Abstract
Terpene synthesis in the majority of bacterial species, together with plant plastids, takes place via the 1-deoxy-d-xylulose 5-phosphate (DXP) pathway. The first step of this pathway involves the condensation of pyruvate and glyceraldehyde 3-phosphate by DXP synthase (Dxs), with one-sixth of the carbon lost as CO2. A hypothetical novel route from a pentose phosphate to DXP (nDXP) could enable a more direct pathway from C5 sugars to terpenes and also circumvent regulatory mechanisms that control Dxs, but there is no enzyme known that can convert a sugar into its 1-deoxy equivalent. Employing a selection for complementation of a dxs deletion in Escherichia coli grown on xylose as the sole carbon source, we uncovered two candidate nDXP genes. Complementation was achieved either via overexpression of the wild-type E. coli yajO gene, annotated as a putative xylose reductase, or via various mutations in the native ribB gene. In vitro analysis performed with purified YajO and mutant RibB proteins revealed that DXP was synthesized in both cases from ribulose 5-phosphate (Ru5P). We demonstrate the utility of these genes for microbial terpene biosynthesis by engineering the DXP pathway in E. coli for production of the sesquiterpene bisabolene, a candidate biodiesel. To further improve flux into the pathway from Ru5P, nDXP enzymes were expressed as fusions to DXP reductase (Dxr), the second enzyme in the DXP pathway. Expression of a Dxr-RibB(G108S) fusion improved bisabolene titers more than 4-fold and alleviated accumulation of intracellular DXP.
INTRODUCTION
Terpenes constitute a very large family of natural products, members of which are produced in virtually all free-living organisms (1). The diverse array of structures within the terpene family is reflected by the variety of applications in society, ranging from nutrition (carotenoids) and medicine (artemisinin, paclitaxel [originally taxol]) to industrial materials (isoprene, linalool) and candidate biofuels (farnesene, bisabolene, pinene) (2). Terpenes can be synthesized via either the mevalonate pathway or the 1-deoxy-d-xylulose 5-phosphate (DXP) pathway, the former predominating in the eukaryotic cytosol and the latter in plastids, while prokaryotes may contain either pathway or, in some cases, both pathways (3).
Commercial-scale terpene production has been demonstrated in a variety of organisms, for example, carotenoids (via the DXP pathway) in algae (4, 5), paclitaxel (via DXP) in Taxus sp. (6), and artemisinin (via mevalonate) in Saccharomyces cerevisiae (7). Key metrics in determining the likelihood of commercial viability, particularly when targeting terpenes valued within the range of commodity chemicals or biofuels, are yield, productivity, and titer (8, 9). Of the two metabolic routes, the DXP pathway is considered the better option from the viewpoint of pathway efficiency—for example, the theoretical maximum yield of isoprene from glucose is around 20% higher when synthesized via DXP instead of mevalonate (9, 10). In considering production of terpenes from hemicellulosic feedstocks, conversion efficiencies of pentoses, predominantly xylose and arabinose, should also be taken into account (11). Although DXP is structurally similar to pentose phosphates, it is synthesized by DXP synthase (Dxs) via condensation of pyruvate and glyceraldehyde 3-phosphate, with a concomitant loss of CO2. Dxs is a key control point in the pathway and has been found to be regulated at the transcriptional and translational levels as well as being feedback inhibited by the prenyl phosphates (12). We reasoned that a novel route from a pentose phosphate to DXP (nDXP) could have several advantages from a pathway-engineering standpoint, including carbon conservation, avoidance of regulatory mechanisms that target Dxs, and a more direct entry point for pentoses into the terpene biosynthetic pathway. However, to our knowledge there are no 1-deoxy sugars made from sugars in nature, and we therefore decided to employ a directed-evolution strategy.
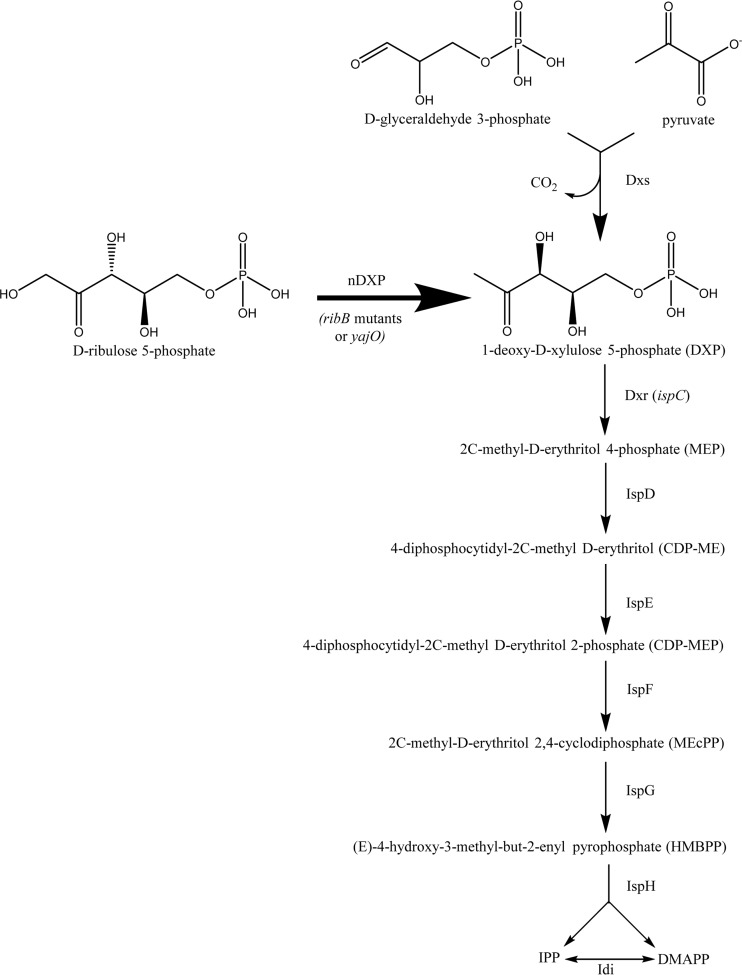
We report the discovery of two nDXP routes following complementation of a dxs deletion in Escherichia coli grown on xylose. One of these arose through spontaneous mutations in the native ribB coding sequence (cds), normally involved in riboflavin biosynthesis (13). The second route was discovered following overexpression of one of several rationally selected candidate nDXP genes, yajO, located next to dxs on the E. coli chromosome and annotated as a putative xylose reductase, although little evidence has been found to support this proposed function (14). Both nDXP enzymes were found to catalyze conversion of ribulose 5-phosphate (Ru5P) to DXP, providing a direct route from pentoses to terpenes (Fig. 1).
FIG 1.
The DXP pathway, together with the alternative nDXP route from Ru5P. IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate.
