Abstract
Proof of concept for the in vivo bacterial production of a polyester resin displaying various customizable affinity protein binding domains is provided. This was achieved by engineering various protein binding domains into a bacterial polyester-synthesizing enzyme. Affinity binding domains based on various structural folds and derived from molecular libraries were used to demonstrate the potential of this technique. Designed ankyrin repeat proteins (DARPins), engineered OB-fold domains (OBodies), and VHH domains from camelid antibodies (nanobodies) were employed. The respective resins were produced in a single bacterial fermentation step, and a simple purification protocol was developed. Purified resins were suitable for most lab-scale affinity chromatography purposes. All of the affinity domains tested produced polyester beads with specific affinity for the target protein. The binding capacity of these affinity resins ranged from 90 to 600 nmol of protein per wet gram of polyester affinity resin, enabling purification of a recombinant protein target from a complex bacterial cell lysate up to a purity level of 96% in one step. The polyester resin was efficiently produced by conventional lab-scale shake flask fermentation, resulting in bacteria accumulating up to 55% of their cellular dry weight as polyester. A further proof of concept demonstrating the practicality of this technique was obtained through the intracellular coproduction of a specific affinity resin and its target. This enables in vivo binding and purification of the coproduced “target protein.” Overall, this study provides evidence for the use of molecular engineering of polyester synthases toward the microbial production of specific bioseparation resins implementing previously selected binding domains.
INTRODUCTION
Affinity chromatography in various forms has been a cornerstone of protein purification for decades. In its simplest form, the inherent interaction between a target enzyme and its substrate is used, for example, purifying amylase by adsorbing it on insoluble starch. However, this technique is limited by the availability of a naturally occurring interaction and the ability of the substrate to be immobilized on an insoluble matrix (1). A more common and versatile technique is to engineer a tag into the target protein and utilize a protein or matrix with affinity toward that tag (2). Though widely used, this approach is often unacceptable for structural studies and in many commercial production systems due to the chance that the affinity tag may affect the structure, function, or other characteristics of the target protein. Furthermore, the presence of tags is commonly not tolerated in the pharmaceutical industry (3). One approach to combat this is to remove the tag after purification by engineering specific protease cleavage sites or self-cleaving sites such as inteins between the target and the tag, but this introduces further processing and purification steps (to remove the protease and/or cleaved tag) and can often result in a “scar” (residual amino acids) as well as negatively impacting process economics (3).
A different approach is to utilize antibodies raised against the target of interest. Antibodies can be generated to have highly specific and strong binding affinities for target proteins. Antibodies can be bound to a common protein A resin, resulting in a custom affinity chromatography resin. Due to the time and relatively high initial cost to generate an antibody, this approach is often not applicable for lab-scale processes unless an antibody already exists but may be appropriate for large-scale production of a high-value protein, for which the initial cost would soon be offset. The main downsides of this approach are the additional steps involved in producing, purifying, and cross-linking the antibody to the resin before the target can be applied and the potential for antibody leaching.
Here we describe the one-step production of an affinity matrix based on the in vivo conjugation of various customizable affinity binding domains to biopolyester beads produced by Escherichia coli. These beads are composed of poly-β-hydroxybutyrate (PHB) and can be naturally produced as intracellular inclusions by a wide range of bacteria and archaea (4). The beads are produced in the bacterial cytosol when the polyhydroxyalkanoate (PHA) synthase, PhaC, catalyzes the polymerization of (R)-3-hydroxyacyl-coenzyme A (CoA). PhaC remains covalently attached to the nascent PHA chain, which self-assembles into beads within the cell (5). These beads have a diameter of 100 to 500 nm and can be simply purified from the cell. The PHA beads can be functionalized by translationally fusing a protein or domain of interest to either terminus of PhaC, which is subsequently displayed at the bead surface (6). Previously, PHA beads have been engineered to display proteins with diverse characteristics such as antibody binding domains, various enzymatic functions, biotin binding, and vaccine antigens (7–11).
It was previously demonstrated that a single-chain variable-fragment antibody (scFv) that had been specifically engineered for soluble production in the bacterial cytoplasm could be functionally produced and displayed on PHA beads in vivo (12). Here we utilize several customizable affinity domain scaffolds with diverse libraries to demonstrate the potential for a bacterially produced custom affinity resin.
Three affinity binding proteins/domain were assessed: VHH domains from camelid antibodies, designed ankyrin repeat proteins (DARPins), and OB-folds (OBodies). Antibodies from camelid species lack light chains and can recognize antigens via their single monomeric variable antibody domain of (also known as VHH domains or nanobodies). VHH domains represent the smallest known natural antigen-binding domain (<15 kDa) and typically are more soluble and stable than multidomain recombinant antibodies. This makes them an attractive target for bacterial production. VHH domains with specific affinities can be derived from immune, nonimmune, or semisynthetic libraries (13). DARPins are customizable synthetic affinity proteins based on a consensus of naturally occurring ankyrin repeat domains. They are very stable and can be produced in the bacterial cytoplasm at high yields. By shuffling specific potentially interacting residues within a repeat and the number of repeat domains themselves, libraries with diversities of up to 1014 can be generated, enabling selection of binding domains with binding affinities in the pM range (14, 15). OBodies are affinity binding proteins derived from OB-fold domains of naturally occurring proteins, typically from Pyrobaculum aerophilum (e.g., tRNA synthetase or translational initiation factor IF5A). Several residues on the binding face of the OB-fold are randomized, and the resulting library is screened for binding to the target of interest by phage display; binding affinities as low as 3 nM have been demonstrated (16, 17).
