exo-α-l-1,3-arabinofuranosidases (EC 3.2.1.55) are encountered in families GH43, GH51, and GH62, and exo-α-l-1,5-arabinofuranosidases (EC 3.2.1.55) are found in GH43 and GH54, whereas endo-α-l-1,5-arabinanases (EC 3.2.1.99) have thus far only been described in GH43. In addition, exo-α-l-1,5-arabinanase (EC 3.2.1.-) has been described in the family GH93. These latter enzymes release arabinobiose as a major product from linear arabinan (11, 12).
In the genomes of biomass-degrading bacteria, notably those present in gut environments, GH-encoding sequences are often grouped together in loci that are dedicated to the breakdown of specific polysaccharides (13). Good examples of such loci, or gene clusters, have been identified in the genomes of Gram-negative bacteria belonging to the genus Bacteroides, which are known to degrade a wide variety of plant polysaccharides (14, 15). Polysaccharide utilization loci (PUL) usually encode cell envelope-associated proteins that confer to bacteria the ability to specifically recognize and hydrolyze polysaccharides and transport the sugar products into the cell for further metabolism. An early and now well-studied example of a PUL is the starch utilization system (Sus) that is comprised of an outer-membrane-associated binding complex, composed of four proteins (SusC, SusD, SusE, and SusF), and a starch hydrolytic machinery, composed of SusG, SusA, and SusB. Together, these Sus components hydrolyze starch to glucose (16). More recently, other Sus-like PUL have been described for the hydrolysis of hemicelluloses and pectins, notably in members of the genus Bacteroides (2, 17).
In recent years, the discovery of new GHs has been accelerated through the use of high-throughput methods, such as metagenomics, which not only reveal individual enzyme-coding sequences but also PUL that provide precious clues as to how complex polysaccharides are degraded by microorganisms that produce large enzyme arrays (18–22). In a recently published study (22), we described the metagenomic analysis of the microbiota of the fungus-growing termite, Pseudacanthotermes militaris. In that study, we identified several GH clusters, including one that is part of a 41-kb metagenomic DNA fragment (designated clone G12) that has been correlated with an arabinofuranosidase-type phenotype when the clone is harbored by Escherichia coli cells. According to sequence analysis, this cluster is composed of two putative GH43 arabinanases or arabinofuranosidases (Abn43A and Abn43B) and two GH51 arabinofuranosidases (Abf51A and Abf51B, with the latter being part of a multimodular enzyme). In addition, the cluster appears to belong to a PUL, which also encodes several putative protein components of a pentose metabolism pathway. Finally, taxonomic assignment of the genes present in clone G12 fragment has led us to postulate that the fragment arises from a bacterium belonging to the genus Bacteroides.
In the present study, we set out to better understand the function of this newly identified PUL candidate, performing biochemical characterization of each of the putative arabinose-active enzyme components in order to determine the specificity and the action of each of these. As a result, we have succeeded in revealing enzyme synergy and provided evidence that suggests that the PUL under study is an arabinan utilization locus, as well as providing a biochemical description of each enzyme.
MATERIALS AND METHODS
The fosmid sequence studied here was previously deposited under GenBank accession number HF548278.
Bioinformatic analysis of the arabinose utilization locus.
Promoter regions and ribosome binding sites (RBS) were tentatively identified by comparing query sequences with previously described consensus sequences (23) and (24). Rho-independent terminators were detected using ARNold Finding Terminators software (http://rna.igmors.u-psud.fr/toolbox/arnold/) and taxonomic assignments were achieved using the MEGAN software (http://ab.inf.uni-tuebingen.de/software/megan/) (22, 25, 26). Putative signal peptide sequence analysis were performed using SignalP software (27). Hemicellulolytic CAZymes and single open reading frames (ORFs) were analyzed both by using blastx (http://blast.ncbi.nlm.nih.gov/), searching against the nonredundant NCBI and Swiss-Prot databases, and by performing another search using the CAZy database (http://www.cazy.org/) as previously described (22).
Gene cloning and standard protein procedures.
The sequences encoding Abf51A (CCO20976.1), Abf51B (CCO20994.1), Abn43A (CCO20984.1) and Abn43B (CCO20993.1), all previously identified within the metagenomic fragment G12 (HF548278), were cloned into the plasmid pET28a and expressed as previously described (22). Abf51B is an enzyme displaying an unusual multimodular configuration of the type CBM4-Abf51B-Abf43C. The sequences encoding CBM4-Abf51Btrunc and Abf43Ctrunc were cloned in the present study according to the same method and using the forward primers 5′-GGGGGGGCTAGCGCGCAAACCAACGAACTGGTG-3′ and 5′-GGGGGGCATATGAAAGAGACCGCTTTCGTAGCCACG-3′, respectively, and the reverse primers 5′-CCCCCCCTCGAGTCATTGTGGCGTAAAGATATAGACTGC-3′ and 5′-CCCCCCCTCGAGCTATATCCCCTCGGAGGTCATC-3′, respectively. The recombinant proteins, all harboring His6 tags at their N-terminal extremities, were purified by applying cell lysates to Talon metal affinity resin (Clontech, USA), with elution of the proteins in 100 mM imidazole being achieved according to the manufacturer's instructions. After purification, Abn43B was concentrated by using an Amicon centrifugal unit (50-kDa cutoff). All purified recombinant proteins were conserved at 4°C in 50 mM phosphate buffer (pH 7.0).
Protein concentrations were determined by measuring the absorbance of purified protein solutions at 280 nm using a Cary 100 Bio UV-visible spectrophotometer (Agilent Technologies, Santa Clara, CA) and applying the Beer-Lambert relationship using extinction coefficient values calculated by using ProtParam software (http://web.expasy.org/protparam/) for each enzyme. Similarly, theoretical protein molecular masses were calculated using ProtParam software.
Enzyme activity determination.
Enzyme activities on synthetic para-nitrophenol (pNP) glycosides were measured by monitoring the release of pNP-OH spectrophotometrically (405 nm) using a microtiter plate absorbance reader (Sunrise; Tecan, Switzerland). Working in triplicate, an aliquot of enzyme was added to 50 mM phosphate buffer (pH 7.0) containing a pNP-glycoside (5 mM) and bovine serum albumin (BSA; 1 mg/ml), which was then incubated at 30°C, removing 100-μl samples at regular intervals. Upon removal, the samples were immediately combined with 500 μl of cold 1 M Na2CO3 and placed on ice before the absorbance was determined. The specific activity was determined by comparing the experimental absorbance values to a pNP-OH standard curve ranging from 0 to 1 mM. One unit of activity was defined as the amount of enzyme required to release 1 μmol of pNP-OH per min. pNP-α-l-arabinofuranoside (pNP-Araf), pNP-β-d-xylopyranoside (pNP-Xylp), pNP-β-d-galactofuranoside (pNP-Galf), and pNP-β-d-glucopyranoside (pNP-Glcp) were purchased from Carbosynth, Ltd. (Berkshire, United Kingdom).
