Abstract
The objective of the study described in this article was, first, to investigate the effect of the simultaneous application of near-infrared (NIR) heating and UV irradiation on inactivation of Escherichia coli O157:H7, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes in ready-to-eat (RTE) sliced ham and as well as its effect on product quality and, second, to elucidate the underlying mechanisms of the synergistic bactericidal action of NIR heating and UV irradiation. With the inoculation amounts used, simultaneous NIR-UV combined treatment for 70 s achieved 3.62, 4.17, and 3.43 log CFU reductions of E. coli O157:H7, S. Typhimurium, and L. monocytogenes, respectively. For all three pathogens, the simultaneous application of both technologies resulted in an additional log unit reduction as a result of their synergism compared to the sum of the reductions obtained after the individual treatments. To investigate the mechanisms of NIR-UV synergistic injury for a particular microorganism in a food base, we evaluated the effect of four types of metabolic inhibitors using the overlay method and confirmed that damage to cellular membranes and the inability of cells to repair these structures due to ribosomal damage were the primary factors related to the synergistic lethal effect. Additionally, NIR-UV combined treatment for a maximum of 70 s did not alter the color values or texture parameters of ham slices significantly (P > 0.05). These results suggest that a NIR-UV combined process could be an innovative antimicrobial intervention for RTE meat products.
INTRODUCTION
Ready-to-eat (RTE) meat products, especially precooked sliced ham, are widely sold in delicatessens and consumed in homes due to their convenience. However, deli meats have been identified to be high-risk products, as they are highly perishable and easily contaminated (1). Escherichia coli O157:H7, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes are the major causes of outbreaks and product recalls associated with contaminated delicatessen meats (2–5). Recent outbreaks of E. coli O157:H7 and S. Typhimurium infections in the United States resulted in a total of 311 cases, which were traced to precooked salami (3). L. monocytogenes is the most relevant pathogen in RTE meat products (6). A significant trend toward an increase in the incidence of listeriosis has been observed in many European countries, with deli meat products having the highest frequency of positive samples (7). L. monocytogenes has been listed in the top 5 highest-ranking pathogens regarding the total cost of foodborne illness in the United States (8), and deli meats have been reported to be the leading vehicle of foodborne listeriosis (6).
In many recent studies, cross contamination during slicing has been suspected to be the mode of transmission of these three pathogens (3, 9–11). In other words, the most important route of sliced deli meat contamination is probably via contact with surfaces. Therefore, an additional superficial decontamination step may become necessary to control pathogenic bacteria on sliced ham products immediately before final packaging or after the products are unwrapped at the delicatessen or other retail outlet.
Infrared (IR) radiation transfers thermal energy in the form of an electromagnetic wave and can be classified into 3 regions, near IR (NIR; 0.76 to 2 μm), medium IR (MIR; 2 to 4 μm), and far IR (FIR; 4 to 1,000 μm). IR radiant heating provides significant advantages over convection and conduction heating, including a higher heat transfer capacity and high energy efficiency, as it heats the product directly without being influenced by the air surrounding the food (12). Several studies have employed infrared heating for surface pasteurization of precooked deli meats (13–15). In our previous study, NIR radiation was investigated as a heat source and showed improved heating and pasteurization efficacy on RTE sliced ham compared to the heating and pasteurization efficacy of conventional convective heating (15).
UV radiation covers part of the electromagnetic spectrum from 100 to 400 nm and is distinguished as UV-A (320 to 400 nm), UV-B (280 to 320 nm), and UV-C (200 to 280 nm) (16). Among them, surface disinfection by 253.7-nm UV irradiation (UV-C) has been widely utilized as an antimicrobial treatment of food surfaces. However, industrial application is still limited because of the low penetration capacity of UV photons into foods. For this reason, UV-C radiation has been used in combination with other antimicrobial processing techniques to produce a hurdle effect on pathogen contaminants (17–19). In our previous studies, the simultaneous use of a combination of NIR radiant heating with UV-C irradiation was found to be markedly effective for reducing E. coli O157:H7, S. Typhimurium, and Cronobacter sakazakii in powdered food systems due to their synergistic effects (20, 21). Thus, the combination of NIR heating with UV-C irradiation could be especially efficient in decontaminating the surfaces of sliced deli meats. In addition, despite being a topic of great interest and importance, the mechanism of bactericidal improvement by simultaneous NIR-UV treatment is not well-known.
The aims of the present study were, first, to investigate the efficacy of the simultaneous use of the combination of NIR heating and UV-C irradiation for inactivating foodborne pathogens, including E. coli O157:H7, S. Typhimurium, and L. monocytogenes, on RTE sliced ham and to determine the effect of the combination treatment on factors related to the quality of the sliced ham product. Second, we tried to elucidate the mechanism of synergistic bacterial inactivation by the simultaneous application of NIR and UV-C radiation.
MATERIALS AND METHODS
Bacterial strains.
Three strains each of E. coli O157:H7 (ATCC 35150, ATCC 43889, and ATCC 43890), S. Typhimurium (ATCC 19585, ATCC 43971, and DT 104), and L. monocytogenes (ATCC 15313, ATCC 19111, and ATCC 19115), obtained from the bacterial culture collection of Seoul National University (Seoul, South Korea), were used in this experiment. Stock cultures were kept frozen at −80°C in 0.7 ml of tryptic soy broth (TSB; Difco, Becton, Dickinson, Sparks, MD, USA) and 0.3 ml of 50% glycerol. Working cultures were streaked onto tryptic soy agar (TSA; Difco), incubated at 37°C for 24 h, and stored at 4°C.
Preparation of pathogen inocula.
All strains of E. coli O157:H7, S. Typhimurium, and L. monocytogenes were cultured individually in 5 ml of TSB at 37°C for 24 h, followed by centrifugation (4,000 × g for 20 min at 4°C) and washing three times with buffered peptone water (BPW; Difco). The final pellets were resuspended in BPW, corresponding to ca. 107 to 108 CFU/ml. Subsequently, suspended pellets of each strain of the three pathogenic species (nine strains total) were combined to construct mixed culture cocktails. These cocktails were used in this inactivation study at a final concentration of approximately 108 CFU/ml. To analyze the mechanism of inactivation, S. Typhimurium strain DT 104 was selected as a model microorganism, and the cell suspension was grown and prepared in the same way.
