Abstract
Although the molecular surveillance network RotaNet-Italy provides useful nationwide data on rotaviruses causing severe acute gastroenteritis in children in Italy, scarce information is available on rotavirus circulation in the general Italian population, including adults with mild or asymptomatic infection. We investigated the genotypes of rotaviruses present in urban wastewaters and compared them with those of viral strains from clinical pediatric cases. During 2010 and 2011, 285 sewage samples from 4 Italian cities were tested by reverse transcription-PCRs (RT-PCRs) specific for rotavirus VP7 and VP4 genes. Rotavirus was detected in 172 (60.4%) samples, 26 of which contained multiple rotavirus G (VP7 gene) genotypes, for a total of 198 G types. Thirty-two samples also contained multiple P (VP4 gene) genotypes, yielding 204 P types in 172 samples. Genotype G1 accounted for 65.6% of rotaviruses typed, followed by genotypes G2 (20.2%), G9 (7.6%), G4 (4.6%), G6 (1.0%), G3 (0.5%), and G26 (0.5%). VP4 genotype P[8] accounted for 75.0% of strains, genotype P[4] accounted for 23.0% of strains, and the uncommon genotypes P[6], P[9], P[14], and P[19] accounted for 2.0% of strains altogether. These rotavirus genotypes were also found in pediatric patients hospitalized in the same areas and years but in different proportions. Specifically, genotypes G2, G9, and P[4] were more prevalent in sewage samples than among samples from patients, which suggests either a larger circulation of the latter strains through the general population not requiring medical care or their greater survival in wastewaters. A high level of nucleotide identity in the G1, G2, and G6 VP7 sequences was observed between strains from the environment and those from patients.
INTRODUCTION
Human group A rotaviruses (RVA) are responsible for severe gastroenteritis in children worldwide and cause ∼450,000 deaths annually, mostly in developing countries (1). Rotavirus remains a common cause of morbidity with a significant economic burden in developed countries (2). Although reinfection may occur during life, most clinically relevant cases involve children <5 years old (3–5).
Rotaviruses are characterized by a double-stranded RNA (dsRNA) genome with 11 segments, which encode six structural proteins (VPs) and five or six nonstructural proteins (NSPs) (4). RVA are commonly classified based on genes encoding the outer capsid proteins, defining G (glycoprotein) (for VP7) and P (protease-cleaved protein) (for VP4) genotypes. Currently, 27 G and 37 P genotypes in humans and animals have been reported (6, 7). However, a limited number of GxP[y] genotype combinations are common in humans, such as G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] (7–9). Uncommon combinations, such as G8P[4], G12P[8], G12P[6], and others, have also been reported recently (10–13).
Rotavirus is transmitted through the fecal-oral route, directly from person to person, or by water and food contaminated with human or animal feces. After replication in the gastrointestinal tract, viruses are shed at very high concentrations (up to 1010 viruses/g) in feces and can persist in the environment for a long time (14–16). Like other enteric viruses, rotavirus is highly resistant to processes used in wastewater treatment plants (WWTPs), which can favor their spread into the environment (17–19), particularly in surface waters. However, RVA have been implicated in waterborne gastroenteritis outbreaks only sporadically (15, 20, 21).
Sewage contains enteric viruses shed by individuals with either overt disease or asymptomatic infection (22, 23), and molecular virus surveillance of urban sewage is therefore useful to assess potential threatening viruses circulating in the population, independent of subjects' age and disease severity.
Two live oral rotavirus vaccines have been used since 2006 in >100 countries worldwide (24), i.e., Rotarix (monovalent G1P[8] vaccine; GlaxoSmithKline) and RotaTeq (pentavalent, containing the G1 to G4 and P[8] genotypes; Sanofi-Pasteur MSD). Surveillance of RVA genotypes in patients is useful to identify viral strains causing residual cases in populations that use mass vaccination, particularly for possible emerging strains of zoonotic origin or imported strains. In Italy, molecular surveillance of RVA gastroenteritis in hospitalized children (RotaNet-Italy) has been conducted since 2007 (25), as part of the EuroRotaNet network (9).
As an aid to clinical surveillance, monitoring of RVA in sewage ahead of WWTPs may provide an additional means to assess genotypes that also circulate in the normal population (26). However, detection and genotyping of RVA in sewage may be affected by the simultaneous presence of several common or uncommon strains, and the segmented nature of the rotavirus genome may preclude the definite identification of the full genome constellation of RVA detected. Possible RNA inhibitors in environmental samples may also interfere with molecular detection (27, 28).
In this study, we investigated the RVA genotypes present in sewage before entry into 10 WWTPs serving 4 cities located in different areas of Italy, comparing environmental strain genotypes with those detected in pediatric gastroenteritis cases occurring in the same cities and years.
MATERIALS AND METHODS
Virus and cell cultures.
Control G1P[8] Wa rotavirus was grown in MA104 cells and titrated in 96-well microcultures after 24 h of infection and immunostaining with a rabbit antirotavirus hyperimmune serum and a peroxidase-labeled anti-rabbit antibody (Bio-Rad, Segrate, Italy), essentially as described previously (29).
Sewage samples.
Inlet sewage was sampled at 10 urban wastewater treatment plants located in four different cities in Italy (Naples, Bari, Palermo, and Sassari), where environmental control is particularly considered due to the high magnitude of either tourism or immigration. The WWTPs of Bari and Naples started to collect samples in the last part of 2010, whereas the WWTPs of Palermo and Sassari arranged sample collection only in 2011. All WWTPs monitored are conventional activated sludge plants, receiving waters from urban areas. Daily flows range from 4,800 to 750,000 m3, with design capacities of 70,000 to 1,000,000 population equivalents. In addition to human waste, Naples WWTPs also treat industrial wastewater occasionally (F. Pennino, University of Naples, personal communication). Sewage samples (1 liter) were taken by using sterile plastic bottles, correlating the number of samples with the sewer-linked population: 1 sample every 15 days for WWTPs serving >300,000 inhabitants and 1 sample every month for populations of <300,000 (Table 1). This sampling schedule was respected in most cases, but fewer samples were collected in some months due to logistic problems.
TABLE 1.