In order to test the utility of the nDXP genes for terpene production in E. coli, we engineered a strain for production of the sesquiterpene bisabolene, a biofuel candidate, by overexpression of pathway enzymes previously found to be limiting. Coexpression of the nDXP genes enhanced bisabolene production 2- to 3-fold, while translational fusions to the succeeding pathway enzyme, DXP reductase (Dxr), increased production up to 4.3-fold.
MATERIALS AND METHODS
Generation of an E. coli dxs knockout.
To permit deletion of dxs, E. coli MG1655 was first transformed with plasmid pMBI (15), harboring four genes (ERG12 [mevalonate kinase], ERG8 [phosphomevaloante kinase], and MVD1 [mevalonate pyrophosphate decarboxylase], all from Saccharomyces cerevisiae, and idi [isopentenyl pyrophosphate {IPP} isomerase], from E. coli) that enable biosynthesis of the isoprenoid precursors IPP and DMAPP when mevalonate is supplied exogenously to cells. Following transformation with pMBI, λ Red recombinase was used to replace the dxs cds with a kanamycin marker cassette (16), generating strain ΔdxsMB (Table 1). After selection on kanamycin, deletion of dxs was confirmed by several diagnostic PCRs, together with verification of mevalonate auxotrophy.
TABLE 1.
Primers used in this study
| Target gene | Primer name | Primer sequence |
|---|---|---|
| dxs knockouta | Dxs-KO-f | ATGAGTTTTGATATTGCCAAATACCCGACCCTGGCACTGGTCGACTCCACGTGTAGGCTGGAGCTGCTTCG |
| Dxs-KO-r | TTCTACGGTGACCAGCGCTTCATGGCTGGCGGCCATTTCCAGAATTAACGATGGGAATTAGCCATGGTCC | |
| ribB | pTrc-ribB-f | GACCATGGCAAATCAGACGCTACTTTCCTCTTTTGGTACG |
| pTrc-ribB-r | CCAAGCTTTCAGCTGGCTTTACGCTCATGTGCGTGAC | |
| yajOb | pTrc-yajO-f | AACCATGGAACAATACAACCCCTTAGGAAAAACC |
| pTrc-yajO-r | AAAAGCTTTTATTTAAATCCTACGACAGGATGCG | |
| rib | pTrc-ribB-S-f | GACCATGGCAAATCAGACGCTACTTTCC |
| rib StrepII tag | pTrc-ribB-S-r | TTAAGCTTTCATTTTTCAAACTGCGGATGGCTCCAGCTGGCTTTACGCTCATGTGCC |
| yajO | pTrc-yajO-S-f | ACATCGATGCAATACAACCCCTTAGGA |
| yajO StrepII tag | pTrc-yajO-S-r | TTAAGCTTTTATTTTTCAAACTGCGGATGGCTCCATTTAAATCCTACGACAGGATGCG |
| AgBIS gene | pBbA1k-BIS-f | AAGAATTCAGTTTTTCCCTACTAGTTCAGGAGGTATTCATGGCGGGTGTTTCTGCGGTTTCTAAAGTTTCTTCTCTGG |
| pBbA1k-BIS-r | TTGGATCCTTACAGCGGCAGCGGTTCGATCAGGCA | |
| ispAc | pBbA1k-ispA-f | AAGGATCCTCTAGAGGAGGTACACTATGGACTTTCCGCAGCAACTCGAAGC |
| pBbA1k-ispA-r | AACTCGAGTTATTTATTACGCTGGATGATGTAG | |
| ispDFd | pBbA1k-ispDF-f | CGCTGCCGCTGTAAGGATCCCCCGGGAATTAACATGGCAA |
| pBbA1k-ispDF-r | CATAATTTCTCACATGTAATTCATTTTGTTGCCTTAATGAGTAGC | |
| idid | pBbA1k-idi-f | TCATTAAGGCAACAAAATGAATTACATGTGAGAAATTATGC |
| pBbA1k-idi-f | CCTGGAGATCCTTACTCGAGTTATTTAAGCTGGGTAAATG | |
| yajO-ispCe | Ptrc-yajO-F-f | GAATTGTGAGCGGATAACAATTTCACACAGGAAACAGACCATGACTGGGGTGAACGAATGCAG |
| ispC-GP-f | AAACCGCATCCTGTCGTAGGATTTAAAGGTCCAGGCCCTAAGCAACTCACCATTCTGGGCT | |
| yajO-GP-r | CGGTCGAGCCCAGAATGFGTGAGTTGCTTAGGGCCTGGACCTTTAAATCCTACGACAGGATGCGGTTT | |
| ispC-PT-f | CCTGTCGTAGGATTTAAACCAACACCAACGCCAACGACACCAACTCCAACAAAGCAACTCACCATTCTGGGC | |
| yajO-PT-r | AGAATGGTGAGTTGCTTTGTTGGAGTTGGTGTCGTTGGCGTTGGTGTTGGTTTAAATCCTACGACAGGATGCGGTT | |
| ispC-Ptrc-r | GGCTGAAAATCTTCTCTCATCCGCCAAAACAGCCAAGCTTTCAGCTTGCGAGACGCATCAC | |
| ribB-ispCe | Ptrc-ribB-F-f | GAATTGTGAGCGGATAACAATTTCACACAGGAAACAGACCATGGCAAATCAGACGCTACTTTCCTCTTTT |
| IspC-G2-F-f | CGTCAGGCACATGAGCGTAAAGCCAGCGGTTCTGGCGGTTCCGGTAAGCAACTCACCATTCTGGGCT | |
| ribB-G2-r | GAGCCGGTCGAGCCCAGAATGGTGAGTTGCTTACCGGAACCGCCAGAACCGCTGGCTTTACGCTCATGTGCCTG | |
| ispC-GP-F-f | CCGTCAGGCACATGAGCGTAAAGCCAGCGGTCCAGGCCCTAAGCAACTCACCATTCTGGGCTCG | |
| ribB-GP-r | GAGCCGGTCGAGCCCAGAATGGTGAGTTGCTTAGGGCCTGGACCGCTGGCTTTACGCTCATGTGCCTGA | |
| ispC-Ptrc-r | GGCTGAAAATCTTCTCTCATCCGCCAAAACAGCCAAGCTTTCAGCTTGCGAGACGCATCAC | |
| ispC-yajOe | ispC-Ptrc-f | GAATTGTGAGCGGATAACAATTTCACACAGGAAACAGACCATGAAGCAACTCACCATTCTGGGCT |
| IspC-G2-r | GGCTGCATTCGTTCACCCCAGTACCGGAACCGCCAGAACCGCTTGCGAGACGCATCACCTCTTTT | |
| yajO-G2-f | AGGTGATGCGTCTCGCAAGCGGTTCTGGCGGTTCCGGTACTGGGGTGAACGAATGCAGC | |
| yajO-Ptrc-r | GGCTGAAAATCTTCTCTCATCCGCCAAAACAGCCAAGCTTTTATTTAAATCCTACGACAGGATGCGGTTTATACG | |
| ispC-ribBe | IspC-Ptrc-f | GAATTGTGAGCGGATAACAATTTCACACAGGAAACAGACCATGAAGCAACTCACCATTCTGGGCT |
| IspC-G2-R-r | AAAAGAGGAAAGTAGCGTCTGATTTGCACCGGAACCGCCAGAACCGCTTGCGAGACGCATCACCTC | |
| ribB-G2-f | TGATGCGTCTCGCAAGCGGTTCTGGCGGTTCCGGTGCAAATCAGACGCTACTTTCCTCTTTTGG | |
| ribB-Ptrc-r | GGCTGAAAATCTTCTCTCATCCGCCAAAACAGCCAAGCTTTCAGCTGGCTTTACGCTCATGTGC |
The 50 nucleotides (nt) at the 5′ end of the primers match the ends of the dxs cds, while the underlined sequence matches the kanamycin resistance cassette. The kanamycin marker cassette was amplified by PCR from plasmid pDK13 (16) using the primers listed. E. coli MG1655 harboring pMBI was transformed with this PCR product together with the pKD46 plasmid, containing the gene for γ Red recombinase (16). Following selection on LB-kanamycin, deletion of dxs was confirmed by several diagnostic PCRs, and loss of the temperature-sensitive pDK46 plasmid was facilitated by growth at 37°C.