Sample preparation and inoculation.
Precooked, vacuum-packaged sliced ham (approximately 90 by 90 by 2 mm) was purchased from a local grocery store (Seoul, South Korea), maintained in a refrigerator (4°C), and used within 2 days. For surface inoculation, 8 ml of the prepared mixed culture cocktail was diluted in 0.8 liter of sterile 0.2% peptone water. Each ham slice was immersed in the mixed pathogen suspension for 3 min at room temperature (22 ± 2°C), drained on a sterilized rack, and dried for 20 min inside a biosafety hood with the fan running. Two ham slices (ca. 25 g; inoculum level, 105 to 106 CFU per sample) were used in each experimental trial.
Near-infrared heating and UV-C irradiation.
A model aluminum chamber (41 by 34 by 29 cm) was used in this study for NIR heating (NIR), UV-C irradiation (UV), and NIR-UV combined treatment (Fig. 1). A quartz halogen infrared heating lamp (350 mm; NS-104; NSTECH, South Korea) with a maximum power of 500 W (radiation intensity at the sample location, 200.36 μW/cm2/nm) at a 230-V input was used as a NIR-emitting source. The maximum wavelength (λm) generated from the infrared heater used in this study was about 1,300 nm, which is within the NIR wave range. A UV germicidal lamp (357 mm; G10T5/4P; Sankyo, Japan) with a nominal output power of 16 W (radiation intensity at the sample location, 1.85 mW/cm2) was used as a UV-C-emitting source. The radiation intensities generated from the NIR and UV lamps were measured and recorded by a NIR fiber optic spectrometer (AvaSpec-NIR256-1.7; Avantes, Eerbeek, Netherlands) and a UV fiber optic spectrometer (AvaSpec-ULS2048; Avantes). Two NIR emitters and two UV-C emitters were arranged horizontally in parallel with the four emitting surfaces facing each other, and four aluminum reflectors were installed behind the emitters to focus as much of the radiation as possible uniformly onto the process line and enhance the efficiency of NIR and UV irradiation (Fig. 1). The total power consumption of the four emitters was approximately 0.9 kW, as measured by a digital power meter (WT-230; Yokogawa, Japan) at the standard voltage (220 V). The vertical distance between the emitters and the sample was 13.5 cm (5.3 in.) on each side. For the subsequent pasteurization experiments (NIR radiant heating, UV-C irradiation, and the simultaneous application of both technologies), the surface-inoculated ham slices were placed side-by-side in the center of a sterilized stainless rack with the long axis parallel to the NIR and UV lamps.
FIG 1.
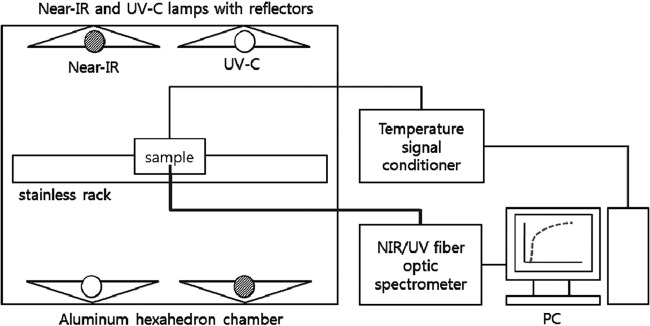
Schematic diagram of the NIR-UV combined treatment system used in this study. Adapted from reference 15.
Temperature measurement.
A fiber optic temperature sensor (FOT-L; FISO Technologies Inc., Quebec, Canada) connected to a signal conditioner (TMI-4; FISO Technologies Inc.) was used to measure the temperatures in the samples in real time during NIR heating and simultaneous NIR-UV treatment. The sensor was placed directly on the surface of the treated ham slices, and the temperature was manually recorded every 5 s. The fiber optic sensors were coated with electrical insulating material. All experiments were replicated three times, and means and standard deviations of sample temperatures for the NIR and NIR-UV combined treatment were compared to determine the rate of heating of the samples.
Bacterial enumeration.
At selected time intervals, each of two treated ham slices (ca. 25 g) was removed, immediately transferred into a sterile stomacher bag (Labplas Inc., Sainte-Julie, Quebec, Canada) containing 225 ml of BPW (detection limit, 10 CFU/g), and homogenized for 2 min with a stomacher (Easy mix; AES Chemunex, Rennes, France). After homogenization, 1-ml aliquots of the sample were 10-fold serially diluted in 9 ml of BPW, and 0.1 ml of sample or diluent was spread plated onto each selective medium. Sorbitol MacConkey agar (SMAC; Difco), xylose lysine desoxycholate agar (XLD; Difco), and Oxford agar base (OAB) with Bacto Oxford antimicrobial supplement (MOX; Difco) were used as selective media for the enumeration of E. coli O157:H7, S. Typhimurium, and L. monocytogenes, respectively. Where low numbers of surviving cells were anticipated, 1 ml of undiluted sample was equally distributed onto four plates to lower the detection limit. All agar media were incubated at 37°C for 24 to 48 h before colonies were counted. To confirm the identity of the pathogens, random colonies were selected from the enumeration plates and subjected to biochemical and serological tests. These tests consisted of the E. coli O157:H7 latex agglutination assay (RIM; Remel, Lenexa, KS), the Salmonella latex agglutination assay (Oxoid, Ogdensburg, NY), and the API Listeria test (bioMérieux, Hazelwood, MO).
Enumeration of injured cells.
