Abstract
Myoviruses and podoviruses that infect cyanobacteria are the two major groups of marine cyanophages, but little is known of how their phylogenetic lineages are distributed in different habitats. In this study, we analyzed the phylogenetic relationships of cyanopodoviruses and cyanomyoviruses based on the existing genomes. The 28 cyanomyoviruses were classified into four clusters (I to IV), and 19 of the 20 cyanopodoviruses were classified into two clusters, MPP-A and MPP-B, with four subclusters within cluster MPP-B. These genomes were used to recruit cyanophage-like fragments from microbial and viral metagenomes to estimate the relative abundances of these cyanophage lineages. Our results showed that cyanopodoviruses and cyanomyoviruses are both abundant in various marine environments and that clusters MPP-B, II and III appear to be the most dominant lineages. Cyanopodoviruses and cluster I and IV cyanomyoviruses exhibited habitat-related variability in their relative levels of abundance, while cluster II and III cyanomyoviruses appeared to be consistently dominant in various habitats. Multivariate analyses showed that reads that mapped to Synechococcus phages and Prochlorococcus phages had distinct distribution patterns that were significantly correlated to those of Synechococcus and Prochlorococcus, respectively. The Mantel test also revealed a strong correlation between the community compositions of cyanophages and picocyanobacteria. Given that cyanomyoviruses tend to have a broad host range and some can cross-infect Synechococcus and Prochlorococcus, while cyanopodoviruses are commonly host specific, the observation that their community compositions both correlated significantly with that of picocyanobacteria was unexpected. Although cyanomyoviruses and cyanopodoviruses differ in host specificity, their biogeographic distributions are likely both constrained by the picocyanobacterial community.
INTRODUCTION
Picocyanobacteria of the genera Synechococcus and Prochlorococcus are the most abundant primary producers in the ocean (1). Cyanobacterial viruses or cyanophages can influence the abundance, diversity, and productivity of cyanobacteria in the ocean (2–7). All known marine cyanophages are tailed double-stranded DNA viruses belonging to three well-defined bacteriophage families, Myoviridae, Podoviridae, and Siphoviridae. Cyanomyoviruses tend to infect a broad range of hosts, often across Prochlorococcus and Synechococcus, while cyanopodoviruses and cyanosiphoviruses are generally host specific (5, 8–13).
Many complete genomes of marine cyanophages have been published for cyanomyoviruses (14–20), cyanopodoviruses (14, 21–23), and cyanosiphoviruses (24, 25). Nearly all known marine cyanomyoviruses (except S-TIM5 [19]) and all known marine cyanopodoviruses are morphologically and genetically similar to the archetypical coliphages T4 and T7, respectively. Cyanophage sequences have commonly been found in both microbial (26–28) and viral metagenomes (29, 30), which underlies the use of existing metagenomic databases to investigate cyanophage communities (20, 21, 25, 31, 32).
Cyanomyoviruses and cyanopodoviruses appear to be much more abundant than cyanosiphoviruses in the ocean (11, 25, 33, 34), and cyanopodoviruses are likely as abundant as cyanomyoviruses (21). The genetic diversity of marine cyanomyoviruses and cyanopodoviruses has been extensively investigated using molecular markers (see reference 35 for a review). Four cyanomyovirus phylogenetic clusters (I, II, III, and PSSM9/11/12 new cluster) have been well defined in earlier studies, using the signature gene g20 that codes for the phage portal protein (36, 37). Metagenomic recruitment of g20 sequences suggested that cluster II may be the most abundant lineage of marine cyanomyoviruses (37). Two discrete marine picocyanobacterial podovirus (MPP) phylogenetic clusters (MPP-A and MPP-B) were recognized based on the viral DNA polymerase gene (12). Recent studies suggested that MPP-B podoviruses are more abundant than MPP-A podoviruses (13, 38, 39). However, the relative abundances and spatial patterns of specific cyanopodovirus and cyanomyovirus phylogenetic lineages over the global ocean are still broadly unclear.
Covariation between cyanophage titers and cyanobacterial abundances has been observed in the sea (5–8, 10, 11, 33). Moreover, both the abundance and genetic diversity of marine cyanomyoviruses were found to covary with those of Synechococcus in a seasonal time series in the Gulf of Aqaba, Red Sea (6). However, another study reported that the cyanomyovirus diversity was not significantly correlated with the abundance or diversity of the co-occurring Prochlorococcus or Synechococcus populations along a North-to-South Atlantic Ocean transect (40). It appears that temporal phage-host correlation is observed more often than spatial correlation. However, investigations of phage-host covariation on large spatial scales are still seldom done, especially for specific phage-host systems. Prochlorococcus and Synechococcus have nearly complementary biogeographies across the global ocean (41). Prochlorococcus dominates the photoautotroph community numerically in the broad oligotrophic open oceans between 40°N and 40°S but is much rarer in nearshore or nutrient-rich waters and generally absent from brackish waters or in high-latitude regions (1, 41, 42). In contrast, Synechococcus is nearly 1 order of magnitude less abundant than Prochlorococcus in open oceans but more abundant in mesotrophic nearshore waters (1, 41, 43) and can be found in the polar seas (44–46). Moreover, Prochlorococcus and Synechococcus are both distinguished by their genetic and physiological diversity, forming closely associated genotypes and ecotypes (47–51) that exhibit remarkable global biogeographic patterns (44, 52–56). Thus, it is interesting to investigate whether the cyanophages have a biogeographic pattern similar to that of picocyanobacteria and to test the correlation between their biogeographic distributions on a large global scale over the world's oceans.
In this study, we analyzed the phylogenetic relationships of 20 marine cyanopodoviruses and 28 T4-like marine cyanomyoviruses based on their genomes. The relative abundances of cyanopodovirus and cyanomyovirus lineages in various marine environments were estimated by recruiting their similar reads from numerous microbial and viral metagenomes. Meanwhile, the read abundances of cyanophage lineages in microbial metagenomes were coanalyzed with those of Prochlorococcus and Synechococcus to test the possible correlation between biogeographic patterns of cyanophages and picocyanobacteria in the sea.
MATERIALS AND METHODS
Phylogenetic analysis.
Genomes from 20 marine cyanopodoviruses and 28 marine cyanomyoviruses were included in the analyses (see Table S1 in the supplemental material). Sequences of previously reported core genes of the 17 marine cyanomyoviruses (57 core genes) (18, 57) and the 12 marine cyanopodoviruses (15 core genes) (21) were extracted from their genomes. The protein sequences of the two core gene sets were compared to all the protein sequences of the 20 cyanopodoviruses and the 28 cyanomyoviruses to reassign core genes, since additional genomes were included in the analyses here. A homologous relationship between sequences was assigned when their reciprocal best hits met the criterion cutoff of an E value of <10−5 and the alignment covered at least 50% of the shorter sequence. Two (T4-GC_4 and T4-GC_250, coding for a DUF1825 domain-containing protein and a hypothetical protein) of the 57 previously identified core genes of marine cyanomyoviruses based on 17 genomes (18, 57) are not shared by all 28 genomes, and thus, these two were excluded in our phylogenetic analysis. Each of the 15 core genes that were identified based on a collection of 12 cyanopodovirus genomes (21) was also shared by all 8 additional genomes. The amino acid sequences of the 15 cyanopodovirus core genes and those of the 55 cyanomyovirus core genes were concatenated. Clustal X2 (58) was used to align the concatenated sequences, and the resulting alignments were trimmed to remove highly divergent regions by using the program Gblocks (59). The maximum-likelihood (ML) phylogenies were built by using RAxML (60, 61) using the JTT protein substitution matrix and the GTRGAMMA+I model to estimate the proportions of invariable sites. A bootstrap test with 100 replicates was used to evaluate the robustness of the trees. The concatenated nucleotide sequences of six single-copy housekeeping genes of 27 picocyanobacteria strains (see the description of these six genes below) were aligned using Clustal X2 and trimmed using Gblocks. The resulting alignment was input to PAUP* 4b10 (62) to construct a distance-based phylogenetic tree and to perform a bootstrap test with 1,000 replicates.
Metagenomic analysis.