The yajO cds was selected according to the GenBank annotation for E. coli MG1655, beginning with the amino acid sequence MQYN. However, the yajO sequence in E. coli H736 is annotated to contain an additional 14 N-terminal amino acids (beginning MTGV) and the corresponding region in MG1655 forms a contiguous open reading frame (ORF), but with a putative valine start codon (reading VTGV). Since it is uncertain whether translation of the MG1655 yajO cds begins at the annotated Met or at the Val codon 14 amino acids upstream, we overexpressed both versions in the ΔdxsMB strain (but substituting Met for Val in the longer version) and found that they both complemented the dxs knockout.
The ispA cds (encoding FPP synthase) was amplified from E. coli MG1655 genomic DNA and cloned between the BamHI and XhoI sites of pBbA1k-AgBIS to make plasmid pBbA1k-AgBIS-ispA (pBbA1k-AF).
The ispDF operon and idi gene were amplified from E. coli MG1655 genomic DNA and cloned between the BamHI and XhoI sites of pBbA1k-AgBIS using SLIC to make plasmid pBbA1k-AgBIS-ispDF-idi (pBbA1k-Aii). Sequences homologous to each other or to the vector backbone are underlined. This fragment was subsequently cloned into the XhoI site of pBbA1k-AgBIS-ispA to make plasmid pBbA1k-AgBIS-ispA-ispDF-idi (pBbA1k-AFii).
nDXP-Dxr fusions were constructed by the use of PCR splicing by overlap extension (SOE). Three fusions for yajO-ispC and three fusions for rib-ispC were constructed and cloned between the NcoI and HindIII sites of pTrc99A using SLIC. Nucleotides encoding fusion linkers (GP, GPGP; PT, PTPTPTTPTPT; G2, GSGGSG) are shown in bold.
Use of selective pressure to isolate spontaneous nDXP mutants.
Subculturing of strain ΔdxsMB was carried out in EZ Rich medium (Teknova, Hollister, CA, USA) containing 1% d-xylose (Sigma-Aldrich) as the carbon source (EZ-X). Selective pressure was applied by reducing the concentration of mevalonate (Sigma-Aldrich) from 1.0 mM to 0.1 mM over the course of three cultures (subcultured at mid-log phase; 1/1,000 dilution) and thereafter maintaining mevalonate at 0.1 mM. At each subculture, aliquots containing approximately 109 cells were plated onto EZ-X agar lacking mevalonate. Colonies were initially ranked in order of size and further prioritized based on growth in liquid medium lacking mevalonate and containing either glucose or xylose.
Genome sequencing of mevalonate-independent ΔdxsMB strains.
Strains isolated under selective pressure that grew well in the absence of mevalonate were submitted to the Joint Genome Institute (JGI) for genome sequencing, using the parent ΔdxsMB strain as a reference. Libraries were generated from 1 μg of genomic DNA using a modified version of Illumina's standard protocol (Illumina Inc., Hayward, CA, USA). DNA was sheared using a sonicator (Covaris, Inc., Wolburn, MA, USA) to generate fragments of 200 to 800 bp in length, and the fragments were then size selected by solid-phase reversible immobilization (SPRI) (Beckman Coulter, Indianapolis, IN, USA) to around 300 bp. Selected fragments were end repaired, phosphorylated, and A-tailed using the polymerase activity of the Klenow fragment of E. coli DNA polymerase I and then ligated with Illumina paired-end sequencing adapters and amplified by 10 cycles of PCR. The prepared sample libraries were quantified using a Kapa next-generation sequencing library quantitative PCR (qPCR) kit (Kapa Biosystems, Wolburn, MA, USA) and run on a LightCycler 480 real-time PCR instrument (Roche, Pleasanton, CA, USA). The libraries of quantified samples were then prepared for sequencing on the Illumina sequencing platform utilizing a paired-end cluster generation kit, v4, and Illumina's cBot instrument to generate clustered flow cells for sequencing. Sequencing of the flow cells was performed on an Illumina GAIIx sequencer using SBS sequencing kits, v4, following a program employing 2 sets of 36 runs.
Generation of additional ribB mutants.
To screen for additional ribB mutations that complement Δdxs, two approaches were taken: chemical mutagenesis by hydroxylamine (Sigma-Aldrich) and error-prone PCR (EP-PCR) mutagenesis. Hydroxylamine mutagenesis was performed as described previously (17) except that DNA was cleaned up using a QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). EP-PCR mutagenesis was performed using the Clontech Diversify approach (Clontech, Mountain View, CA, USA) and conditions 2 and 5 in the manufacturer's protocol, predicted to generate 2.3 and 4.6 mutations per kb, respectively. The ribB cds was cloned between the NcoI and HindIII sites of pTrc99A under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible trc promoter (Table 1). Mutant libraries were transformed into strain ΔdxsMB and grown under selective pressure as described above except that 50 μM IPTG (Calbiochem/EMD Millipore, Billerica, MA, USA) was included in media to induce ribB expression. Plasmids were recovered from mevalonate-independent strains, which were isolated from EZ-X agar plates and ranked in order of growth, and the ribB cds was sequenced.
Expression of other candidate nDXP genes.
Several genes or gene combinations were investigated for their ability to complement a dxs knockout, and overexpression of one of these candidates, yajO from E. coli, enabled mevalonate-independent growth. The yajO cds was cloned between the NcoI and HindIII sites of pTrc99A (Table 1) and transformed into strain ΔdxsMB, and the resulting strain was then grown under selective pressure in the presence of 50 μM IPTG. EP-PCR mutagenesis of yajO was carried out as described for ribB above.
