Abstract
Using populations of two sympatric Peromyscus species, we characterized the importance of the host species, physiology, environment, diet, and other factors in shaping the structure and dynamics of their gut microbiota. We performed a capture-mark-release experiment in which we obtained 16S rRNA gene sequence data from 49 animals at multiple time points. In addition, we performed 18S rRNA gene sequencing of the same samples to characterize the diet of each individual. Our analysis could not distinguish between the two species of Peromyscus on the basis of the structures of their microbiotas. However, we did observe a set of bacterial populations that were found in every animal. Most notable were abundant representatives of the genera Lactobacillus and Helicobacter. When we combined the 16S and 18S rRNA gene sequence analyses, we were unable to distinguish the communities on the basis of the animal's diet. Furthermore, there were no discernible differences in the structure of the gut communities based on the capture site or their developmental or physiological status. Finally, in contrast to humans, where each individual has a unique microbiota when sampled over years, among the animals captured in this study, the uniqueness of each microbiota was lost within a week of the original sampling. Wild populations provide an opportunity to study host-microbiota interactions as they originally evolved, and the ability to perform natural experiments will facilitate a greater understanding of the factors that shape the structure and function of the gut microbiota.
INTRODUCTION
The mechanisms that give rise to differences in the structure of communities associated with hosts are poorly understood. Among humans, it is widely accepted that the bacterial species composition within the intestines (i.e., the gut microbiota) is unique to each person and that the composition of the microbiota is the product of various factors, including the individual's genetics, diet, immune system, and behaviors (1). The amount that each of these contributes to shaping of the microbiota is unclear. Regardless, it is apparent that individuals within a family tend to harbor a more similar microbiota than others, leading to the suggestion that a person's microbiota is obtained horizontally as it is seeded by those in the environment (e.g., parents and siblings) and shaped via selection by a common culture, environment, and diet (2–5). This process of host selection and transmission has led to the hypothesis that the structure of the human gut microbiota and animal-associated communities, in general, is the result of coevolutionary processes (6).
Given that evolutionary divergence among hosts results in differences in physiology, diet, immunity, and behavior, it is reasonable to expect host-specific signatures within their microbiota. One approach has been the reciprocal transplants of gut contents between germfree animals (e.g., zebrafish to mice, mice to zebrafish) in which the altered niche space of the host environment results in a remodeling of the inoculated community (7–9). Another approach has been to characterize the gut microbiotas of diverse animals, largely sampled in zoos and reared on artificial diets, to show that these gut communities cluster according to broad dietary classifications (e.g., herbivore, carnivore, omnivore) (10–12). However, both approaches represent artifactual and extreme comparisons across the evolutionary history of animals. Others have performed natural experiments of sympatric and nonsympatric populations of insects (13, 14), primates (15–17), and bats (18). By studying animals within a common habitat, studies utilizing sympatric populations have shown that interindividual variations within a host species are greater than the variation between host species. That these studies frequently fail to find convincing concordance between host evolution and the similarity of their gut communities suggests that host genetics may have a limited role in the shaping of the microbiota.
A hallmark of the human gut microbiota is that the structure of an individual's gut microbiota represents a signature that is unique to that individual over periods of time spanning years (3, 19, 20). In essence, every person has a highly individualized gut microbiota. Again, it is unclear whether this is a product of the unique genetics of each individual or differences in diet, life history, or behaviors. In contrast to humans, chimpanzees do not appear to harbor individualized gut community structures (16). In a laboratory setting, while individual Mus musculus mice living with other mice do not harbor individualized gut community structures, littermates, cagemates, and mice from the same breeding colony or vendor still tend to have more similar microbiota than others (21–23). Such results again suggest that nongenetic factors such as diet and life history have a more significant impact on the shaping the structure of an individual's microbiota.
We sought to evaluate these observations by using sympatric populations of Peromyscus leucopus and P. maniculatus gracilis. These species are thought to have diverged from their common ancestor approximately 500 thousand years ago (24). Both species are widely distributed throughout North America, with P. leucopus being more commonly associated with deciduous forests and P. m. gracilis being more common in boreal forests (25). Northern Michigan is unique because it is part of a tension zone where there is a transition between boreal and deciduous habitats. As a result, P. leucopus and P. m. gracilis are sympatric in this environment, despite occupying apparently identical niches (26). This framework provided a unique opportunity to study the contributions of host phylogeny, environment, and other factors to the shaping of the mammalian gut microbiota. Using 16S and 18S rRNA gene sequencing, we were able to characterize the gut community structures of these animals and the composition of their diets. Furthermore, because the animals were captured live and released, we were able to characterize the change in the microbiota of the animals over the course of several months. Thus, it was possible to assess inter- and intraindividual variations in the context of interspecies differences while accounting for the diet, habitat, and physiological characteristics of the individuals.