The metagenome data sets tested in this study (see Table S2 in the supplemental material) were downloaded from the CAMERA database (63). The microbial metagenomes included the whole database of the Global Ocean Sampling (GOS) Expedition Project (64, 65) and data sets from the Hawaii Ocean Time-series (HOT) station samples (26, 66, 67). The viral metagenomes were comprised of data sets from MarineVirome projects (29) and BroadPhage projects (http://www.broadinstitute.org). The GOS and HOT microbial samples targeted microbial fractions of 0.1 to 0.8 μm and 0.22 to 1.6 μm, respectively, while the viral metagenome projects sampled particles smaller than 0.2 μm. Both the microbial and viral samples analyzed in this study covered broad geographic regions and diverse marine environments, such as estuary, coastal water, open ocean, and reef (the habitat types of samples or locations were obtained from the CAMERA database).
Microbial and viral metagenomic reads were recruited by each entire genome of the 20 cyanopodoviruses and 28 cyanomyoviruses (see Table S1 in the supplemental material). In order to minimize the bias caused by nonspecific recruitment, especially from phage-host shared genes, the following steps were carried out. First, reads were retrieved from metagenomes by local BLASTN runs, using the following parameters: E value of ≤1e−3, gap open penalty of −1, gap extend penalty of −1, mismatch penalty of −3, and reward of 1. Second, each of the recruited metagenomic reads was compared against the NCBI RefSeq database, and the most similar RefSeq sequence of each read was exported and filtered to remove repeat redundancy. We then built a data set that encompassed the exported RefSeq sequences and all gene sequences (DNA) from the genomes of the 20 cyanopodoviruses, 28 cyanomyoviruses, and 27 picocyanobacteria (see Table S3 in the supplemental material). Each of the metagenomic reads was again compared against this data set, and reads that had best hits to non-cyanopodovirus or non-cyanomyovirus sequences were removed. Third, the retained reads were mapped onto one of the 20 cyanopodoviruses or 28 cyanomyoviruses by using BLASTN comparison. In order to test the possible correlation between the distributions of cyanophages and cyanobacteria, the relative abundances of Prochlorococcus and marine Synechococcus sequences in each GOS station were calculated using six single-copy housekeeping genes as metrics (recA, atpD, gyrB, rpoB, tuf, and nrdJ) (see Table S3). These single-copy genes have been applied to assess the microbial diversity (68) or to estimate the relative abundances of other genes as metrics (69, 70) when analyzing the GOS metagenomic data sets. Reference DNA sequences of these genes were retrieved from the complete or draft genomes of 12 Prochlorococcus and 15 marine Synechococcus strains (see Table S3). Recruitments of these gene sequences from the GOS database also followed the three steps described above. Perl scripts were used to parse BLAST outputs and to calculate the G+C content of reads.
Statistical analyses.
The t test was carried out using SPSS software version 13. CANOCO version 4.5 was used to perform the multivariate analysis, based on the recruitment results against the GOS metagenomes. The environmental parameters (temperature, latitude, depth, and concentration of chlorophyll a) of GOS sampling sites were obtained from the CAMERA database. Chlorophyll data at a GOS site were represented by the average chlorophyll a density in the sampling month. To perform constrained ordination, environmental parameters were treated as explanatory variables, and the cyanopodovirus, cyanomyovirus, Synechococcus, and Prochlorococcus read abundances were treated as response variables. Redundancy analysis (RDA; based on a linear model) rather than canonical correspondence analysis (CCA; based on a unimodal model) was chosen because the maximum gradient length of a priori detrended correspondence analysis (DCA) is shorter than 3.0 (71). A subset of explanatory variables was forward selected using partial Monte Carlo permutation tests (999 permutations), in order to reduce cross-correlation. The variables chlorophyll a density and latitude failed to pass the test (P value > 0.05) and were excluded in subsequent steps. The null hypothesis that the response is independent of the explanatory variables was tested using a Monte Carlo permutation test (999 permutations). Nonmetric multidimensional scaling (NMDS) based on the Bray-Curtis similarity matrix was performed using PRIMER version 5 (Primer-E, Lutton, United Kingdom). One-way analysis of similarity (ANOSIM) with 999 permutations was used to test the significance of grouping of cyanophage or picocyanobacterial lineages on the NMDS diagrams. The Mantel test with 999 permutations was used to estimate the correlations between cyanophage community composition and cyanobacterial community composition and between cyanophage community composition and environmental parameters. Cyanobacteria-based Bray-Curtis similarity between GOS samples was calculated based on the relative abundances of the reads that were recruited to each of the cyanobacterial genotypes. Cyanophage-based Bray-Curtis similarity was calculated based on the relative abundances of the reads that were recruited to each of the cyanophage isolates and to each of the cyanophage genotypes, respectively. Environmental parameters (temperature, depth, latitude, and chlorophyll a) were z-score transformed, and the Euclidean distance between GOS samples was calculated based on the four parameters. PRIMER was used to calculate the matrixes and perform the tests.
RESULTS
Phylogenetic lineages of cyanopodoviruses, cyanomyoviruses, and picocyanobacteria.
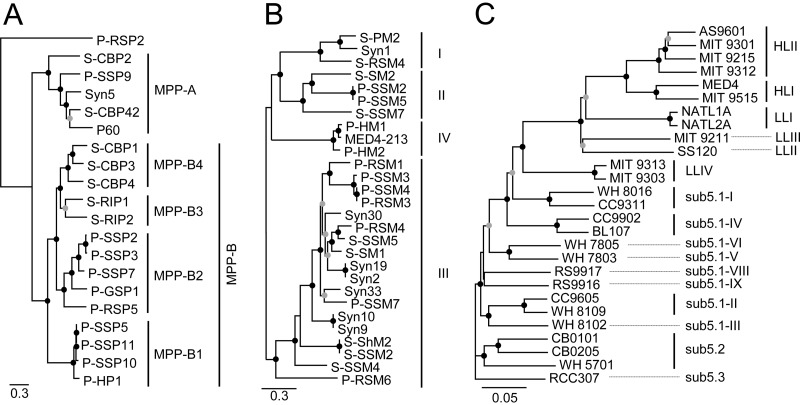
The whole-genome phylogeny divided cyanopodoviruses into two clusters, MPP-A (5 phages) and MPP-B (14 phages), and a distant outgroup, Prochlorococcus podovirus P-RSP2 (Fig. 1A). Two subclusters, MPP-B3 and MPP-B4, which both comprised Synechococcus podoviruses, were added to the MPP-B cluster, which previously consisted of the two Prochlorococcus podovirus subclusters MPP-B1 and MPP-B2 (21). Two Synechococcus podovirus strains were added into the MPP-A cluster, which previously contained one Prochlorococcus phage and two Synechococcus phages (21). Thus, among the 20 cyanopodovirus isolates analyzed here, most (four of five) MPP-A phages infect Synechococcus bacteria, while MPP-B phages infect either Synechococcus or Prochlorococcus (Fig. 1A). A recent study also reported a similar trend of most MPP-A phage isolates infecting Synechococcus (13). The average G+C content of cyanopodoviruses infecting Synechococcus (ca. 49.7%) is much higher than that of those infecting Prochlorococcus (ca. 38.6%) (see Table S1 in the supplemental material), with the difference being significant (t test, P < 0.01).
FIG 1.
Phylogenetic relationships of marine cyanopodoviruses (A), marine cyanomyoviruses (B), and Prochlorococcus and marine Synechococcus strains (C). The concatenated protein sequences of the 15 core genes of 20 cyanopodoviruses and the 55 core genes of 28 cyanomyoviruses were aligned and used to construct the maximum-likelihood trees. The concatenated nucleotide sequences of six single-copy housekeeping genes of 27 picocyanobacterial strains were aligned and used to construct a distance-based phylogenetic tree. Black and gray circles at branch nodes represent bootstrap support of 100% and 75 to 99%, respectively.
Four cyanomyovirus clusters (I to IV) were grouped on the whole-genome tree (Fig. 1B). These clusters are consistent with those that resulted from phylogenetic analysis using the portal gene g20 (36) (cluster IV was previously denoted as “PSSM9/11/12 new cluster” [37]). Although clusters I and IV were comprised solely of phages isolated on Synechococcus and Prochlorococcus, respectively, in this study, they indeed contain phages isolated on both Synechococcus and Prochlorococcus (37). In cluster II or III, cyanomyoviruses that originally infect Prochlorococcus and Synechococcus were nearly numerically equal (Fig. 1B). The average G+C content of myoviruses isolated on Synechococcus (40.3%) is slightly higher than that of myoviruses isolated on Prochlorococcus (37.5%) (see Table S1 in the supplemental material), with the difference being significant (t test, P < 0.01).