Detection of rotavirus in sewage samples from four WWTPs in Italy in 2010 and 2011
| City | WWTPa | No. of inhabitants | No. of samples collectedb |
No. (%) of positive samples | ||
|---|---|---|---|---|---|---|
| 2010 | 2011 | Total | ||||
| Bari | BF | 300,000 | 15 | 24 | 39 | 27 (69.2) |
| MdB | 300,000 | 15 | 24 | 39 | 32 (82.1) | |
| J | 300,000 | 14 | 24 | 38 | 28 (73.7) | |
| All | 44 | 72 | 116 | 87 (75.0) | ||
| Palermo | AdC | 130,000 | NC | 24 | 24 | 14 (58.3) |
| FV | 70,000 | NC | 20 | 20 | 13 (65.0) | |
| JH | 70,000 | NC | 10 | 10 | 7 (70.0) | |
| All | 0 | 54 | 54 | 34 (62.9) | ||
| Naples | NCu | 1,000,000 | NC | 30 | 30 | 12 (40.0) |
| NTe | 700,000 | 9 | 24 | 33 | 13 (39.4) | |
| NE | 500,000 | 8 | 20 | 28 | 7 (25.0) | |
| All | 17 | 74 | 91 | 32 (35.1) | ||
| Sassari | SCa | 120,000 | NC | 24 | 24 | 19 (79.2) |
| All | 61 | 224 | 285 | 172 (60.4) | ||
Abbreviations: BF, Bari Fesca; MdB, Mola di Bari; J, Japigia; AdC, Acqua dei Corsari; FD, Fondo Verde; JH, Jolly Hotel; NCu, Naples Cuma; NTed, Naples Teduccio; NE, Naples Est; SCa, Sassari Caniggia.
NC, not collected.
An automated 24-h sampling system was present in Palermo and Naples (Naples Est and Cuma), where a timer-operated valve allows collection of the total sample volume at regular intervals during 24 h. In Sassari, Bari, and Naples (San Giovanni a Teduccio) WWTPs, manual sampling was performed at selected sites during peak hours. Samples were kept at −20°C until processing was performed.
Rotavirus concentration in sewage.
Sixty-five milliliters of sewage was clarified by centrifugation at 1,200 × g for 20 min at 4°C, and supernatants were centrifuged in a Beckman L8-80M ultracentrifuge using a Ti45 rotor at 126,000 × g for 2 h at 4°C, as described previously by Fumian and coworkers (30). Pellets were suspended in 1 ml of phosphate-buffered saline (PBS), and 140 μl was used for RNA extraction by using the Viral RNeasy minikit (Qiagen, Milan, Italy). Extracted RNAs were eluted in 50 μl of RNase-free water and stored at −80°C.
Rotavirus detection and G and P genotyping.
Rotavirus RNA was amplified by reverse transcription-PCR (RT-PCR) using primers GEN_VP6_F and GEN_VP6_R (31). To increase sensitivity, a nested PCR was occasionally performed by using internal primers VP6-F and VP6-R (32). VP6-positive samples were genotyped for both VP7 and VP4 by nested PCR, as described previously (33, 34). The molecular size of genotyping PCR products was determined by agarose gel electrophoresis, using a Gel Doc XR molecular imager with Quantity-One software (Bio-Rad, Segrate, Italy).
Rotaviruses in clinical samples.
A total of 343 fecal samples were collected from children with rotavirus diarrhea admitted to hospitals in Naples, Bari, Palermo, and Sassari in 2011 within the Italian RVA AGE surveillance program. Rotavirus infection was diagnosed by using a commercial immunochromatographic enzyme-linked immunosorbent assay (ELISA) or latex agglutination methods in use in each hospital. For strain characterization, stool samples were diluted 10% in distilled water, and rotavirus RNA was extracted with the Viral RNeasy minikit and directly subjected to G and P genotyping, performed as described above for sewage samples (33, 34). All samples failing both G and P genotyping were confirmed to be rotavirus positive by RT-PCR amplification of the VP6 gene (32).
Sequencing and phylogenetic analysis.
A subgroup of samples was selected randomly for each city, and the respective G and P amplicons were characterized by nucleotide sequencing using PCR primers at Macrogen Inc. (Seoul, South Korea). Chromatograms were analyzed with Chromas Pro 2.23 (Tecnelysium) and aligned with SeqMan II (DNAstar). The phylogenetic dendrograms were constructed with the neighbor-joining method, using the Kimura two-parameter model with MEGA5.1 software (35). The robustness of each node was assessed by 1,000 bootstrap replications. All relevant sequences from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) were used in comparisons. RVA VP7 and VP4 genotypes were defined according to guidelines of the Rotavirus Classification Working Group (RCWG) (31).
Evaluation of rotavirus concentration protocols.
To control the efficiency of ultracentrifugation for concentrating rotaviruses from sewage, virus recovery was determined by testing 65-ml sewage samples spiked with 1 ml of a viral stock suspension containing 3 × 105 focus-forming units (FFU)/ml of human Wa rotavirus (∼1 × 104 genome PCR units/ml). Ultracentrifugation pellets were serially diluted (1:1.5 and 1:2 steps) and analyzed by VP6 RT-PCR using primers VP6-F and VP6-R (32). The last dilution yielding a discernible PCR-amplified DNA band was considered to contain a rotavirus genome PCR unit. The ratios between molecular titers before and those after the concentration procedure were used to calculate the recovery efficiency.
Testing for inhibitors of RT-PCR and rotavirus in sewage.
Preliminary experiments were conducted to investigate the possible presence of chemical inhibitors of RT-PCR in sewage. RNA extracts from concentrated sewage samples that were negative by RT-PCR were spiked with serial dilutions of RNA extracted from RVA-positive fecal samples and retested by VP6-specific RT-PCR. The results of the test were compared with results for control RNA samples diluted in RNase-free water.
Separate experiments were also performed to exclude possible rotavirus damage following protracted presence in sewage. To simulate field conditions, untreated samples of rotavirus-negative sewage from the different plants or distilled water were spiked with 3 × 105 FFU/ml of human Wa rotavirus and left to stand at room temperature (RT) for 24 h. Titration of residual rotavirus persisting in each sample was then performed by endpoint VP6-specific RT-PCR (see above), and the ratios between viral titers in sewage and those in clean water were calculated.
Statistical assays.
The Z-test (36), Fisher's exact test (http://www.langsrud.com/fisher.htm), and Pearson's correlation and Lin's correlation concordance coefficients (37, 38) were used to evaluate possible differences and correlations between the distributions of RVA genotypes in sewage and clinical samples.
Nucleotide sequence accession numbers.
The nucleotide sequence data obtained in this study have been submitted to GenBank under accession numbers KF414532 to KF414624.
RESULTS
Efficiency of rotavirus recovery by ultracentrifugation.
To evaluate the efficiency of ultracentrifugation in recovery of virus, 20 sewage samples were collected from each WWTP, at the start and at the end of the sampling period, and were spiked with Wa rotavirus. After analysis of serial dilutions of the ultracentrifugation pellets by VP6 RT-PCR, it was calculated that between ≥30 and <67% of spike virus RNA was recovered from the different samples.
Since the sewage sample was eventually concentrated 65-fold, a 21-fold virus concentration or higher was attained throughout the process.
Rotavirus detection and distribution of G and P genotypes in sewage samples.