MATERIALS AND METHODS
Sample collection.
The samples used for this study were collected over a 7-week period during the months of May, June, and July of 2010 in the Pigeon River State Forest in northern Michigan (latitude, 45.24; longitude, −84.48). The trapping area was a 400-by-400-m plot with 400 traps set 20 m apart in a 20-by-20 grid (Fig. 1). Small folding aluminum Sherman traps (2 by 2.5 by 6.5 in.; H. B. Sherman Traps, Inc., Tallahassee, FL) were baited with sunflower seeds each night and opened in the morning. For each catch, the mouse's weight, sex, reproductive condition, approximate age, and other notes were collected along with the trapping coordinates, a tissue sample for identification to the species level (25), and a fresh stool sample for microbiota and dietary analyses. This study was approved by the University Committee on Use and Care of Animals at the University of Michigan.
FIG 1.
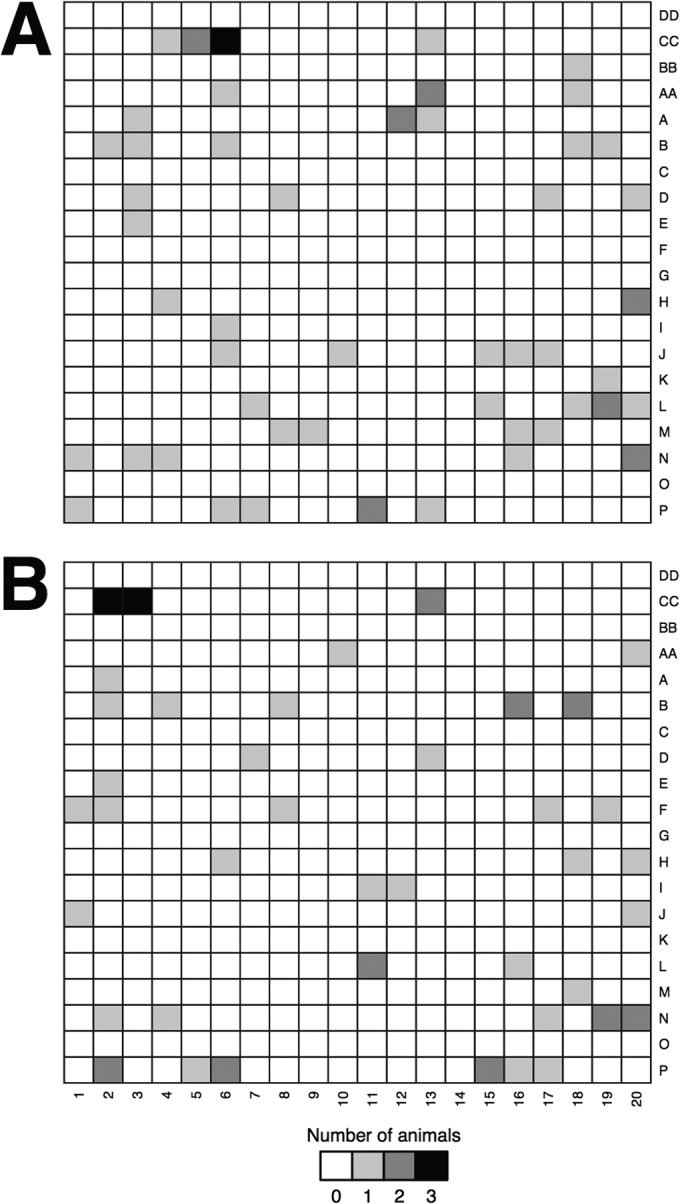
Spatial sampling of P. leucopus (A) and P. m. gracilis (B) indicates that their habitats overlap.
16S and 18S rRNA gene sequencing and curation.
Microbial genomic DNA was extracted from fecal samples with the PowerSoil-htp 96-well soil DNA isolation kit (MO BIO Laboratories) and an EpMotion 5075 automated pipetting system. The V4 regions of the 16S and 18S rRNA genes from each sample were amplified with custom barcoded primers and sequenced on an Illumina MiSeq sequencer (27). To sequence the 18S rRNA gene, we adapted the approach used to sequence the V4 region by using the TAReuk454FWD1 and TAReukREV3 primers, which target the V4 region of the 18S rRNA gene (28). The 16S and 18S rRNA gene sequences were processed separately with the mothur software package (29) as described previously (27). Briefly, paired-end reads were assembled into contigs and aligned with 16S and 18S reference sequences from the SILVA small-subunit rRNA sequence database (30). Aligned sequences were screened for chimeras with UCHIME (31). The actual commands used in mothur are available at http://www.mothur.org/wiki/MiSeq_SOP. To assess the error rate after curating the sequences, we included mock community samples in each sequencing run and processed these samples in parallel with the fecal samples. The observed error rate was 0.03%.