Here, examples of these affinity domains, previously derived from immune and synthetic libraries, were genetically fused to a polyester synthase gene (phaC) as a proof of concept for the in vivo production of biopolyester-based customizable affinity resins. The expression of the disulfide-containing VHH domain constructs was optimized to allow efficient cytosolic production of functional protein. Furthermore, we demonstrated the application of this technique to coproduce both the affinity matrix and the target protein within the same cell. This allowed for the rapid in vivo “one-step” production and purification of an untagged protein of interest.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. All E. coli strains were grown in LB at 37°C or 25°C unless otherwise stated. When required, antibiotics were used at the following concentrations: ampicillin, 75 μg/ml; chloramphenicol, 34 μg/ml. For PHA bead and protein production, E. coli cultures were grown in LB with 1% glucose at 25°C to an optical density at 600 nm (OD600) of 0.4 to 0.5, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and allowed to grow for approximately 48 h.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description or sequence | Source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| SHuffle T7 Express | Chromosomal copy of dsbC trxB gor mutant | NEB |
| Plasmids | ||
| pMCS69 | pBBR1MCS with phaA and phaB | 22 |
| pETC | pET14b containing wild-type phaC | 11 |
| pET14b:M-PhaC | pET14b encoding an Mpl-PhaC fusion protein | 6 |
| pET14b:PhaC-linker-MalE | pET14b encoding a PhaC-linker-MalE fusion protein | 6 |
| pET14b:M-PhaC-linker-MalE | pET14b encoding an MPL-PhaC-linker-MalE fusion protein | 6 |
| pUC57:GFPVHH_opt | Plasmid carrying the cAbGFP4-coding region (optimized for expression in E. coli) | This study |
| pUC57:TNFaVHH_opt | Plasmid carrying the B4 anti-TNF-α VHH-coding region (optimized for expression in E. coli) | This study |
| pUC57:off7 | Plasmid carrying the off7 DARPin-coding region (optimized for expression in E. coli) | This study |
| pMCS69E | pMCS69 with Erv1P ORF inserted upstream of phaA and phaB | This study |
| pET14b:GFPVHH-PhaC | pET14b encoding the GFPVHH-PhaC fusion protein | This study |
| pET14b:PhaC-GFPVHH | pET14b encoding the PhaC-GFPVHH fusion protein | This study |
| pET14b:GFPVHH-PhaC-GFPVHH | pET14b encoding the GFPVHH-PhaC-GFPVHH fusion protein | This study |
| pET14b:off7-PhaC | pET14b encoding the off7-PhaC fusion protein | This study |
| pET14b:PhaC-off7 | pET14b encoding the PhaC-off7 fusion protein | This study |
| pET14b:OB-PhaC | pET14b encoding the OBody-PhaC fusion protein | This study |
| pET14b:PhaC-OB | pET14b encoding the PhaC-OBody fusion protein | This study |
| pET14b:TNFaVHH-PhaC | pET14b encoding the TNFaVHH-PhaC fusion protein | This study |
| pET14b:PhaC-TNFaVHH | pET14b encoding the PhaC-TNFaVHH fusion protein | This study |
| pET14b:GFPVHH-PhaC-GFPVHH_GFP | pET14b encoding the GFPVHH-PhaC-GFPVHH fusion protein and GFP to be expressed polycistronically | This study |
| pET14b:PhaC_GFP | pET14b encoding the PhaC fusion protein and GFP to be expressed polycistronically | This study |
| Primers | ||
| GFPVHH-atg_F_XhoI_(C) | GCTCTCGAGCAGGTCCAACTGGTCGAATCAGGTG | This study |
| GFPVHH+taa_R_BamHI_(C) | GCTGGATCCTTACGAAGACACCGTGACTTGCGTGCCC | This study |
| TNFaB5VHH-atg_F_XhoI_(C) | GCTCTCGAGCAAGCGGAAGTCCAACTGCAAGAATC | This study |
| TNFaB5VHH+taa_R_BamHI_(C | GCTGGATCCTTACGCTGCCGGTTGCGGTTTCGGGG | This study |
| off7-atg_F_XhoI_(C) | ACTCTCGAGTCGGACCTGGGCCGTAAACTGCTGG | This study |
| off7+taa_R_BamHI_(C) | CGAGGATCCTTATTGCAGGATTTCAGCCAGGTCTTCG | This study |
| OB_F_SpeI | CCGACTAGTGTGTATCCTAAAAAGACCCACTGGACC | This study |
| OB_R_SpeI | ATAACTAGTGTCTATTGGAAGCGGCTTGGCCTTG | This study |
| OB_F_SmaI | GATACCCGGGGTGTATCCTAAAAAGACCCACTGGACC | This study |
| OB_R_BamHI | TATGGATCCGTCTATTGGAAGCGGCTTGGCCTTG | This study |
| GFP_polycis_F_(BamHI-RBS) | CGATGGATCCAATAATTTTGTTTAACTTTAAGAAGGAGATATACCCATATGAGTAAAGGAGAAGAACTTTTC | This study |
| GFP_polycis_R_(StuI-BclI) | CATCTGATCAAGTAGGCCTTCATTTGTATAGTTCATCCATGCCATGTGTAATCCCAG | This study |
Plasmids, DNA constructs, and oligonucleotides.
All plasmids and oligonucleotides used in this study are listed in Table 1. DNA primers, deoxynucleoside triphosphate, Taq and Platinum Pfx polymerases, and T4 DNA ligase were purchased from Invitrogen. DNA sequencing was performed by the Massey University Genome Service.
The amino acid sequence for the anti-green fluorescent protein (GFP) VHH domain cAbGFP4 was obtained from Saerens et al. (19). The amino acid sequence for the anti-ovine tumor necrosis factor alpha (TNF-α) VHH domain B5 was kindly provided by Anton Pernthaner, AgResearch New Zealand (20). The amino acid sequence for the maltose binding protein (MBP) DARPin off7 was obtained from Binz et al. (15). The VHH and DARPin affinity domains were codon optimized, synthesized, and inserted into pUC57 by Genscript USA Inc. with the addition of XbaI sites followed by ribosomal binding site (RBS) and start codons on the 5′ end and a 48-bp region encoding a serine-glycine linker, S(G4)3S, followed by SpeI sites, on the 3′ end. Plasmids encoding fusion of the VHH domains or DARPin to the N terminus of the PHA synthase were generated by excising the respective domains from the pUC57 backbone with XbaI and SpeI and ligating the fragments into the corresponding sites of pET-14b:M-PhaC (6) (replacing the Mpl-encoding region), resulting in plasmids pET-14b:GFPVHH-SG-PhaC, pET-14b:TNFaVHH-SG-PhaC, and pET-14b:off7-SG-PhaC.
To create plasmids encoding the fusion of the C terminus of PhaC to the VHH or DARPin domains, the respective domains were amplified from the synthesized pUC57 templates using the following primers: GFPVHH-atg_F_XhoI_(C) and GFPVHH+taa_R_BamHI_(C); TNFaB5VHH-atg_F_XhoI_(C) and TNFaB5VHH+taa_R_BamHI_(C); and off7-atg_F_XhoI_(C) and off7+taa_R_BamHI_(C). The resulting fragments were cleaved with XhoI and BamHI and ligated into the corresponding sites on the plasmid pET-14b:PhaC-linker-MalE (6) (replacing the MalE-encoding region), resulting in plasmids pET-14b:PhaC-linker-GFPVHH, pET-14b:PhaC-linker-TNFaVHH, and pET-14b:PhaC-linker-off7.
To create the constructs pET-14b:GFPVHH-SG-PhaC-linker-DsbC, pET-14b:TNFaVHH-SG-PhaC-linker-DsbC, pET-14b:Tub1VHH-SG-PhaC-linker-DsbC, and pET-14b:PhaC-linker-DsbC, a fragment encoding DsbC was amplified from pBADΔSSdsbC (21) using the primers DsbC-ss_F_XhoI_(C) and DsbC-ss_R_BamHI_(C). The resulting fragment was cleaved with XhoI and BamHI and ligated into the corresponding sites of pET-14b:M-PhaC-linker-MalE and pET-14b:M-PhaC-linker-MalE (6) (replacing the MalE-encoding region), resulting in the plasmids pET-14b:M-PhaC-linker-DsbC and pET-14b:PhaC-linker-DsbC. The VHH domains were inserted upstream of the PhaC-coding region of pET-14b:M-PhaC-linker-DsbC via XbaI and SpeI as described above.