In vitro analysis of RibB(G108S) and YajO.
Coding sequences for RibB(G108S) and YajO were amplified by PCR using reverse primers that added sequence encoding C-terminal StrepII peptide tags ([WSHPQFEK]) and cloned into pTrc99A (Table 1). Protein was expressed in E. coli BLR(DE3) grown in LB medium containing 100 μM IPTG and purified using a StrepTactin SpinPrep kit (EMD Millipore). Protein purity was determined to be around 95% by SDS-PAGE, and protein was directly used for assays. In vitro reactions were carried out by combining 13.2 μg purified protein and 5 mM substrate in assay buffer (50 mM TrisCl [pH 7.5], 150 mM NaCl, 10 mM MgCl2, 5 mM dithiothreitol [DTT]) and incubating at 37°C for 30 min (18). Following addition of methanol (high-performance liquid chromatography [HPLC] grade; Sigma-Aldrich) to reach a concentration of 50% (vol/vol), samples were analyzed by liquid chromatography-mass spectrometry (LC-MS) alongside a DXP standard (Echelon Inc., Salt Lake City, UT, USA). Chromatographic separation was achieved using an Agilent 1200 Rapid Resolution HPLC system and the hydrophilic interaction liquid chromatography (HILIC) method from Baidoo et al. (19), and metabolites were detected by the use of an Agilent 6210 TOF MS system as described previously (20).
Terpene biosynthesis in E. coli.
Amorphadiene production levels were compared using the original ΔdxsMB parent strain and an equivalent strain harboring a single mutation encoding RibB(G92D). Strains were transformed with the pADS plasmid, encoding amorphadiene synthase from Artemisia annua (15), and grown at 37°C in M9 medium (Sigma-Aldrich) containing 1% xylose, 0.5 mM IPTG, and a 10% (vol/vol) dodecane (Sigma-Aldrich) overlay (to capture amorphadiene), with or without 5 mM mevalonate. Amorphadiene production was measured by gas chromatography-mass spectrometry (GC-MS) as described previously (21).
Bisabolene production was tested in E. coli BL21(DE3) harboring a plasmid containing an nDXP gene, or containing rfp (encoding red fluorescent protein [RFP]) as a control, and a second plasmid harboring the Abies grandis bisabolene synthase (AgBIS) gene, amplified from pRSLeu2d-BISopt (22), coupled with E. coli DXP pathway genes ispD, ispF, and idi (Fig. 1; see Table 1 for cloning details). The nDXP genes were expressed on pTrc99A plasmids as described above, while the AgBIS and the DXP pathway genes were expressed on pBbA1k (23). Strains were grown at 37°C in EZ-X medium containing a 20% (vol/vol) dodecane overlay to an optical density at 600 nm (OD600) of around 1.0, at which point IPTG (200 μM) was added and growth was continued at 30°C; bisabolene was measured by GC-MS as described previously (22).
Vectors encoding translation fusions between Dxr (encoded by the E. coli ispC gene) and either RibB(G108S) or YajO were constructed using the sequence-and-ligation-independent-cloning (SLIC) approach (24). A combination of fusions was made with Dxr located at either the N or the C terminus and employing peptide linker G2 (GSGGSG), GP (GPGP), or PT (PTPTPTTPTPT) (Table 1). Fusions were expressed on pTrc99A plasmids, while the AgBIS gene combined with E. coli DXP pathway genes ispD, ispF, idi, and/or ispA was expressed on pBbA1k. Strains were grown in EZ-X medium and bisabolene levels quantified as described above.
Quantification of DXP pathway intermediates.
DXP pathway intermediates were extracted from E. coli by brief centrifugation (3,000 × g for 3 min at room temperature) of each 5-ml culture, removal of medium, addition of 1 ml ice-cold 50% (vol/vol) methanol (in water) to the cell pellet, vortex mixing for 1 min, centrifugation (15,000 × g for 10 min at 4°C), and removal of high-molecular-weight (MW) material from the supernatant by centrifugation (14,000 × g for 45 min at 4°C) through a 3,000-MW-cutoff (MWCO) Amicon Ultra 0.5-ml filter (Millipore, Carrigtwohill, Cork, Ireland). Separation of DXP pathway intermediates and quantification using authentic standards (Echelon Inc.) were performed using LC-MS as described above.
RESULTS
Use of selective pressure to isolate spontaneous nDXP mutations.
The ΔdxsMB strain was constructed to screen for nDXP routes by transforming E. coli MG1655 with the pMBI plasmid and deletion of the dxs gene. pMBI, which encodes the lower half of the mevalonate pathway, enables growth of strain ΔdxsMB in the presence of exogenously supplied mevalonate. A selection for spontaneous nDXP mutants was carried out by subculturing in defined medium containing xylose (EZ-X), coupled with reduction of the mevalonate concentration to growth-limiting levels. Aliquots of each subculture were plated on EZ-X agar lacking mevalonate, and colonies were picked for further analysis. Since mutations in the native aceE gene have been reported to complement a dxs knockout (25), the aceE gene was sequenced in the first 50 isolates, but no mutations were found.
Sequencing of mevalonate-independent ΔdxsMB strains.
The genomes of eight strains that were isolated at the beginning of the selection process were sequenced, and all were found to have a single nucleotide mutation in the ribB gene, namely, S89R (three isolates), G92D (three isolates), or T106I (two isolates). The wild-type RibB enzyme is responsible for synthesis of the riboflavin precursor 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) from Ru5P (13, 26). We also sequenced the genomes of four strains isolated at the end of the screening process (following 3 weeks of subculturing under selective pressure), and all three were found to harbor mutations in the gatC gene, encoding D296A. GatC functions as a galactitol phosphotransferase permease and has also been found to contribute to xylose uptake (27, 28). Since the strains harboring ribB mutations had grown considerably faster than those harboring gatC mutations on EZ-X agar lacking mevalonate (3 versus 6 days, respectively), the RibB mutants were investigated further as the most promising nDXP candidates.
Generation of additional ribB mutants.
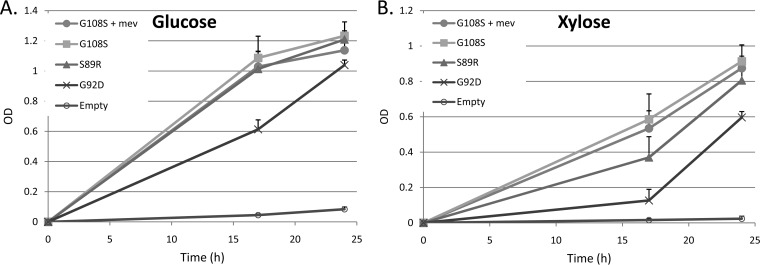
Following chemical and EP-PCR mutagenesis of the ribB cds, mutant libraries were constructed in pTrc99A and transformed into strain ΔdxsMB. Selection in EZ-X was carried out as before, and the 44 fastest-growing isolates were sequenced and found to comprise five discrete mutations: G108S, T88I, V109I, M182I, and G92D (see Table S1 in the supplemental material). A total of 16 mutants that formed colonies within the first 24 h were represented by two mutations, G108S and T88I. The efficiencies of complementation of Δdxs by three of the ribB mutants were compared by expression on pTrc99A in strain ΔdxsMB and culturing in the absence of mevalonate in either EZ-X or EZ-G (EZ-Rich medium containing 1% d-glucose) (Fig. 2). Expression of ribB(G108S) facilitated growth on either glucose or xylose in the absence of mevalonate at rates similar to those seen in media containing mevalonate. The growth rate of the ribB(G92D) strain was appreciably lower, while negligible growth was observed in the strain containing an empty vector.