In 2010 and 2011, 285 sewage samples were collected from the WWTPs of the four cities monitored (Table 1). Altogether, 172 (60.2%) samples tested positive by rotavirus VP6 RT-PCR. The proportion of RVA-positive samples ranged between 58.3 and 82.1% in 7 WWTPs serving three cities and was lower (25.0 to 40.0%) for the three WWTPs of Naples.
The RVA G and P genotypes identified in sewage samples are shown in Table 2. The VP7 genotype detected more frequently was G1 (130/172 sewage samples), followed by G2 (40 cases), G9 (15 cases), G4 (9 cases), and G3 (1 case). Genotypes G6 and G26 were detected in 2 samples and 1 sample, respectively. In 32 cases, 2 different genotypes were detected in the same sample. These genotypes were mostly G1 with either G2 or G4 (26 and 3 cases, respectively). Genotypes P[8] and P[4] were found in 153 and 47 sewage samples, respectively. Multiple P genotypes were detected in 38 samples, 4 of which contained the uncommon P[6]+P[8], P[9]+P[4], P[14], and P[19] genotypes, respectively. Twelve samples contained G1+G2-P[4]+P[8] virus.
TABLE 2.
Distribution of rotavirus G and P genotypes in inlet wastewater in four Italian cities in 2010 and 2011
| Genotype | No. (%) of samples |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2010 |
2011 |
2010 total | 2011 total | 2010–2011 total | |||||
| Naples | Bari | Naples | Bari | Palermo | Sassari | ||||
| G types | |||||||||
| G1 | 8 (88.8) | 40 (81.6) | 17 (56.7) | 28 (53.8) | 19 (50.0) | 18 (90.0) | 48 (82.7) | 82 (58.6) | 130 (65.6) |
| G2 | 1 (11.2) | 6 (12.2) | 6 (20.0) | 14 (26.9) | 12 (31.6) | 1 (10.0) | 7 (12.1) | 33 (23.6) | 40 (20.2) |
| G3 | 0 | 0 | 1 (3.3) | 0 | 0 | 0 | 0 | 1 (0.7) | 1 (0.5) |
| G4 | 0 | 3 (6.2) | 3 (10.0) | 2 (3.9) | 1 (2.6) | 0 | 3 (5.2) | 6 (4.3) | 9 (4.6) |
| G9 | 0 | 0 | 2 (6.7) | 6 (11.5) | 6 (15.8) | 1 (10.0) | 0 | 15 (10.7) | 15 (7.6) |
| G6 | 0 | 0 | 0 | 2 (3.9) | 0 | 0 | 0 | 2 (1.4) | 2 (1.0) |
| G26 | 0 | 0 | 1 (3.3) | 0 | 0 | 0 | 0 | 1 (0.7) | 1 (0.5) |
| Total | 9 | 49 | 30 | 52 | 38 | 20 | 58 | 140 | 198 |
| P types | |||||||||
| P[8] | 8 (72.7) | 41 (87.2) | 20 (74.0) | 35 (63.6) | 30 (66.7) | 19 (100.0) | 49 (84.4) | 104 (71.2) | 153 (75.0) |
| P[4] | 3 (27.3) | 6 (12.8) | 5 (18.5) | 18 (32.8) | 15 (33.3) | 0 | 9 (15.6) | 38 (26.0) | 47 (23.0) |
| P[6] | 0 | 0 | 1 (3.7) | 0 | 0 | 0 | 0 | 1 (0.7) | 1 (0.5) |
| P[9] | 0 | 0 | 0 | 1 (1.8) | 0 | 0 | 0 | 1 (0.7) | 1 (0.5) |
| P[14] | 0 | 0 | 0 | 1 (1.8) | 0 | 0 | 0 | 1 (0.7) | 1 (0.5) |
| P[19] | 0 | 0 | 1 (3.7) | 0 | 0 | 0 | 0 | 1 (0.7) | 1 (0.5) |
| Total | 11 | 47 | 27 | 55 | 45 | 19 | 58 | 146 | 204 |
Absence of RT-PCR inhibitors and rotavirus inactivation in sewage samples.
To verify if the lower RVA detection rate obtained consistently in the WWTPs of Naples could be due to RT-PCR inhibitors being present in sewage, negative RNA samples extracted from all WWTPs were retested by RT-PCR after spiking with rotavirus RNA. No difference between spiked sewage and RNase-free water samples was observed (see Table S1 in the supplemental material).
Possible rotavirus destruction by a prolonged stay in sewage was also investigated by spiking two raw sewage samples that had tested RVA negative in each WWTP with RVA Wa. The residual virus after 24 h of incubation at RT was titrated by endpoint VP6 RT-PCR. No decrease in virus titer was observed for any sample, excluding the occurrence of significant rotavirus damage in any of the WWTP samples investigated (see Table S2 in the supplemental material).
Prevalence of rotavirus G and P genotypes in clinical samples.
Three hundred forty-three rotaviruses from stool samples of clinical pediatric cases in Bari, Naples, Palermo, and Sassari were genotyped by RT-PCR. Altogether, G1 was the predominant genotype (74.6% of strains investigated), followed by G2 (10.3%), G9 (5.8%), G3 (2.0%), and G4 (2.5%) (Table 3). Uncommon G8, G10, and G12 rotaviruses were observed in a single case each. A G genotype could not be defined for 14 samples, although the presence of RVA was confirmed by either P genotyping or VP6 RT-PCR. The common G types G1, G3, G4, and G9 and the common G type G2 were associated mostly with the common P types P[8] (87.0%) and P[4] (11.5%), respectively (Table 3).
TABLE 3.