To create the constructs pET-14b:DsbC-SG-PhaC-linker-GFPVHH, pET-14b:DsbC-SG-PhaC-linker-TNFaVHH, and pET-14b:DsbC-SG-PhaC-linker-Tub1VHH, a fragment encoding Dsbc was amplified from pBADΔSSdsbC (21), using the primers DsbC-ss+RBS+atg_F_XbaI_(N) and DsbC-taa_R_KpnI_(N). This fragment was cleaved with XbaI and KpnI and ligated into the corresponding sites of pUC57:GFPVHHopt (replacing the GFPVHH domain) to introduce an SG linker after the DsbC-encoding region, and the fragment encoding DsbC followed by an SG linker was excised via XbaI and SpeI from the resulting plasmid and ligated into the corresponding sites of pET-14b:M-PhaC-linker-MalE and pET-14b:M-PhaC. The VHH domains were inserted downstream of the PhaC-coding region and the linker via XhoI and BamHI as described above.
pET-14b:GFPVHH-SG-PhaC-linker-GFPVHH was generated by replacing the DsbC-coding region from pET-14b:GFPVHH-SG-PhaC-linker-DsbC with the GFPVHH domain from pET-14b:PhaC-linker-GFPVHH via XhoI and BamHI as described above.
The lysozyme-specific OBody-coding region was obtained from the plasmid pProExHtb-L200EP-06 (16). The region was amplified for fusion to the N or C terminus of PhaC using the primer set OB_F_SpeI_(N) and OB_R_SpeI_(N) and the primer set OB_F_SmaI_(C) and OB_R_BamHI_(C), respectively. The products were hydrolyzed with SpeI or SmaI and BamHI and ligated into the corresponding sites of pET14b:GFP-phaC (for the N-terminal fusion) or pET14b:phaC-linker-SG-GFP (for the C-terminal fusion), resulting in pET14b:OB-PhaC and pET14b:PhaC-OB.
The plasmids for the coexpression of GFPVHH beads and GFP were generated by inserting the GFP open reading frame (ORF) behind the VHH-PhaC-VHH ORF of the plasmid pET-14b:GFPVHH-SG-PhaC-linker-GFPVHH (also pET-14b:PhaC as a negative control). The GFP ORF was amplified (with the addition of an RBS) from the plasmid pET14b:PhaC-GFP (6) with the primers GFP_polycis_F_(BamHI-RBS) and GFP_polycis_R_(StuI-BclI). This product was cut with BamHI and BclI and ligated into the BamHI site of pET-14b:GFPVHH-PhaC-GFPVHH and pET-14b:PhaC, resulting in pET-14b:GFPVHH-PhaC-GFPVHH_GFP and pET-14b:PhaC_GFP. Orientation and sequence were confirmed by sequencing.
To generate the sulfhydryl oxidase-expressing plasmid pMSC69E, the amino acid sequence of Erv1p was obtained from NCBI (NP_011547.2) and codon optimized by Geneart (Life technologies) with the addition of HindIII and an RBS on the 5′ end and PstI on the 3′ end, and then this product was cut with HindIII and PstI and ligated into the corresponding sites of pMCS69 (22) (upstream of phaA and phaB before the promoter).
Production and isolation of beads.
The strains used to produce beads were E. coli BL21(DE3)(pMCS69) or SHuffle T7 express(pMCS69E) containing the pET14b-derived phaC construct of interest. Beads were produced and harvested essentially as described elsewhere (6). Briefly, cells were harvested by centrifugation and mechanically disrupted via a French press or a similar process; the insoluble (bead-containing) fraction was collected and washed in phosphate-buffered saline (PBS; 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4) before being subjected to centrifugation at 100,000 × g on a glycerol density gradient (88% cushion, 44% layer, bead-containing mixture). The bead-containing interface between the 44% and 88% layers was collected and washed twice in PBS. For the lysozyme-binding OBody beads, no lysozyme was added during cell lysis or processing. For the coexpression experiments, the glycerol gradient step was omitted and Bugbuster (Merck Milipore) was added to the lysis buffer according to the manufacturer's instructions. When wet weights of PHA beads are reported, they represent the weights of the beads after centrifugation at 5,000 × g for 5 min and removal of the supernatant.
Preparation of GFP- or MBP-containing lysate.
The GFP open reading frame was amplified and inserted into pET-14B under the control of the T7 promoter. The plasmid pMAL-c2g (NEB) was used to express MBP. These plasmids were transformed into E. coli BL21(DE3). The resulting transformants were grown as described above, harvested, and lysed using Bugbuster according to the manufacturer's instructions. Lysates were cleared by centrifugation at 9,000 × g and filtered with a 0.2-μm filter.
GFP and MBP binding assay.
Beads were prewashed once in Bugbuster lysis buffer. Approximately 50 mg of beads was added to 1 ml of cleared GFP or MBP lysate. The beads were resuspended and incubated on a rotary mixer for 20 min, after which the beads were sedimented by centrifugation at 6,000 × g. The supernatant was removed, and the beads were washed three times in PBS with 0.05% Tween 20 (each wash consisting of resuspension of the beads, centrifugation, and removal of supernatant). To elute the bound protein, the beads were resuspended in 500 μl of 50 mM glycine (pH 2.7); the beads were removed by centrifugation at 16,000 × g, and the supernatant was neutralized with 50 μl of 1 M K2HPO4. The protein content of the resulting elution fractions was assessed using the Bio-Rad protein assay reagent according to the manufacturer's instructions (bovine serum albumin [BSA] was used as a standard for protein concentration) and by SDS-PAGE. Purification stringency was estimated by pixel densitometry.
Lysozyme OBody binding assay.
Beads were prewashed once in TBST (150 mM NaCl, 50 mM Tris-Cl [pH 8.4], 0.1% Tween 20). Approximately 50 mg of beads was added to 1 ml of TBST containing 2 mg/ml BSA, 2 mg/ml skim milk powder, and 1 mg/ml lysozyme. The beads were resuspended and incubated on a rotary mixer for 20 min, after which the beads were sedimented by centrifugation at 6,000 × g. The supernatant was removed, and the beads were washed three times (each wash consisting of resuspension of the beads, centrifugation, and removal of supernatant) in TBST with 0.05% Tween 20. To elute the bound protein, the beads were resuspended in 500 μl of 50 mM glycine (pH 2.0); the beads were removed by centrifugation at 16,000 × g, and the supernatant was neutralized with 50 μl of 1 M K2HPO4. The protein content of the resulting elution fractions was assessed as above.
Sandwich ELISA for ovine TNF-α.