The phylogenetic tree built based on concatenated nucleotide sequences of six single-copy housekeeping genes from 27 picocyanobacterial strains resolved these strains into six Prochlorococcus clades (HLI, HLII, LLI, LLII, LLIII, and LLIV, where HL and LL indicate high-light-adapted and low-light-adapted clades, respectively) and eight clades in marine Synechococcus subcluster 5.1 (I, II, III, IV, V, VI, VIII, and IX) and marine Synechococcus subclusters 5.2 and 5.3 (Fig. 1C). The affiliations of these strains agree with those in a previous phylogenetic analysis based on the 16S rRNA gene sequence (72).
Assessing metagenomic fragment recruitment.
We searched and retrieved cyanopodovirus-like and cyanomyovirus-like reads from the microbial (GOS and HOT) and the viral (MarineVirome and BroadPhage) metagenome databases, which cover a broad habitat range and geographic scale (see Table S2 in the supplemental material), filtered away noises, binned the reads into cyanopodovirus and cyanomyovirus genotypes, and calculated the frequency of each genotype. Similar approaches have been used to estimate the abundances of newly isolated phage types in the ocean (19, 73–75). We also retrieved Synechococcus-like and Prochlorococcus-like reads from the GOS microbial metagenome database and estimated the relative abundances of picocyanobacterial genotypes. The numbers of nonredundant metagenomic reads that were retrieved and the numbers of the fractions of reads that passed our quality filtration are shown in Table 1.
TABLE 1.
Metagenomic fragment recruitment summary statistics
| Database, analytical stage | Cyanopodovirus | Cyanomyovirus | Cyanobacteria |
|---|---|---|---|
| GOS | |||
| No. of reads recruiteda | 19,119 | 251,620 | 47,395 |
| No. of reads after filtrationb | 11,874 | 174,683 | 8,111 |
| HOT | |||
| No. of reads recruited | 1,047 | 7,510 | NAc |
| No. of reads after filtration | 479 | 2,518 | NA |
| MarineVirome | |||
| No. of reads recruited | 4,484 | 5,818 | NA |
| No. of reads after filtration | 4,230 | 4,116 | NA |
| BroadPhage | |||
| No. of reads recruited | 18,682 | 86,033 | NA |
| No. of reads after filtration | 11,577 | 70,814 | NA |
Numbers of nonredundant raw reads that were recruited by cyanopodoviruses, cyanomyoviruses, or cyanobacteria.
Numbers of reads that passed the quality filtration. The filtration process is described in detail in Materials and Methods.
NA, not analyzed.
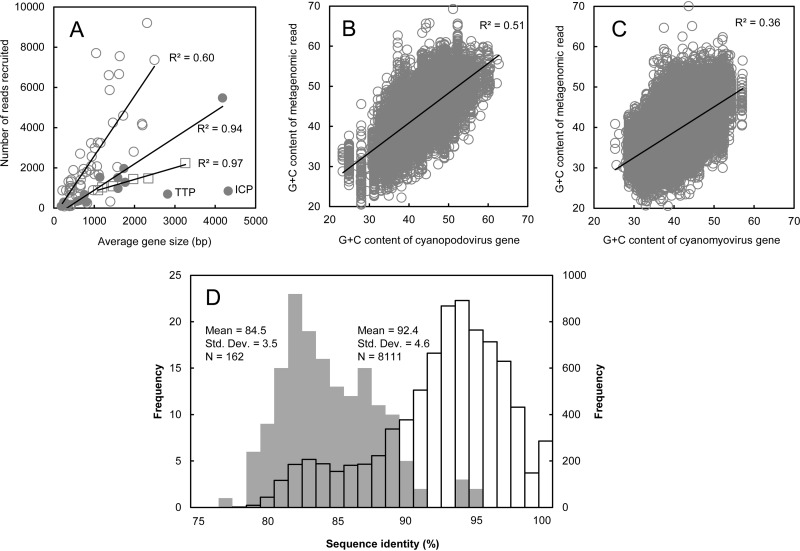
The abundance of reads that were recruited by and then binned to a gene depended on three factors: rate of presence in genomes, gene size, and level of sequence conservation (or divergence). Many genes are shared between phages and bacteria or between different bacterial groups. Miscategorizing the origins of reads could lead to a significant bias in metagenomic fragment recruitment. To minimize the bias caused by nonspecific recruitments, we filtered the recruited reads by comparing them to the NCBI RefSeq database and removed those hitting unrelated organisms. This strategy has been widely used in viral metagenomics (20, 21, 73, 74). Our assessment suggests that the metagenomic analysis procedures approximately prevent nonspecific recruitments, based on the following factors. (i) When the number of reads recruited to a gene was plotted against its gene size, a linear correlation was observed for most core genes of cyanopodoviruses (R2 = 0.94) and cyanomyoviruses (R2 = 0.6) and the six single-copy housekeeping genes of cyanobacteria (R2 = 0.97) (Fig. 2A). (ii) The reads that map to cyanopodoviruses (Fig. 2B) and cyanomyoviruses (Fig. 2C) have G+C contents similar to those of the genes to which these reads were hitting, also indicated by the linear regression (R2 = 0.51 for cyanopodoviruses and R2 = 0.36 for cyanomyoviruses). (iii) The identities between picocyanobacterium-like metagenomic reads and the reference sequences most similar to them (mean = 92.4% ± 4.6%, n = 8,111) were higher than the maximum identities between reference sequences from different picocyanobacterial clades (mean = 84.5% ± 3.5% [mean ± standard deviation], n = 162) (Fig. 2D; see also Table S3 in the supplemental material), suggesting that metagenomic read recruitment using these six reference genes can distinguish different picocyanobacterial clades.
FIG 2.
Assessing metagenomic read recruitment. (A) The numbers of reads yielded for cyanopodovirus core genes (filled circles), cyanomyovirus core genes (empty circles), and six single-copy cyanobacterial housekeeping genes (empty squares) were plotted against the sizes of these genes. Linear regression is shown for each set of genes except for two genes coding for the tail tube B (TTB) protein and internal core protein (ICP) of cyanopodoviruses. (B and C) The G+C content of a cyanopodovirus gene (B) or a cyanomyovirus gene (C) was plotted against those of metagenomic reads recruited to this gene. (D) The gray chart (left y axis) shows the frequencies of maximum identities between sequences from picocyanobacterial strains in different clades. A sequence of one of the six housekeeping genes from a picocyanobacterial strain was compared to the sequences of this gene from strains in the other clades, and the maximum identity was recorded (see Table S3 in the supplemental material). The mean of the maximum sequence identities between different picocyanobacterial clades is 84.5% ± 3.5% (n = 162). The open chart (right y axis) shows the frequencies of identities between recruited GOS metagenomic reads and the reference sequences of the picocyanobacterial strains to which the reads were binned. The mean of the sequence identities between recruited GOS reads and the reference sequences most similar to them is 92.4% ± 4.6% (n = 8,111).
Relative abundances of cyanopodoviruses and cyanomyoviruses.
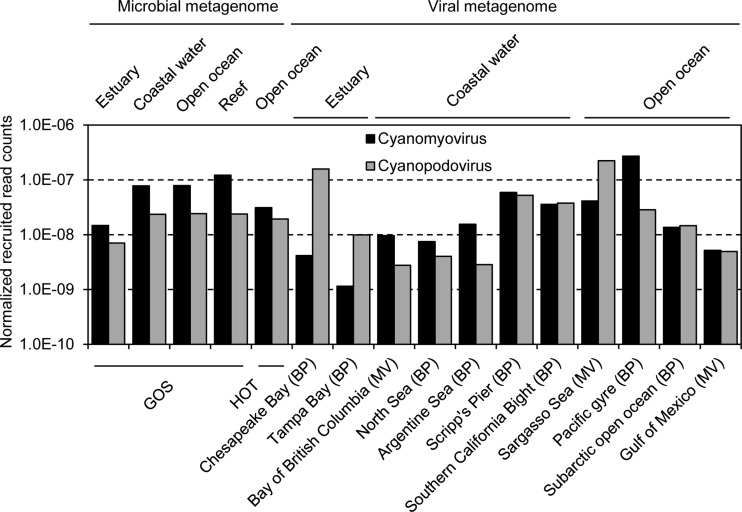
The comparisons between the abundances of cyanopodovirus-like and cyanomyovirus-like reads resulted in greater variation in the viral metagenomes than in the microbial metagenomes (Fig. 3). In the microbial metagenomes, the rates of detection of cyanomyoviruses (normalized against database size and phage genome size) consistently exceeded those of cyanopodoviruses in all of the types of environments (Fig. 3), consistent with the results from previous recruitments against the GOS database (25, 27) or other microbial metagenome data sets (21). However, the overabundance of cyanomyovirus-like reads compared to the amounts of cyanopodovirus-like reads was not obvious in the viral metagenomes (Fig. 3). In fact, the frequencies of cyanopodovirus-like reads could be lower than (four data sets), similar to (four data sets), or higher than (three data sets) those of cyanomyovirus-like reads in viral metagenomes (Fig. 3). In addition, cyanopodovirus, cyanomyovirus, and picocyanobacterial reads were detected at the rates of 2.5 ×10−8, 7.5 ×10−8, and 6 ×10−8, respectively, per base pair in the microbial metagenomes (Fig. 4). It can be roughly estimated that once one picocyanobacterial cell was sampled, nearly 0.4 and 1.2 copies of cyanopodovirus and cyanomyovirus genomes were cotrapped, respectively. Together, these results suggest that cyanopodoviruses and cyanomyoviruses are both abundant in the sea.