The overlay (OV) method was used to enumerate injured cells of S. Typhimurium and L. monocytogenes (22). TSA was used as a nonselective medium to repair injured cells. One hundred microliters of the appropriate dilutions was spread plated onto TSA medium, and the plates were incubated at 37°C for 2 h to allow injured cells to resuscitate (23). The plates were then overlaid with 7 to 8 ml of selective medium (XLD or OAB). After solidification, the plates were further incubated for an additional 22 to 46 h at 37°C. Following incubation, typical black colonies were counted. In the case of E. coli O157:H7, it is not appropriate to overlay with SMAC medium. Instead, phenol red agar base with 1% sorbitol (SPRAB; Difco) was used to enumerate injured cells of this pathogen (24). After incubation at 37°C for 24 h, presumptive colonies of E. coli O157:H7 with typical white colonies were enumerated. Isolates randomly selected from SPRAB plates were subjected to serological confirmation as E. coli O157:H7 (E. coli O157:H7 latex agglutination assay; RIM; Remel, Lenexa, KS), because SPRAB is not typically used as a selective agar for enumerating E. coli O157:H7.
Investigation of the bactericidal mechanism.
To investigate sites of cellular damage in S. Typhimurium DT 104 (the strain selected as a model microorganism) caused by NIR, UV, and simultaneous NIR-UV treatments, four kinds of metabolic inhibitors, chloramphenicol (28.0 μg/ml; Sigma-Aldrich, St. Louis, MO), nalidixic acid (1.2 μg/ml; Sigma-Aldrich), penicillin G (350.0 μg/ml; Sigma-Aldrich), and rifampin (5.0 μg/ml; Sigma-Aldrich), were utilized. The concentrations of the metabolic inhibitors applied were chosen after preliminary experiments were performed, and the synthesis inhibition targets of these antibiotics are presented in Table 1.
TABLE 1.
Concentrations of metabolic inhibitors incorporated into TSA medium and targets of synthesis inhibition
| Metabolic inhibitor | Concna (μg/ml) | Target of synthesis inhibition |
|---|---|---|
| Chloramphenicol | 28.0 | Protein (ribosome) |
| Nalidixic acid | 1.2 | DNA |
| Penicillin G | 350.0 | Cell wall |
| Rifampin | 5.0 | RNA (RNA polymerase) |
The maximum concentration at which selective antibiotics had no effect on colony formation of intact Salmonella Typhimurium DT 104 cells.
Two ham slices (ca. 25 g) inoculated with S. Typhimurium DT 104 were treated with NIR, UV, and NIR-UV for 70 s and homogenized in BPW as described above. One hundred microliters of the appropriate dilutions was spread plated onto TSA medium (with or without metabolic inhibitors incorporated at the concentrations listed above), and the plates were incubated at 37°C for 2 h to allow injured cells to resuscitate. The plates were then overlaid with 7 to 8 ml of selective medium (XLD). After solidification, the plates were further incubated for an additional 22 h at 37°C. Following incubation, typical black colonies were enumerated. The quantitative levels of recovery inhibition were calculated by subtracting the population numbers on medium containing each antibiotic from the population numbers obtained on nonselective controls that did not contain a selective reagent in the TSA medium and were thus used to probe the site of injury corresponding to the particular metabolic inhibitor.
Color and texture measurement.
In order to determine the effect of NIR-UV treatment on the color of the ham slices, a Minolta colorimeter (model CR400; Minolta Co., Osaka, Japan) was used to measure changes to the color of the treated samples. The color attributes were quantified from the values of L*, a*, and b*, which indicate the color lightness, redness, and yellowness of the sample, respectively, and which were measured at random locations on the ham slices. All measurements were taken in triplicate.
Changes in the texture of the NIR-UV-treated ham slices were evaluated with a Brookfield texture analyzer (CT3-10k; Brookfield Engineering Laboratories, Inc., MA, USA) with a blade set probe (TA7; knife edge, 60 mm wide). After the treated samples were cooled, four stacked slices (45 by 90 mm) were placed onto the press holder, and a blade was moved down at 2 mm/s. The maximum force required to cut the sample was recorded using TexturePro CT software (version 1.2; Brookfield Engineering Laboratories, Inc.). The peak force required to shear the samples was utilized as an indicator of hardness. All experiments were replicated three times.
Statistical analysis.
All experiments were repeated three times with duplicate samples. Triplicate data were analyzed by analysis of variance (ANOVA) and Duncan's multiple-range test of a statistical analysis system (SAS Institute, Cary, NC, USA). A P value of <0.05 was used to indicate significant differences.
RESULTS
Inactivation of pathogenic cells by simultaneous NIR-UV treatment.
Reductions in the viable counts of E. coli O157:H7, S. Typhimurium, and L. monocytogenes cells on ham slice surfaces during NIR radiant heating (NIR), UV-C irradiation (UV), and the simultaneous application of both technologies (NIR-UV) are summarized in Tables 2, 3, and 4, respectively. Significant (P < 0.05) log reductions of the three pathogens were observed after 10 s of UV irradiation alone and the NIR-UV combined treatment, whereas for NIR radiant heating, the time to initiation of a significant (P < 0.05) reduction was delayed about 30 to 40 s (Tables 2 to 4). In addition, the UV sensitivities (irradiation dose based) of the treated pathogens were consistent with those presented in another report describing S. Typhimurium and L. monocytogenes inactivation by UV-C light in RTE sliced ham (25). The simultaneous NIR-UV combined treatment for 70 s led to mean reductions of 3.62, 4.17, and 3.43 log CFU/g in E. coli O157:H7, S. Typhimurium, and L. monocytogenes, respectively. For all three pathogens, the sums of the reductions for NIR and UV inactivation were lower than the values reached by the simultaneous application of both technologies. In other words, the existence of a synergistic effect against all pathogens was confirmed for all treatment times. However, statistically significant (P < 0.05) differences between the sums of NIR and UV inactivation and the inactivation obtained with the combination treatment were observed for the three pathogens only after treatment times of 70 s (Tables 2 to 4). The reductions in the E. coli O157:H7, S. Typhimurium, and L. monocytogenes counts resulting from the synergistic effect occurring after 70 s of treatment, calculated by subtracting the sums of the NIR and UV reductions from those achieved during simultaneous NIR-UV treatment, were 0.64, 0.76, and 0.43 log units, respectively.
TABLE 2.