The amount of reducing sugars released from polysaccharides during enzyme-catalyzed hydrolysis was determined discontinuously by using 3,5-dinitrosalicylic acid (DNS) reagent. Sugar beet arabinan (SBA), debranched arabinan (DA), wheat arabinoxylan (WAX), and larch arabinogalactan (LAG), all purchased from Megazyme International (Bray, Ireland), were used as substrates at a concentration of 10 mg/ml. Reactions, performed in triplicate, were incubated for 1 h at 30°C in 50 mM phosphate buffer (pH 7.0) containing BSA (1 mg/ml). At regular intervals, 100 μl of the reaction were added to 100 μl of DNS and kept on ice until all samples were ready to be placed at 95°C. After the samples were heated for 5 min and cooled on ice, 50-μl portions were diluted in 250 μl of deionized water for absorbance measurement at 540 nm. A standard curve was prepared using l-arabinose. This was used to calculate the specific activities, which were expressed as the quantity of arabinose released (in μmol) per liter per min and per mg of protein.
Determination of pH and temperature optima.
The apparent optimal temperature for activity of the different enzymes was determined by measuring the enzyme activity on the preferred substrate in the temperature range from 25 to 55°C. Reactions were conducted in a solution of phosphate buffer 50 mM (pH 7.0) supplemented with 1 mg/ml BSA and an appropriate amount of substrate and enzyme. Although the actual pH of the phosphate buffer was lower (−0.14 pH units) at 55°C compared to the temperature at which the buffer was prepared (21.5°C), this was not taken into account, because the pH optima were generally estimated with an accuracy of less than ±0.5 pH units. For Abf51A and Abf51Btrunc, reactions were performed using 93 and 9 μM enzyme, respectively, with 5 mM pNP-Araf. For Abn43A (92 μM) and Abn43B (0.5 μM), SBA and DA (at 0.5% [wt/vol]), respectively, were used as the substrates. All enzymatic assays were carried out in triplicate, and at least seven temperatures were assayed for each enzyme.
The apparent optimal pH for the activity of each enzyme was determined in an identical manner, except that all reactions were performed at 30°C in the pH range from 4.0 to 8.0 (measurements were performed using at least seven pH values). For assays performed at pH 4.0 to 6.0, the 50 mM phosphate buffer was replaced by 50 mM citrate buffer.
Enzyme stability assays.
To ascertain the effects of temperature and pH on enzyme stability, triplicate samples of enzymes were incubated at least in three different buffer (pH) conditions and at three different temperatures for between 15 min and 24 h before determining the enzyme activity using the previously described protocol and the enzyme's preferred substrate and optimal pH or temperature. The measured activities were expressed as a percentage of the normal enzyme activity (i.e., the activity measured with no incubation).
Determination of kinetic parameters.
To determine Michaelis-Menten constants, the initial rate of enzyme activity was measured under optimal operating conditions (pH and temperature) using a range of substrate concentrations (0.125 to 6% [wt/vol] for polysaccharides and 0.2 to 6 mM for pNP-glycosides) and collecting at least seven data points in triplicate in each case. The data analysis was performed using the enzyme kinetics module embedded in the SigmaPlot 11.0 software (Systat Software, USA). This provided values for kcat and Km. In the case of reactions involving polysaccharides, the computed Km value was necessarily only an apparent value.
In addition, Abn43B catalytic parameters were determined on arabino-oligosaccharides (AOS) arabinobiose (A2) to arabinohexaose (A6) (Megazyme International, Bray, Ireland). Abn43B (0.45 μM) was incubated with individual AOS at concentrations ranging from 0.1 to 5 mM in 1 ml of 50 mM phosphate buffer (pH 6.0) supplemented with 1 mg/ml BSA, in duplicate in each case. At regular time intervals (i.e., at 0, 2, 5, 7, 10, 15, 20, 30, and 60 min), aliquots were removed and analyzed using high-performance anionic-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using an ICS 3000 dual (Dionex, France) equipped with Carbo-Pac PA-100 analytical and guard columns (250 by 2 mm and 50 by 2 mm, respectively). To perform HPAEC analyses, 5 μl of sample was injected, and analytes were separated in 500 mM sodium acetate–150 mM NaOH at a 1-ml/min flow rate. Calibration was achieved using arabinose and AOS (A2, A3, A4, A5, and A6) at concentrations from 5 to 60 mg/liter. The catalytic constants kcat and Km were determined using nonlinear regression from the enzyme kinetics module embedded in the SigmaPlot 11.0 software.
Determination of the mode of action of the enzymes alone and in combined reactions.
To procure a preliminary understanding of the linkage preferences of the four arabinose-active enzymes, the activity of each one was evaluated on SBA and on linear arabinan (LA), a polymer of α-1,5-linked l-arabinosyl residues. These tests were achieved by adding 2 μg of enzyme to a 1 ml of 50 mM phosphate buffer (pH 6.5) containing 1 mg/ml BSA and 0.5% (wt/vol) polysaccharide substrate, followed by incubation at 30°C. After mixing, 100-μl aliquots of the reaction mixture were removed at regular intervals (at 0, 1, 2, 4, 8, and 24 h), and the reaction was stopped by the immediate addition of the DNS reagent, followed by boiling for 5 min, cooling on ice, and dilution in 50 μl in 250 μl of deionized water. Afterward, the absorbance was measured at 540 nm in order to quantify free reducing ends. The reaction mixture remaining at the end of the kinetics study was heated at 95°C for 15 min to stop the reaction, and then the products were analyzed by HPAEC-PAD (see above).
Having investigated the action of single enzymes on arabinans, similar experiments were performed using enzyme combinations in which enzymes were deployed pairwise with three enzymes or all four enzymes at concentrations of 2 μg/ml each. The overall experimental protocol was similar to that described above. Both DNS reducing-sugar and HPAEC-PAD analyses were performed. For each reaction, a synergistic index was calculated by determining the ratio between experimentally derived activities and theoretical results obtained by the addition of individual activities.
RESULTS
Analysis of the G12 fosmid: sequence, organization, and annotation.