FIG 3.
Rates of occurrence of cyanopodovirus-like and cyanomyovirus-like reads in metagenomes, showing the relative abundances of cyanopodoviruses and cyanomyoviruses. Rates for the reads recruited from each type of habitat (GOS project) or each location (HOT, MarineVirome [MV], and BroadPhage [BP] projects) are shown. The absolute numbers of retrieved reads were normalized against the data sizes of metagenomes and the average genome size of cyanopodoviruses or cyanomyoviruses.
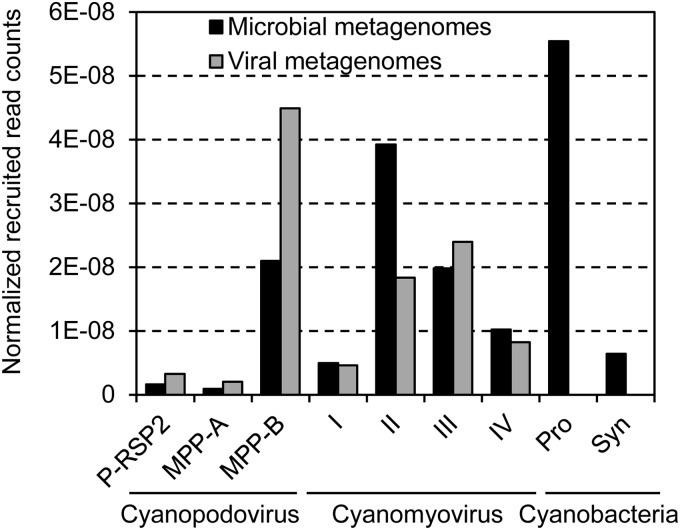
FIG 4.
Relative abundances of cyanopodovirus and cyanomyovirus genotypes and cyanobacteria of the genera Prochlorococcus and Synechococcus. The read number was normalized against the data sizes of metagenomes and the average genome size of each specific cyanopodovirus or cyanomyovirus genotype or the average size of the concatenated six reference genes of Prochlorococcus or Synechococcus.
Relative abundances of cyanopodovirus and cyanomyovirus genotypes.
Cyanopodoviruses in cluster MPP-B yielded far more reads than those in cluster MPP-A (Fig. 4), consistent with previous findings that MPP-B phages were detected at much higher frequencies than MPP-A phages either in clone libraries (38, 39) or among phage isolates (13). Cyanomyoviruses in clusters II and III recruited significantly more reads than those in clusters I and IV (Fig. 4). The high abundance of cluster II cyanomyovirus-like sequences in the GOS database has been reported based on g20 gene recruitment (37). Here, our results showed that cluster III cyanomyoviruses can be numerically abundant, too. It is noticeable that the numbers of genomes used in recruitment varied significantly among cyanophage clusters. It was shown that the abundance of cyanopodoviruses can be greatly underestimated if a low number of genomes are used for recruitment (21). Thus, fewer reference genomes can also potentially cause underestimation of some specific clusters, such as cyanopodovirus cluster MPP-A, which only comprised five genomes here (Fig. 1). Nevertheless, cyanomyovirus cluster II was still among the most abundant cyanophage lineages (Fig. 4), though it only had four genomes analyzed (Fig. 1). In addition, the recruitment abundances of each of the clusters were consistent between microbial and viral metagenomes (Fig. 4).
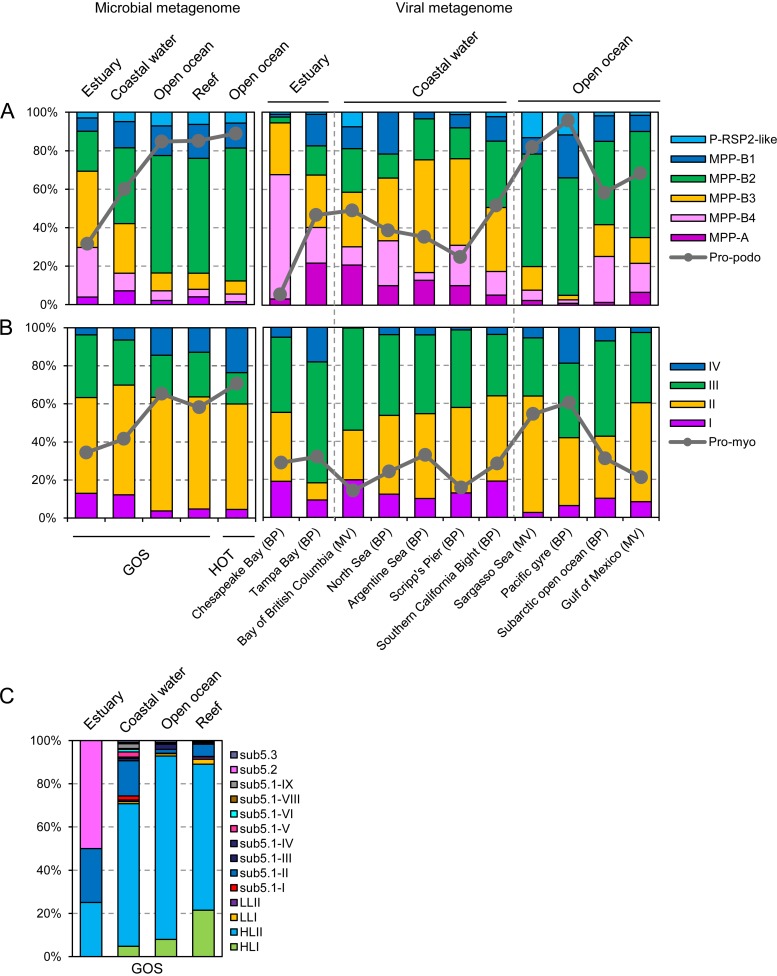
We further examined the relative abundances of these cyanophage clusters or subclusters in various types of marine habitats. In coastal and estuarine waters, the cyanopodovirus communities were quite diverse and their compositions varied greatly among different sampling locations (Fig. 5A). In general, subclusters MPP-B2, -B3, and -B4 made up the majority of the communities. In the open oceans, the communities were likely less diverse, and subcluster MPP-B2 was strikingly dominant, accounting for up to 80% of the total recruited reads (Fig. 5A). In addition, these cyanopodovirus lineages appeared to occur preferentially in specific habitats. For instance, the MPP-A cluster and the MPP-B3 and -B4 subclusters contributed far more recruited reads in the estuarine and coastal waters than in the open oceans, while the MPP-B2 subcluster exhibited a contrasting trend (Fig. 5A). For cyanomyoviruses, the community structure on the cluster level is quite consistent among different habitats and was generally dominated by clusters II and III (Fig. 5B). These two major lineages appeared to exhibit not a habitat-related distribution pattern but a trend in which cluster III contributed less to the recruited reads in microbial metagenomes (20 to 35%) than in viral metagenomes (35 to 60%). In contrast, the two minor lineages, clusters I and IV, likely had habitat-related relative abundances, with the former being more abundant in estuarine and coastal waters and the latter being more abundant in open oceans (Fig. 5B). Such a pattern may be related to the host range of the phages analyzed in each of the clusters, as clusters I and IV only contained phages isolated on either Synechococcus or Prochlorococcus while clusters II and III contained phages isolated on both host genera.