Reductions in numbers of viable Escherichia coli O157:H7 cells on ham slice surfaces treated with UV, NIR, and NIR-UV
| Treatment time (s) | Log reduction (log10
n0 − log10
n) by treatment type and selective mediuma |
|||
|---|---|---|---|---|
| UV (SMAC) | NIR (SMAC) | NIR-UV |
||
| SMAC | SPRAB | |||
| 0 | 0.00 ± 0.00A | 0.00 ± 0.00A | 0.00 ± 0.00Aa | 0.00 ± 0.00Aa |
| 10 | 1.22 ± 0.27B | 0.06 ± 0.07A | 1.57 ± 0.33Ba | 1.49 ± 0.01Ba |
| 20 | 1.24 ± 0.26B | 0.11 ± 0.07A | 1.88 ± 0.24BCa | 1.54 ± 0.02Ba |
| 30 | 1.29 ± 0.22B | 0.21 ± 0.11A | 2.00 ± 0.38BCDa | 1.62 ± 0.08BCa |
| 40 | 1.43 ± 0.23B | 0.25 ± 0.11A | 2.14 ± 0.30CDa | 1.81 ± 0.34BCa |
| 50 | 1.46 ± 0.21B | 0.69 ± 0.27B | 2.46 ± 0.19DEa | 1.96 ± 0.25Ca |
| 60 | 1.50 ± 0.24B | 1.05 ± 0.23C | 2.68 ± 0.16Ea | 2.46 ± 0.30Da |
| 70b | 1.52 ± 0.15B | 1.46 ± 0.09D | 3.62 ± 0.26Fa | 3.23 ± 0.23Ea |
The values are means ± standard deviations from three replications. Means with the same uppercase letter in the same column are not significantly different (P > 0.05). Within the NIR-UV columns, values in the same row followed by the same lowercase letter are not significantly different (P > 0.05). SMAC, sorbitol MacConkey agar; SPRAB, phenol red agar base with 1% sorbitol; n0, initial population; n, population after treatment.
A statistically significant (P < 0.05) difference between the sum of NIR and UV inactivation and inactivation achieved with combination treatment was observed.
TABLE 3.
Reductions in numbers of viable Salmonella Typhimurium cells on ham slice surfaces treated with UV, NIR, and NIR-UV
| Treatment time (s) | Log reduction (log10
n0 − log10
n) by treatment type and selective mediuma |
|||
|---|---|---|---|---|
| UV (XLD) | NIR (XLD) | NIR-UV |
||
| XLD | OV-XLD | |||
| 0 | 0.00 ± 0.00A | 0.00 ± 0.00A | 0.00 ± 0.00Aa | 0.00 ± 0.00Aa |
| 10 | 1.16 ± 0.17B | 0.03 ± 0.03A | 1.55 ± 0.23Ba | 1.38 ± 0.16Ba |
| 20 | 1.22 ± 0.16B | 0.06 ± 0.00A | 1.68 ± 0.37Ba | 1.46 ± 0.19Ba |
| 30 | 1.48 ± 0.22C | 0.06 ± 0.04A | 1.71 ± 0.34Ba | 1.55 ± 0.23Ba |
| 40 | 1.56 ± 0.14C | 0.11 ± 0.12A | 1.96 ± 0.35BCa | 1.83 ± 0.39BCa |
| 50 | 1.57 ± 0.15C | 0.43 ± 0.30B | 2.31 ± 0.30Ca | 2.25 ± 0.24Ca |
| 60 | 1.63 ± 0.12C | 1.19 ± 0.20C | 3.05 ± 0.32Da | 2.78 ± 0.27Da |
| 70b | 1.73 ± 0.03C | 1.68 ± 0.03D | 4.17 ± 0.11Ea | 3.66 ± 0.34Ea |
The values are means ± standard deviations from three replications. Means with the same uppercase letter in the same column are not significantly different (P > 0.05). Within the NIR-UV columns, values in the same row followed by the same lowercase letter are not significantly different (P > 0.05). XLD, xylose lysine desoxycholate agar; OV-XLD, overlay XLD on TSA; n0, initial population; n, population after treatment.
A statistically significant (P < 0.05) difference between the sum of NIR and UV inactivation and inactivation achieved with combination treatment was observed.
TABLE 4.
Reductions in numbers of viable Listeria monocytogenes cells on ham slice surfaces treated with UV, NIR, and NIR-UV
| Treatment time (s) | Log reduction (log10
n0 − log10
n) by treatment type and selective mediuma |
|||
|---|---|---|---|---|
| UV (OAB) | NIR (OAB) | NIR-UV |
||
| OAB | OV-OAB | |||
| 0 | 0.00 ± 0.00A | 0.00 ± 0.00A | 0.00 ± 0.00Aa | 0.00 ± 0.00Aa |
| 10 | 1.00 ± 0.21B | 0.06 ± 0.10A | 1.37 ± 0.42Ba | 1.26 ± 0.03Ba |
| 20 | 1.08 ± 0.28B | 0.11 ± 0.14A | 1.66 ± 0.26BCa | 1.36 ± 0.15Ba |
| 30 | 1.11 ± 0.28BC | 0.23 ± 0.26AB | 1.73 ± 0.20BCa | 1.52 ± 0.21BCa |
| 40 | 1.18 ± 0.18BC | 0.48 ± 0.31BC | 1.96 ± 0.05Ca | 1.83 ± 0.34Ca |
| 50 | 1.27 ± 0.16BCD | 0.67 ± 0.21CD | 2.45 ± 0.25Da | 2.26 ± 0.20Da |
| 60 | 1.46 ± 0.16CD | 0.87 ± 0.20D | 2.73 ± 0.36Da | 2.62 ± 0.32Da |
| 70b | 1.55 ± 0.17D | 1.45 ± 0.21E | 3.43 ± 0.10Ea | 3.16 ± 0.18Ea |
The values are means ± standard deviations from three replications. Means with the same uppercase letter in the same column are not significantly different (P > 0.05). Within the NIR-UV columns, values in the same row followed by the same lowercase letter are not significantly different (P > 0.05). OAB, Oxford agar base; OV-OAB, overlay OAB on TSA; n0, initial population; n, population after treatment.
A statistically significant (P < 0.05) difference between the sum of NIR and UV inactivation and inactivation achieved with combination treatment was observed.