The G12 metagenomic clone described by Bastien et al. was initially isolated by using the chromogenic substrate 5-bromo-4-chloro-3-indolyl-α-l-arabinofuranoside, which primarily reveals α-l-arabinofuranosidase activity. This finding was corroborated by the fact that a cell lysate of clone G12 was able to hydrolyze pNP-Araf and by the results of a bioinformatic analysis that revealed the presence of four putative arabinofurano-hydrolases (Fig. 1), including two putative GH51 α-l-arabinofuranosidases, one of which forms a larger, multidomain enzyme (22).
FIG 1.

Genetic map of the 41-kb fosmid containing arabinose and arabinan utilization genes from the G12 metagenomic clone. The letters P and Ω indicate, respectively, the putative promoter rho-independent transcription termination sites. Black segments represent ORFs encoding glycoside hydrolases, while gray segments correspond to ORFs encoding putative pentose metabolism enzymes. White boxes represent ORFs encoding proteins putatively involved in arabinan and arabinose recognition and utilization. No hypothetical function could be attributed to ORF16 and ORF17.
Pursuing previous bioinformatics work that revealed the four putative arabinofurano-hydrolases and allowed the assignment of the 41-kb metagenomic fragment to the genus Bacteroides (22), we determined in the present study that the DNA fragment contains a total of 19 ORFs, as well as four promoter sites, 18 ribosome binding sites (RBS), and two rho-independent terminator regions (Fig. 1). Overall, these results imply that the fragment bears three gene operons that encode a total of 19 putative proteins. The results of the BLAST analysis of the individual protein sequences revealed that most of these, except those encoded by ORF16 and ORF17, displayed strong similarities to protein components of known polysaccharide utilization operons, including proteins involved in polysaccharide recognition, degradation, or monosaccharide utilization (Table 1). The araL-araK and araJ-araI pairs of ORFs found in the operon araLKJIH-abnA-araGFEDCB-araA-abfA encode proteins that share more than 80% identity with the susC-susD pair found in numerous Bacteroides species, especially the one from B. thetaiotaomicron (15). On the same operon, araGFEDCB encodes proteins that share between 30 and 79% identity with proteins involved in l-arabinose metabolic pathways. Finally, araA corresponds to a putative GH97 enzyme, a family of proteins that has previously been observed in sus multigene clusters (28), and the abfA gene encodes an enzyme that is classified as a GH51 arabinofuranosidase. The second operon is composed of orf16, orf17, and araM. The latter (araM) encodes a protein that shares 66% identity with an annotated two-component sensor histidine kinase/response regulator (15). Significantly, a similar protein has been shown to be responsible for the expression of a polysaccharide utilization locus in Bacteroides thetaiotaomicron (2). Regarding ORF16 and ORF17, BLAST analysis failed to reveal any significant homology with known protein sequences. The third operon, abnB-abfB, encodes Abn43B and Abf51B, which have been shown to be an arabinanase and an arabinofuranosidase, respectively (22) (Fig. 1).
TABLE 1.
Sequence identity scores of G12 ORFs and proposed biological function for encoded proteins
| Process(es) and ORF | Gene | Homologous protein(s) (highest % identity) identified by BLAST | Putative generic function |
|---|---|---|---|
| Arabinose binding, transport, and utilization | |||
| 4 | araB | Ribulose kinase from Salmonella enterica (52) | Hypothetical arabinose metabolism |
| 5 | araC | l-Arabinose isomerase from Bacteroides vulgatus (79) | Arabinose metabolism |
| 6 | araD | l-Ribulose-5-phosphate 4-epimerase from Geobacillus stearothermophilus (65) | Hypothetical arabinose metabolism |
| 7 | araE | ADP-ribose pyrophosphatase from Methanococcus jannaschii (35) | Hypothetical arabinose metabolism |
| 8 | araF | l-Deoxy-d-xylulose-5-phosphate synthase from Bacteroides thetaiotaomicron (30%) | Hypothtical arabinose metabolism |
| 9 | araG | Uronate isomerase from Bacteroides fragilis (32) | Hypothetical arabinose metabolism |
| 11 | araH | Hypothetical protein from Bacteroides intestinalis (74) | None |
| 12 | araI | SusD from B. intestinalis (89) | Substrate binding |
| 13 | araJ | SusC/RagA family TonB-linked protein from Bacteroides oleiciplenus (85) | Substrate transport |
| 14 | araK | SusD from Bacteroides intestinalis (80) | Substrate binding |
| 15 | araL | SusC/RagA family TonB-linked protein from Bacteroides oleiciplenus (83) | Substrate transport |
| 18 | araM | Two-component system sensor/regulator from Bacteroides thetaiotaomicron (66) | Hypothetical histidine kinase regulator |
| Arabinan and arabino-oligosaccharide hydrolysis | |||
| 2 | abfA | GH51 from Bacteroides ovatus (81) | α-l-Arabinofuranosidase |
| 3 | araA | GH97 from Bacteroides intestinalis (73) | Glycoside hydrolase |
| 10 | abnA | GH43 from Bacteroides intestinalis (77) | endo-Arabinanase |
| 19 | abnB | GH43 from Bacteroides gallinarum (74) | endo-Arabinanase |
| 20 | abfBa | GH51 from Bacteroides intestinalis (84), GH43 from Bacteroides sp. (87) | α-l-Arabinofuranosidase |
abfB encodes two GH domains.
In summary, when combined with the knowledge that clone G12 expresses arabinofuranosidase activity, these data suggest a Sus-like organization and an arabinose utilization function for the three gene operons. Regarding the cellular localization of the GHs, the presence of putative peptide signals was detected, suggesting that the proteins might be secreted. However, the results of the analysis were insufficiently precise to predict the exact cellular localization.
Characterization of the four arabinofurano-hydrolases: Abf51A, Abf51B, Abn43A, and Abn43B.
In order to characterize the PUL, the sequences encoding Abf51A, Abn43A, and Abn43B were successfully cloned and expressed in E. coli to permit the production of soluble proteins. Unlike Abf51A, Abf51B, is part of a multimodular enzyme CBM4-GH51B-GH43C. Therefore, to analyze the activity of each of the catalytic modules of this enzyme, the entire protein and two truncated forms, Abf51Btrunc (CBM4-GH51B) and truncAbn43C (GH43C), were cloned into E. coli (Fig. 2). Unfortunately, the overexpression of the entire protein and truncAbn43C could not be achieved, although it was possible to express CBM4-Abf51B, here referred to as Abf51Btrunc.
FIG 2.