Rotavirus G/P genotypes in samples from children hospitalized with acute gastroenteritis in four Italian cities in 2011
| Genotype | No. (%) of samples |
||||
|---|---|---|---|---|---|
| Naples | Bari | Palermo | Sassari | Total | |
| Common | |||||
| G1P[8] | 37 (56.1) | 77 (79.5) | 62 (68.1) | 79 (88.8) | 255 (74.4) |
| G2P[4] | 5 (7.6) | 5 (5.1) | 10 (11.0) | 3 (3.4) | 23 (6.7) |
| G3P[8] | 4 (6.1) | 2 (2.1) | 0 | 0 | 6 (1.7) |
| G4P[8] | 0 | 8 (8.2) | 0 | 0 | 8 (2.3) |
| G9P[8] | 10 (15.2) | 2 (2.1) | 0 | 2 (2.2) | 14 (4.1) |
| Total common | 56 (84.8) | 94 (97.0) | 72 (79.1) | 84 (94.4) | 306 (89.2) |
| Uncommon | |||||
| G3P[9] | 0 | 0 | 1 (1.1) | 0 | 1 (0.3) |
| G10P[8] | 0 | 0 | 1 (1.1) | 0 | 1 (0.3) |
| G12P[8] | 0 | 1 (1.0) | 0 | 0 | 1 (0.3) |
| G8P[4] | 0 | 0 | 0 | 1 (1.1) | 1 (0.3) |
| G1P[4] | 0 | 0 | 1 (1.1) | 0 | 1 (0.3) |
| Total uncommon | 0 | 1 (1.0) | 3 (3.3) | 1 (1.1) | 5 (1.4) |
| Mixed types | |||||
| G1,G2P[4,8] | 2 (3.0) | 0 | 0 | 4 (4.5) | 6 (1.7) |
| G2,G9P[4,8] | 4 (6.1) | 0 | 0 | 0 | 4 (1.2) |
| G2,G9P[8] | 0 | 1 (1.0) | 0 | 0 | 1 (0.3) |
| G1,G4P[8] | 0 | 1 (1.0) | 0 | 0 | 1 (0.3) |
| G1,G9P[8] | 2 (3.0) | 0 | 0 | 0 | 2 (0.6) |
| G1,G2P[4] | 0 | 0 | 2 (2.2) | 0 | 2 (0.6) |
| Total mixed types | 8 (12.1) | 2 (2.0) | 2 (2.2) | 4 (4.5) | 16 (4.7) |
| Untypeable | |||||
| GNtP[8] | 1 (1.5) | 0 | 7 (7.7) | 0 | 8 (2.3) |
| GNtP[4] | 0 | 0 | 4 (4.4) | 0 | 4 (1.2) |
| GNtP[9] | 0 | 0 | 1 (1.1) | 0 | 1 (0.3) |
| G1P[Nt] | 0 | 0 | 1 (1.1) | 0 | 1 (0.3) |
| G2P[Nt] | 1 (1.5) | 0 | 0 | 0 | 1 (0.3) |
| GNtP[Nt] | 0 | 0 | 1 (1.1) | 0 | 1 (0.3) |
| Total untypeable | 0 | 0 | 14 (15.4) | 0 | 16 (4.7) |
| Total | 66 (100.0) | 97 (100.0) | 91 (100.0) | 89 (100.0) | 343 (100.0) |
Sixteen (4.7%) and ten samples presented dual G and P genotypes, respectively, indicating mixed rotavirus infection.
Comparison of rotavirus genotypes from sewage samples and samples from gastroenteritis cases.
Overall, the relative distribution of the G and P RVA genotypes detected in sewage sample (Table 2) was similar to that detected in acute gastroenteritis (AGE) patients (Table 3), with G1 and P[8] accounting for most of the genotypes identified in both cases. In particular, comparison of the distributions of different genotypes in sewage samples and patient samples by correlative statistic tools yielded high concordance, i.e., Pearson's correlation coefficient of 0.957 and Lin's correlation concordance coefficient of 0.933 (not shown).
Although it is likely that the viruses found in the WWTPs reflect the strains circulating in the population, comparisons of the two sets of data have some limitations. In fact, the detection of particular G and P types may indicate either a higher concentration or a higher stability of these strains in sewage, whereas the data from patients correspond to the actual numbers of children infected with each RVA strain.
The proportions of sewage samples and samples from children where specific RVA genotypes were detected in 2011 presented some differences between cities. In particular, in Bari and Palermo, genotype G1 was detected in 80% and 68% of patients, respectively, whereas this genotype was present in 54% and 50% of sewage samples, respectively. In contrast, G2 RVA infected 5% and 11% of children with AGE in Bari and Palermo, respectively, and was detected in 27% and 32% of the corresponding sewage samples (Tables 2 and 3). Equivalent ratios were also found for comparison of the P[8] and P[4] genotypes in Bari and Palermo. In the other two cities, the ratios between genotypes G1 and G2 in patient samples were very similar to those determined in sewage samples.
G9 RVA was found for between 7% (Naples) and 16% (Palermo) of sewage samples but was detected in a high percentage (15%) of patient samples only in Naples, being rare elsewhere (<2.2%).
The uncommon G6 genotype was detected in two sewage samples from Bari, one of which also contained the rare P[14] genotype. Other uncommon genotypes included G8 and G10 strains in samples from two patients in Sassari and Palermo, respectively, and three P[9] strains in samples from two patients in Palermo (Table 3) and in one sewage sample from Bari (Table 2).
The two rare P[6] and P[19] genotypes detected in Naples were present only in sewage (1 sample each), despite sewage samples in this city being positive for RVA less frequently than elsewhere. This observation may suggest that these uncommon RVA circulate in the population more extensively than is indicated by patient strain genotyping.
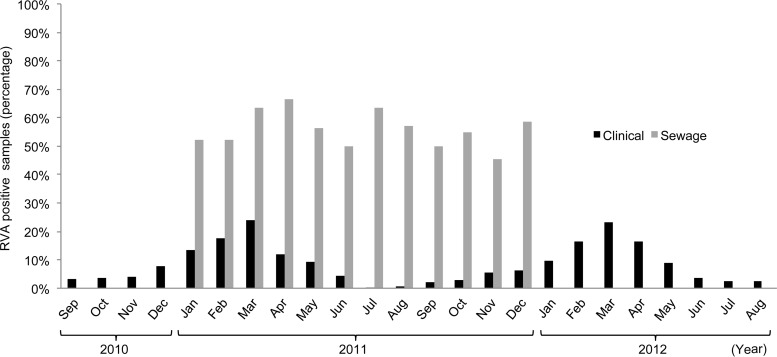
Figure 1 reports the average proportions of RVA-positive sewage samples by month in 2011, when samples were taken from all four cities of the study, showing the occurrence of RVA throughout the year. For a better comparison with the marked seasonal distribution of rotavirus infections, data on pediatric patients were reported for the period from September 2010 to August 2012, including two sequential winter-spring epidemic peaks. Whereas patients presented a marked seasonal distribution, with cases peaking in March and being almost absent during summer, the percentage of RVA-positive sewage samples ranged between 45 and 67% throughout the year, showing no specific seasonal trend.
FIG 1.
Distribution of RVA-positive sewage samples (2011) and samples from pediatric AGE cases (2010 to 2012) in Naples, Bari, Palermo, and Sassari, by month.
Phylogenetic analysis of rotavirus G and P genotypes.