Resuscitation of NIR-UV-injured cells.
The levels of sublethally injured E. coli O157:H7, S. Typhimurium, and L. monocytogenes cells on ham slice surfaces following simultaneous NIR-UV treatment are presented in Tables 2, 3, and 4, respectively. At the maximum treatment time of 70 s, injured cell levels of 0.39, 0.51, and 0.27 log CFU/g were detected for E. coli O157:H7, S. Typhimurium, and L. monocytogenes, respectively. Overall, the reductions in the numbers of the three pathogens observed at whole treatment time intervals using the agar OV method (SPRAB in the case of E. coli O157:H7) were slightly less than the reductions observed by direct plating on selective agar. However, statistically significant (P > 0.05) differences between the inactivation levels enumerated on the appropriate selective agar (SMAC, XLD, and OAB) versus the agar used for recovery (SPRAB, OV-XLD, and OV-OAB) were not observed for any treatment time interval (Tables 2 to 4).
Average temperature-time histories of ham slices.
The average surface temperatures of the ham slices during NIR heating and simultaneous NIR-UV treatment are shown in Fig. 2. The surface temperature rose immediately in response to infrared waves when the ham slice samples were exposed to NIR radiation, and the heating rate of the simultaneous NIR-UV treatment was not significantly (P > 0.05) different from that of the NIR treatment alone (Fig. 2). The surface temperature of the ham slices increased from room temperature (22 ± 2°C) and reached ca. 74°C after 70 s of NIR-UV treatment. The difference in temperature between treatment with NIR alone and NIR-UV treatment at 70 s was under 1°C.
FIG 2.

Average temperature-time histories of ham slice surfaces during NIR heating and simultaneous NIR-UV treatment. The error bars indicate standard deviations calculated from triplicates.
Determination of injury sites in NIR-UV-treated cells.
To gain further insight into the basis of NIR-UV injury, the effect of metabolic inhibitors on repair of S. Typhimurium DT 104 cells injured by UV, NIR, and NIR-UV was examined. Table 5 shows the quantitative levels of recovery inhibition of S. Typhimurium DT 104 after each treatment. The degree of recovery inhibition of intact S. Typhimurium DT 104 cells (untreated control) was close to zero; namely, the resuscitated populations of untreated S. Typhimurium DT 104 cells were not significantly different in the presence or absence of the selective antibiotics (chloramphenicol, nalidixic acid, penicillin G, and rifampin). Penicillin G did not inhibit the recovery of UV- or NIR-treated cells at all. However, significant (P < 0.05) inhibition of repair (ca. 0.27 log unit) was detected following the NIR-UV combined treatment. In the presence of chloramphenicol, the degree of recovery inhibition of NIR-UV-treated cells (ca. 0.58 log unit) was significantly (P < 0.05) higher than the degree of recovery inhibition of UV- or NIR-treated cells (ca. 0.02 or 0.29 log unit, respectively). Nalidixic acid only slightly inhibited the repair of injuries induced by UV or NIR-UV and did not affect the viability of NIR-treated cells, whereas in the presence of rifampin, equal ca. 0.65 log unit of resuscitation of NIR- or NIR-UV-treated cells, respectively, was inhibited. Therefore, S. Typhimurium DT 104 cells subjected to simultaneous NIR-UV treatment showed significantly (P < 0.05) greater values of recovery inhibition than cells subjected to the other treatments in the presence of chloramphenicol or penicillin G.
TABLE 5.
Effect of metabolic inhibitors on resuscitation of UV-, NIR-, and NIR-UV-injured Salmonella Typhimurium DT 104 cells
| Treatment type | Level of recovery inhibition (log10 CFU/g) by selective antibioticsa |
|||
|---|---|---|---|---|
| Chloramphenicol | Nalidixic acid | Penicillin G | Rifampin | |
| Untreated control | 0.09 ± 0.01A | −0.01 ± 0.07AB | 0.01 ± 0.09A | 0.02 ± 0.05A |
| UV | 0.02 ± 0.04A | 0.07 ± 0.06B | −0.01 ± 0.06A | 0.23 ± 0.04B |
| NIR | 0.29 ± 0.17A | −0.04 ± 0.01A | −0.01 ± 0.07A | 0.65 ± 0.11C |
| NIR-UV | 0.58 ± 0.23B | 0.06 ± 0.04B | 0.27 ± 0.03B | 0.65 ± 0.08C |
Values are means of three replications ± standard deviations. Values followed by the same letters within the column for each metabolic inhibitor are not significantly different (P > 0.05). The quantitative levels of recovery inhibition were calculated by subtracting the populations on medium containing each antibiotic from the populations obtained on nonselective medium that did not contain a selective reagent (control).
Effect of simultaneous NIR-UV treatment on product quality.
The color and texture parameters of the ham slices after simultaneous NIR-UV treatment are shown in Table 6. The color (L*, a*, and b*) values for NIR-UV-treated (70 s) ham slices were not significantly (P > 0.05) different from those for nontreated samples. Although the values of a* and b* (redness and yellowness, respectively) slightly increased and the values of L* (lightness) slightly decreased, in accordance with the prolonged treatment time, statistically significant differences were not observed during the entire treatment interval (Table 6). Also, the NIR-UV combined treatment for 70 s did not significantly (P > 0.05) change the maximum load values of the texture measurements. Thus, the simultaneous application of NIR and UV treatment for 70 s did not significantly alter the quality of the ham slice products.
TABLE 6.
Surface color values and maximum load values for quantifying texture of ham slices simultaneously treated with NIR-UVa
| Treatment time (s) | Color value for parameter |
Maximum load (g) | ||
|---|---|---|---|---|
| L* | a* | b* | ||
| 0 | 63.54 ± 0.57A | 15.69 ± 0.10A | 9.60 ± 0.55A | 1,015.67 ± 43.66A |
| 10 | 63.43 ± 0.55A | 15.91 ± 0.18A | 9.59 ± 1.02A | 1,001.00 ± 72.33A |
| 30 | 63.54 ± 0.96A | 15.81 ± 0.27A | 9.88 ± 0.98A | 1,012.67 ± 52.00A |
| 50 | 62.82 ± 0.56A | 16.00 ± 0.53A | 10.21 ± 0.40A | 1,007.33 ± 73.93A |
| 70 | 62.31 ± 0.29A | 16.19 ± 0.41A | 10.79 ± 0.67A | 1,022.33 ± 71.63A |
Values are means from three replications ± standard deviations. Values followed by the same letters within each column are not significantly different (P > 0.05). L*, lightness; a*, redness; b*, yellowness.