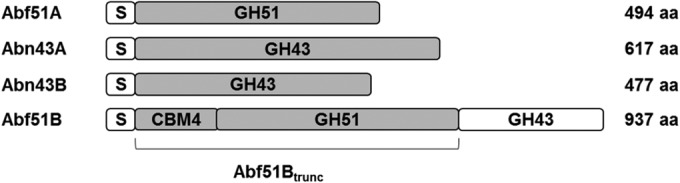
Schematic representation of the four ORFs encoding the glycoside hydrolases studied. Enzymatic modules (gray rectangles) were successfully cloned and expressed in E. coli, while attempts to express other modules (white rectangles) failed. The protein size in number of amino acids does not account for signal peptide sequences.
(i) Optimal parameters.
Abf51A, Abn43B, and Abf51Btrunc showed optimal activity in a pH range of 5.5 to 7.0, while Abn43A was more active in a pH range of 4.5 to 6 (Table 2). The apparent optimum temperatures of the different enzymes were in the range 45 to 55°C for Abf51A, Abf51Btrunc, and Abn43A, whereas Abn43B showed a maximum activity at lower temperature, with the optimum being at 37°C, and exhibited rapid precipitation above 40°C. Once the optimal conditions for the measurement of enzyme activities was established, it was possible to investigate enzyme stability. In particular, these analyses revealed that Abf51Btrunc was quite unstable, losing up to 90% of its activity after being held at 37°C for 24 h, although this activity loss was attenuated at 30°C. Similarly, at 30°C and pH 6.5, the activities of the three other enzymes remained quite stable over a 24-h period.
TABLE 2.
Biochemical properties of the GH51 and GH43 encoded by the Bacteroides sp. strain G12 metagenomic cluster
| Enzyme | Final yield of purified protein (mg/liter) | Theoretical pI | Mol mass (kDa) | Apparent optimum rangea |
|
|---|---|---|---|---|---|
| pH | Temp (°C) | ||||
| Abf51A | 11 | 5.61 | 55.68 | 5.5–7.0 (6.5) | 45–55 (50) |
| Abn43A | 7.41 | 4.72 | 68.51 | 4.5–6.0 (4.5) | 45–47 (47) |
| Abn43B | 26.07 | 5.63 | 56.05 | 5.5–6.5 (6.0) | 30–40 (37) |
| Abf51Btrunc | 2.87 | 5.42 | 72.01 | 6.0–7.0 (7.0) | 45–55 (55) |
Values indicate that the relative enzyme activity was >80% of the activity observed at optimum conditions. Values in parentheses are the optimal conditions defined for biochemical characterization.
(ii) Characterization of enzyme specificity and kinetics.
Both Abf51A and Abf51Btrunc were active on pNP-Araf, but only Abf51Btrunc displayed activity on the 5-hydroxymethyl analog, pNP-Galf, although this activity was lower than that observed on pNP-Araf. Abf51A was also shown to release reducing sugars from both SBA and WAX (Table 3), unlike Abf51Btrunc that only released reducing sugars from DA. SBA and DA are pectic polymers, both consisting of main chains composed of α-1,5-linked l-arabinosyl residues. However, the main chain of SBA is also decorated by single or double l-arabinosyl substitutions that are linked via α-1,2 and/or α-1,3 bonds (6, 29). Similarly, Abn43A displayed a slightly higher (∼1.2-fold) catalytic performance on SBA than on DA, while Abn43B was inactive on SBA but highly active on DA and α-1,5-linked l-arabino-oligosaccharides. These observations were corroborated by complementary assays performed using SBA and LA as substrates, which showed that Abf51Btrunc displays a marked preference for linear (unbranched) polysaccharides (data not shown), which is in sharp contrast to the behavior of Abf51A. Regarding Abn43A and Abn43B, no activity was detected on pNP-Araf, and the results obtained clearly indicated that these can be classified as arabinanases (Table 3), with Abn43A acting more or less indiscriminately on linear and branched arabinan, whereas Abn43B can apparently only hydrolyze bonds in linear substrates (Fig. 3).
TABLE 3.
Activities of Abf51A, Abf51Btrunc, Abn43A, and Abn43Ba
| Enzyme | Substrate | Mean ± SD |
|||
|---|---|---|---|---|---|
| SA (U/mg) | Km (g liter−1 or mmol liter−1)b | kcat (103 min−1) | kcat/Km (103 g/liter min−1 or 103 mmol/liter min−1) | ||
| Abf51A | pNP-Araf | 118.26 ± 2.16 | 0.58 ± 0.02 | 7.48 ± 0.01 | 12.83 ± 0.21 |
| WAX | 8.71 ± 0.53 | ND | ND | ND | |
| SBA | 9.44 ± 0.13 | ND | ND | ND | |
| DA | 0 | ND | ND | ND | |
| Abf51Btrunc | pNP-Araf | 38.4 ± 0.45 | 0.42 ± 0.02 | 2.99 ± 0.03 | 7.13 ± 0.09 |
| pNP-Galf | 0.27 ± 0.02 | ND | ND | ND | |
| WAX | 0 | ND | ND | ND | |
| LAG | 0 | ND | ND | ND | |
| SBA | 0.63 ± 0.04 | ND | ND | ND | |
| DA | 13.01 ± 0.15 | ND | ND | ND | |
| Abn43A | WAX | 0 | ND | ND | ND |
| LAG | 0 | ND | ND | ND | |
| SBA | 21.21 ± 0.06 | 5.95 ± 0.31 | 2.41 ± 0.04 | 0.41 ± 0.01 | |
| DA | 9.58 ± 0.64 | 2.58 ± 0.21 | 0.87 ± 0.02 | 0.34 ± 0.01 | |
| Abn43B | A2 | 0 | ND | ND | ND |
| A3 | 17.16 ± 2.15 | 3.65 ± 1.29 | 1.84 ± 0.35 | 0.50 ± 0.27 | |
| A4 | 14.06 ± 1.60 | 0.55 ± 0,15 | 1.03 ± 0.08 | 1.88 ± 0.53 | |
| A5 | 12.98 ± 0.92 | 0.64 ± 0.20 | 0.97 ± 0.09 | 1.50 ± 0.45 | |
| A6 | 11.77 ± 2.16 | 1.26 ± 0.38 | 0.91 ± 0.10 | 0.73 ± 0.28 | |
| WAX | 0 | ND | ND | ND | |
| LAG | 0 | ND | ND | ND | |
| SBA | 0 | ND | ND | ND | |
| DA | 13.90 ± 0.20 | 1.28 ± 0.09 | 0.87 ± 0.02 | 0.68 ± 0.02 | |
All the substrates were assayed. Data are reported only when activity was detected. ND, not determined due to no or extremely weak activity to calculate the value.