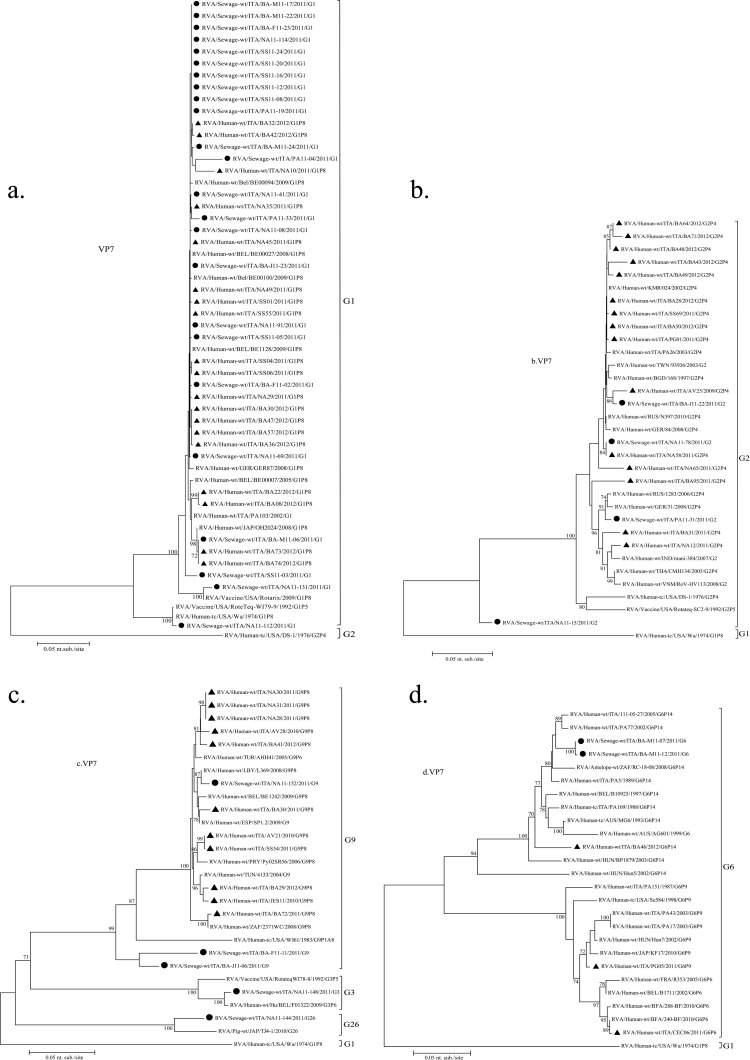
Seventy-one strains with common G types (G1 to G3 and G9) detected in either wastewater (32) or clinical (39) samples in the cities investigated in 2011 were selected randomly and subjected to VP7 and VP4 gene sequencing and phylogenetic analysis.
All G1 strains sequenced presented high nucleotide identity (97.5 to 100%), regardless of the source and city, except for strain NA11-112, identified in Naples (Fig. 2a), which coclustered with the Wa (G1P[8]) prototype strain. Sewage sample strain NA11-112 also contained a P[8] sequence that coclustered with the P[8] Wa strain separately from the other P[8] sequences analyzed in this study.
FIG 2.
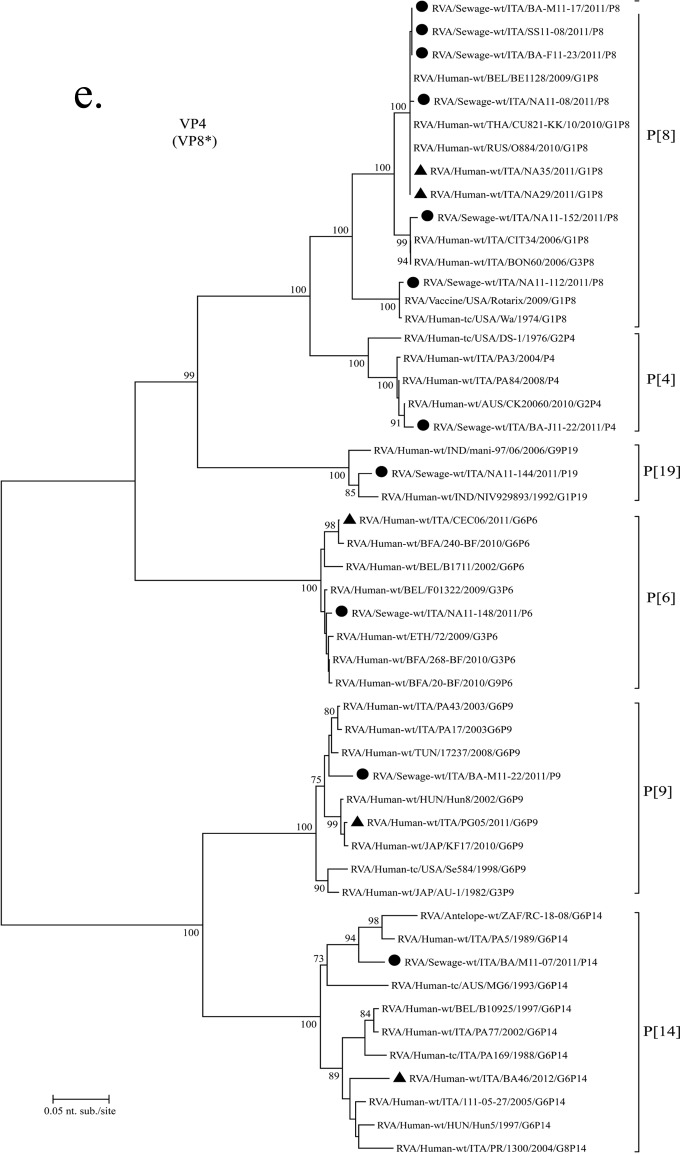
Phylogenetic dendrograms based on partial VP7 sequences of genotypes G1 (a), G2 (b), G3 and G9 (c), and G6 (d) and on partial VP4 nucleotide sequences of genotypes P[8], P[4], P[14], P[19], P[6], and P[9] (e). RVA strains from Italy were detected in sewage and clinical samples. All sequences obtained from GenBank are named as described previously by Matthijnssens et al. (7), and G and P genotypes are indicated on the right. Environmental samples are marked with filled circles; AGE samples are marked with filled triangles. The scale bar at the bottom of the tree indicates the number nucleotide substitutions/site. Bootstrap values (2,000 replicates) are shown at the branch nodes; values of <70 are not shown.
The nucleotide identities were also very high (97 to 99%) among the G2 strains, except for strain NA11-15, which presented a >10% difference from all other G2 sequences (Fig. 2b). Similarly, most G9 clinical strains and the G9 sewage strain NA11-152 from Naples coclustered within the G9-III lineage, except for sewage strains J11-06 and F11-11 from Bari (Fig. 2c).
The G3 VP7 gene sequence from a sewage sample from Bari was closely related to a G3P[6] strain reported in Belgium in 2009 (Fig. 2c). A second strain formerly genotyped as G3 by PCR was eventually assigned to the rare G26 genotype after sequencing, showing strict relatedness to swine G26 strain TJ4-1, detected in Japan in 2010 (Fig. 2c). The generation of a G3-sized amplicon was likely due to the 74% sequence identity found between nucleotides 250 and 268 of the G26 gene and the G3 primer.
The uncommon G6 strains M11-07 and M11-12 detected in the Bari WWTP in April and June (Fig. 2d) were identical and presented a 96% similarity to human G6P[14] strain BA46 detected in Bari 1 year later and to other G6P[14] RVA strains identified in Italy during 1988 to 2005. The P[14] genotype found in Bari sewage sample strain M11-07 showed 93.5% nucleotide identity with G6P[14] clinical strain BA46 (Fig. 2e).