FIG 5.
Relative abundances of cyanopodovirus (A), cyanomyovirus (B), and picocyanobacterial (C) genotypes in different types of habitats. It is noteworthy that the GOS reef samples were mainly collected from reefs located in open oceans. Abbreviations: Pro-podo and Pro-myo (gray line charts in panels A and B), the fractions of reads that were recruited to the genomes of cyanopodoviruses or cyanomyoviruses that were originally isolated on Prochlorococcus; HL, high-light-adapted Prochlorococcus clade; LL, low-light-adapted Prochlorococcus clade; sub5.1, sub5.2, and sub5.3, marine Synechococcus subclusters 5.1, 5.2, and 5.3; BP, BroadPhage; MV, MarineVirome.
Relation between the distribution patterns of cyanophages and cyanobacteria.
We compared the numbers of reads recruited by genomes of cyanophages isolated on Prochlorococcus and Synechococcus (Fig. 5A and B, gray lines). A clear pattern showed that reads recruited by the Prochlorococcus-infecting cyanophages contributed significantly higher proportions in the open-ocean metagenomes (the proportions ranged from 60 to 95% for cyanopodovirus and from 20 to 70% for cyanomyovirus) than in the estuarine and coastal metagenomes (from 5 to 60% for cyanopodovirus and from 15 to 40% for cyanomyovirus), while the recruitments by Synechococcus-infecting cyanophages resulted in an opposite trend (t tests: P < 0.01 for cyanopodovirus, n = 16; P < 0.05 for cyanomyovirus, n = 16; P < 0.01 for cyanopodovirus and cyanomyovirus, n = 32).
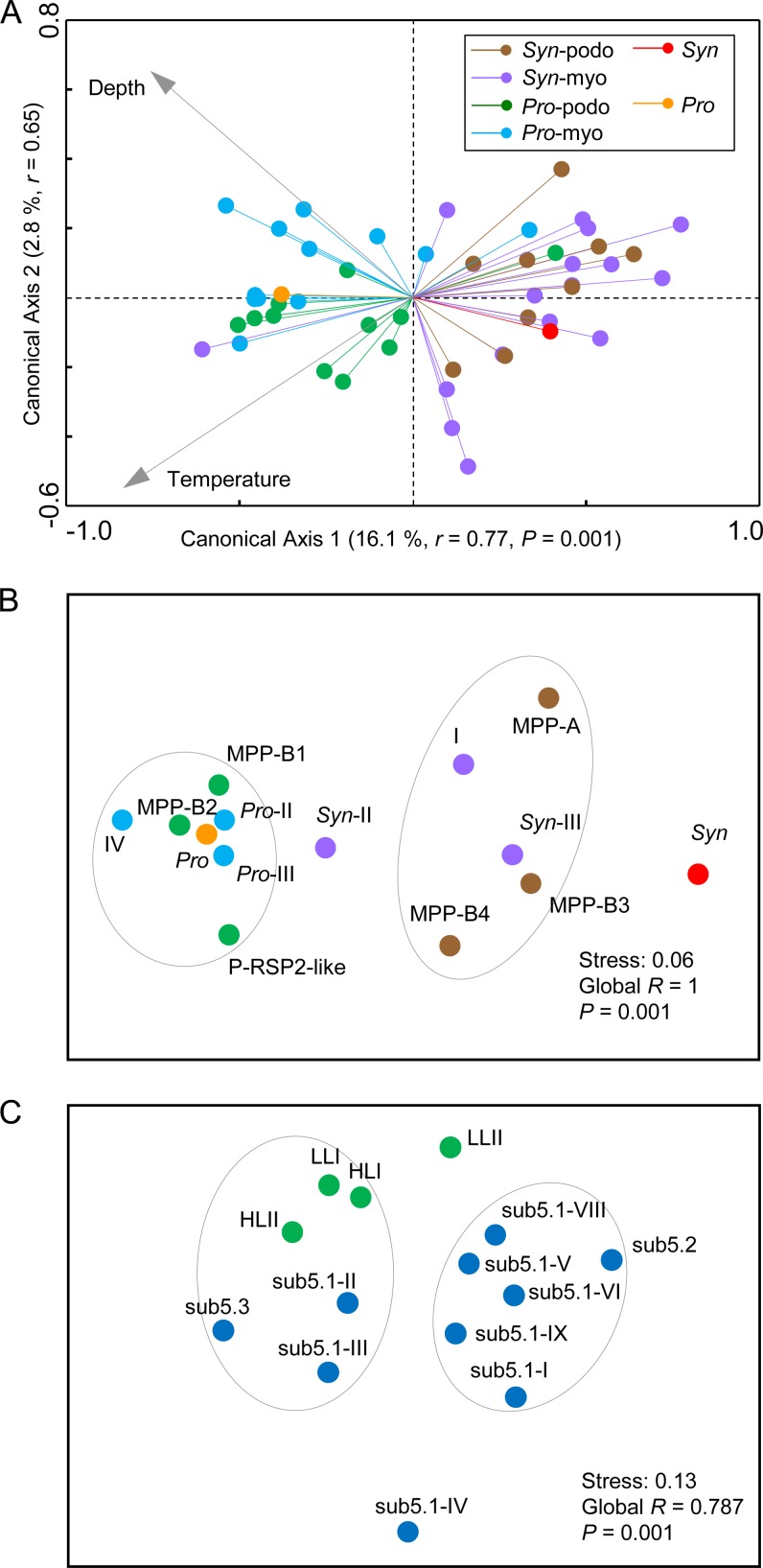
We further performed RDA based on the relative abundances of reads that were recruited to each of the cyanophage isolates and to Prochlorococcus and Synechococcus at 65 GOS sampling sites (see Fig. S1 in the supplemental material). Note that here we were describing the pattern observed among the metagenomic reads that were recruited by genomes of cyanophages that were originally isolated on different hosts but not assigning hosts to metagenomic reads. The RDA diagram showed that the cyanophages originally isolated on Synechococcus and those originally isolated on Prochlorococcus separated into two groups, approximately along the first canonical axis, which explained 16.1% of the total variability in community composition and revealed a significant species-environment correlation (r = 0.77, P < 0.01) (Fig. 6A). Interestingly, the distributions of reads recruited to the cyanophages that were originally isolated on Prochlorococcus and Synechococcus were significantly correlated with those recruited to Prochlorococcus and Synechococcus, respectively (Fig. 6A). The RDA also showed that Prochlorococcus and the cyanophages that were isolated on Prochlorococcus were both positively correlated with temperature and water depth, two environmental parameters that had significance in explanation of the species-environment relationship (P < 0.05, partial Monte Carlo permutation test), while Synechococcus and cyanophages that were isolated on Synechococcus were both negatively correlated with them (Fig. 6A). We also performed NMDS based on the relative abundances of reads that were recruited to each of the cyanophage genotypes and to Prochlorococcus and Synechococcus at those GOS sites (Fig. 6B). Note that the cyanomyovirus clusters II and III were each artificially divided into two subclusters (Pro-II, Syn-II, Pro-III, and Syn-III). For example, in the analysis, Pro-II represented the relative abundances of reads that were recruited to the genomes of cluster II cyanomyoviruses that were originally isolated on Prochlorococcus. The NMDS diagram showed that cyanopodovirus lineages MPP-B1, MPP-B2, and P-RSP2-like and cyanomyovirus cluster IV and subclusters Pro-II and Pro-III were clustered together with Prochlorococcus (Fig. 6B). The analyzed phages in these genotypes or defined groups were all isolated on Prochlorococcus. Cyanopodovirus lineages MPP-A, MPP-B3, and MPP-B4 and cyanomyovirus cluster I and subcluster Syn-III also converged in the diagram. The original hosts of most of the analyzed phage genomes in these genotypes or defined groups were Synechococcus strains. Such groupings had statistical significance (global R = 1, P < 0.01) (Fig. 6B).
FIG 6.