For reactions involving pNP-glycosides the Km was expressed in molar terms using mmol liter−1, whereas for reactions involving complex polysaccharides the Km was expressed as g liter−1.
FIG 3.
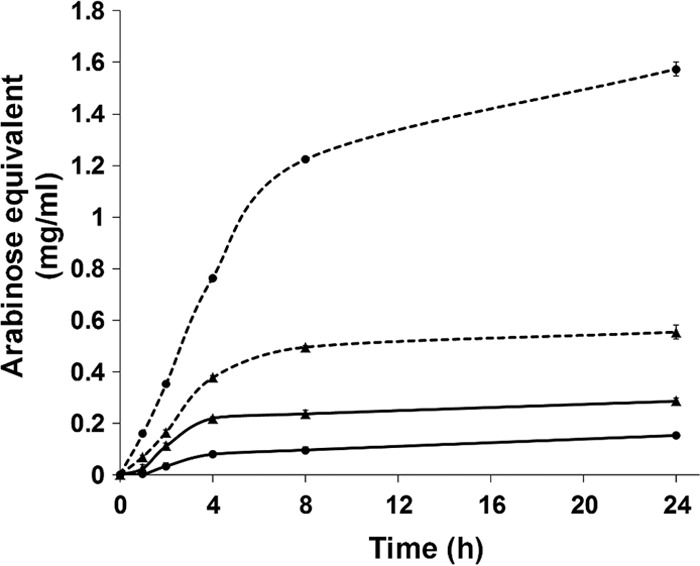
Degradation of arabinans by Abn43A and Abn43B. Time-dependent assays were performed using the DNS protocol to quantify reducing sugars at different time points. Solid and dotted lines represent SBA and LA hydrolysis, respectively, by Abn43A (▲) or Abn43B (●).
HPAEC-PAD analysis of the reaction end products of SBA and LA hydrolysis showed that both Abf51A and Abf51Btrunc released only arabinose, whereas Abn43A generated a range of AOSs, with arabinotetraose being the major product (48%), followed by arabinopentaose (23%), when LA was the substrate. Moreover, when acting on SBA, Abn43A released other unidentified oligosaccharides, which might correspond to branched AOS. Abn43B produced both arabinose and A2 as products, although the latter was a minor product being 15-fold less abundant on LA hydrolysis, suggesting that Abn43B displays exo-α-l-1,5-arabinanase activity. In this respect, further investigation of the activity of Abn43B on linear AOSs (A2 to A6) revealed that the shortest oligosaccharide that is hydrolyzed is arabinotriose, with A2 remaining intact even after a 60-min incubation (Table 3). Moreover, time course analyses revealed that Abn43B hydrolyzed A3 to A6 in exo-mode, releasing arabinose and A(n − 1) as sole products (Fig. 4A). The hydrolysis of 1 mol of A3 was concomitant with the equimolar liberation of A2 and arabinose, and the hydrolysis of A6 (Fig. 4B) proceeded through the successive production of A5 to A2, the reaction being complete after 1 h. In terms of relative efficiency, Abn43B appeared to be more active on A4 and A5 (3.5- and 2.5-fold, respectively) than on A3 or A6 (Table 3), perhaps suggesting that the active site is composed of four or five subsites.
FIG 4.
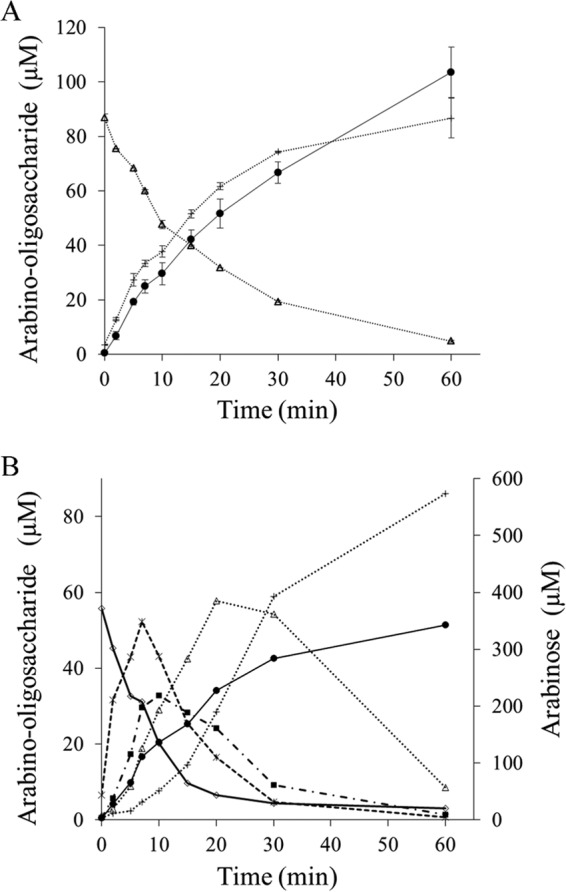
Hydrolysis of arabino-oligosaccharides by Abn43B. The time-dependent degradation of arabinotriose (A) and arabinohexaose (B) by Abn43B was analyzed using HPAEC-PAD. Symbols: ●, arabinose; +, arabinobiose; △, arabinotriose; □, arabinotetraose; ×, arabinopetaose; ◇, arabinohexaose.
Synergistic effects revealed by enzyme combinations.