16S rRNA gene sequences were clustered into operational taxonomic units (OTUs) by using the average-neighbor algorithm with a 3% distance cutoff. In addition, the 16S rRNA gene sequences were classified by using a naive Bayesian classifier with a training set (version 9) made available through the Ribosomal Database Project (http://rdp.cme.msu.edu) (32). Taxonomic assignments were made on the basis of the lowest level that provided a confidence score of at least 80%. The taxonomic assignment of each OTU was made by using the majority consensus taxonomy of each sequence affiliated with the OTU. The taxonomic assignments were also used to bin each sequence into a phylotype. To minimize the detrimental effects of uneven sampling on community metrics, all samples were subsampled or rarefied to 4,465 per sample.
18S rRNA gene sequences were also classified by using the naive Bayesian classifier with a training set derived from the SILVA SSU reference taxonomy database (30). The reference taxonomy used to classify the 18S rRNA sequences was manually curated to include only eukaryotic taxa whose members would make up a meaningful portion of the host's diet. That is, only sequences mapping to metazoa, embryophyta, ascomycota, or basidiomycota were included. For practical reasons, we did not attempt to remove all of the microscopic members of these taxa; however, downstream analysis showed that microscopic members accounted for only a small portion of the total sequences. The remaining sequences, presumed to be the major constituents of the Peromyscus diet, were then assigned to phylotypes at the genus level. To minimize the detrimental effects of uneven sampling on community metrics, all samples were subsampled or rarefied to 500 sequences per sample. Because of the prevalence of host DNA in many samples, we were only able to characterize the animals' diet in 87 samples by using 18S rRNA gene sequences.
Microbial community and diet analysis.
We used several community analysis tools to help analyze the abundance distributions of the bacterial OTUs and phylotypes and the diet-related phylotypes. The number of observed OTUs and Shannon diversity were calculated by rarefaction with 1,000 randomizations. Comparisons of rank-transformed α-diversity metrics were performed by analysis of variance (ANOVA) accounting for the presence of repeated measures for some animals. To calculate distances between the structures of communities, we used the Yue and Clayton (33) distance metric (θYC), which takes into account the membership and abundance of each OTU, and the weighted UniFrac distance, which takes into account the phylogenetic relationships between the populations in the communities (34). These distances were calculated in mothur on the basis of an average of 100 randomizations. Where appropriate, results based on the θYC metric are denoted with a θ subscript and those based on the weighted UniFrac metric are denoted with a W subscript. Tests to determine whether the communities overlapped were performed by analysis of molecular variance (AMOVA) and analysis of homogeneity of variance, again accounting for repeated measures (35, 36). In addition, we used the Wilcoxon nonparametric test to compare the median distance of each sample with that of every other sample. Correlations between matrices were performed with the Mantel test by using 10,000 randomizations. After a single subsampling, samples were assigned to community types by using Dirichlet multinomial mixture (DMM) models based on their phylotype abundance (37); comparisons of DMM models generated after multiple subsamplings were not meaningfully different. To identify OTUs that were significantly different between samples, we applied two approaches to data derived from a single subsampling of the data. Again, repeated analyses with multiple subsamplings did not meaningfully affect the results. First, we used the random-forest algorithm as implemented in the randomForest R package with 10,000 trees (38). Second, we simplified the data set by selecting those OTUs that were identified in at least half of the samples and then used a rank-transformed ANOVA that corrected for repeated measures to identify OTUs that differed significantly between the two groups. In all of the analyses, P values were corrected for multiple comparisons by using the Benjamini-Hochberg correction with an experiment-wide significance threshold of 0.05 (39).
Nucleotide sequence accession numbers and source code availability.
The sequence data obtained in this study and the MIMARKS metadata are available through the NCBI Sequence Read Archive under accession number SRP044050 and BioProject accession number PRJNA254334. A literate programming document prepared with the knitr R package is available as a GitHub repository (https://github.com/SchlossLab/wild_mice). This document contains the source code that was used to generate results and the figures.
RESULTS
Field capture of Peromyscus spp.
Between 25 May and 14 July 2010, we used live traps to capture and release P. leucopus and P. m. gracilis from sites within a 400-by-400-m grid (at 20-m intervals) within Pigeon River State Forest in Cheboygan County, MI. Because animals were given ear tags at the initial capture event, we were able to document the animals that were captured multiple times. We obtained 58 fecal samples from 26 P. leucopus animals and 53 fecal samples from 23 P. m. gracilis animals. Among the samples from animals captured multiple times (n = 29 animals), the median distance between successive resampling events was 45 m (minimum = 0 m, maximum = 362 m). When we overlaid the species of Peromyscus on the sampling grid, we were unable to detect variation in the range of the mice, supporting the notion that the habitats of these mice overlapped (Fig. 1A and B).