Five of the common P[8] sequences from Naples, Bari, and Sassari WWTPs exhibited high nucleotide identity to two clinical P[8] sequences identified in Naples and to other strains from GenBank. The only sewage P[4] strain sequenced presented 98% identity to Italian strains PA84/2008 and PA3/2004.
A rare P[19] genotype was detected in sewage sample strain NA11-144 (Fig. 2e), which also yielded the uncommon G26 genotype (Fig. 2c). This P[19] sequence was 97 to 98% identical to two human G1P[19] and G9P[19] strains previously identified in India.
The P[6] genotype from sewage sample strain NA11-148 (also containing G3) presented 98% identity with pediatric strain RVA/Human-wt/ITA/CEC06/2011/G6P[6], reported in central Italy in 2011. The P[9] genotype from sewage sample strain BA-M11-22 (also containing genotype G1) showed only 95% nucleotide identity to the contemporary strain RVA/Human-wt/ITA/PG05/2011/G6P[9] but coclustered with older G6P[9] Italian strains.
DISCUSSION
We investigated the correlation between rotavirus genotypes G and P in strains from inflowing water in the WWTPs of four Italian cities and the RVA strains isolated from children with severe gastroenteritis, identified by the RotaNet-Italy surveillance network in the same cities, in 2011.
Combined ultracentrifugation and molecular methods permitted efficient rotavirus recovery and RNA detection in sewage samples, altogether yielding a 20-fold rotavirus concentration or higher with removal of PCR inhibitors. This method of viral concentration in sewage samples was described previously by Fumian and coworkers (30) and was demonstrated to work better than adsorption-elution protocols. In our hands, this method allowed a rate of recovery of the original virus in spiked samples of at least 30% to between 50 and 67%, which is in the same order as that reported by Fumian and coworkers (45%), with some differences between samples.
Besides allowing rapid and sensitive rotavirus identification, the molecular methods applied offer the additional advantage of being easily extended to the simultaneous detection of other enteric viruses.
Although molecular detection may not assess infectious RVA in sewage, the detection of viral RNA is meaningful, since wild RVA strains are normally resistant in the environment (39).
The finding of RVA in a large fraction of the sewage samples examined indicates both that considerable and continuous virus circulation occurs in the populations of the cities investigated and that RVA is shed with feces in large amounts, permitting its detection despite extensive dilution in sewage.
Moreover, detection of rotavirus in sewage also points out possible risks for human health, particularly in the case of wastewater treatment failures, as may occur during heavy raining and flooding.
Only a few other studies compared rotavirus strains in samples from pediatric AGE patients and sewage in the same location and year (40–43). In this study, RVA genotyping and phylogenetic analysis of common VP7 genotype G1, G2, G3, and G9 sequences showed high similarity between clinical and environmental strains. These findings indicate that rotaviruses released into urban sewage largely match the RVA strains that cause severe disease in children, implying that common RVA genotypes are also involved in asymptomatic or mild infection of adults and children not requiring hospitalization.
It should be considered that coverage with rotavirus vaccine in Italy, particularly in the period considered in this study, did not exceed 5% of the pediatric population, including the cities monitored. Therefore, the genotypes found to be circulating were not receiving any selective pressure, which Matthijnssens et al. (44) hypothesized was induced by mass vaccination, favoring specific viral strains. However, other studies disagree with any occurrence of genotype replacement in largely vaccinated populations (5, 45), rather highlighting that common genotypes, particularly G1P[8], remain predominant despite the overall decrease in the number of cases.
A high prevalence of genotypes G2 and G9 was observed in sewage samples from Palermo and Bari, while these genotypes were infrequently associated with cases of disease in children in this study. The prevalence of genotype G2 in sewage was confirmed by the correspondingly high rate of detection of genotype P[4], normally associated with G2P[4] strains. It is possible that the higher rate of detection of G2 and P[4] merely reflects temporal fluctuations in the circulation of different genotypes in the population. Nevertheless, this finding might otherwise indicate a different persistence of distinct viruses in sewage or higher rates of intestinal replication of individual strains, which would result in different viral concentrations in wastewater. In fact, fewer sewage samples were found to be positive for rotavirus in Naples, where a higher dilution of fecal viruses in the city WWTPs is very likely, since these WWTPs also collect industrial wastewaters in addition to urban waste.
Uncommon G6 and P[14] sequences were detected in sewage twice in 2011 in Bari. Interestingly, these RVA strains were related phylogenetically to a genotype G6P[14] strain detected in the stool samples of a patient in the same city in 2012. This findings may suggest that a rare G6P[14] RVA strain circulated in the city population for at least a year, being shed into the sewer system at high concentrations. It is tempting to believe that environmental surveillance may help predict the circulation of emerging RVA strains before symptomatic cases are detected, as observed with other enteric viruses (46).
Although related, the G6 strains from sewage samples in 2011 and the patient in 2012 in Bari were not identical, suggesting that the same autochthonous strain evolved obviously between 2011 and 2012. All other G6 sequences from other cities belonged to completely separate clusters.
The rare G26 strain NA11-144 detected in sewage samples from Naples showed high sequence similarity to a Japanese strain of apparent swine origin. The sewage sample also contained a rare P[19] genotype, previously detected in a G1P[19] strain from a patient in India, which was proposed to represent a human/swine reassortant (47). The simultaneous presence of typically swine genotypes G26 and P[19] in the same sewage sample suggests that animal feces may have been disposed of into the urban wastewater sewer system of Naples. Although animal waste should not merge with human drainage, we cannot exclude that unauthorized dumping from a nearby swine farm or slaughterhouse may have occurred.
The initial erroneous identification of G26 RVA as G3 was likely due to sequence conservation of the G3 primer-binding region in the G26 strain, generating a typical G3-like amplicon by nested PCR. Because this may in principle lead to wrong genotyping and a lack of detection of other uncommon emerging RVAs, the G3-specific primer may need to be redesigned. At present, whenever an uncommon G or P genotype is identified in association with an apparently common P or G type, sequencing of both genes may be recommended to avoid mistyping.
Interestingly, whereas most hospitalized rotavirus cases clustered between January and April 2011 and cases were virtually absent in July and August, RVA was constantly found in sewage samples in all months investigated. Similar observations of the persistence of RVA in sewage have been reported previously, mostly independent from the seasonal pattern of hospitalized RVA cases (40, 48, 49), and indicate that the population maintains a high level of virus shedding throughout the year, independent of pediatric clinical disease. The lack of seasonality in virus discharge in sewage suggests that rotaviruses might circulate through asymptomatic or subclinical reinfections of children and adults, before a new epidemic starts among susceptible young children.