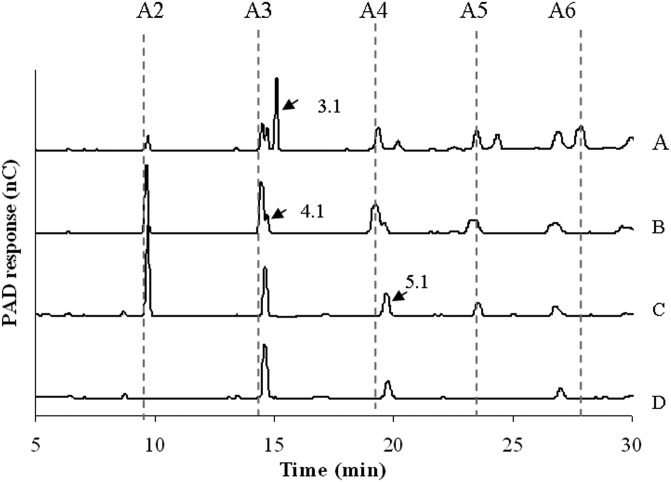
To reveal synergistic effects, the combined activities of Abf51A, Abf51Btrunc, Abn43A, and Abn43B were tested using SBA and LA as substrates and different enzyme combinations (two, three, or four enzymes). Irrespective of the enzyme mixture, a synergistic effect was observed, with the maximum synergistic index always being >1.5 (Fig. 5A). In reactions performed in the absence of Abf51A (i.e., in the presence of the three other enzymes), the final amount of arabinose released was only 21% of that obtained when all four enzymes were present, demonstrating the essential contribution of Abf51A. Moreover, although hydrolysis of SBA was slower when only Abf51A and Abn43B were deployed, the final yield (after 24 h) of arabinose nevertheless reached 1.89 ± 0.01 mg (i.e., 91.5% of the maximum yield). It is also noteworthy that HPAEC-PAD analysis revealed that the profile of the reaction end products was progressively modified as the complexity of the enzyme mixture was increased (Fig. 6). Indeed, as mentioned earlier, Abn43A alone released a range of linear and perhaps branched AOS (Fig. 6A), but when this enzyme was used pairwise with Abf51A, peaks corresponding to putative branched structures disappeared (e.g., peak 3.1, Fig. 6B). Similarly, when Abn43B was added, forming a three-enzyme mixture, the yield of arabinose and A2 was increased, while the peaks corresponding to longer linear AOS disappeared (Fig. 6C). Finally, deployment of the complete cocktail procured the highest yield of arabinose, although some unidentified species persisted (e.g., peaks 4.1 and 5.1), even after 24 h (Fig. 6D).
FIG 5.
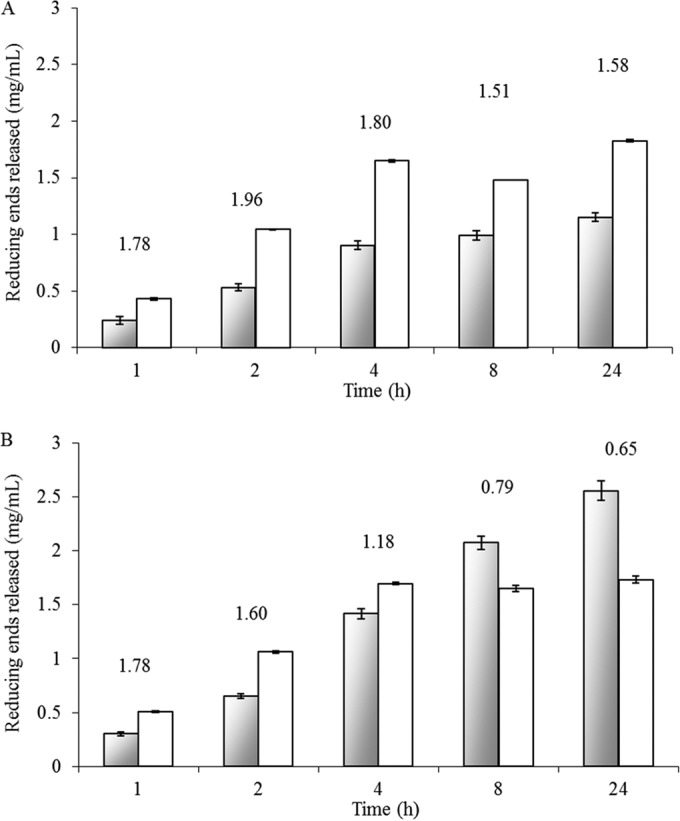
Synergistic effect of enzymes in cocktail. The time-dependent release of reducing sugars from sugar beet arabinan (A) and α-1,5 linear arabinan (B) was quantified by using the DNS assay. Gray bars represent the theoretical sum of the individual action of the four enzymes, while white bars represent the experimentally measured impact of the four enzymes working together (2 μg/ml of each enzyme). The numbers above the bars indicate the synergistic index.
FIG 6.
Degradation of sugar beet arabinan. HPAEC-PAD elution patterns of AOS after degradation of sugar beet arabinan by Abn43A (A), Abn43A + Abf51A (B), Abn43A + Abf51A + Abn43B (C), and Abn43A + Abf51A + Abn43B + Abf51Btrunc (D) during a 24-h reaction at 30°C and pH 6.5 are shown.
Testing the different enzyme combinations on LA provided similar results, although logically the presence of Abf51A was rendered superfluous, since this enzyme had already been shown to be inactive on LA. Monitoring the time-dependent release of reducing sugars from LA revealed that a core cocktail composed of Abf51Btrunc, Abn43A, and Abn43B provided a clear synergistic effect during the initial stages of the reaction. However, after reaching a maximum, the synergistic index diminished to <1 after 8 h (Fig. 5B). Presumably, this is because in the latter stages of the reaction, competition between the three enzymes for the soluble linear AOS becomes strong. Assuming that Abn43B might display catalytic processivity, its prolonged interaction with its substrate could hamper access to AOS by the other two enzymes. Finally, after long incubation periods, it is also possible that end product (arabinose) inhibition becomes an issue. Interestingly, when Abn43B and Abf51Btrunc were paired, their combined action led to a final arabinose yield that was close to that achieved using all three enzymes (94%). However, the presence of Abn43A speeded up the reaction, since its presence ensured that the reaction was almost (97% of total reducing sugar released) complete after 4 h, whereas without it the yield of arabinose at this time point was only 70%. Finally, combining Abf51Btrunc with either Abn43A or Abn43B to hydrolyze LA procured arabinose as the sole end product (data not shown), thus providing a clear indication that Abf51Btrunc can hydrolyze the AOS (DP2 to DP6) produced by Abn43A. Interestingly, the unidentified peaks (e.g., 4.1 and 5.1) were absent at the end of LA hydrolysis by the four enzymes.
DISCUSSION
Several studies have revealed the presence of PUL in members of Bacteroidetes, a bacterial phylum that is represented among the inhabitants of termite guts (18, 30). In a previous study, we isolated a Bacteroidetes-affiliated metagenomic DNA fragment from the gut microbiota of the fungal-growing termite Pseudacanthotermes militaris and revealed that this DNA bears four arabinofuranohydrolase-encoding ORFs (22). This discovery led us to postulate that the metagenomic DNA fragment encodes a PUL that is probably responsible for the degradation of an arabinose-containing polymer such as arabinan, a function that would be consistent with the diet of the animal host, known to feed on young plants such as sugarcane, which contains significant levels of pectins (9).