Recombinant ovine TNF-α was kindly supplied by Anton Penthaner, AgResearch New Zealand, and prepared as described elsewhere (20). Tubulin was obtained from Cytoskeleton, Inc. Mouse monoclonal antibodies against bovine TNF-α (CC328), secondary anti-mouse antibodies conjugated to horseradish peroxidase (HRP), were obtained from Abcam. Beads were diluted to a concentration of 800 μg wet bead mass/ml in enzyme-linked immunosorbent assay (ELISA) buffer (PBS, 0.05% Tween 20, pH 7.4), and 100 μl of this bead suspension was added to the wells of a high-binding microtiter plate (Greiner bio-one) and incubated at 4°C overnight. The wells were then rinsed twice with ELISA buffer before blocking the wells with 200 μl of 1% BSA in ELISA buffer for 1 h. The blocking buffer was removed, and 100 μl of antigen in blocking buffer (10 μg/ml recombinant TNF-α) was added to the wells and incubated for 2 h, followed by washing of the wells four times with ELISA buffer. The primary anti-TNF-α antibody was diluted to 2.5 μg/ml in ELISA buffer, and 100 μl was added to wells and incubated for 2 h. Wells were washed twice in ELISA buffer before addition of 100 μl of secondary antibody at a 5,000-fold dilution and incubation for 2 h. The ELISA was also conducted with the omission of the TNF-α antigen as a control for unspecific binding of the antibodies by the beads. Wells were finally washed four times with ELISA buffer before adding 100 μl of o-phenylenediamine dihydrochloride developing solution. After 15 min, the reaction was stopped with the addition of 50 μl 1 N H2SO4 and the absorbance of the wells at 490 nm was read.
PHA quantification.
PHA content of the lyophilized cells was quantified using gas chromatography-mass spectrometry (GC/MS) after conversion of the PHA into 3-hydroxymethylester by acid-catalyzed methanolysis. GC/MS was performed by The New Zealand Institute for Plant & Food Research (Palmerston North, New Zealand).
RESULTS
Generation of DARPin-based MBP affinity beads.
A synthetic DARPin domain was selected as the first affinity binding domain for proof of concept due to its demonstrated production and solubility in the E. coli cytosol. A previously generated DARPin (off7) raised against MBP was selected (23). Two fusion proteins were generated, off7-PhaC and PhaC-off7, and fusions to the N terminus of PhaC were done via an unstructured serine-glycine linker to allow the domains to move and function independently. Fusions to the C terminus of PhaC utilized the linker described by Jahns and Rehm (6); this linker allows fusion of proteins to the conserved hydrophobic C terminus of PhaC. Recombinant production of these fusion proteins in E. coli BL21(DE3) harboring pMCS69 (pMCS69 carries the β-ketothiolase [phaA] and the acetoacetyl-CoA reductase [phaB)]genes, which are required for the formation of the precursor R-3-hydroxybutyryl-CoA, the substrate of the polyester synthase PhaC) produced PHB at levels lower than those obtained with strain BL21(DE3) expressing native PhaC: approximately 2 times less for the PhaC-off7 fusion and 4 times less for the off7-PhaC and off7-PhaC-off7 fusions (see Table S1 in the supplemental material). The isolated PHA beads had a protein coat density similar to that of PhaC beads (see Fig. S1 in the supplemental material).
The isolated fusion protein-displaying PHA beads were used to purify MBP from a cleared E. coli lysate containing recombinant MBP (Fig. 1A). One major band corresponding to MBP (50.9 kDa) is apparent in the elution fraction of both the N- and C-terminal DARPin fusions; this band is not apparent in the PhaC control (Fig. 1A). The purity of the elution fraction as estimated by gel densitometry was similar, as was the amount of protein eluted from the beads, whereas the binding of only PhaC beads was negligible (Fig. 1A and Table 2). These DARPin-displaying beads showed minimal nonspecific products in the elution fraction that were not present in the PhaC beads. It should be noted that significant amounts of MBP remained bound to the beads after a low-pH elution. Various different pH buffers, salt concentrations, and detergents were tested during elution as well as prolonged incubation in elution buffer in an attempt to elute this MBP from the beads, but no further protein could be eluted from the beads (data not shown).
FIG 1.
(A) Use of DARPin-based PHA beads for the purification of MBP from lysed E. coli cells expressing MBP. (B) Use of OBody-based PHA beads for the purification of lysozyme from a solution of skim milk, BSA, and lysozyme. (C) Use of VHH-based PHA beads produced in BL21(pMCS69) for the purification of GFP from lysed E. coli cells expressing GFP. (D) Use of VHH-based PHA beads produced in SHuffle(pMCS69E) for the purification of GFP from lysed E. coli cells expressing GFP. MW, molecular weight ladder (Life Technologies Mark 12 protein ladder); molecular weights (in thousands) are shown on the right.
TABLE 2.
Binding capacity and purification power of various affinity domain-displaying PHA beadsa
| Construct | Affinity domain type | Strain | Amt of target protein eluted (mg protein per g bead ± SD) | Amt of target protein (nmol per g bead) | % purity (based on SDS-PAGE) |
|---|---|---|---|---|---|
| PhaC | NA | B | 0.41 ± 0.20 | 8 | NA |
| off7-PhaC | DARPin | B | 4.71 ± 1.04 | 93 | 91 |
| PhaC-off7 | DARPin | B | 4.48 ± 0.56 | 88 | 90 |
| PhaC | NA | B | 0.32 ± 0.11 | 22 | NA |
| OB-PhaC | OBody | B | 6.65 ± 0.17 | 465 | 95 |
| PhaC-OB | OBody | B | 5.13 ± 0.20 | 360 | 72 |
| PhaC | NA | B | 0.21 ± 0.18 | 8 | NA |
| GFPVHH-PhaC | VHH | B | 6.19 ± 0.17 | 230 | 88 |
| PhaC-GFPVHH | VHH | B | 4.31 ± 0.31 | 160 | 78 |
| GFPVHH-PhaC-GFPVHH | VHH | B | 6.36 ± 0.19 | 236 | 90 |
| GFPVHH-PhaC | VHH | S | 9.07 ± 0.37 | 337 | 91 |
| PhaC-GFPVHH | VHH | S | 7.82 ± 0.18 | 291 | 90 |
| GFPVHH-PhaC-GFPVHH | VHH | S | 16.07 ± 0.31 | 598 | 96 |
Abbreviations: B, BL21(DE3)(pMCS69); S, SHuffle T7 Express(pMCS69E); SD, standard deviation; NA, not applicable.
Generation of OBody-based lysozyme affinity beads.
A synthetic OBody domain raised against hen white egg lysozyme was used as a second proof of concept (16). Two fusion proteins were generated; OB-PhaC and PhaC-OB. Expression of these fusions in E. coli BL21(DE3)(pMCS69) produced PHA at levels equivalent to that obtained with PhaC alone (see Table S1 in the supplemental material). The isolated PHA beads had a protein coat density similar to that of PhaC beads (see Fig. S1 in the supplemental material). The PHA beads were used to purify lysozyme from a solution containing BSA, skim milk powder, and lysozyme. Both N- and C-terminal OBody fusions could be used to purify lysozyme (14.3 kDa) from the solution, whereas the control beads containing PhaC only did not. These OBody-displaying beads showed low levels of nonspecific binding (Fig. 1B and Table 2). Based on densitometry, the purity of the eluted products was higher and the amount of lysozyme eluted was higher for the N-terminal fusions than for the C-terminal fusions, whereas the binding of the PhaC-only beads was negligible. Both the N- and C-terminal fusions bound at least 3.5 mol more target protein than the DARPin-based beads (Fig. 1B and Table 2).