(A) RDA based on metagenomic fragment recruitments against the GOS database. Prochlorococcus, Synechococcus, and each of the 20 cyanopodovirus and 28 cyanomyovirus isolates are indicated by colored lines ending with filled circles. The environmental parameters (temperature and depth, indicated by gray arrows) shown on this ordination diagram were those that passed the partial Monte Carlo permutation test (P < 0.05). The angle (cosine) between lines expresses the correlation, and the length of a line expresses how well the values are approximated by the ordination diagram. Canonical axes 1 and 2 explained 16.1% and 2.8% of the total variability in community composition, respectively, and both showed significant species-environment correlation (r = 0.77 and r = 0.65; Monte Carlo permutation test P values for the first axis and for all the axes were both <0.01). (B and C) NMDS analyses for cyanophages (B) and cyanobacteria (C) based on metagenomic fragment recruitments against the GOS database. Pro-II, Pro-III, Syn-II, and Syn-III represent the fractions of reads that were recruited to the genomes of cluster II or III cyanomyoviruses that were originally isolated on Prochlorococcus or Synechococcus. The significance of grouping (indicated by gray circles) was tested by ANOSIM.
We also estimated the relative abundances of Prochlorococcus and Synechococcus genotypes (see Table S3 in the supplemental material) based on read recruitment against the GOS metagenomes. HLII Prochlorococcus dominated the picocyanobacterial communities in coastal waters, open oceans, and reefs (Fig. 5C). Despite this, the genetic community composition still varied among those GOS sites (see Fig. S1A). Moreover, the NMDS analysis resulted in a pattern where Prochlorococcus genotypes HLI, HLII, and LLI likely co-occurred with Synechococcus genotypes sub5.1-II (Synechococcus clade II in marine subcluster 5.1), sub5.1-III, and sub5.3, while most of the other Synechococcus genotypes formed a discrete guild (Fig. 6C). This pattern agrees approximately with the known biogeography scenario of picocyanobacterial lineages, such as the fact that Synechococcus lineages sub5.1-II, sub5.1-III, and sub5.3 and Prochlorococcus lineages were mostly confined to tropical and subtropical oceans (44, 46, 52, 54). Mantel tests, which estimate the correlation between matrices, indicated tighter correlations between cyanophage and cyanobacterial community structures (most r > 0.5, P < 0.01) than between cyanophage community structure and environmental factors (most r > 0.4, P < 0.01) (Table 2). The cyanobacterial community structure correlated to environmental factors at a level (r = 0.475, P < 0.01) similar to the correlation between cyanophage and environmental factors. However, the community composition of cyanomyoviruses at the cluster level correlated only moderately to that of cyanobacteria or to environmental factors, differing from the results for cyanopodoviruses (Table 2). This may be in part due to the more homogeneous cyanomyovirus communities among the GOS samples, which were consistently dominated by clusters II and III (see Fig. S1C).
TABLE 2.
Mantel test summary statistics
| Test description | R (cyanobacteriac,e) | R (environmentd,e) |
|---|---|---|
| Matrix based on cyanophage isolatea | ||
| Podovirus + myovirus | 0.530* | 0.475* |
| Podovirus | 0.522* | 0.481* |
| Myovirus | 0.516* | 0.432* |
| Matrix based on cyanophage genotypeb | ||
| Podovirus + myovirus | 0.523* | 0.494* |
| Podovirus | 0.539* | 0.488* |
| Myovirus | 0.263* | 0.300* |
| Matrix based on cyanobacterial genotypec | ||
| Cyanobacteria | - | 0.475* |
The Bray-Curtis similarity matrix was calculated based on the relative abundances of the reads that were recruited to each of the cyanophage isolates.
The Bray-Curtis similarity matrix was calculated based on the relative abundances of the reads that were recruited to each of the cyanophage genotypes (MPP-A, MPP-B1, -B2, -B3, and -B4, and P-RSP2-like for cyanopodoviruses and I, II, III, and IV for cyanomyoviruses).
The Bray-Curtis similarity matrix was calculated based on the relative abundances of the reads that were recruited to each of the cyanobacterial genotypes (HLI, HLII, LLI, and LLII for Prochlorococcus and sub5.1-I, -II, -III, -IV, -V, -VI, -VIII, and -IX and sub5.2 and sub5.3 for marine Synechococcus).
The Euclidean distance matrix was calculated based on the z-score transformed environmental parameters of temperature, depth, latitude, and chlorophyll a.
*, P < 0.01.
DISCUSSION
We estimated the relative abundances of cyanopodovirus and cyanomyovirus lineages using metagenomic read recruitment against the existing microbial and viral metagenomes in the public domain. The cyanophage lineages were defined based on the phylogenetic relationship of sequenced cyanophage genomes (Fig. 1), which are many fewer than the previously defined cyanomyovirus genotypes (36, 40) and cyanopodovirus genotypes (13, 38, 39), respectively. Despite this, the lineage classifications used here are broadly accepted, such as clusters I to IV for cyanomyoviruses (36, 37) and clusters MPP-A and MPP-B for cyanopodoviruses (12, 13, 21, 38, 39).
Our results suggest equally high abundances of cyanopodoviruses and cyanomyoviruses in the sea overall (Fig. 3), although a discrepancy was observed when comparing the results from recruitments against microbial and viral metagenomes. A few previous studies also observed a common trend of cyanomyovirus-like reads often outnumbering cyanopodovirus-like reads in microbial metagenomes (21, 25, 27). It was suggested that the overabundance of cyanomyoviruses may be related to their large particle size, which would cause them to be more likely to be retained by filters when sampling the microbial fraction in seawater (21). Alternatively, if the majority of the cyanophage-like sequences identified in the microbial fraction of data originated from cyanophage DNA within infected host cells (26, 27), the differences in burst size and latent period between cyanopodoviruses and cyanomyoviruses may also have an impact on this issue. Irrespective of this discrepancy, our data revealed similar ranges of abundance of cyanophage-like reads and cyanobacterium-like reads in the GOS microbial metagenomes (Fig. 4), suggestive of both numerical abundance and physiological activity of marine cyanophages. This observation is in agreement with the extensive literature in which cyanomyoviruses and cyanopodoviruses were both commonly found in marine environments during the last 2 decades (4, 5, 8–11, 13, 33, 76–78).
Cyanopodovirus cluster MPP-B and cyanomyovirus clusters II and III outcompete other lineages in relative abundance remarkably (Fig. 4). Cyanopodovirus cluster MPP-A and subclusters MPP-B1, -B2, -B3, and -B4 and cyanomyovirus clusters I and IV exhibit habitat-related variability in relative abundance, whereas cyanomyovirus clusters II and III appear to be ubiquitous and consistently dominant in various marine environments (Fig. 5A and B). This pattern indicates strong heterogeneity existing in the biogeographic distribution of different cyanopodovirus and cyanomyovirus genotypes, suggestive of the presence of rare and abundant viral taxa. This pattern contrasts with the observation that different viral morphotypes (i.e., myovirus, podovirus, siphovirus, and nontailed virus) in the global ocean exhibit little variance in relative abundances (79). However, on the other hand, a comparative viral metagenomic analysis showed that whole viral communities may have highly structured genetic compositions in the ocean, likely driven by factors such as depth and proximity to shore (80). Temporal and spatial variations of marine cyanophage communities have been delineated for both cyanomyoviruses (6, 10, 40, 77, 81) and cyanopodoviruses (38, 39). These studies also suggested that the variations of cyanophage communities follow certain temporal and spatial patterns. Our results further support the idea that closely related viral genotypes within a fairly narrow group may not be randomly distributed in the world's oceans but, instead, may follow specific patterns (37, 77). Biotic or/and abiotic factors may impose pressures on viruses that infect hosts within a specific taxonomic unit, shaping the composition of viral genotypes.
Our analysis suggests a relation of co-occurrence of lineages of cyanophages and picocyanobacteria in the sea. First, the recruited metagenomic reads that map to reference cyanophages isolated on Prochlorococcus or Synechococcus tend to have greater representation in the habitats where Prochlorococcus or Synechococcus presumably thrives (Fig. 5A and B). The RDA ordination further suggests that the reads recruited to Prochlorococcus phages commonly prevailed in warmer and deeper oceans, while the reads recruited to Synechococcus phages were more prevalent in shallower or cooler areas, mirroring the overall biogeographic features of Prochlorococcus and Synechococcus in the ocean (Fig. 6A) (1, 41, 42, 82). Second, we observed strong correlations between the genotype compositions of cyanophages and picocyanobacteria for both cyanopodoviruses and cyanomyoviruses (Table 2). Cyanopodoviruses are highly host strain-specific, while cyanomyoviruses can coinfect hosts in different genotypes and can even cross-infect Prochlorococcus and Synechococcus (11). We therefore thought that cyanopodoviruses and picocyanobacteria would have a stronger correlation than cyanomyoviruses and picocyanobacteria when coanalyzing the community compositions of cyanophages and picocyanobacteria. However, the two correlations appear to be equally strong, with similar correlation coefficients (Table 2). This suggests a cohesive biogeographic pattern between some lineages of cyanophages and picocyanobacteria and further suggests phylogenetic specificity of cyanophages. Prochlorococcus and Synechococcus both encompass numerous genotypes that are associated with physiological and ecological traits, forming corresponding ecotypes (47–51). These genotypes are marked by their biogeographic patterns that are bounded to specific horizontal and vertical profiles (44, 52–56). Our results demonstrate that this highly structured community composition of picocyanobacteria on the global scale correlates with that of cyanophages, suggesting a role of the host in structuring viral communities.