FIG 2.
Growth of E. coli ΔdxsMB expressing various ribB mutants on pTrc99A on glucose (A) or xylose (B) as the sole carbon source. Supplementation of the RibB(G108S) strain with 1 mM mevalonate made no significant difference in the growth rate. The ΔdxsMB strain harboring an empty vector exhibited minimal growth within 24 h.
Rational selection of candidate nDXP genes.
We selected candidate genes from several species on the basis of genetic or biochemical evidence of possible connections to both C5 sugars and DXP and investigated their capacity to complement a dxs knockout. One of these genes, yajO from E. coli, was found to support growth on EZ-X agar in the absence of mevalonate when overexpressed in strain ΔdxsMB. Two lines of evidence had led to the selection of yajO, originally annotated to encode a putative xylose reductase, as a candidate nDXP gene. First, yajO was identified in a screen for genes linked to thiamine biosynthesis that employed growth-inhibiting thiamine analog precursors (29). Although the mechanism for resistance to these analogs has not been elucidated, one of the possibilities is increased synthesis of DXP, a precursor to thiamine. Second, yajO and the isoprenoid biosynthetic genes dxs and ispA (the latter encodes farnesyl pyrophosphate synthase) comprise an operon in the genomes of E. coli and related species.
Colony formation for strain ΔdxsMB harboring pTrc99A-yajO on EZ-X agar plates lacking mevalonate usually took 4 to 5 days. Efforts to make this route more efficient through mutagenesis of yajO followed by mevalonate selective pressure were unsuccessful. In addition, growth in defined media lacking mevalonate was observed when xylose but not glucose was provided as the sole carbon source (see Fig. S1 in the supplemental material).
Reactions catalyzed by RibB(G108S) and YajO.
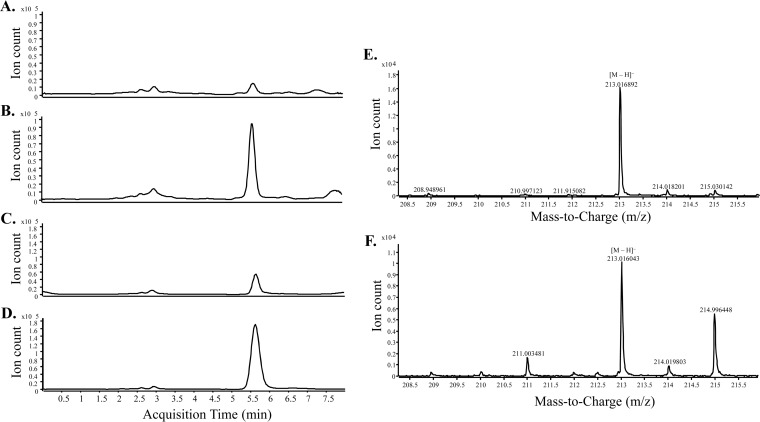
To elucidate the reactions catalyzed by the nDXP enzymes, RibB(G108S) and YajO containing C-terminal StrepII tags were overexpressed in E. coli and purified. In vitro assays were conducted using candidate substrates xylose, xylulose, xylulose 5-phospate, and Ru5P, followed by analysis by LC-MS for potential products DXP and 1-deoxy-d-xylulose (DX). Production of DX was considered a possible nDXP route, since DX can be phosphorylated to DXP by the endogenous E. coli xylulokinase XylB (30). In the case of both enzymes, DXP was detected only when Ru5P was included as the substrate (Fig. 3). The addition of a cofactor, NADH, NADPH, or ATP, to the reaction mixtures (at 1 mM) did not yield an increase in DXP levels, and no product was detected when the wild-type RibB enzyme was used instead of RibB(G108S) (data not shown).
FIG 3.
In vitro production of DXP (5.7 min) from Ru5P by RibB(G108S) and YajO. (A to D) Chromatograms for DXP produced in vivo by yajO (A) and RibB(G108S) (C); the data in panels B and D, respectively, represent the results determined with the same samples spiked with 4 μM DXP to confirm the retention time for DXP. (E and F) Mass spectra for a DXP standard (E) and DXP produced by RibB(G108S) (F).
Utility of nDXP routes for terpene production in E. coli.
The use of an nDXP route for isoprenoid production was initially evaluated by transforming strain ΔdxsMB harboring a genomic ribB(G92D) mutation and the corresponding parent strain with pADS, encoding amorphadiene synthase from A. annua (15). As expected, growth and amorphadiene production were negligible in the parent strain in the absence of mevalonate whereas the corresponding ribB(G92D) strain grew normally and generated 2 mg/liter amorphadiene (see Fig. S2 in the supplemental material). When supplemented with 5 mM mevalonate, strains grew to equivalent ODs, but the amorphadiene titer (5.4 mg/liter or 17 mg/g dry cell weight [DCW]) was over 4-fold higher in the ribB(G92D) mutant than in the control strain.
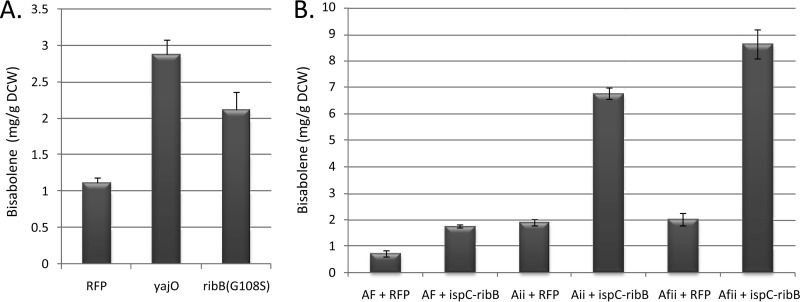
In order to evaluate the use of nDXP enzymes for terpene pathway engineering alongside native levels of Dxs, the host strain was changed from strain ΔdxsMB to E. coli BL21(DE3). In addition, we selected the candidate biofuel α-bisabolene as the target sesquiterpene product. To assess synergy between DXP pathway engineering and nDXP routes, we constructed vectors for expression of bisabolene synthase (AgBIS) together with DXP pathway enzymes that have previously been shown to limit pathway flux, namely, those encoded by ispD, ispF, idi, and ispA (31). Expression of yajO or ribB(G108S) alongside plasmid pBbA1k-AgBIS-ispDF-idi yielded a 2-to-3-fold improvement in bisabolene titers compared to those seen with an rfp control vector (Fig. 4A). Having previously tested fusions of Dxs/Dxr for DXP pathway engineering and found no benefit in terms of the bisabolene titer (data not shown), we constructed nDXP/Dxr fusions, using the native ispC gene encoding Dxr, to determine if pathway flux could be enhanced. In comparisons of several fusions in which different orders of the proteins and the peptide linkers were used, ispC-G2-ribB(G108S), where G2 corresponds to a GSGGSG linker, yielded the highest increase in the bisabolene titer (see Fig. S3 in the supplemental material).