In conclusion, this paper suggests that environmental monitoring of sewage provides a good assessment of RVA genotypes circulating in the local human population, with minor differences with respect to the strains causing severe infantile diarrhea. Also, uncommon RVA strains might be detected in sewage earlier than in patients, which might help in preparation for the future spread of emerging strains. Environmental monitoring of WWTPs might be complementary to molecular RVA surveillance of clinical cases within vaccine monitoring programs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michele Labianca (Osservatorio Epidemiologico Regione Puglia, Bari, Italy) and Francesca Pennino (University of Naples, Naples, Italy) for providing sewage samples and Anna Morea (University of Bari, Bari, Italy) for sequencing of rotavirus clinical strains from Bari.
This study was supported by grants from the Ministry of Health, Italy, to L.F. (CCM Epidemiologia Molecolare di Rotavirus in Atà Pediatrica in Italia, Creazione di una Rete di Sorveglianza per Monitorare la Diffusione e l'Evoluzione di Genotipi Virali, 2007 to 2009) and to F.M.R. (Italy/United States, Investigating the Evolution of Zoonotic Norovirus and Rotavirus Strains from Swine, 2011) and from EuroRotaNet (2007) (http://www.eurorota.net).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02695-14.
REFERENCES
- 1.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. 2012. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 12:136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- 2.Charles MD, Holman RC, Curns AT, Parashar UD, Glass RI, Bresee JS. 2006. Hospitalizations associated with rotavirus gastroenteritis in the United States, 1993-2002. Pediatr Infect Dis J 25:489–493. doi: 10.1097/01.inf.0000215234.91997.21. [DOI] [PubMed] [Google Scholar]
- 3.Kapikian AZ. 1996. Overview of viral gastroenteritis. Arch Virol Suppl 12:7–19. [DOI] [PubMed] [Google Scholar]
- 4.Estes MK, Kapikian AZ. 2007. Rotaviruses, p 1917–1974. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Dennehy PH. 2013. Treatment and prevention of rotavirus infection in children. Curr Infect Dis Rep 15:242–250. doi: 10.1007/s11908-013-0333-5. [DOI] [PubMed] [Google Scholar]
- 6.Abe M, Ito N, Morikawa S, Takasu M, Murase T, Kawashima T, Kawai Y, Kohara J, Sugiyama M. 2009. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res 144:250–257. doi: 10.1016/j.virusres.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 9.Iturriza-Gomara M, Dallman T, Banyai K, Bottiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Koopmans M, Korsun N, Koukou D, Kroneman A, Laszlo B, Lappalainen M, Maunula L, Marques AM, Matthijnssens J, Midgley S, Mladenova Z, Nawaz S, Poljsak-Prijatelj M, Pothier P, Ruggeri FM, Sanchez-Fauquier A, Steyer A, Sidaraviciute-Ivaskeviciene I, Syriopoulou V, Tran AN, Usonis V, Van Ranst M, De Rougemont A, Gray J. 2011. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol Infect 139:895–909. doi: 10.1017/S0950268810001810. [DOI] [PubMed] [Google Scholar]
- 10.Pietsch C, Petersen L, Patzer L, Liebert UG. 2009. Molecular characteristics of German G8P[4] rotavirus strain GER1H-09 suggest that a genotyping and subclassification update is required for G8. J Clin Microbiol 47:3569–3576. doi: 10.1128/JCM.01471-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg GA, Payne DC, Teel EN, Mijatovic-Rustempasic S, Bowen MD, Wikswo M, Gentsch JR, Parashar UD. 2012. First reports of human rotavirus G8P[4] gastroenteritis in the United States. J Clin Microbiol 50:1118–1121. doi: 10.1128/JCM.05743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauchan P, Nakagomi T, Sherchand JB, Yokoo M, Pandey BD, Cunliffe NA, Nakagomi O. 2013. Continued circulation of G12P[6] rotaviruses over 28 months in Nepal: successive replacement of predominant strains. Trop Med Health 41:7–12. doi: 10.2149/tmh.2012-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oluwatoyin Japhet M, Adeyemi Adesina O, Famurewa O, Svensson L, Nordgren J. 2012. Molecular epidemiology of rotavirus and norovirus in Ile-Ife, Nigeria: high prevalence of G12P[8] rotavirus strains and detection of a rare norovirus genotype. J Med Virol 84:1489–1496. doi: 10.1002/jmv.23343. [DOI] [PubMed] [Google Scholar]
- 14.Carter MJ. 2005. Enterically infecting viruses: pathogenicity, transmission and significance for food and waterborne infection. J Appl Microbiol 98:1354–1380. doi: 10.1111/j.1365-2672.2005.02635.x. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins RS, Gaspard GB, Williams FP Jr, Karlin RJ, Cukor G, Blacklow NR. 1984. A community waterborne gastroenteritis outbreak: evidence for rotavirus as the agent. Am J Public Health 74:263–265. doi: 10.2105/AJPH.74.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansari SA, Springthorpe VS, Sattar SA. 1991. Survival and vehicular spread of human rotaviruses: possible relation to seasonality of outbreaks. Rev Infect Dis 13:448–461. doi: 10.1093/clinids/13.3.448. [DOI] [PubMed] [Google Scholar]
- 17.Espinosa AC, Mazari-Hiriart M, Espinosa R, Maruri-Avidal L, Mendez E, Arias CF. 2008. Infectivity and genome persistence of rotavirus and astrovirus in groundwater and surface water. Water Res 42:2618–2628. doi: 10.1016/j.watres.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Petrinca AR, Donia D, Pierangeli A, Gabrieli R, Degener AM, Bonanni E, Diaco L, Cecchini G, Anastasi P, Divizia M. 2009. Presence and environmental circulation of enteric viruses in three different wastewater treatment plants. J Appl Microbiol 106:1608–1617. doi: 10.1111/j.1365-2672.2008.04128.x. [DOI] [PubMed] [Google Scholar]
- 19.Villena C, El-Senousy WM, Abad FX, Pinto RM, Bosch A. 2003. Group A rotavirus in sewage samples from Barcelona and Cairo: emergence of unusual genotypes. Appl Environ Microbiol 69:3919–3923. doi: 10.1128/AEM.69.7.3919-3923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timenetsky MC, Gouvea V, Santos N, Alge ME, Kisiellius JJ, Carmona RC. 1996. Outbreak of severe gastroenteritis in adults and children associated with type G2 rotavirus. Study Group on Diarrhea of the Instituto Adolfo Lutz. J Diarrhoeal Dis Res 14:71–74. [PubMed] [Google Scholar]