Host species does not affect microbiota structure.
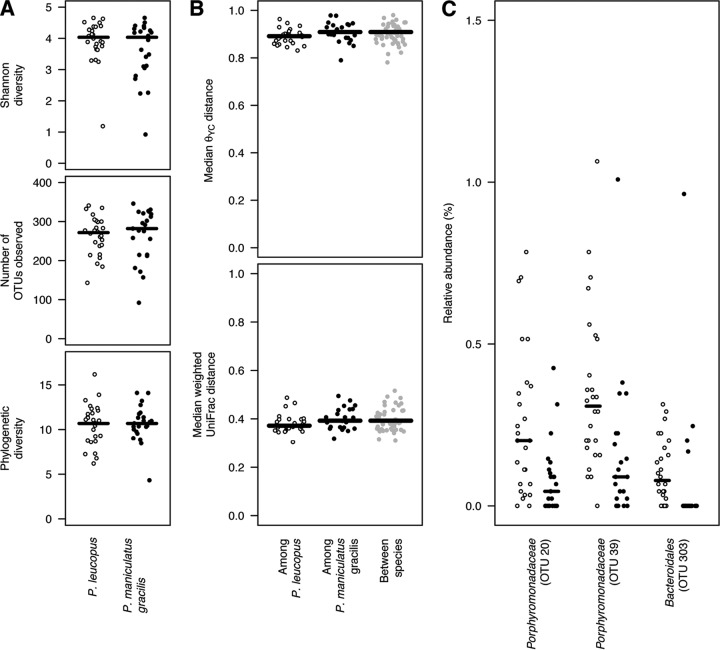
We compared the fecal communities of the two host species to test whether the genetic differences between the two animal species would translate into differences in their microbiotas. Using a variety of statistical tests, we were unable to detect a meaningful difference between the two species. First, there was no significant difference in the richness, Shannon diversity, or phylogenetic diversity between the two species of mice (Fig. 2A). Second, there was not a significant difference in the community structures of the characterized microbiotas of the two species when the θYC or weighted UniFrac community structure metric was used. Third, there was no significant difference in the variation of their community structure within the characterized microbiotas of the two species when either of the community structure metrics was used. Finally, when we attempted to fit the OTU abundance data to DMM models, there was no support for more than one community type. In each of these comparisons, the level of variation across mice within either species was considerable (Fig. 2B).
FIG 2.
The microbiotas of P. leucopus and P. m. gracilis could not be differentiated on the basis of 16S rRNA gene sequences. Open and closed circles represent the results for P. leucopus and P. m. gracilis, respectively. The alpha diversity of the two species was calculated on the basis of the number of observed OTUs, Shannon diversity, and phylogenetic diversity; the metrics did not differ significantly between the species of mice (A). The median θYC and weighted UniFrac distances among mice within the same species and across species were large, and the difference was not significant (B). The relative abundances of two OTUs were significantly different between the two species; however, the effect of the size and relative abundance of those OTUs was small (C).
We observed one OTU that was significantly different between the two species. This OTU was affiliated with the unclassified members of the family Porphyromonadaceae (Fig. 2C). Furthermore, among all of the OTUs, this one provided the greatest mean decrease in accuracy (MDA = 21.0) when we used the random-forest machine-learning algorithm to distinguish between the Peromyscus species. The two OTUs that resulted in the next greatest MDAs were associated with Prevotella and Clostridium (MDAs = 20.5 and 17.7, respectively). For the random-forest feature selection procedure, we observed an out-of-bag error rate of 33.3%, suggesting that it was not possible to use these OTUs to reliably and correctly classify the communities found in the two species of mice. Thus, there was a lack of clear support for the differentiation of specific members of the Peromyscus species' microbiotas that parallels that of the host species.