DISCUSSION
Recently, to meet the needs of food industries, new designs of combined antimicrobial processes that are applied at lower intensities but that exhibit degrees of microbial inactivation equivalent to or even higher than that of either treatment used alone are in demand. Therefore, in order to prevent excessive heating of sliced ham without compromising decontamination ability, we explored the effect of decreasing the input energy of NIR heating while combining it with UV-C irradiation. As a result, the simultaneous NIR-UV treatment resulted in a level of inactivation of E. coli O157:H7, S. Typhimurium, and L. monocytogenes significantly greater than that achieved with either treatment used alone as a result of synergism (Tables 2 to 4) without causing any deterioration in product quality due to the lower intensity of NIR (Table 6). Many researchers have emphasized the importance of the presence of sublethally injured cells after bactericidal treatments, since they are able to repair themselves and resume growth under favorable conditions (26). In the present study, the extent to which sublethally injured pathogens survived after NIR-UV treatment was evaluated by plating on selective agars with and without a resuscitation step, yet there were no significant (P > 0.05) differences at any of the treatment time intervals (Tables 2 to 4).
One of the purposes of the current study was to examine the mechanism of the synergistic lethal effect induced by simultaneous NIR and UV treatment. On the basis of the extent of synergistic inactivation of the three pathogens detected and the treatment interval at which statistically significant (P < 0.05) synergism was observed, S. Typhimurium strain DT 104 and a 70-s treatment time were chosen as model factors for investigation of the mechanism (Tables 2 to 4). We postulated that UV-, NIR-, and NIR-UV-injured cells have different sensitivities to antibiotics which promote specific inhibitory actions on bacteria, and four types of metabolic inhibitors (antibiotics), of which primary synthesis inhibitions are well-known, were employed in order to specify the character of the injury in the target microorganism. Antibiotic-induced cell death has been well studied and predominantly falls into four classes: inhibition of DNA synthesis, inhibition of protein synthesis, inhibition of cell wall synthesis, and inhibition of RNA synthesis (27). Antibiotics can be also classified on the basis of whether they induce cell death with an efficiency of >99.9% (bactericidal) or merely inhibit cell growth (bacteriostatic) (28). In this study, we employed chloramphenicol, nalidixic acid, penicillin G, and rifampin as the protein, DNA, cell wall, and RNA-specific synthesis inhibitors, respectively, since each is bacteriostatic. The bacteriostatic antibiotics block a particular cellular process and do not produce highly deleterious hydroxyl radicals, whereas in Gram-negative and Gram-positive bacteria, all classes of bactericidal antibiotics stimulate the production of hydroxyl radicals, which ultimately contribute to cell death (28), and in those instances it may be difficult to determine the specific site of injury in cells.
Several research studies have used these metabolic inhibitors to investigate the cellular metabolic activities and the site of damage in treated cells (29–34). In all previous studies, however, liquid suspensions of bacterial cells were treated and then subjected to analysis of the mechanism. It is well-known that the physicochemical state and composition of the treatment medium may affect the bactericidal efficacy of most food preservation technologies, and more specifically, sites of cellular damage or the extent of damage at a particular cellular site could be different (31). Therefore, to accurately study the mechanism of NIR-UV-induced injury for a particular microorganism on ham slices, the studies should be performed in the same food system. For this purpose, we combined the metabolic inhibitors with the overlay method (TSA-XLD). TSA medium containing the antibiotics was used as the selective medium. S. Typhimurium DT 104 cells treated on ham slices were spread plated onto selective medium and nonselective medium that did not contain an antibiotic. Colonies of target Salmonella cells could be readily enumerated on the overlaid XLD medium. The difference in colony counts obtained with and without the addition of selective reagents can thus be taken as a measure of injury resulting from each treatment. This proposed method could be utilized in a food base as a simple and effective means of detecting and classifying sites of cellular damage to bacterial cells induced by physical or chemical antimicrobial technologies.
Since the number of colonies of noninjured cells on selective medium (TSA-antibiotic) has to be same as that on nonselective medium, we investigated the effects of various concentrations of metabolic inhibitors in TSA medium on inactivation of intact S. Typhimurium DT 104 cells to find the maximum concentrations at which selective antibiotics had no quantitative effect on the formation of colonies. The applied maximum concentrations, especially those of chloramphenicol and penicillin G, were relatively high due to the multidrug resistance of S. Typhimurium strain DT 104 (35). As a result, exposure of intact S. Typhimurium DT 104 cells to the selective antibiotic agents at the concentrations listed in Table 1 caused no substantial effects on colony formation (Table 5). These concentrations are adequate to allow detection of any slight damage to bacterial cells (resulting in a reduction in colony numbers). In the present study, there were no significant (P > 0.05) differences in the level of recovery inhibition between NIR- and NIR-UV-treated cells in the presence of rifampin or between UV- and NIR-UV-treated cells in the presence of nalidixic acid. Rifampin acts on RNA polymerase to arrest DNA-dependent RNA synthesis, and nalidixic acid targets DNA replication and repair by binding DNA gyrase complexed with DNA (27). These results indicate that RNA or DNA damage did not affect the synergistic effect of the NIR-UV combined treatment. On the other hand, simultaneous NIR-UV treatment markedly enhanced the level of recovery inhibition in the presence of penicillin G or chloramphenicol. Moreover, the extent of recovery inhibition induced by the simultaneous application of NIR and UV was significantly higher than the sum of the recovery inhibition levels obtained by separate NIR and UV treatments (Table 5). From this synergistic tendency, the main cellular damage contributing to the synergistic lethal effect of the NIR-UV combined treatment could be inferred. The mode of inhibition for penicillin G is interference with cell wall synthesis by blocking transpeptidation on peptidoglycan strands, and that for chloramphenicol is disruption of protein-synthesizing systems (blockage of protein translation) depending on the reaction with the 50S subunit of ribosomes (27). Typically, structural proteins are necessary for the normal development of the cell membrane or cell wall. Especially, during restoration of injured cell envelopes, ribosomal damage can significantly affect membrane integrity owing to the inhibition of membrane protein synthesis (36). Therefore, the mechanism of synergistic bacterial inactivation by simultaneous NIR-UV treatment might be related to damage to cellular envelopes and the inability of cells to repair these structures due to ribosomal damage. Thus, more than one type of damage may be related to the synergistic lethal effect.