It has been known that there is a tight coupling between picocyanobacterial abundance and cyanophage titer, either temporally or spatially (4, 5, 7, 11, 33). A monthly temporal survey at the same sampling site in the Gulf of Aqaba showed that the diversity and abundance of cyanomyoviruses were correlated to those of co-occurring Synechococcus populations (6). However, such correlations were barely observed between cyanomyoviruses and picocyanobacteria in a spatial survey across the North and South Atlantic basins (47°N to 28°S) (40). The authors attributed this to the temporally and spatially discrete sampling and/or to the viral molecular marker g20 they used that lacks easily detectable functional and evolutionary phage-host connections (40). On the other hand, our analysis, which involved the whole-genome information of cyanopodoviruses and cyanomyoviruses and covered a broad range of marine habitats and geographic regions, showed a spatial pattern of co-occurrence of cyanophages and picocyanobacteria. A recent network study that investigated the temporal covariation of T4-like myoviruses and bacteria found a dominance of positive over negative correlations between bacterial and viral operational taxonomic units (OTUs) and suggested that, on the monthly and seasonal time scale, it is more likely that viral taxa are controlled by host availability than that they control host abundance or, in other words, that viruses are following the hosts (83). Here, we also demonstrated a strong correlation between the biogeographic patterns of cyanophages and cyanobacteria in the ocean for both cyanopodoviruses and cyanomyoviruses, irrespective of their different host specificities. This correlation may reflect either the ecological roles of cyanophages in influencing the community of cyanobacteria or, more apparent here, the roles of cyanobacteria in constraining the community of cyanophages.
The cyanophage-cyanobacteria system has been emerging as one of the model virus-host systems in marine microbiology and microbial ecology studies (15, 18, 84), probably the most matured one, and future investigation of this system will continuously contribute to our understanding of the ecological roles of both virus and host for each other.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NSFC grants 41206131 (S.H.) and 41230962 (S.Z.) and the Xiamen University 111 Program (F.C.). We also thank the Hanse-Wissenschaftskolleg for a fellowship (F.C.) supporting the collaborative study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02483-14.
REFERENCES
- 1.Partensky F, Hess WR, Vaulot D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63:106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proctor LM, Fuhrman JA. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60–62. doi: 10.1038/343060a0. [DOI] [Google Scholar]
- 3.Suttle CA, Chan AM, Cottrell MT. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467–469. doi: 10.1038/347467a0. [DOI] [Google Scholar]
- 4.Suttle CA, Chan AM. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol 60:3167–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterbury JB, Valois FW. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol 59:3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muhling M, Fuller NJ, Millard A, Somerfield PJ, Marie D, Wilson WH, Scanlan DJ, Post AF, Joint I, Mann NH. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ Microbiol 7:499–508. doi: 10.1111/j.1462-2920.2005.00713.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, Wommack KE, Chen F. 2011. Abundance and distribution of Synechococcus and cyanophages in the Chesapeake Bay. Appl Environ Microbiol 77:7459–7468. doi: 10.1128/AEM.00267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suttle CA, Chan AM. 1993. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar Ecol Prog Ser 92:99–109. doi: 10.3354/meps092099. [DOI] [Google Scholar]
- 9.Lu J, Chen F, Hodson RE. 2001. Distribution, isolation, host specificity, and diversity of cyanophages infecting marine Synechococcus spp. in river estuaries. Appl Environ Microbiol 67:3285–3290. doi: 10.1128/AEM.67.7.3285-3290.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marston MF, Sallee JL. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl Environ Microbiol 69:4639–4647. doi: 10.1128/AEM.69.8.4639-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan MB, Waterbury JB, Chisholm SW. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 12.Wang K, Chen F. 2008. Prevalence of highly host-specific cyanophages in the estuarine environment. Environ Microbiol 10:300–312. doi: 10.1111/j.1462-2920.2007.01452.x. [DOI] [PubMed] [Google Scholar]
- 13.Dekel-Bird NP, Avrani S, Sabehi G, Pekarsky I, Marston MF, Kirzner S, Lindell D. 2013. Diversity and evolutionary relationships of T7-like podoviruses infecting marine cyanobacteria. Environ Microbiol 15:1476–1491. doi: 10.1111/1462-2920.12103. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann NH, Clokie MR, Millard A, Cook A, Wilson WH, Wheatley PJ, Letarov A, Krisch HM. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J Bacteriol 187:3188–3200. doi: 10.1128/JB.187.9.3188-3200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, King J, Hatfull GF, Lawrence JG, Hendrix RW. 2007. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol 9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- 17.Millard AD, Zwirglmaier K, Downey MJ, Mann NH, Scanlan DJ. 2009. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ Microbiol 11:2370–2387. doi: 10.1111/j.1462-2920.2009.01966.x. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, DeFrancesco AS, Kern SE, Thompson LR, Young S, Yandava C, Fu R, Krastins B, Chase M, Sarracino D, Osburne MS, Henn MR, Chisholm SW. 2010. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol 12:3035–3056. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabehi G, Shaulov L, Silver DH, Yanai I, Harel A, Lindell D. 2012. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc Natl Acad Sci U S A 109:2037–2042. doi: 10.1073/pnas.1115467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly L, Ding H, Huang KH, Osburne MS, Chisholm SW. 2013. Genetic diversity in cultured and wild marine cyanomyoviruses reveals phosphorus stress as a strong selective agent. ISME J 7:1827–1841. doi: 10.1038/ismej.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labrie S, Frois-Moniz K, Osburne M, Kelly L, Roggensack S, Sullivan M, Gearin G, Zeng Q, Fitzgerald M, Henn M. 2013. Genomes of marine cyanopodoviruses reveal multiple origins of diversity. Environ Microbiol 15:1356–1376. doi: 10.1111/1462-2920.12053. [DOI] [PubMed] [Google Scholar]
- 22.Pope WH, Weigele PR, Chang J, Pedulla ML, Ford ME, Houtz JM, Jiang W, Chiu W, Hatfull GF, Hendrix RW, King J. 2007. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. J Mol Biol 368:966–981. doi: 10.1016/j.jmb.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F, Lu J. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl Environ Microbiol 68:2589–2594. doi: 10.1128/AEM.68.5.2589-2594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D, Chisholm SW. 2009. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome’. Environ Microbiol 11:2935–2951. doi: 10.1111/j.1462-2920.2009.02081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S, Wang K, Jiao N, Chen F. 2012. Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage-host genetic exchanges. Environ Microbiol 14:540–558. doi: 10.1111/j.1462-2920.2011.02667.x. [DOI] [PubMed] [Google Scholar]
- 26.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 27.Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, Frazier M, Venter JC. 2008. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghai R, Martin-Cuadrado AB, Molto AG, Heredia IG, Cabrera R, Martin J, Verdu M, Deschamps P, Moreira D, Lopez-Garcia P, Mira A, Rodriguez-Valera F. 2010. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J 4:1154–1166. doi: 10.1038/ismej.2010.44. [DOI] [PubMed] [Google Scholar]
- 29.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H. 2006. The marine viromes of four oceanic regions. PLoS Biol 4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, Wommack KE. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl Environ Microbiol 73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuno CM, Rodriguez-Valera F, Garcia-Heredia I, Martin-Cuadrado AB, Ghai R. 2013. Reconstruction of novel cyanobacterial siphovirus genomes from Mediterranean metagenomic fosmids. Appl Environ Microbiol 79:688–695. doi: 10.1128/AEM.02742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y, Allen LZ, Palenik B. 27 April 2014. Diversity and genome dynamics of marine cyanophages using metagenomic analyses. Environ Microbiol Rep doi: 10.1111/1758-2229.12160. [DOI] [PubMed] [Google Scholar]
- 33.Millard AD, Mann NH. 2006. A temporal and spatial investigation of cyanophage abundance in the Gulf of Aqaba, Red Sea. J Mar Biol Assoc UK 86:507–515. doi: 10.1017/S0025315406013415. [DOI] [Google Scholar]