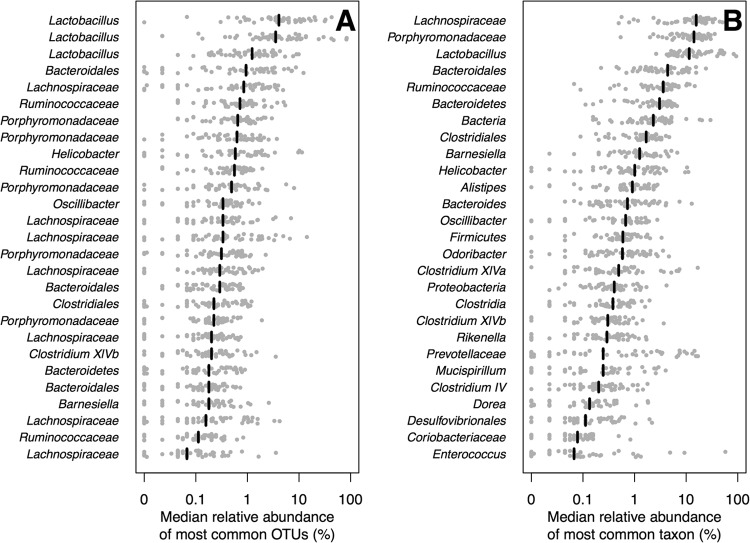
One striking feature of the Peromyscus sp. microbial communities was the large amount of variation in the composition of their gut microbiota. We observed a total of 3,021 OTUs among all of the mice; however, the median number of OTUs observed in each mouse was 277 (95% confidence interval = 112.5 to 369.75). Across all of the OTUs, 6 were present in every sample but 23 were observed in at least 90% of the mice (Fig. 3A). The latter set of OTUs were affiliated with 10 different taxa and represented 33.1% of the 16S rRNA gene sequences sampled from the mice (Fig. 3A). These OTUs did not overlap the one that was identified as being significantly different between the two species of mice. We next considered the taxonomic diversity of bacterial genera. Among the 358 genera that were observed, 22 were observed in at least 90% of the mice and 12 were observed in every mouse sampled (Fig. 3B). Those genera found in at least 90% of the mice represented 86.3% of the sequences sampled, and those found in every animal represented 78.1% of the sequences sampled. Within each animal, we identified the dominant taxa and observed that Lachnospiraceae was the dominant family in 34.7% of the mice, followed by Porphyromonadaceae (30.6%) and the genus Lactobacillus (24.5%). These results demonstrate that although there was considerable variation among the Peromyscus sp. animals that we sampled, there was a conserved taxonomic core.
FIG 3.
A core microbiota exists across mice of both Peromyscus species. The relative abundance of the OTUs (A) and genera and other taxa (B) that were found in at least 90% of the mice. The limit of detection was 0.02%.
Effects of host physiology on diversity and richness.
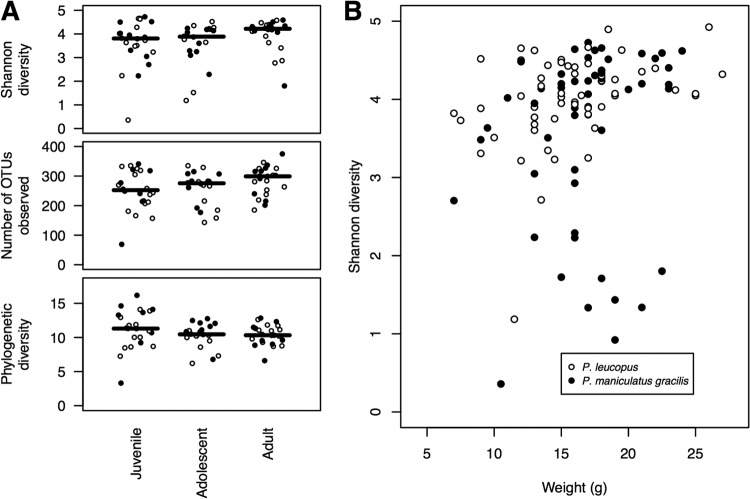
We next tested how differences in sex, reproductive condition, and age affected the gut microbiota of the Peromyscus sp. animals. We were unable to detect a statistically significant sex-based difference in the richness, Shannon diversity, phylogenetic diversity, or community structure of the Peromyscus sp. microbiota. To investigate possible sex-based differences further, we focused on sexually mature adult males and females, which had descended testes and emerged nipples, respectively; however, we were unable to identify a sex-based effect on the richness, Shannon diversity, phylogenetic diversity, or community structure. Furthermore, none of the OTUs that appeared in at least half of the samples were differentially enriched in either sex. Stratification of the mice into the age categories of juvenile (n = 30), adolescent (n = 30), and adult (n = 51) did reveal small but significant differences in the richness and diversity between the cohorts; the observed differences in their phylogenetic diversity was not statistically significant (Fig. 4A). The overall trend was for older mice to have a richer and more complex community. This was paralleled by a modest correlation between the animals' weight and their Shannon diversity (Spearman's ρ = 0.31; Fig. 4B). Although there did appear to be some effect of age on the richness and diversity of the communities, it was difficult to ascribe much biological significance to these differences, considering that the differences in overall community structure were not significantly different from each other and there were no OTUs that differed in abundance among the three age categories. Overall, by the parameters we were able to measure, we were unable to detect a robust effect of host physiology on their microbiota.
FIG 4.
Effects of age (A) and weight (B) on richness and diversity.
Location of sampling was not associated with variation in microbiota.