Generation of VHH-based GFP affinity beads.
A VHH domain against GFP was selected due to the ability to easily visualize GFP binding and the relative ease of recombinantly producing GFP. The GFP-specific VHH domain used here is the domain cAbGFP4, which was initially generated from an immune alpaca library after immunization with GFP (24). GFPVHH-PhaC and PhaC-GFPVHH were generated as described for the DARPin fusions. One additional fusion that had GFPVHH domains on both termini of PhaC was made. When expressed in E. coli BL21(DE3)(pMCS69), both constructs with N-terminal VHH domains (GFPVHH-PhaC and GFPVHH-PhaC-GFPVHH) produced PHA at levels at least 4 times lower than those of the construct containing only PhaC, whereas the C-terminal fusion (PhaC-GFPVHH) produced PHA at levels equivalent to that obtained with PhaC alone (see Table S1 in the supplemental material). The isolated PHA beads had a protein coat density similar to that of PhaC beads (see Fig. S1 in the supplemental material). Beads were isolated from these strains and used to purify GFP from a cleared E. coli lysate expressing GFP. All three VHH fusions could purify GFP from the cell lysate (Fig. 1C). The purity levels of the eluted products were similar for the three fusion proteins. The binding capacity was higher for the N-terminal fusion than for the C-terminal fusions, whereas the binding of the PhaC-only beads was negligible. Both the N- and C-terminal fusions bound approximately one-half the moles of target protein bound by the OBody-based beads (Table 2). Interestingly, the double-VHH fusion (GFPVHH-PhaC-GFPVHH) beads had a GFP binding capacity similar to that of the single-fusion GFPVHH-PhaC beads (Fig. 1C and Table 2). This suggests that there could be a limitation in the correct folding of the disulfide-containing VHH domains in the E. coli BL21(DE3) cytosol.
Binding capacity of VHH-based beads can be increased by coexpression of the beads with DsbC and Erv1P in an oxidizing cytosol.
VHH domains derived from alpacas contain at least one disulfide bond, required for stability. The fact that the VHH double-fusion GFPVHH-PhaC-GFPVHH did not have a GFP binding capacity equal to the sum of its parts (Table 2) suggested that a subset of the VHH domains might not be folding correctly in the reducing cytosol. The PHA bead platform is based on the production of the VHH-PhaC fusion proteins in the bacterial cytosol and cannot be shifted to the oxidizing periplasm. To combat this, we initially adapted a method previously used for the cytosolic expression of VHH and scFv proteins in E. coli (25, 26). Briefly, leaderless disulfide bond isomerase (DsbC) was fused to PhaC at the opposing terminus of the VHH domain (i.e., GFPVHH-PhaC-DsbC and DsbC-PhaC-GFPVHH). Though both PHA yields and the overall binding capacities were generally increased in both cases, cofusion with DsbC increased the unspecific binding of the material and subsequently decreased the purity of the eluted protein (data not shown). This was presumably due to DsbC interacting with nontarget proteins via its chaperone function.
An alternative approach was based on the findings that coexpression of DsbC and the sulfhydryl oxidase Erv1p can increase the cytosolic expression of VHH fusion proteins (18). To do this, we used the E. coli strain SHuffle T7 express, which constitutively expresses a cytosolic form of DsbC and also has an oxidizing cytosol due to the trxB gor mutations. In addition to this, we modified the PhaA- and PhaB-expressing plasmid pMCS69 to also express Erv1P (generating the plasmid pMCS69E). Expression of GFPVHH-PhaC, PhaC-GFPVHH, and GFPVHH-PhaC-GFPVHH fusions in the E. coli SHuffle T7 Express(pMSC69E) background resulted in a significant increase both in the PHA yields and in the binding capacities. All constructs produced PHA at levels at least equivalent to that of the PhaC-only-mediated PHA synthesis (see Table S1 in the supplemental material). The binding capacity of PHA resin mediated by all constructs increased with GFPVHH-PhaC 1.47-fold; PhaC-GFPVHH increased 1.81-fold; and the double fusion GFPVHH-PhaC-GFPVHH increased 2.53-fold, to 16.07 mg (598 nmol GFP) per g wet beads (Fig. 1D and Table 2). The fact that the dual fusion gives a binding capacity very close to the sum of the two independent fusions suggests that VHH folding is no longer a problem in these strains. Along with the increases in binding capacity, there was an apparent increase in the purity of the eluted product, with minimal additional bands visible on a Coomassie blue-stained gel (Fig. 1D and Table 2).
TNF-α VHH affinity beads for use in ELISA.
To demonstrate the versatility of the affinity PHA beads, a second VHH domain against a more physiologically relevant target was used. A VHH domain raised from an immune alpaca library against ovine TNF-α was initially selected (20). N- and C-terminal fusions to PhaC were generated as described above. Production of these fusion proteins in E. coli BL21(DE3) mediated lower levels of PHA production, contributing to about 25% and 75% less PHA per cellular dry weight for PhaC-TNF-αVHH and TNF-α VHH-PhaC, respectively, than for PhaC only, whereas expression in E. coli SHuffle T7 Express(pMCS69E) resulted in higher levels, similar to what was seen with PhaC alone.
As TNF-α cannot be easily obtained naturally or be recombinantly produced at quantities required to conduct batch purification assays as described above for MBP, lysozyme, and GFP, a sandwich ELISA was performed to assess function. Due to the nature of the assay, binding capacities could not be accurately determined. A clear increased signal was observed in the samples containing beads displaying TNF-αVHH-PhaC and PhaC-TNF-αVHH compared to the PhaC-only sample. Also, the relative results were similar to those for the GFPVHH beads, with the material produced in E. coli SHuffle T7 Express(pMCS69E) performing at least 1.78-fold better in both cases (see Fig. S2 in the supplemental material).
Coproduction of an affinity resin and a target protein in the same cell.