FIG 4.
Bisabolene production via the DXP pathway in E. coli using nDXP genes. (A) Expression of rfp, yajO, or ribB(G108S) combined with pBbA1k-AgBIS-ispDF-idi. (B) Expression of rfp or an ispC-(GSG)2-ribB(G108S) fusion (ispC-ribB) coupled with pBbA1k-AgBIS-ispA (AF), pBbA1k-AgBIS-ispDF-idi (Aii), or pBbA1k-AgBIS-ispA-ispDF-idi (Afii).
To further investigate the ispC-G2-ribB(G108S) fusion, we first coupled it with three different vectors harboring AgBIS and DXP pathway genes (Fig. 4B). The overall bisabolene titer as well as the increase over control levels became progressively higher as the ispC-G2-ribB(G108S) fusion was coupled with progressively engineered DXP pathway vectors. Coupling pTrc99A-ispC-G2-ribB(G108S) with pBbA1k-AgBIS-ispA-ispDF-idi (Afii) yielded a 4.3-fold increase over the levels of a control strain in which Afii was coupled with rfp. To determine how this affected DXP pathway flux, we measured pathway intermediates in these two strains (Table 2). In the RFP control strain, only two metabolites were detected, DXP and MEcPP (2-C-methyl-d-erythritol 2,4-cyclophosphate), the latter being the substrate for IspG, which is known to be kinetically slow (32). In the equivalent strain harboring pTrc99A-ispC-G2-ribB(G108S), DXP did not accumulate to detectable levels whereas MEcPP levels were almost 3-fold higher than those seen with the rfp strain. Together, these data suggest that fusion of Dxr to RibB increases flux through the pathway, circumvents accumulation of DXP, and results in increased accumulation of MEcPP, implicating IspG as a suitable target for further engineering studies.
TABLE 2.
Measurement of DXP pathway intermediates in two E. coli strains engineered for bisabolene productiona
| E. coli strain | Concn (μM) |
|||||
|---|---|---|---|---|---|---|
| DXP | MEP | CDP-ME | MEcPP | HMBPP | IPP/DMAPP | |
| Afii + RFP | 1.13 (0.62) | 0.00 | 0.00 | 13.9 (5.2) | 0.00 | 0.00 |
| Afii + Dxr-RibB | 0.00 | 0.15 (0.01) | 0.43 (0.06) | 38.7 (13.3) | 0.00 | 0.00 |
Vectors harboring rfp or an ispC-G2-ribB(G108S) fusion (Dxr-RibB) were coupled with pBbA1k-AgBIS-ispA-ispDF-idi (Afii). Concentrations are normalized to culture OD; standard deviations from three cultures are shown in parentheses. CDP-ME, 4-diphosphocytidyl-2-C-methyl-d-erythritol; HMBPP, (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate.
DISCUSSION
Metabolic engineering of terpene production in microbes or plants usually entails overexpression of key enzymes in the mevalonate or DXP precursor pathways (33). Dxs is an important control point in the DXP pathway as demonstrated by the fact that overexpression has increased terpene titers in several species (34, 35). However, the benefit gained by increasing Dxs levels is somewhat limited by regulatory mechanisms that can occur posttranscriptionally (36, 37), for example, via feedback inhibition by prenyl phosphates (12, 38). The nDXP enzymes described here provide alternative routes to DXP accumulation that may facilitate terpene pathway engineering by circumventing regulatory issues and also carbon loss associated with the Dxs-catalyzed reaction (Fig. 1). Additional benefits may arise depending on the carbon source provided to the production host; for example, nDXP expression may benefit E. coli grown on mixed sugars derived from hemicellulosic feedstocks by shunting the pentose fraction more directly to the DXP pathway following conversion to Ru5P. Alternatively, engineering nDXP in plants and algae could provide a more direct link from carbon fixation (Ru5P in the Calvin cycle) to the terpene pathway.
Calculation of the stoichiometric pathway efficiency, accounting for the cost of NADPH and ATP utilized together with the molar product yield (10), reveals that the nDXP route is more efficient not only from pentoses but also from glucose. Conversion of glucose 6-phosphate to Ru5P via the oxidative phase of the pentose phosphate pathway generates 2 NADPHs, which may serve as cofactors in the DXP pathway. The resulting theoretical pathway efficiency of production of terpenes from glucose via the nDXP route is close to the maximum theoretical biochemical yield (0.309 g bisabolene per gram of glucose versus 0.324 g bisabolene per gram of glucose, respectively).
Our approach to screening for nDXP genes entailed growth on xylose as the sole carbon source. Interestingly, mutations in the aceE gene, previously shown to convert pyruvate and glyceraldehyde to DX (25), were not encountered in any of the first 50 mevalonate prototrophs isolated, perhaps indicating that the mutant RibB route for synthesis of DXP is more efficient than the mutant AceE route in the presence of high Ru5P levels. Overexpression of ribB(G108S) also enabled normal growth in the absence of mevalonate of strain ΔdxsMB on glucose, while overexpression of yajO enabled growth only on xylose, suggesting that YajO may require higher Ru5P levels to support growth due to kinetic limitations.
The remaining genomic mutation found to complement growth in strain ΔdxsMB, encoding GatC(D296A), was not investigated further. However, since GatC can function as an ATP-independent xylose transporter, it seems likely that the main benefit from the mutation may be increased uptake of xylose (27), perhaps increasing the Ru5P concentration to the point where native levels of YajO can generate sufficient DXP to sustain a low level of growth.
Two mutations in ribB have been recently found to complement Δdxs in E. coli grown on LB medium (20); here we have identified one of the same mutations (G108S) along with 6 others (T88I, S89R, G92D, T106I, V109I, and M182I) that complement Δdxs in E. coli grown on xylose and have elucidated the reaction catalyzed. Nuclear magnetic resonance (NMR) and crystal structures have been solved for RibB from E. coli and other species, and a reaction mechanism has been proposed for conversion of Ru5P to the riboflavin precursor DHBP (39–42). Several of the RibB mutations that enable production of DXP are at or close to catalytically important residues; for example, G108 is conserved among RibB proteins from diverse sources and is suggested by NMR studies to play a role in substrate binding (40), while crystal structure analysis suggests that the presence of a nonglycine residue at this critical turn would result in a sterically strained conformation (41). Since the reaction mechanism proposed for wt RibB involves the generation of 1-deoxy intermediates, one possibility is that the mutations result in termination of the reaction prior to the proposed carbon rearrangement and elimination of formate (26, 41). However, the exact mechanism and means of reduction remain unknown and require further investigation.