Next, we hypothesized that animals collected from the same environment would be more likely to have similar microbiotas than animals collected from different environments. For this analysis, we used the sampling coordinates to define the animals' environment (Fig. 1). When we performed a Mantel test to examine the association between the distance between the sampling points and the β diversity of the gut microbiotas, we failed to detect a significant association (Mantel correlation coefficient for θYC distances [Mθ] = −0.03; Mantel correlation coefficient for weighted UniFrac distances [MW] = ∼−0.03). To test for a more localized habitat effect, we compared the distances collected at the same site to those collected at different sites and again failed to detect a statistically significant effect. These results suggest the lack of a geographic or habitat-based effect on the Peromyscus sp. microbiota.
Diet was not associated with variation in microbiota.
To test the role of diet in the shaping of the Peromyscus species gut microbiota, we characterized the dietary contents of each P. leucopus and P. m. gracilis sample by 18S rRNA sequencing. The diets of P. leucopus and P. m. gracilis consisted of plant material (e.g., seeds, nuts, fruits, and green vegetation), arthropods, and fungi, which was consistent with previous results (26). We removed sequences outside these broad taxonomic groups to minimize the contributions of microeukaryotic members of the gut microbiota. First, we tested for a difference in diet composition based on the two species of Peromyscus. We failed to find a significant difference between the structures of the diets when using AMOVA with correction for repeated measures. Second, using the Mantel test, we failed to identify a significant association between the distances between samples based on their bacterial community structure and the distances based on the structure of their diet (Mθ = 0.07, MW = 0.11). Both analyses suggest the lack of an association between the structures of the diet and microbiota.
To further characterize the relationship between diet and the microbiota, we attempted to correlate the relative abundance of OTUs that were found in more than half of the samples with the relative abundance of dietary components that appeared in more than half of the samples. We were unable to detect any statistically significant Spearman correlation coefficients. Next, we used DMM models to identify clusters within the diet data. This approach identified two clusters, a cluster of samples dominated by plant material (n = 51) and a cluster of samples with a more diverse mixture of plants, arthropods, and fungi (n = 36). When we used these clusters to test for an association within the community structure distance matrix between the microbiotas of the mice, we did not observe a significant effect of diet on community structure. We then used the random-forest algorithm to identify features in the microbiota that could distinguish the two dietary groups; however, the out-of-bag error rate was 37.9%, which indicates that it was unable to correctly classify the samples. As these analyses demonstrate, we were unable to find associations between specific members of the microbiota and the dietary contents of each sample. This confirms our observation that the weak association between the distances between the microbiota and diet structures was unlikely to be biologically relevant.
A transient microbiota.
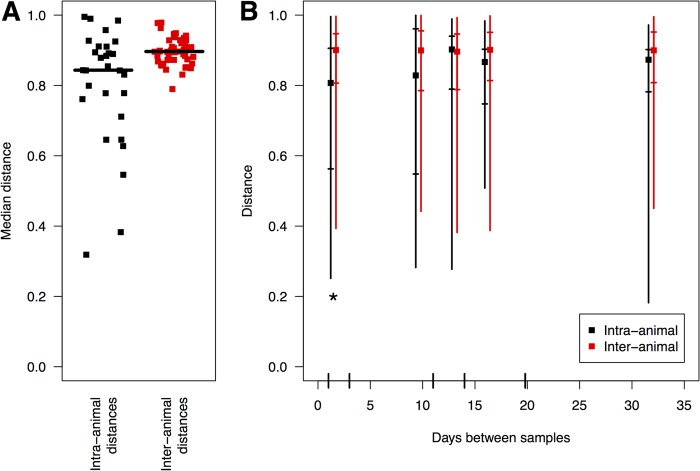
A striking characteristic of the microbiotas characterized in this study was the high intra- and interindividual variation. Studies of other animals have observed less variation in microbiota structure within an individual over time than between individuals. We sought to determine whether nondomesticated animals also harbor personalized microbiotas. To investigate this in Peromyscus spp., we took advantage of the ability to catch and release the same animal multiple times. By the OTU-based approach, the median intraindividual distance was significantly less than the median interindividual distance (Fig. 5A); however, by the weighted UniFrac-based approach, the difference was not significant. To investigate this further, we calculated the number of days that elapsed between recapturing events and created five time windows such that each window had approximately the same number of mice that were recaptured within that time period (i.e., 1 to 3 [n = 30], 4 to 11 [n = 22], 12 to 14 [n = 24], 15 to 19 [n = 21], and ≥20 [n = 24] days apart). We then compared the intra-animal distances within these time periods to the interanimal distances of different mice captured within the same intervals. Surprisingly, only the mice captured between 1 and 3 days apart were more similar to themselves than to other mice (Fig. 5B). In none of the other time windows was there a significant difference between the samples collected from the same animal and the samples collected from different animals. These results suggest that any individualization of an animal's microbiota is quickly lost.