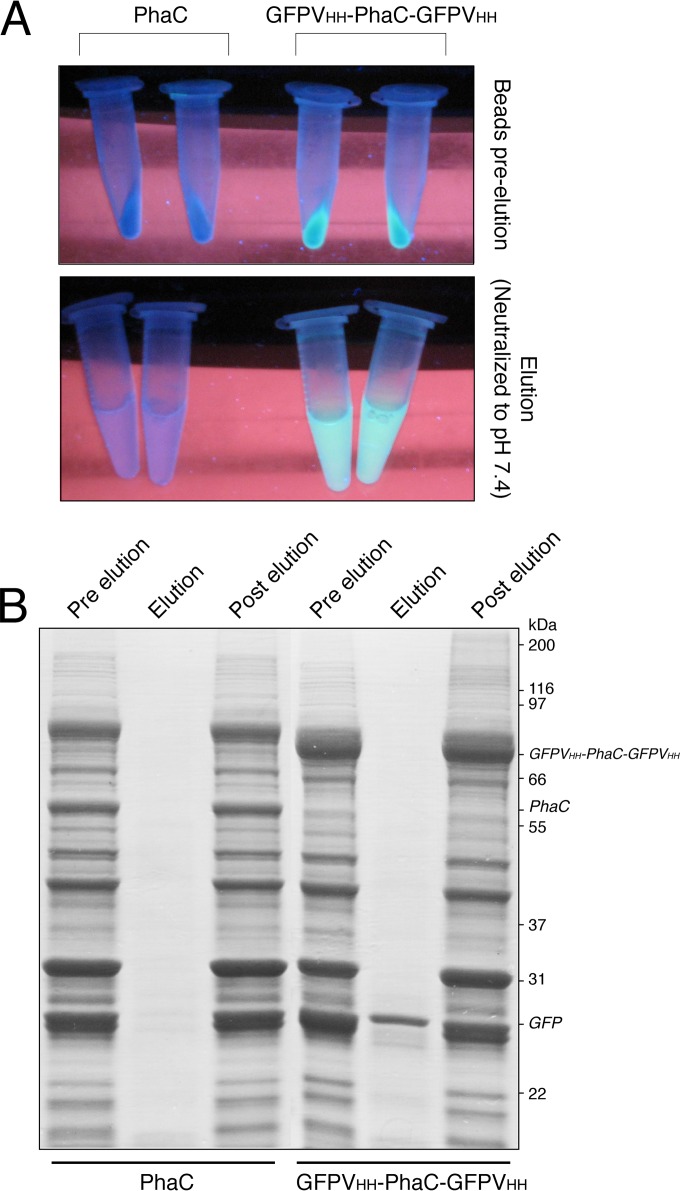
The ability to produce an affinity resin in vivo led to the idea of generating a system whereby a recombinant product and an affinity resin binding that product are produced in a single step within a single cell. As the dual VHH-GFP fusion produced in E. coli SHuffle T7 Express(pMCS69E) yielded material with the highest binding capacity and had high PHB yields (Fig. 1D; see also Table S1 in the supplemental material), we used this construct to assess the coproduction. A plasmid that allowed the polycistronic expression of GFP and the VHH-PhaC-VHH fusion protein was generated. PHA beads were produced based on the optimized conditions described above for the production of the GFPVHH domain fusions. In order to minimize the potential of loss of bound GFP during the isolation and washing steps and to simplify the purification steps, a simplified PHA bead isolation method was used. Briefly, the glycerol density gradient step was omitted and the detergent mixture Bugbuster was added to the lysis buffer. When GFP was coproduced with GFPVHH-PhaC-GFPVHH, GFP associated with the beads, whereas when GFP was coexpressed with PhaC alone, no GFP was found to be associated with the beads (Fig. 2A). The GFP could be eluted from the beads with a low-pH wash, yielding 13.47 mg (503 nmol GFP) per g wet PHA beads with an estimated purity of 81%, whereas the amount of protein eluted from the PhaC beads was negligible (Fig. 2B). The overall protein profile of the isolated PHA beads was not as clean with this simplified purification method, but as little nonspecific protein appeared in the elution profile, this does not appear to be a problem (Fig. 2; see also Fig. S1 in the supplemental material). Using this method under conventional lab-scale growth conditions (LB in a shake flask), a purified target protein yield of approximately 16 mg GFP per liter of culture could be obtained. It should be noted that some level of GFP remained in the soluble fraction of the cell lysate, indicating that not all the GFP was bound. This could be addressed by fine-tuning the expression of the VHH-PhaC fusion and the target (GFP) independently. Alternatively, reusing the postelution beads to bind the remaining soluble target protein contained in the lysate could provide a solution. The beads appeared to retain much of their activity postelution after several washes in PBS (data not shown).
FIG 2.
Coexpression of VHH affinity PHA bead and its target. (A) GFP can be seen associating with and eluted from the GFPVHH beads but not the PhaC beads. (B) GFP can be eluted from the GFPVHH material; lanes 1 and 4 contain the insoluble PHA-containing material obtained from cells coexpressing PHA beads and GFP; lanes 2 and 5 contain the low-pH elution from the isolated PHA material; lanes 3 and 6 contain the crude PHA bead material after the low-pH elution.
DISCUSSION
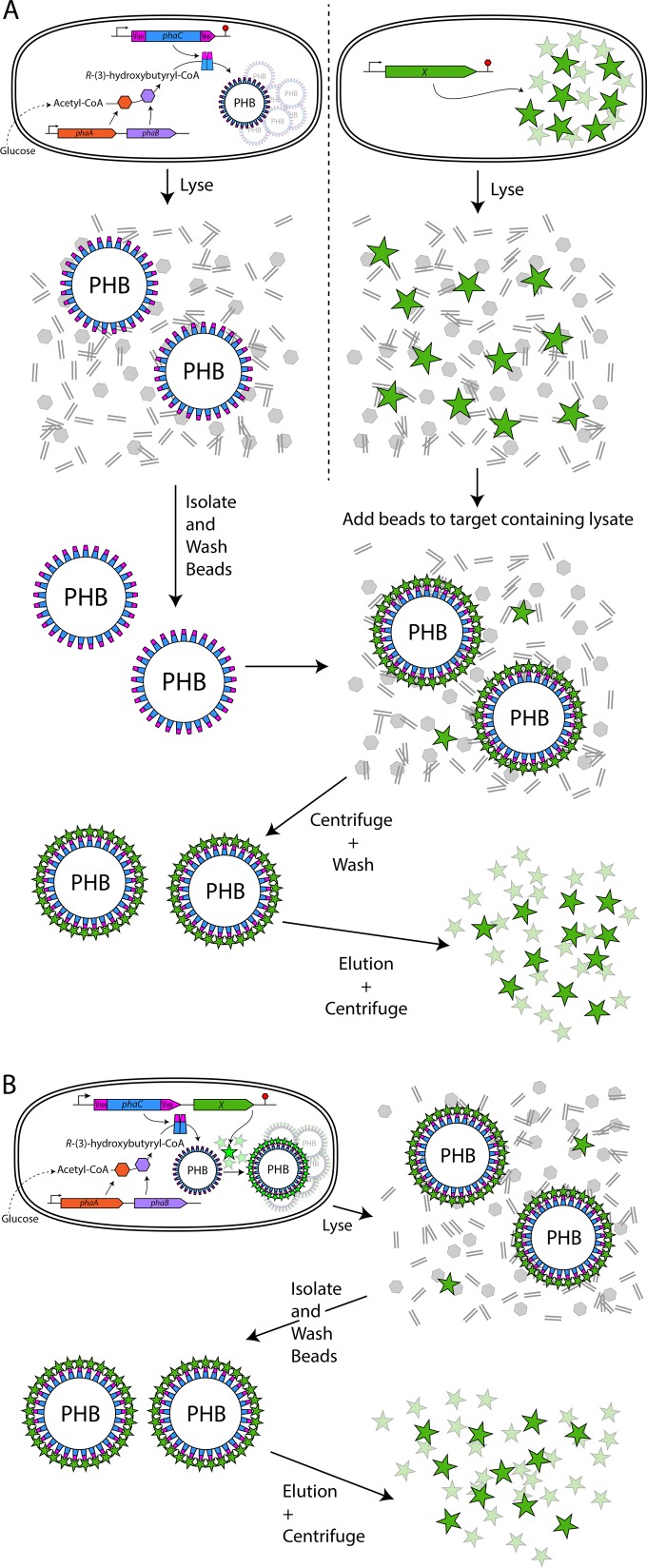
Here we have demonstrated the ability to generate affinity resins in vivo by utilizing and combining several existing technologies: 3 different customizable affinity binding domains, i.e., camelid VHH domains, DARPins, and OBodies, and the microbial production of intracellular biopolyester beads. The ability to recombinantly produce these affinity binding domains covalently attached to a matrix/support material in a single step could expand their attractiveness for bioseparation applications. These resins could be used to isolate target proteins from complex biological samples (e.g., total bacterial cell lysates). Furthermore, as demonstrated here, the ability to produce a desired target protein simultaneously with a defined affinity matrix inside the same cell provides a fast and simple alternative to conventional purification processes (Fig. 3A and B). As well as demonstrating the proof of principle for in vivo-produced customizable affinity resins, we generated usable affinity matrices to proteins commonly used as tags (MBP and GFP) (27, 28), thus providing a cheap and easily produced affinity resin to proteins currently used in many research labs.