The first indication that our hypothesis is correct was supplied by the results of a thorough bioinformatics study, which revealed a Sus-like organization among the genetic elements of the 41-kb metagenomic fragment (G12), a configuration that is common in Bacteroides species but has thus far not been reported in bacterial inhabitants of termite guts. A key indicator of sus-like gene clusters is the susC-susD pair (15, 31), which encodes proteins that are responsible for cell surface substrate binding and transport to the periplasmic space, respectively (16). Therefore, the identification of two susC and susD homologues (i.e., either araL and araK or araJ and araI) is significant, as is the presence of araM, which is highly similar to BT0366. The latter has been shown to be a two-component system sensor histidine kinase/response regulator that controls the expression of the PUL BT0348-69 from B. thetaiotaomicron (2). Therefore, by analogy it is possible to assume that araM encodes the protein that regulates the expression of the G12-associated PUL. Finally, in addition to the fact that the different elements of G12 share high identity with those of the PUL BT0348-69, it is noteworthy these two DNAs display conserved synteny, which implies that the G12 and BT0348-69 PUL might originate from a common ancestor.
Regarding the four arabinofuranohydrolases, our results clearly indicated that these enzymes are able to degrade arabinan in a cooperative manner, with each enzyme contributing its specific catalytic attributes. Significantly, both so-called arabinanases, Abn43A and Abn43B, appear to share significant identity with Bt0360 and Bt0367, respectively, two B. thetaiotaomicron GH43 enzymes that also display arabinanase activity (32). Nevertheless, the exact mode of action of Abn43B appears to be different from that of Bt0367 because, unlike Abn43B, the latter did display activity on SBA. In this respect, the mode of action of Abn43B appears to be unique in family 43, since exo-α-l-1,5-arabinanase activity has thus far only been reported for enzymes belonging to the family GH93, although GH93 enzymes release arabinobiose, and not arabinose, as the major product from linear arabinan. Therefore, the mode of action of Abn43B is apparently quite original and is perhaps closer to that of an exo-α-l-1,5-arabinofuranosidase activity. In this respect, it is interesting that the active site of Abn43B appears to possess at least three subsites (−1 to +2), with interactions in subsite +2 being indispensable for the formation of a productive enzyme-substrate complex. The fact that Abn43B is more active on A4 or A5 than A3 further suggests the presence of a subsite +3, and even a subsite +4.
We also detected very distinctive arabinofuranosidase activities for Abf51A and Abf51Btrunc, the former hydrolyzing both α-1,2 and α-1,3 linkages (in SBA and WAX). This is consistent with the fact that Abf51A shares quite good identity with Abf2 from Bacillus subtilis, an enzyme that displays highly similar catalytic properties (33). That Abf51Btrunc was also able to hydrolyze pNP-Galf (i.e., displays α-d-galactofuranosidase activity) is unsurprising, since such catalytic promiscuity has already been observed in other GH51 enzymes (34) and is due to tolerance in subsite −1 of the d-galactofuranosyl moiety, which can be considered to be a C5-hydroxymethylated analog of l-arabinofuranosyl.
Taken together, the results presented here allow us to propose a scheme describing how the G12-encoded PUL might degrade arabinan. Clearly, one might expect Abn43A to be the key depolymerizing enzyme in the process, releasing branched and linear AOS of DP4 and DP5 from the ramified polymer. Branched AOS would then constitute substrates for Abf51A, which would remove arabinose side chains and create debranched AOS substrates for Abn43B. The exo-action of the latter enzyme would mainly release arabinose from linear AOS, as would that of Abf51Btrunc, which could mop up any residual arabinobiose left by Abn43B. Of course, this scheme is rather simplistic and certainly does not account for all of the complexity of the system, including the role of the uncharacterized Abf43C, which is the second catalytic domain of the Abf51B-Abf43C enzyme. In this regard, it is tempting to speculate that Abf43C is active on double-substituted (i.e., α-1,2 and α-1,3) arabinosyl moieties and allows hydrolysis of arabinan to go even further toward completion. Two observations support this speculation. First, Abf43C shares 75% of identity with Bt0369, a family GH43 enzyme from B. thetaiotaomicron that can remove O2-linked arabinose from both mono- and disubstituted arabinosyl moieties in arabinan (32). Second, the action of a family GH43 Chrysosporium lucknowense arabinanase on SBA (35) has been shown to generate among its products two AOS bearing one double-substituted (i.e., α-1,2 and α-1,3) arabinosyl moiety each, whose HPAEC-PAD elution profiles coincide well with those species (peaks 4.1 and 5.1 in Fig. 6) produced in the present study by the combined action of the four enzymes on SBA. This implies that none of the tested enzymes are able to act on these doubly substituted products and provides the rationale for attributing this missing activity to Abf43C.
Originally, the fosmid clone G12 was selected on the basis of its ability to hydrolyze the synthetic substrate 5-bromo-4-chloro-3-indolyl-α-l-arabinofuranoside, but not the chromogenic substrate azurine-cross-linked (AZCL) linear arabinan, implying that only arabinofuranosidase was expressed. In the present study, this assumption was corroborated by reverse transcription-quantitative PCR analysis, which showed that only Abn43B and Abf51B were expressed in E. coli (see the supplemental material). This finding adds further support to the claim that the genetic regulatory elements of members of Bacteroides are not always well recognized in E. coli (36) and further underlines the importance of cloning large metagenomic fragments, a strategy which offers the possibility to identify whole gene clusters, even when only one activity is expressed.
Currently, next-generation sequencing coupled to powerful metagenomics is increasingly providing the means to probe the complexity of complex gut-based microbial communities (15, 18) and thus shed light upon the intricate, multienzyme systems that are deployed by gut bacteria to hydrolyze complex substrates such as plant cell walls. Nevertheless, as exemplified in the present study, to acquire a complete understanding of these enzyme processes, it is necessary to obtain hard biochemical data, because even very sequence-related enzymes, such as Abn43B and Bt0367, can display subtle catalytic differences, which can modify the overall interpretation of the processes involved. In this regard, within the P. militaris metagenomic library we have identified several fosmids that bear gene clusters of Bacteroides origin, which appear to be responsible for functions, such as xylan and glucan degradation and utilization. Therefore, in future work we intend to pursue our functional approach in order to dissect more multienzymatic systems and begin to form a more complex picture of how the P. militaris gut microbiota deal with the polysaccharide-rich diet of its host.
Supplementary Material
ACKNOWLEDGMENTS
Financial support for this study was provided by OSEO Innovation and the Institut National de la Recherche Agronomique, the latter supplying Ph.D. stipends to G.A. and G.B. A.A. was funded by Midi-Pyrénées regional authorities (program R2390070/12053333).