Although the reaction catalyzed by the mutant RibB enzyme is unusual, it is not unique; YajO from E. coli can also catalyze formation of DXP from Ru5P, albeit less efficiently. YajO was originally annotated as a putative xylose reductase, but little evidence was found for this activity in vitro, and E. coli is known to utilize the alternative xylose isomerase route for xylose assimilation (14). YajO is now most often annotated as a 2-carboxybenzaldehyde reductase based on detection of this activity in vitro, but since E. coli is not known for polyaromatic hydrocarbon degradation, the true biological function for the enzyme remains unknown (14). It is tempting to speculate that YajO may play a role in biosynthesis of DXP (which, besides its role in the isoprenoid pathway, serves as a precursor to thiamine) under conditions of thiamine starvation. Since Dxs requires thiamine pyrophosphate as a cofactor, thiamine starvation would leave cells in a state where synthesis of DXP for de novo thiamine (and isoprenoid) biosynthesis would not be possible. YajO could serve a role in low-level DXP biosynthesis from Ru5P to kick-start thiamine biosynthesis and, consequently, Dxs. As mentioned, yajO is located next to dxs on the E. coli genome, and thiL, which encodes the final enzyme in the thiamine biosynthetic pathway, is situated less than 1 kb downstream.
Having demonstrated the utility of nDXP genes in engineering terpene production in E. coli, we investigated the use of protein fusions to further increase yields of the candidate biofuel bisabolene. The use of fusions or scaffolds has been demonstrated to increase pathway flux in several situations, including terpene pathway engineering (43, 44). Having found that Dxs/Dxr fusions (in either orientation, with both using GSGGSG linkers) did not yield an increase in the bisabolene titer, we tested Dxr/nDXP fusions where the gene corresponding to nDXP was either yajO or ribB(G108S) and found that ispC-G2-ribB(G108S) yielded a 4.3 improvement in bisabolene production over the level seen with the control strain. The fact that DXP accumulation in this strain was below the limit of detection suggests that the metabolic flux from Ru5P to MEP (2C-methyl-d-erythritol 4-phosphate) is efficient. However, extensive further engineering of the DXP pathway will be required to achieve terpene yields close to the theoretical maximum in E. coli. Our analysis of DXP pathway intermediates, together with previously published studies, suggests that IspG is a key pathway bottleneck that should be addressed together with the genes expressed here (45, 46). We are also currently investigating the utility of the nDXP genes and fusions in plant hosts where a direct link from carbon fixation to biofuel production presents an attractive opportunity.
Supplementary Material
ACKNOWLEDGMENTS
J.D.K. has a financial interest in Amyris and Lygos.
The work conducted through the Joint BioEnergy Institute was supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. The work conducted through the University of California at Berkeley was funded through the U.S. Department of Energy ARPA-E PETRO program, under grant no. DE-AR0000209.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02920-14.
REFERENCES
- 1.Pérez-Gil J, Rodríguez-Concepción M. 2013. Metabolic plasticity for isoprenoid biosynthesis in bacteria. Biochem J 452:19–25. [DOI] [PubMed] [Google Scholar]
- 2.Tippmann S, Chen Y, Siewers V, Nielsen J. 2013. From flavors and pharmaceuticals to advanced biofuels: production of isoprenoids in Saccharomyces cerevisiae. Biotechnol J 8:1435–1444. doi: 10.1002/biot.201300028. [DOI] [PubMed] [Google Scholar]
- 3.Boucher Y, Doolittle WF. 2000. The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol Microbiol 37:703–716. doi: 10.1046/j.1365-2958.2000.02004.x. [DOI] [PubMed] [Google Scholar]
- 4.Skjånes K, Rebours C, Lindblad P. 2013. Potential for green microalgae to produce hydrogen, pharmaceuticals and other high value products in a combined process. Crit Rev Biotechnol 33:172–215. doi: 10.3109/07388551.2012.681625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye VM, Bhatia SK. 2012. Pathway engineering strategies for production of beneficial carotenoids in microbial hosts. Biotechnol Lett 34:1405–1414. doi: 10.1007/s10529-012-0921-8. [DOI] [PubMed] [Google Scholar]
- 6.Onrubia M, Cusido RM, Ramirez K, Hernandez-Vazquez L, Moyano E, Bonfill M, Palazon J. 2013. Bioprocessing of plant in vitro systems for the mass production of pharmaceutically important metabolites: paclitaxel and its derivatives. Curr Med Chem 20:880–891. doi: 10.2174/0929867311320070004. [DOI] [PubMed] [Google Scholar]
- 7.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. 2013. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 8.Du J, Shao Z, Zhao H. 2011. Engineering microbial factories for synthesis of value-added products. J Ind Microbiol Biotechnol 38:873–890. doi: 10.1007/s10295-011-0970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Cann AF, Liao JC. 2010. Biofuels: biomolecular engineering fundamentals and advances. Annu Rev Chem Biomol Eng 1:19–36. doi: 10.1146/annurev-chembioeng-073009-100938. [DOI] [PubMed] [Google Scholar]
- 10.Dugar D, Stephanopoulos G. 2011. Relative potential of biosynthetic pathways for biofuels and bio-based products. Nat Biotechnol 29:1074–1078. doi: 10.1038/nbt.2055. [DOI] [PubMed] [Google Scholar]
- 11.Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu Rev Plant Biol 61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee A, Wu Y, Banerjee R, Li Y, Yan H, Sharkey TD. 2013. Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J Biol Chem 288:16926–16936. doi: 10.1074/jbc.M113.464636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter G, Volk R, Krieger C, Lahm HW, Rothlisberger U, Bacher A. 1992. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J Bacteriol 174:4050–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko J, Kim I, Yoo S, Min B, Kim K, Park C. 2005. Conversion of methylglyoxal to acetol by Escherichia coli aldo-keto reductases. J Bacteriol 187:5782–5789. doi: 10.1128/JB.187.16.5782-5789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose MD, Fink GR. 1987. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- 18.Schlösser T, Schmidt G, Stahmann KP. 2001. Transcriptional regulation of 3,4-dihydroxy-2-butanone 4-phosphate synthase. Microbiology 147:3377–3386. [DOI] [PubMed] [Google Scholar]