FIG 3.
Schematic representations of the two approaches used to protein purification in this study. (A) Independent expression of an affinity bead (blue, PhaC domain; pink, affinity domain) and a target (green) in separate cultures. (B) Coexpression of the affinity bead and target (green) in a single cell.
Although the ligand density was similar on all beads (see Fig. S1 in the supplemental material), the binding capacities varied. DARPins initially appeared to be the most suitable candidate for the in vivo immobilization of an affinity domain on PHA due to their great stability and ability to be expressed at high levels in the bacterial cytosol (30% total cell protein or milligrams per liter) (14, 29), but the DARPin-based material performed the worst in this study. In terms of binding capacity, the amount of DARPin-based material was 3 to 6 times lower than the OBody- or optimized VHH domain-based material. Furthermore, the presence of the MBP-binding DARPin appeared to have an inhibitory effect on the function of the polyester synthase activity of PhaC, as observed by the 4-fold reduction in PHB content of the cells (see Table S1 in the supplemental material). It is unclear why fusion of the DARPin had a detrimental effect on the polyester synthase. The reduced target yield observed when using the DARPIN-based PHA beads is presumably due to the inability to completely elute the bound MBP from the beads. As the off7 DARPin domain has a dissociation constant of 4.4 nM, which is intermediate between that of the cABGFP VHH domain (0.23 nM) and that of the OBody domain (612.8 nM), it remains unclear why the target couldn't be eluted efficiently using standard elution conditions (15, 16, 24). However, it is not uncommon that for new high-affinity binding resin an elution protocol needs to be developed and optimized.
VHH proteins have commonly been produced in the oxidizing bacterial periplasm; here they are expressed in the reducing bacterial cytosol. A review of the literature reveals three other examples of the recombinant production of VHH molecules in the cytosol of E coli, of which two were achieved through fusion with DsbC or thioredoxin 1 (26, 30). In the third example, the VHH fusion protein was coproduced with DsbC and the sulfhydryl oxidase Erv1P in a tor grx mutant background. Here, coproduction with DsbC and the sulfhydryl oxidase Erv1P in a tor grx mutant background significantly enhanced the activity of the VHH domain-displaying beads, but it was not absolutely required for production of functional VHH domains. This is in contrast to the above-mentioned study, in which the fusion to DsbC required functional expression of the VHH domain (26, 31, 32).
Recently, a similar approach to produce immobilized binding domains in vivo was used with a red fluorescent protein (RFP)-specific VHH domain on the surface of magnetosomes in Magnetospirillum gryphiswaldense, but no data were supplied regarding the yield or binding capacity (31).
Previously, an in vivo-produced IgG binding bead based on fusion of a modified protein A domain to PHA synthase had been reported; the binding capacity of this bead was reported to be 100 mg of IgG per g of beads or approximately 660 nmol IgG per g beads (32). A similar binding capacity was observed in this study, in which the best-performing beads bound 16.07 mg of GFP per g of wet beads, corresponding to 598 nmol GFP per g beads. Currently, BAC/Life Technologies produces affinity resins based on Lama VHH binding domains against various targets (CaptureSelect). The respective VHHs are recombinantly produced by yeast and after purification cross-linked to agarose beads. The binding capacities of the various commercially available CaptureSelect products are quoted as between 2 and >15 mg of protein (depending on the specific target protein) per ml of resin; this equates to between 90 and 250 nmol per ml resin (Life Technologies CaptureSelect Pub. No. 4486257 Rev. B, 28 June 2013). Thus, all of the recombinant affinity polyester beads described here have at least as high a binding capacity, with the highest GFP binding VHH affinity PHA resin outperforming these agarose bead-based resins more than 2-fold and without the need to purify the VHH protein and to cross-link it to agarose. The higher binding capacity may be due to the smaller size (and thus increased surface area) of the bacterially produced polyester beads (typically up to around 600 nm) compared to the agarose beads used in the commercially available product (35 μm to 100 μm). It should be noted that as previously shown with enzyme-immobilized PHA beads (33), these PHA-based beads could be reused several times without obvious loss of binding capacity (data not shown).
The ability to coproduce an affinity resin and a target protein within a single cell has been demonstrated. This allows for the convenient production and isolation of the target protein. Expanding on this idea, it is widely accepted that the immobilization of proteins can have a beneficial effect on the stability and solubility of proteins, and thus this method may help in the soluble production of problematic (i.e., prone to inclusion body formation) proteins. One potential application for this finding is that the in vivo immobilization of the affinity domains and subsequent physical separation of the target protein on the surface of the polyester beads may aid in its folding or in preventing its incorrect aggregation. Indeed, VHH domains have been used to aid in the stability, solubility, and conformation of problematic proteins and to allow their crystallization (34, 35). We have also demonstrated that this system is compatible with other factors commonly used to combat problematic proteins (e.g., oxidizing cytosol mutant background or expression of sulfhydryl oxidase or disulfide bond isomerase).
Overall, in this study we have provided experimental evidence that PHA synthase engineering can be applied to implement various library-derived binding domains for the in vivo one-step production of disposable bioseparation resins with commercially relevant binding capacities and purification power. Furthermore, this method can be adapted to immobilize a target protein in vivo, allowing the single-step isolation of the target protein with mg yields per liter of culture with high purities.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02595-14.