- 34.Clokie MR, Millard AD, Mehta JY, Mann NH. 2006. Virus isolation studies suggest short-term variations in abundance in natural cyanophage populations of the Indian Ocean. J Mar Biol Assoc UK 86:499–505. doi: 10.1017/S0025315406013403. [DOI] [Google Scholar]
- 35.Adriaenssens EM, Cowan DA. 2014. Using signature genes as tools to assess environmental viral ecology and diversity. Appl Environ Microbiol 80:4470–4480. doi: 10.1128/AEM.00878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong Y, Chen F, Wilhelm SW, Poorvin L, Hodson RE. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl Environ Microbiol 68:1576–1584. doi: 10.1128/AEM.68.4.1576-1584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan MB, Coleman ML, Quinlivan V, Rosenkrantz JE, Defrancesco AS, Tan G, Fu R, Lee JA, Waterbury JB, Bielawski JP, Chisholm SW. 2008. Portal protein diversity and phage ecology. Environ Microbiol 10:2810–2823. doi: 10.1111/j.1462-2920.2008.01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen F, Wang K, Huang S, Cai H, Zhao M, Jiao N, Wommack KE. 2009. Diverse and dynamic populations of cyanobacterial podoviruses in the Chesapeake Bay unveiled through DNA polymerase gene sequences. Environ Microbiol 11:2884–2892. doi: 10.1111/j.1462-2920.2009.02033.x. [DOI] [PubMed] [Google Scholar]
- 39.Huang S, Wilhelm SW, Jiao N, Chen F. 2010. Ubiquitous cyanobacterial podoviruses in the global oceans unveiled through viral DNA polymerase gene sequences. ISME J 4:1243–1251. doi: 10.1038/ismej.2010.56. [DOI] [PubMed] [Google Scholar]
- 40.Jameson E, Mann NH, Joint I, Sambles C, Muhling M. 2011. The diversity of cyanomyovirus populations along a North-South Atlantic Ocean transect. ISME J 5:1713–1721. doi: 10.1038/ismej.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partensky F, Blanchot J, Vaulot D. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull Inst Oceanogr Monaco NS 19:457–475. [Google Scholar]
- 42.Scanlan DJ, West NJ. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol 40:1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 43.Olson RJ, Chisholm SW, Zettler ER, Armbrust EV. 1990. Pigments, size, and distribution of Synechococcus in the North Atlantic and Pacific Oceans. Limnol Oceanogr 35:45–58. doi: 10.4319/lo.1990.35.1.0045. [DOI] [Google Scholar]
- 44.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O, Scanlan DJ. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol 10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 45.Waleron M, Waleron K, Vincent WF, Wilmotte A. 2007. Allochthonous inputs of riverine picocyanobacteria to coastal waters in the Arctic Ocean. FEMS Microbiol Ecol 59:356–365. doi: 10.1111/j.1574-6941.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- 46.Huang S, Wilhelm S, Harvey H, Taylor K, Jiao N, Chen F. 2012. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J 6:285–297. doi: 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore LR, Rocap G, Chisholm SW. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 48.Moore LR, Chisholm SW. 1999. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol Oceanogr 44:628–638. doi: 10.4319/lo.1999.44.3.0628. [DOI] [Google Scholar]
- 49.West NJ, Scanlan DJ. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl Environ Microbiol 65:2585–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocap G, Distel DL, Waterbury JB, Chisholm SW. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol 69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwirglmaier K, Heywood JL, Chamberlain K, Woodward EM, Zubkov MV, Scanlan DJ. 2007. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ Microbiol 9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- 53.Garczarek L, Dufresne A, Rousvoal S, West NJ, Mazard S, Marie D, Claustre H, Raimbault P, Post AF, Scanlan DJ, Partensky F. 2007. High vertical and low horizontal diversity of Prochlorococcus ecotypes in the Mediterranean Sea in summer. FEMS Microbiol Ecol 60:189–206. doi: 10.1111/j.1574-6941.2007.00297.x. [DOI] [PubMed] [Google Scholar]
- 54.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 55.Bouman HA, Ulloa O, Scanlan DJ, Zwirglmaier K, Li WK, Platt T, Stuart V, Barlow R, Leth O, Clementson L, Lutz V, Fukasawa M, Watanabe S, Sathyendranath S. 2006. Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science 312:918–921. doi: 10.1126/science.1122692. [DOI] [PubMed] [Google Scholar]
- 56.Fuller NJ, Tarran GA, Yallop M, Orcutt KM, Scanlan DJ. 2006. Molecular analysis of picocyanobacterial community structure along an Arabian Sea transect reveals distinct spatial separation of lineages. Limnol Oceanogr 51:2515–2526. doi: 10.4319/lo.2006.51.6.2515. [DOI] [Google Scholar]
- 57.Ignacio-Espinoza J, Sullivan M. 2012. Phylogenomics of T4 cyanophages: lateral gene transfer in the ‘core’ and origins of host genes. Environ Microbiol 14:2113. doi: 10.1111/j.1462-2920.2012.02704.x. [DOI] [PubMed] [Google Scholar]
- 58.Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 59.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 60.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 61.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 62.Swofford DL. 2002. PAUP: Phylogenetic Analysis Using Parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 63.Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. 2007. CAMERA: a community resource for metagenomics. PLoS Biol 5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W. 2007. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol 5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, DeLong EF. 2008. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A 105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez A, Tyson GW, DeLong EF. 2010. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol 12:222–238. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- 68.Biers EJ, Sun S, Howard EC. 2009. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl Environ Microbiol 75:2221–2229. doi: 10.1128/AEM.02118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howard EC, Sun S, Biers EJ, Moran MA. 2008. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol 10:2397–2410. doi: 10.1111/j.1462-2920.2008.01665.x. [DOI] [PubMed] [Google Scholar]
- 70.Yutin N, Suzuki MT, Teeling H, Weber M, Venter JC, Rusch DB, Béjà O. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling Expedition metagenomes. Environ Microbiol 9:1464–1475. doi: 10.1111/j.1462-2920.2007.01265.x. [DOI] [PubMed] [Google Scholar]
- 71.Lepš J, Šmilauer P. 2003. Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 72.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, Ellisman M, Deerinck T, Sullivan MB, Giovannoni SJ. 2013. Abundant SAR11 viruses in the ocean. Nature 494:357–360. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- 74.Kang I, Oh H-M, Kang D, Cho J-C. 2013. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci U S A 110:12343–12348. doi: 10.1073/pnas.1219930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holmfeldt K, Solonenko N, Shah M, Corrier K, Riemann L, VerBerkmoes NC, Sullivan MB. 2013. Twelve previously unknown phage genera are ubiquitous in global oceans. Proc Natl Acad Sci U S A 110:12798–12803. doi: 10.1073/pnas.1305956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson WH, Joint IR, Carr NG, Mann NH. 1993. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl Environ Microbiol 59:3736–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marston MF, Taylor S, Sme N, Parsons RJ, Noyes TJ, Martiny JB. 2013. Marine cyanophages exhibit local and regional biogeography. Environ Microbiol 15:1452–1463. doi: 10.1111/1462-2920.12062. [DOI] [PubMed] [Google Scholar]
- 78.Fuller NJ, Wilson WH, Joint IR, Mann NH. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl Environ Microbiol 64:2051–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brum JR, Schenck RO, Sullivan MB. 2013. Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses. ISME J 7:1738–1751. doi: 10.1038/ismej.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hurwitz BL, Westveld AH, Brum JR, Sullivan MB. 2014. Modeling ecological drivers in marine viral communities using comparative metagenomics and network analyses. Proc Natl Acad Sci U S A 111:10714–10719. doi: 10.1073/pnas.1319778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sandaa RA, Larsen A. 2006. Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp. Appl Environ Microbiol 72:4610–4618. doi: 10.1128/AEM.00168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WK, Lomas MW, Veneziano D. 2013. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci U S A 110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chow C-ET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. 2014. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J 8:816–829. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan MB. 2014. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513:242–245. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.