- 19.Baidoo EE, Xiao Y, Dehesh K, Keasling JD. 2014. Metabolite profiling of plastidial deoxyxylulose-5-phosphate pathway intermediates by liquid chromatography and mass spectrometry. Methods Mol Biol 1153:57–76. doi: 10.1007/978-1-4939-0606-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Gil J, Uros EM, Sauret-Gueto S, Lois LM, Kirby J, Nishimoto M, Baidoo EE, Keasling JD, Boronat A, Rodriguez-Concepcion M. 2012. Mutations in Escherichia coli aceE and ribB genes allow survival of strains defective in the first step of the isoprenoid biosynthesis pathway. PLoS One 7:e43775. doi: 10.1371/journal.pone.0043775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paradise EM, Kirby J, Chan R, Keasling JD. 2008. Redirection of flux through the FPP branch-point in Saccharomyces cerevisiae by down-regulating squalene synthase. Biotechnol Bioeng 100:371–378. doi: 10.1002/bit.21766. [DOI] [PubMed] [Google Scholar]
- 22.Peralta-Yahya PP, Ouellet M, Chan R, Mukhopadhyay A, Keasling JD, Lee TS. 2011. Identification and microbial production of a terpene-based advanced biofuel. Nat Commun 2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. 2011. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng 5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 25.Sauret-Güeto S, Urós EM, Ibáñez E, Boronat A, Rodríguez-Concepción M. 2006. A mutant pyruvate dehydrogenase E1 subunit allows survival of Escherichia coli strains defective in 1-deoxy-D-xylulose 5-phosphate synthase. FEBS Lett 580:736–740. doi: 10.1016/j.febslet.2005.12.092. [DOI] [PubMed] [Google Scholar]
- 26.Bacher A, Eberhardt S, Fischer M, Kis K, Richter G. 2000. Biosynthesis of vitamin b2 (riboflavin). Annu Rev Nutr 20:153–167. doi: 10.1146/annurev.nutr.20.1.153. [DOI] [PubMed] [Google Scholar]
- 27.Nduko JM, Matsumoto K, Ooi T, Taguchi S. 2014. Enhanced production of poly(lactate-co-3-hydroxybutyrate) from xylose in engineered Escherichia coli overexpressing a galactitol transporter. Appl Microbiol Biotechnol 98:2453–2460. doi: 10.1007/s00253-013-5401-0. [DOI] [PubMed] [Google Scholar]
- 28.Nobelmann B, Lengeler JW. 1996. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J Bacteriol 178:6790–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawhorn BG, Gerdes SY, Begley TP. 2004. A genetic screen for the identification of thiamin metabolic genes. J Biol Chem 279:43555–43559. doi: 10.1074/jbc.M404284200. [DOI] [PubMed] [Google Scholar]
- 30.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. 2001. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc Natl Acad Sci U S A 98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan LZ, Rouviere PE, Larossa RA, Suh W. 2006. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab Eng 8:79–90. doi: 10.1016/j.ymben.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Xu W, Lees NS, Adedeji D, Wiesner J, Jomaa H, Hoffman BM, Duin EC. 2010. Paramagnetic intermediates of (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE/IspG) under steady-state and pre-steady-state conditions. J Am Chem Soc 132:14509–14520. doi: 10.1021/ja101764w. [DOI] [PubMed] [Google Scholar]
- 33.Vickers CE, Bongers M, Liu Q, Delatte T, Bouwmeester H. 2014. Metabolic engineering of volatile isoprenoids in plants and microbes. Plant Cell Environ 37:1753–1775. doi: 10.1111/pce.12316. [DOI] [PubMed] [Google Scholar]
- 34.Kim SW, Keasling JD. 2001. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng 72:408–415. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Muñoz-Bertomeu J, Arrillaga I, Ros R, Segura J. 2006. Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol 142:890–900. doi: 10.1104/pp.106.086355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordoba E, Salmi M, Leon P. 2009. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot 60:2933–2943. doi: 10.1093/jxb/erp190. [DOI] [PubMed] [Google Scholar]
- 37.Wright LP, Rohwer JM, Ghirardo A, Hammerbacher A, Ortiz-Alcaide M, Raguschke B, Schnitzler JP, Gershenzon J, Phillips MA. 2014. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in Arabidopsis. Plant Physiol 165:1488–1504. doi: 10.1104/pp.114.245191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghirardo A, Wright LP, Bi Z, Rosenkranz M, Pulido P, Rodriguez-Concepcion M, Niinemets U, Bruggemann N, Gershenzon J, Schnitzler JP. 2014. Metabolic flux analysis of plastidic isoprenoid biosynthesis in poplar leaves emitting and nonemitting isoprene. Plant Physiol 165:37–51. doi: 10.1104/pp.114.236018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinbacher S, Schiffmann S, Richter G, Huber R, Bacher A, Fischer M. 2003. Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with divalent metal ions and the substrate ribulose 5-phosphate: implications for the catalytic mechanism. J Biol Chem 278:42256–42265. doi: 10.1074/jbc.M307301200. [DOI] [PubMed] [Google Scholar]
- 40.Kelly MJ, Ball LJ, Krieger C, Yu Y, Fischer M, Schiffmann S, Schmieder P, Kuhne R, Bermel W, Bacher A, Richter G, Oschkinat H. 2001. The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site. Proc Natl Acad Sci U S A 98:13025–13030. doi: 10.1073/pnas.231323598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao DI, Calabrese JC, Wawrzak Z, Viitanen PV, Jordan DB. 2001. Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase of riboflavin biosynthesis. Structure 9:11–18. doi: 10.1016/S0969-2126(00)00550-5. [DOI] [PubMed] [Google Scholar]
- 42.Volk R, Bacher A. 1991. Biosynthesis of riboflavin. Studies on the mechanism of L-3,4-dihydroxy-2-butanone 4-phosphate synthase. J Biol Chem 266:20610–20618. [PubMed] [Google Scholar]
- 43.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. 2009. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 44.Brodelius M, Lundgren A, Mercke P, Brodelius PE. 2002. Fusion of farnesyldiphosphate synthase and epi-aristolochene synthase, a sesquiterpene cyclase involved in capsidiol biosynthesis in Nicotiana tabacum. Eur J Biochem 269:3570–3577. doi: 10.1046/j.1432-1033.2002.03044.x. [DOI] [PubMed] [Google Scholar]
- 45.Rivasseau C, Seemann M, Boisson AM, Streb P, Gout E, Douce R, Rohmer M, Bligny R. 2009. Accumulation of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate in illuminated plant leaves at supraoptimal temperatures reveals a bottleneck of the prokaryotic methylerythritol 4-phosphate pathway of isoprenoid biosynthesis. Plant Cell Environ 32:82–92. doi: 10.1111/j.1365-3040.2008.01903.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhou K, Zou R, Stephanopoulos G, Too HP. 2012. Metabolite profiling identified methylerythritol cyclodiphosphate efflux as a limiting step in microbial isoprenoid production. PLoS One 7:e47513. doi: 10.1371/journal.pone.0047513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.