These results are in agreement with earlier findings from transmission electron microscopy (TEM) analysis and the propidium iodide (PI) uptake test in our previous study (20). Based on TEM images and PI uptake values, we confirmed that disruption of the bacterial cell membrane was the main factor contributing to the synergistic inactivation by the NIR-UV combined treatment. In addition, the outputs involving NIR radiant heating do not contradict the findings of other research studies which dealt with the metabolic requirements involved in the thermal injury and repair of bacteria (30, 31, 37). Tomlins and Ordal (37) reported that rifamycin, an inhibitor of RNA synthesis, blocked the repair of heat-injured S. Typhimurium. Thus, in S. Typhimurium, RNA is degraded during heat treatment and resynthesized during repair. The function of RNA polymerase in cells is very sensitive to temperature change since it plays an important role in induction of heat shock proteins (38). Therefore, in NIR and NIR-UV combined treatment, there were equal levels of recovery inhibition by rifampin (Table 5), which demonstrated that similar degrees of thermal damage occurred in both NIR- and NIR-UV-treated cells due to the same patterns of temperature growth (Fig. 2).
In conclusion, the simultaneous application of NIR and UV-C radiation was especially effective in inactivating three major pathogenic bacteria (E. coli O157:H7, S. Typhimurium, and L. monocytogenes) on RTE sliced ham owing to their aforementioned synergistic mechanisms without affecting product quality. Furthermore, the NIR-UV processing technique can easily be utilized in industrial applications on a continuous basis, and the effectiveness of this decontaminating system can be further improved by refining the procedure, such as rearranging the radiation intensities of NIR and UV-C emitters and adjusting the treatment intervals.
ACKNOWLEDGMENTS
This research was supported by the Public Welfare and Safety research program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT, and Future Planning (NRF-2012M3A2A1051679). This research was also supported by a grant (14162MFDS973) from the Ministry of Food and Drug Safety in 2014.
REFERENCES
- 1.Food Safety and Inspection Service, US Department of Agriculture. 2010. FSIS comparative risk assessment for Listeria monocytogenes in ready-to-eat meat and poultry deli meats. US Department of Agriculture, Washington, DC: http://www.fsis.usda.gov/wps/wcm/connect/c2ac97d0-399e-4c4a-a2bc-d338c2e201b3/Comparative_RA_Lm_Report_May2010.pdf?MOD=AJPERES. Accessed 30 April 2014. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2014. Foodborne outbreak online database. Investigation update: multistate outbreak of listeriosis, E. coli, and human Salmonella infections. Centers for Disease Control and Prevention, Atlanta, GA: http://wwwn.cdc.gov/foodborneoutbreaks/. Accessed 30 April 2014. [Google Scholar]
- 3.Chen D, Zhao T, Doyle MP. 2014. Transfer of foodborne pathogens during mechanical slicing and their inactivation by levulinic acid-based sanitizer on slicers. Food Microbiol 38:263–269. doi: 10.1016/j.fm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Bravo D, de Alba M, Medina M. 2014. Combined treatments of high-pressure with the lactoperoxidase system or lactoferrin on the inactivation of Listeria monocytogenes, Salmonella Enteritidis and Escherichia coli O157:H7 in beef carpaccio. Food Microbiol 41:27–32. doi: 10.1016/j.fm.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Food Safety and Inspection Service, US Department of Agriculture. 2014. FSIS recalls. US Department of Agriculture, Washington, DC: http://www.fsis.usda.gov/fsis_recalls/Recall_Case_Archive. Accessed 30 April 2014. [Google Scholar]
- 6.Zhang L, Moosekian SR, Todd EC, Ryser ET. 2012. Growth of Listeria monocytogenes in different retail delicatessen meats during simulated home storage. J Food Prot 75:896–905. doi: 10.4315/0362-028X.JFP-11-491. [DOI] [PubMed] [Google Scholar]
- 7.European Food Safety Authority. 2007. The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. EFSA J 130:1–352. [Google Scholar]
- 8.Ruckman SA, Rocabayera X, Borzelleca JF, Sandusky CB. 2004. Toxicological and metabolic investigations of the safety of N-α-lauroyl-l-arginine ethyl ester monohydrochloride (LAE). Food Chem Toxicol 42:245–259. doi: 10.1016/j.fct.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 9.Lin CM, Takeuchi K, Zhang L, Dohm CB, Meyer JD, Hall PA. 2006. Cross-contamination between processing equipment and deli meats by Listeria monocytogenes. J Food Prot 69:71–79. [DOI] [PubMed] [Google Scholar]
- 10.Berzins A, Hellström S, Silins I, Korkeala H. 2010. Contamination patterns of Listeria monocytogenes in cold-smoked pork processing. J Food Prot 73:2103–2109. [DOI] [PubMed] [Google Scholar]
- 11.Chaitiemwong N, Hazeleger WC, Beumer RR, Zwietering MH. 2014. Quantification of transfer of Listeria monocytogenes between cooked ham and slicing machine surfaces. Food Control 44:177–184. doi: 10.1016/j.foodcont.2014.03.056. [DOI] [Google Scholar]
- 12.Krishnamurthy K, Khurana HK, Jun SJ, Irudayaraj J, Demirci A. 2008. Infrared heating in food processing: an overview. Compr Rev Food Sci Saf 7:2–12. doi: 10.1111/j.1541-4337.2007.00024.x. [DOI] [Google Scholar]
- 13.Huang L, Sites J. 2004. Infrared surface pasteurization of turkey frankfurters. Innov Food Sci Emerg Technol 5:345–351. doi: 10.1016/j.ifset.2004.03.007. [DOI] [Google Scholar]