FIG 5.
The individualized structure of the Peromyscus microbiota is transient. The median intra-animal distances between samples were significantly lower than the median interanimal distances (A). To compare the change in distance with time (B), time windows were selected so that there were approximately the same number of distances between the same animals per window. The position along the x axis was determined by the mean number of days between samples for the distances within the window and was jittered so the points for the two comparisons did not overlap; the vertical bars on the x axis indicate the boundaries of each window. The squares represent the median distance, and the lengths of the error bars reflect the 95% confidence intervals, with short lines representing the interquartile range. The asterisk indicates that intraindividual distances were significantly smaller than interindividual differences during the first 3 days of sampling.
DISCUSSION
An ongoing question within the field of microbiota research is the degree to which host species, physiology, diet, and environment contribute to shape the structure and function of the microbiota. Laboratory- and zoo-based studies are able to control for these factors; however, they do so in an artificial manner by studying the animals in artificial environments with an unnatural diet. The result is a tightly controlled experimental system that does not allow one to actually study the structure of the microbiota as it likely coevolved with its host. Here we were able to measure the effects of these sources of variation on the microbiota in the animals' actual environment by performing a natural experiment. We characterized the microbiota structures of two species of Peromyscus that had overlapping habitats and similar diets, and we were unable to identify biologically meaningful effects of any of these factors. These results support a previous field-based study of Peromyscus spp. that could not identify a significant effect of environmental contamination or habitat type on cecal diversity (40). They also support recent studies of wild European M. musculus populations that observed that large geographical distances were the most significant factor in explaining microbiota variation and that host genetics and diet had relatively minor effects on variation (41, 42). Other natural experiments investigating the structure of the microbiota in insects (13, 14), primates (15–17), and bats (18) also support our results.
The inability to differentiate between the two species of Peromyscus suggests that the genetic differences that have accumulated in the 500 thousand years since these species differentiated have not been sufficient to differentiate their microbiotas beyond other sources of variation. These results support previous studies of closely related animal species, including primates, bats, and insects, where it has not been possible to associate host evolutionary history with their microbiota (14–18). It remains to be determined whether there is a minimum evolutionary time period required to differentiate the microbiotas of two species. We hypothesize that insufficient mutations have accumulated since host speciation to impact the boundaries of possible gut community structures for these species (43, 44). For example, fixation of mutations affecting the immune system, diet preferences, digestion, or preferred habitat would be likely to differentiate the composition of these communities.
Previous studies have argued that eating a similar diet can streamline the composition of the microbiota (10, 12). In this study, we observed two types of diets in the mice—one dominated by plant material and one that was a mixture of plants, arthropods, and fungi. That we were unable to identify a Peromyscus sp. preference or distinguish between the microbiotas of animals consuming the two diet types indicates that the two species shared a niche and that their diets did not have a major impact on the variation in the observed community structures. This result is not unlike what is observed in humans, where there are immediate shifts in the gut microbiota with an abrupt change in diet; however, the microbiotas of different people do not converge to a common structure (45).
One distinct difference between the microbiotas of humans and the animals we characterized is the lack of individualized microbiotas. Similar results have been observed in wild populations of primates (16). Although the Peromyscus mice we sampled did show signs of individualization of the samples collected within a week of each other, this signal was lost over longer periods of time. The contrast between the individuality of human microbiotas and the lack of individuality in wild populations suggests that there are behavioral or cultural factors (e.g., hygiene, segregated families) that drive individualization. A better understanding of the processes that maintain or reduce individualization will help in designing microbiota-focused therapies for humans.
A product of the individuality observed in humans is the lack of any OTUs that are conserved across all people (3). In spite of the significant variation among animals in this study, there was strong evidence of a core set of taxa that were observed across all of the animals in this study (Fig. 3). At the genus and OTU levels, there were many taxa present that are commonly found in laboratory populations of M. musculus. Most striking, however, was the presence of multiple different OTUs affiliated with Lactobacillus in every animal we sampled. Furthermore, these populations were among the most abundant populations in the Peromyscus gut microbiota (median = 11.2%, range = 3.0 to 93.0%). Although Lactobacillus populations are commonly observed in M. musculus, they tend to be rare (e.g., <3%) (23). It was also interesting that nearly all of the animals we captured harbored OTUs affiliated with Helicobacter, which is commonly associated with colitis in immunodeficient mice (46). Our result confirms the assertion that Helicobacter bacteria are normal symbionts of rodents that can be commensals or pathogens in different contexts. Although Peromyscus and Mus are distantly related, these stark differences in gut community composition suggest the need for further exploration of the natural Mus microbiota and immune system (47).