- 21.Rasanen S, Lappalainen S, Halkosalo A, Salminen M, Vesikari T. 2011. Rotavirus gastroenteritis in Finnish children in 2006-2008, at the introduction of rotavirus vaccination. Scand J Infect Dis 43:58–63. doi: 10.3109/00365548.2010.508462. [DOI] [PubMed] [Google Scholar]
- 22.Koopmans M, von Bonsdorff CH, Vinje J, de Medici D, Monroe S. 2002. Foodborne viruses. FEMS Microbiol Rev 26:187–205. doi: 10.1111/j.1574-6976.2002.tb00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metcalf TG, Melnick JL, Estes MK. 1995. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu Rev Microbiol 49:461–487. doi: 10.1146/annurev.mi.49.100195.002333. [DOI] [PubMed] [Google Scholar]
- 24.Jiang V, Jiang B, Tate J, Parashar UD, Patel MM. 2010. Performance of rotavirus vaccines in developed and developing countries. Hum Vaccin 6:532–542. doi: 10.4161/hv.6.7.11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monini M, Biasin A, Valentini S, Cattoli G, Ruggeri FM. 2011. Recurrent rotavirus diarrhoea outbreaks in a stud farm, in Italy. Vet Microbiol 149:248–253. doi: 10.1016/j.vetmic.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Bosch A, Guix S, Sano D, Pinto RM. 2008. New tools for the study and direct surveillance of viral pathogens in water. Curr Opin Biotechnol 19:295–301. doi: 10.1016/j.copbio.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Bartolo I, Monini M, Losio MN, Pavoni E, Lavazza A, Ruggeri FM. 2011. Molecular characterization of noroviruses and rotaviruses involved in a large outbreak of gastroenteritis in Northern Italy. Appl Environ Microbiol 77:5545–5548. doi: 10.1128/AEM.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira FF, Guimaraes FR, Fumian TM, Victoria M, Vieira CB, Luz S, Shubo T, Leite JP, Miagostovich MP. 2009. Environmental dissemination of group A rotavirus: P-type, G-type and subgroup characterization. Water Sci Technol 60(3):633–642. doi: 10.2166/wst.2009.413. [DOI] [PubMed] [Google Scholar]
- 29.Giammarioli AM, Mackow ER, Fiore L, Greenberg HB, Ruggeri FM. 1996. Production and characterization of murine IgA monoclonal antibodies to the surface antigens of rhesus rotavirus. Virology 225:97–110. doi: 10.1006/viro.1996.0578. [DOI] [PubMed] [Google Scholar]
- 30.Fumian TM, Leite JP, Castello AA, Gaggero A, Caillou MS, Miagostovich MP. 2010. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J Virol Methods 170:42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iturriza Gomara M, Wong C, Blome S, Desselberger U, Gray J. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence of independent segregation. J Virol 76:6596–6601. doi: 10.1128/JVI.76.13.6596-6601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iturriza-Gomara M, Kang G, Gray J. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol 31:259–265. doi: 10.1016/j.jcv.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 30:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zar JH. 1996. Biostatistical analysis, 3rd ed. Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- 37.Pearson K. 1909. Determination of the coefficient of correlation. Science 30:23–25. doi: 10.1126/science.30.757.23. [DOI] [PubMed] [Google Scholar]
- 38.Lin LI. 1989. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45:255–268. doi: 10.2307/2532051. [DOI] [PubMed] [Google Scholar]
- 39.Xue B, Jin M, Yang D, Guo X, Chen Z, Shen Z, Wang X, Qiu Z, Wang J, Zhang B, Li J. 2013. Effects of chlorine and chlorine dioxide on human rotavirus infectivity and genome stability. Water Res 47:3329–3338. doi: 10.1016/j.watres.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 40.Barril PA, Giordano MO, Isa MB, Masachessi G, Ferreyra LJ, Castello AA, Glikmann G, Nates SV. 2010. Correlation between rotavirus A genotypes detected in hospitalized children and sewage samples in 2006, Cordoba, Argentina. J Med Virol 82:1277–1281. doi: 10.1002/jmv.21800. [DOI] [PubMed] [Google Scholar]
- 41.Kamel AH, Ali MA, El-Nady HG, Aho S, Pothier P, Belliot G. 2010. Evidence of the co-circulation of enteric viruses in sewage and in the population of Greater Cairo. J Appl Microbiol 108:1620–1629. doi: 10.1111/j.1365-2672.2009.04562.x. [DOI] [PubMed] [Google Scholar]
- 42.Arraj A, Bohatier J, Aumeran C, Bailly JL, Laveran H, Traore O. 2008. An epidemiological study of enteric viruses in sewage with molecular characterization by RT-PCR and sequence analysis. J Water Health 6:351–358. doi: 10.2166/wh.2008.053. [DOI] [PubMed] [Google Scholar]
- 43.Sdiri-Loulizi K, Hassine M, Aouni Z, Gharbi-Khelifi H, Chouchane S, Sakly N, Neji-Guediche M, Pothier P, Aouni M, Ambert-Balay K. 2010. Detection and molecular characterization of enteric viruses in environmental samples in Monastir, Tunisia between January 2003 and April 2007. J Appl Microbiol 109:1093–1104. doi: 10.1111/j.1365-2672.2010.04772.x. [DOI] [PubMed] [Google Scholar]
- 44.Matthijnssens J, Nakagomi O, Kirkwood CD, Ciarlet M, Desselberger U, Van Ranst M. 2012. Group A rotavirus universal mass vaccination: how and to what extent will selective pressure influence prevalence of rotavirus genotypes? Expert Rev Vaccines 11:1347–1354. doi: 10.1586/erv.12.105. [DOI] [PubMed] [Google Scholar]
- 45.Hemming M, Vesikari T. 2013. Genetic diversity of G1P[8] rotavirus VP7 and VP8* antigens in Finland over a 20-year period: no evidence for selection pressure by universal mass vaccination with RotaTeq vaccine. Infect Genet Evol 19:51–58. doi: 10.1016/j.meegid.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 46.Ganesh A, Lin J. 2013. Waterborne human pathogenic viruses of public health concern. Int J Environ Health Res 23:544–564. doi: 10.1080/09603123.2013.769205. [DOI] [PubMed] [Google Scholar]
- 47.Chitambar SD, Arora R, Chhabra P. 2009. Molecular characterization of a rare G1P[19] rotavirus strain from India: evidence of reassortment between human and porcine rotavirus strains. J Med Microbiol 58:1611–1615. doi: 10.1099/jmm.0.012856-0. [DOI] [PubMed] [Google Scholar]
- 48.Green DH, Lewis GD. 1999. Comparative detection of enteric viruses in wastewaters, sediments and oysters by reverse transcription-PCR and cell culture. Water Res 33:1195–1200. doi: 10.1016/S0043-1354(98)00313-3. [DOI] [Google Scholar]
- 49.Bosch A, Pinto RM, Jofre J. 1988. Non-seasonal distribution of rotavirus in Barcelona raw sewage. Zentralbl Bakteriol Mikrobiol Hyg B 186:273–277. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.