- 14.Huang L, Sites J. 2009. Elimination of Listeria monocytogenes on cooked chicken breast meat surfaces by near-infrared surface pasteurization prior to final packaging. J Food Process Eng 35:1–15. doi: 10.1111/j.1745-4530.2009.00551.x. [DOI] [Google Scholar]
- 15.Ha JW, Ryu SR, Kang DH. 2012. Evaluation of near-infrared pasteurization in controlling Escherichia coli O157:H7, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes in ready-to-eat sliced ham. Appl Environ Microbiol 78:6458–6465. doi: 10.1128/AEM.00942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gayán E, Serrano MJ, Raso J, Álvarez I, Condón S. 2012. Inactivation of Salmonella enterica by UV-C light and by its combinations with mild temperatures. Appl Environ Microbiol 78:8353–8361. doi: 10.1128/AEM.02010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walkling-Ribeiro M, Noci F, Cronin DA, Riener J, Lyng JG, Morgan DJ. 2008. Reduction of Staphylococcus aureus and quality changes in apple juice processed by ultraviolet irradiation, preheating and pulsed electric fields. J Food Eng 89:267–273. doi: 10.1016/j.jfoodeng.2008.05.001. [DOI] [Google Scholar]
- 18.Char CD, Mitilinaki E, Guerrero SN, Alzamora SM. 2010. Use of high-intensity ultrasound and UV-C light to inactivate some microorganisms in fruit juices. Food Bioprocess Technol 3:797–803. doi: 10.1007/s11947-009-0307-7. [DOI] [Google Scholar]
- 19.Ukuku DO, Geveke DJ. 2010. A combined treatment of UV-light and radio frequency electric field for the inactivation of Escherichia coli K-12 in apple juice. Int J Food Microbiol 138:50–55. doi: 10.1016/j.ijfoodmicro.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Ha JW, Kang DH. 2013. Simultaneous near-infrared radiant heating and UV radiation for inactivating Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in powdered red pepper (Capsicum annuum L.). Appl Environ Microbiol 79:6568–6575. doi: 10.1128/AEM.02249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha JW, Kang DH. 2014. Synergistic bactericidal effect of simultaneous near-infrared radiant heating and UV radiation against Cronobacter sakazakii in powdered infant formula. Appl Environ Microbiol 80:1858–1863. doi: 10.1128/AEM.03825-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S-Y, Kang D-H. 2001. Suitability of overlay method for recovery of heat-injured Listeria monocytogenes and Salmonella Typhimurium. Food Sci Biotechnol 10:323–326. http://www.dbpia.co.kr/Article/1631765. [Google Scholar]
- 23.Kang DH, Siragusa GR. 1999. Agar underlay method for recovery of sublethally heat-injured bacteria. Appl Environ Microbiol 65:5334–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhee MS, Lee SY, Hillers VN, McCurdy SM, Kang DH. 2003. Evaluation of consumer-style cooking methods for reduction of Escherichia coli O157:H7 in ground beef. J Food Prot 66:1030–1034. [DOI] [PubMed] [Google Scholar]
- 25.Chun HH, Kim JY, Chung KS, Won MS, Song KB. 2009. Inactivation kinetics of Listeria monocytogenes, Salmonella enterica serovar Typhimurium, and Campylobacter jejuni in ready-to-eat sliced ham using UV-C irradiation. Meat Sci 83:599–603. doi: 10.1016/j.meatsci.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Wu VCH. 2008. A review of microbial injury and recovery methods in food. Food Microbiol 25:735–744. doi: 10.1016/j.fm.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 29.Flowers RS, Adams DM. 1976. Spore membrane(s) as the site of damage within heated Clostridium perfringens spores. J Bacteriol 125:429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez RF, Blais KD, Herrero A, Sinskey AJ. 1976. Effects of inhibitors of protein, RNA and DNA synthesis on heat-injured Salmonella Typhimurium LT2. J Gen Microbiol 97:19–27. doi: 10.1099/00221287-97-1-19. [DOI] [PubMed] [Google Scholar]
- 31.Restaino L, Jeter WS, Hill WM. 1980. Thermal injury of Yersinia enterocolitica. Appl Environ Microbiol 40:939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeChevallier MW, Schiemann DA, McFeters GA. 1987. Factors contributing to the reduced invasiveness of chlorine-injured Yersinia enterocolitica. Appl Environ Microbiol 53:1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawai J, Sagara K, Igarashi H, Hashimoto A, Kokugan T, Shimizu M. 1995. Injury of Escherichia coli in physiological phosphate-buffered saline induced by far-infrared irradiation. J Chem Eng Jpn 28:294–299. doi: 10.1252/jcej.28.294. [DOI] [Google Scholar]
- 34.Sawai J, Kojima H, Igarashi H, Hashimoto A, Shoji S, Sawaki T, Hakoda A, Kawada E, Kokugan T, Shimizu M. 2000. Antibacterial characteristics of magnesium oxide powder. World J Microbiol Biotechnol 16:187–194. doi: 10.1023/A:1008916209784. [DOI] [Google Scholar]
- 35.Threlfall EJ, Frost JA, Ward LR, Rowe B. 1996. Increasing spectrum of resistance in multiresistant Salmonella Typhimurium. Lancet 347:1053–1054. doi: 10.1016/S0140-6736(96)90199-3. [DOI] [PubMed] [Google Scholar]
- 36.Tsuchido T, Aoki I, Takano M. 1989. Interaction of the fluorescent dye 1-N-phenylnaphthylamine with Escherichia coli cells during heat stress and recovery from heat stress. J Gen Microbiol 135:1941–1947. [DOI] [PubMed] [Google Scholar]
- 37.Tomlins RI, Ordal ZJ. 1971. Requirements of Salmonella Typhimurium for recovery from thermal injury. J Bacteriol 105:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yura T, Nagai H, Mori H. 1993. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol 47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]