Natural experiments with wild animals provide a unique opportunity to study host-microbiota relationships that are not affected by artifacts introduced into domesticated animals. Our ability to leverage a capture-mark-release design with sympatric populations afforded us the ability to study animals living in the same natural habitat that were consuming similar diets. Exploration of the microbiota of natural animal populations will help us to better understand the factors that define the human microbiota and, more importantly, what makes the microbiota of each individual so unique.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes for Health to P.D.S. (R01GM099514, R01HG005975, and P30DK034933) and a graduate research fellowship from the National Science Foundation to N.T.B.
REFERENCES
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schloss PD, Iverson KD, Petrosino JF, Schloss SJ. 2014. The dynamics of a family's gut microbiota reveal variations on a theme. Microbiome 2:25 doi: 10.1186/2049-2618-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108(Suppl 1):S4578–S4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg E, Sharon G, Zilber-Rosenberg I. 2009. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol 11:2959–2962. doi: 10.1111/j.1462-2920.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 7.Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpert C, Sczesny S, Gruhl B, Blaut M. 2008. Long-term stability of the human gut microbiota in two different rat strains. Curr Issues Mol Biol 10:17–24. [PubMed] [Google Scholar]
- 10.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher J S, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders JG, Powell S, Kronauer DJ, Vasconcelos HL, Frederickson ME, Pierce NE. 2014. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol 23:1268–1283. doi: 10.1111/mec.12611. [DOI] [PubMed] [Google Scholar]
- 14.Wong AC, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J 7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moeller AH, Peeters M, Ndjango JB, Li Y, Hahn BH, Ochman H. 2013. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res 23:1715–1720. doi: 10.1101/gr.154773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, Ochman H. 2012. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci U S A 109:13034–13039. doi: 10.1073/pnas.1110994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P. 2010. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips CD, Phelan G, Dowd SE, McDonough MM, Ferguson AW, Hanson JD, Siles L, Ordonez-Garza N, San Francisco M, Baker RJ. 2012. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol Ecol 21:2617–2627. doi: 10.1111/j.1365-294X.2012.05568.x. [DOI] [PubMed] [Google Scholar]
- 19.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov II, de Llanos Frutos R, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Schubert AM, Zackular JP, Iverson KD, Young VB, Petrosino JF. 2012. Stabilization of the murine gut microbiome following weaning. Gut Microbes 3:383–393. doi: 10.4161/gmic.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibbard CW. 1968. Paleontology, p 6-24. In King JA. (ed), Biology of Peromyscus (Rodentia). American Society of Mammalogists, Stillwater, OK. doi: 10.5962/bhl.title.39510. [DOI] [Google Scholar]
- 25.Myers P, Lundrigan BL, Hoffman SMG, Haraminac AP, Seto SH. 2009. Climate-induced changes in the small mammal communities of the Northern Great Lakes Region. Glob Change Biol 15:1434–1454. doi: 10.1111/j.1365-2486.2009.01846.x. [DOI] [Google Scholar]
- 26.Wolff JO, Dueser RD, Berry KS. 1985. Food-habits of sympatric Peromyscus leucopus and Peromyscus maniculatus. J Mammal 66:795–798. doi: 10.2307/1380812. [DOI] [Google Scholar]
- 27.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, Richards TA. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19(Suppl 1):S21–S31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 29.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Commun Stat Theory Methods 34:2123–2131. doi: 10.1080/STA-200066418. [DOI] [Google Scholar]
- 34.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 36.Schloss PD. 2008. Evaluating different approaches that test whether microbial communities have the same structure. ISME J 2:265–275. doi: 10.1038/ismej.2008.5. [DOI] [PubMed] [Google Scholar]
- 37.Holmes I, Harris K, Quince C. 2012. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breiman L. 2001. Random forests. Mach Learn Int Conf Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 40.Coolon JD, Jones KL, Narayanan S, Wisely SM. 2010. Microbial ecological response of the intestinal flora of Peromyscus maniculatus and P. leucopus to heavy metal contamination. Mol Ecol 19(Suppl 1):S67–S80. doi: 10.1111/j.1365-294X.2009.04485.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Linnenbrink M, Kunzel S, Fernandes R, Nadeau MJ, Rosenstiel P, Baines JF. 2014. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci U S A 111:E2703–10. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linnenbrink M, Wang J, Hardouin EA, Kunzel S, Metzler D, Baines JF. 2013. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol 22:1904–1916. doi: 10.1111/mec.12206. [DOI] [PubMed] [Google Scholar]
- 43.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A 107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moeller AH, Ochman H. 2013. Factors that drive variation among gut microbial communities. Gut Microbes 4:403–408. doi: 10.4161/gmic.26039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. 1998. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun 66:5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung H, Pamp SJ, Hill J A, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]