We thank the staff of the ICEO facility (part of the IBiSA and GenoToul core facility PICT), who provided access to HPLC equipment and robots. Finally, we thank J. G. Berrin and M. Arlat for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02257-14.
REFERENCES
- 1.Bayer EA, Belaich JP, Shoham Y, Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol 58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 2.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1011221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu Rev Plant Biol 61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 4.Caffall KH, Mohnen D. 2009. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344:1879–1900. doi: 10.1016/j.carres.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Ridley BL, O'Neill MA, Mohnen DA. 2001. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57:929–967. doi: 10.1016/S0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 6.Mohnen D. 2008. Pectin structure and biosynthesis. Curr Opin Plant Biol 11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Zykwinska AW, Ralet MCJ, Garnier CD, Thibault JFJ. 2005. Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol 139:397–407. doi: 10.1104/pp.105.065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumon C, Song LT, Bozonnet S, Fauré R, O'Donohue MJ. 2012. Progress and future prospects for pentose-specific biocatalysts in biorefining. Process Biochem 47:346–357. doi: 10.1016/j.procbio.2011.06.017. [DOI] [Google Scholar]
- 9.de Souza AP, Leite DCC, Pattathil S, Hahn MG, Buckeridge MS. 2013. Composition and structure of sugarcane cell wall polysaccharides: implications for second-generation bioethanol production. Bioenergy Res 6:564–579. doi: 10.1007/s12155-012-9268-1. [DOI] [Google Scholar]
- 10.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carapito R, Imberty A, Jeltsch JM, Byrns SC, Tam PH, Lowary TL, Varrot A, Phalip V. 2009. Molecular basis of arabino-bio-hydrolase activity in phytopathogenic fungi: crystal structure and catalytic mechanism of Fusarium graminearum GH93 exo-α-l-arabinanase. J Biol Chem 284:12285–12296. doi: 10.1074/jbc.M900439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhnel S, Hinz SWA, Pouvreau L, Wery J, Schols HA, Gruppen H. 2010. Chrysosporium lucknowense arabinohydrolases effectively degrade sugar beet arabinan. Bioresource Technol 101:8300–8307. doi: 10.1016/j.biortech.2010.05.070. [DOI] [PubMed] [Google Scholar]
- 13.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 14.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 16.Shipman JA, Berleman JE, Salyers AA. 2000. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol 182:5365–5372. doi: 10.1128/JB.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pope PB, Mackenzie AK, Gregor I, Smith W, Sundset MA, McHardy AC, Morrison M, Eijsink VGH. 2012. Metagenomics of the Svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 7:e38571. doi: 10.1371/journal.pone.0038571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernandez M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 19.Tasse L, Bercovici J, Pizzut-Serin S, Robe P, Tap J, Klopp C, Cantarel BL, Coutinho PM, Henrissat B, Leclerc M, Doré Jl, Monsan P, Remaud-Simeon M, Potocki-Veronese G. 2010. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res 20:1605–1612. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeAngelis K, Gladden J, Allgaier M, D'haeseleer P, Fortney J, Reddy A, Hugenholtz P, Singer S, Vander Gheynst J, Silver W, Simmons B, Hazen T. 2010. Strategies for enhancing the effectiveness of metagenomic-based enzyme discovery in lignocellulolytic microbial communities. Bioenergy Res 3:146. doi: 10.1007/s12155-010-9089-z. [DOI] [Google Scholar]
- 21.Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 22.Bastien G, Arnal G, Bozonnet S, Laguerre S, Ferreira F, Fauré R, Henrissat B, Lefevre F, Robe P, Bouchez O, Noirot C, Dumon C, O'Donohue M. 2013. Mining for hemicellulases in the fungus-growing termite Pseudacanthotermes militaris using functional metagenomics. Biotechnol Biofuels 6:78. doi: 10.1186/1754-6834-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayley DP, Rocha ER, Smith CJ. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol Lett 193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- 24.Tribble GD, Parker AC, Smith CJ. 1999. Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plasmid 42:1–12. doi: 10.1006/plas.1999.1401. [DOI] [PubMed] [Google Scholar]
- 25.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res 17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra S, Klar B, Huson DH. 2009. Visual and statistical comparison of metagenomes. Bioinformatics 25:1849–1855. doi: 10.1093/bioinformatics/btp341. [DOI] [PubMed] [Google Scholar]
- 27.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 28.Kitamura M, Okuyama M, Tanzawa F, Mori H, Kitago Y, Watanabe N, Kimura A, Tanaka I, Yao M. 2008. Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron. J Biol Chem 283:36328–36337. doi: 10.1074/jbc.M806115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beldman G, Schols HA, Pitson SM, Searle-van Leeuwen MF, Voragen AGJ. 1997. Arabinans and arabinan degrading enzymes, p 1–64. Advances in macromolecular carbohydrate research, vol 1 JAI Press, London, United Kingdom. [Google Scholar]
- 30.Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Vongkaluang C, Noparatnaraporn N, Kudol T. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol 71:6590–6599. doi: 10.1128/AEM.71.11.6590-6599.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cartmell A, McKee LS, Pena MJ, Larsbrink J, Brumer H, Kaneko S, Ichinose H, Lewis RJ, Vikso-Nielsen A, Gilbert HJ, Marles-Wright J. 2011. The structure and function of an arabinan-specific α-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem 286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inacio JM, Correia IL, de Sa-Nogueiral I. 2008. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 154:2719–2729. doi: 10.1099/mic.0.2008/018978-0. [DOI] [PubMed] [Google Scholar]
- 34.Remond C, Ferchichi M, Aubry N, Plantier-Royon R, Portella C, O'Donohue MJ. 2002. Enzymatic synthesis of alkyl arabinofuranosides using a thermostable α-l-arabinofuranosidase. Tetrahedron Lett 43:9653–9655. doi: 10.1016/S0040-4039(02)02381-X. [DOI] [Google Scholar]
- 35.Westphal Y, Kuhnel S, de Waard P, Hinz SWA, Schols HA, Voragen AGJ, Gruppen H. 2010. Branched arabino-oligosaccharides isolated from sugar beet arabinan. Carbohydr Res 345:1180–1189. doi: 10.1016/j.carres.2010.03.042. [DOI] [PubMed] [Google Scholar]
- 36.Taupp M, Mewis K, Hallam SJ. 2011. The art and design of functional metagenomic screens. Curr Opin Biotechnol 22:465–472. doi: 10.1016/j.copbio.2011.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.