REFERENCES
- 1.Mondal K, Gupta MN. 2006. The affinity concept in bioseparation: evolving paradigms and expanding range of applications. Biomol Eng 23:59–76. doi: 10.1016/j.bioeng.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Terpe K. 2003. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 3.Arnau J, Lauritzen C, Petersen GE, Pedersen J. 2006. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr Purif 48:1–13. doi: 10.1016/j.pep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Rehm BH. 2010. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol 8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 5.Rehm BH. 2006. Genetics and biochemistry of polyhydroxyalkanoate granule self-assembly: the key role of polyester synthases. Biotechnol Lett 28:207–213. doi: 10.1007/s10529-005-5521-4. [DOI] [PubMed] [Google Scholar]
- 6.Jahns AC, Rehm BH. 2009. Tolerance of the Ralstonia eutropha class I polyhydroxyalkanoate synthase for translational fusions to its C terminus reveals a new mode of functional display. Appl Environ Microbiol 75:5461–5466. doi: 10.1128/AEM.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockelbank JA, Peters V, Rehm BH. 2006. Recombinant Escherichia coli strain produces a ZZ domain displaying biopolyester granules suitable for immunoglobulin G purification. Appl Environ Microbiol 72:7394–7397. doi: 10.1128/AEM.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parlane NA, Grage K, Lee JW, Buddle BM, Denis M, Rehm BH. 2011. Production of a particulate hepatitis C vaccine candidate by engineered Lactococcus lactis. Appl Environ Microbiol 77:8516–8522. doi: 10.1128/AEM.06420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parlane NA, Wedlock DN, Buddle BM, Rehm BH. 2009. Bacterial polyester inclusions engineered to display vaccine candidate antigens for use as a novel class of safe and efficient vaccine delivery agents. Appl Environ Microbiol 75:7739–7744. doi: 10.1128/AEM.01965-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasiah IA, Rehm BH. 2009. One-step production of immobilized alpha-amylase in recombinant Escherichia coli. Appl Environ Microbiol 75:2012–2016. doi: 10.1128/AEM.02782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters V, Rehm BH. 2008. Protein engineering of streptavidin for in vivo assembly of streptavidin beads. J Biotechnol 134:266–274. doi: 10.1016/j.jbiotec.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Grage K, Rehm BH. 2008. In vivo production of scFv-displaying biopolymer beads using a self-assembly-promoting fusion partner. Bioconjug Chem 19:254–262. doi: 10.1021/bc7003473. [DOI] [PubMed] [Google Scholar]
- 13.Harmsen MM, De Haard HJ. 2007. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol 77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stumpp MT, Binz HK, Amstutz P. 2008. DARPins: a new generation of protein therapeutics. Drug Discov Today 13:695–701. doi: 10.1016/j.drudis.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grutter MG, Pluckthun A. 2004. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol 22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 16.Steemson JD. 2011. Directed evolution and structural analysis of an OB-fold domain towards a specific binding reagent. Ph.D. thesis University of Waikato, Hamilton, New Zealand. [Google Scholar]
- 17.Steemson JD, Baake M, Rakonjac J, Arcus VL, Liddament MT. 2014. Tracking molecular recognition at the atomic level with a new protein scaffold based on the OB-fold. PLoS One 9:e86050. doi: 10.1371/journal.pone.0086050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veggiani G, de Marco A. 2011. Improved quantitative and qualitative production of single-domain intrabodies mediated by the co-expression of Erv1p sulfhydryl oxidase. Protein Expr Purif 79:111–114. doi: 10.1016/j.pep.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Saerens D, Conrath K, Govaert J, Muyldermans S. 2008. Disulfide bond introduction for general stabilization of immunoglobulin heavy-chain variable domains. J Mol Biol 377:478–488. doi: 10.1016/j.jmb.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Maass DR, Sepulveda J, Pernthaner A, Shoemaker CB. 2007. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). J Immunol Methods 324:13–25. doi: 10.1016/j.jim.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bessette PH, Aslund F, Beckwith J, Georgiou G. 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci U S A 96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amara AA, Rehm BH. 2003. Replacement of the catalytic nucleophile cysteine-296 by serine in class II polyhydroxyalkanoate synthase from Pseudomonas aeruginosa-mediated synthesis of a new polyester: identification of catalytic residues. Biochem J 374:413–421. doi: 10.1042/BJ20030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binz HK, Amstutz P, Pluckthun A. 2005. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol 23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 24.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, Leonhardt H. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods 3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 25.Jurado P, Ritz D, Beckwith J, de Lorenzo V, Fernandez LA. 2002. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J Mol Biol 320:1–10. doi: 10.1016/S0022-2836(02)00405-9. [DOI] [PubMed] [Google Scholar]
- 26.Olichon A, Surrey T. 2007. Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in Escherichia coli. J Biol Chem 282:36314–36320. doi: 10.1074/jbc.M704908200. [DOI] [PubMed] [Google Scholar]
- 27.Tsien RY. 1998. The green fluorescent protein. Annu Rev Biochem 67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 28.Waugh DS. 2005. Making the most of affinity tags. Trends Biotechnol 23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Tamaskovic R, Simon M, Stefan N, Schwill M, Pluckthun A. 2012. Designed ankyrin repeat proteins (DARPins) from research to therapy. Methods Enzymol 503:101–134. doi: 10.1016/B978-0-12-396962-0.00005-7. [DOI] [PubMed] [Google Scholar]
- 30.Rahbarizadeh F, Rasaee MJ, Forouzandeh-Moghadam M, Allameh AA. 2005. High expression and purification of the recombinant camelid anti-MUC1 single domain antibodies in Escherichia coli. Protein Expr Purif 44:32–38. doi: 10.1016/j.pep.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Pollithy A, Romer T, Lang C, Muller FD, Helma J, Leonhardt H, Rothbauer U, Schuler D. 2011. Magnetosome expression of functional camelid antibody fragments (nanobodies) in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 77:6165–6171. doi: 10.1128/AEM.05282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis JG, Rehm BH. 2009. ZZ polyester beads: an efficient and simple method for purifying IgG from mouse hybridoma supernatants. J Immunol Methods 346:71–74. doi: 10.1016/j.jim.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Hooks DO, Blatchford PA, Rehm BH. 2013. Bioengineering of bacterial polymer inclusions catalyzing the synthesis of N-acetylneuraminic acid. Appl Environ Microbiol 79:3116–3121. doi: 10.1128/AEM.03947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domanska K, Vanderhaegen S, Srinivasan V, Pardon E, Dupeux F, Marquez JA, Giorgetti S, Stoppini M, Wyns L, Bellotti V, Steyaert J. 2011. Atomic structure of a nanobody-trapped domain-swapped dimer of an amyloidogenic beta2-microglobulin variant. Proc Natl Acad Sci U S A 108:1314–1319. doi: 10.1073/pnas.1008560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koide S. 2009. Engineering of recombinant crystallization chaperones. Curr Opin Struct Biol 19:449–457. doi: 10.1016/j.sbi.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.