Abstract
Nitrification, mediated by ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA), is important in global nitrogen cycling. In estuaries where gradients of salinity and ammonia concentrations occur, there may be differential selections for ammonia-oxidizer populations. The aim of this study was to examine the activity, abundance, and diversity of AOA and AOB in surface oxic sediments of a highly nutrified estuary that exhibits gradients of salinity and ammonium. AOB and AOA communities were investigated by measuring ammonia monooxygenase (amoA) gene abundance and nitrification potentials both spatially and temporally. Nitrification potentials differed along the estuary and over time, with the greatest nitrification potentials occurring mid-estuary (8.2 μmol N grams dry weight [gdw]−1 day−1 in June, increasing to 37.4 μmol N gdw−1 day−1 in January). At the estuary head, the nitrification potential was 4.3 μmol N gdw−1 day−1 in June, increasing to 11.7 μmol N gdw−1 day−1 in January. At the estuary head and mouth, nitrification potentials fluctuated throughout the year. AOB amoA gene abundances were significantly greater (by 100-fold) than those of AOA both spatially and temporally. Nitrosomonas spp. were detected along the estuary by denaturing gradient gel electrophoresis (DGGE) band sequence analysis. In conclusion, AOB dominated over AOA in the estuarine sediments, with the ratio of AOB/AOA amoA gene abundance increasing from the upper (freshwater) to lower (marine) regions of the Colne estuary. These findings suggest that in this nutrified estuary, AOB (possibly Nitrosomonas spp.) were of major significance in nitrification.
INTRODUCTION
Nitrification is central to the global nitrogen cycle, coupling ammonia production from mineralization of organic matter with denitrification. In estuaries, discharge of domestic and industrial waste as well as runoff from mineral fertilizers and nitrogen fixation may contribute to ammonium enrichment. Ammonia oxidation is considered to be the rate-limiting step of nitrification and is catalyzed by ammonia monooxygenase (AMO), which is encoded by the amoA gene. It was previously considered that autotrophic ammonia oxidation is carried out solely by ammonia-oxidizing bacteria (AOB). However, the discovery of a marine archaeon belonging to the thaumarchaea which also oxidizes ammonia showed that this is not the case (1, 2). Ammonia-oxidizing archaea (AOA) belonging to the phylum Thaumarchaeota (AOA) are widely distributed in terrestrial and aquatic environments (2, 3) and thus may be more important contributors to nitrification than was previously considered. While previous studies of marine sediments showed different patterns of either AOA or AOB dominance, in estuarine environments where gradients of salinity and ammonia concentrations occur, there may be a differential selection for ammonia-oxidizer populations along these gradients (4–9). It has been previously suggested that AOA are significant in estuarine nitrogen cycling (6) and that AOA were more abundant than AOB along an estuarine salinity gradient (8).
The focus of the current study was the River Colne estuary, a macrotidal, hypernutrified estuary on the east coast of the United Kingdom (Fig. 1) which has very high inorganic nitrogen levels in the upper estuary from inputs from the River Colne and a major sewage treatment works, with decreasing gradients of both ammonium and nitrate downstream (10, 11). Benthic denitrification can remove ∼44% of the total oxidized nitrogen load (25% of total inorganic nitrogen) from the estuary before it enters the North Sea; coupled nitrification-denitrification accounts for about 25% of the total denitrification (12). However, little is known about the benthic AOA and AOB communities along the estuary and their potential links with biogeochemical function (10).
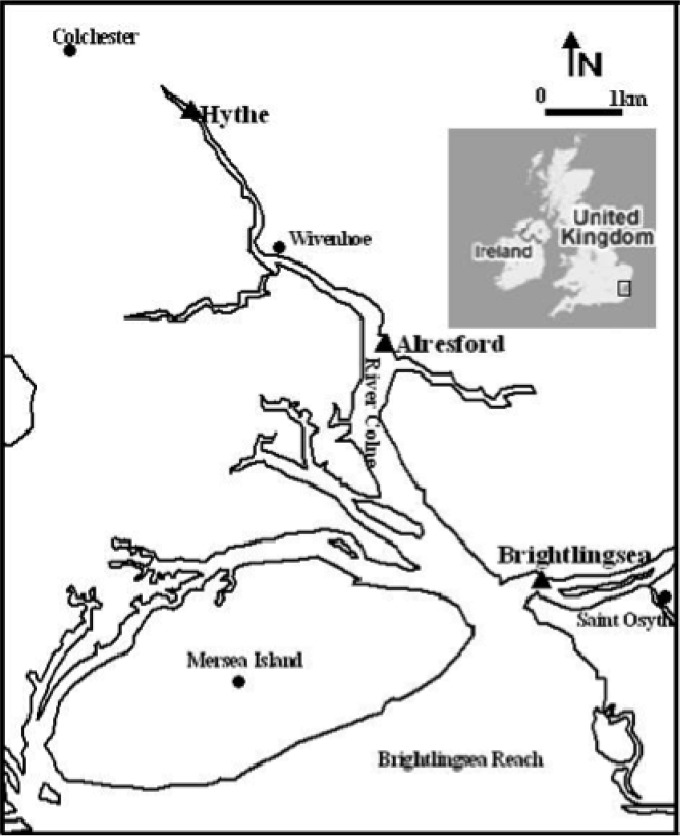
FIG 1.
Map of the Colne estuary showing sample locations at Hythe, Alresford, and Brightlingsea (modified from reference 50).
It has been suggested that AOA and AOB niches are defined by the concentrations of ammonium present (13), with AOA dominating in low-ammonium environments such as some soils (14) and the open ocean (15) and in some estuaries (6, 16). In the present study, we hypothesized that in the upper estuary of the River Colne at the Hythe, where very high levels of inorganic N occur (12), nitrification is driven by AOB, which predominate over AOA. In contrast, as ammonium concentrations in the water column decline downstream, the significance of AOA with respect to nitrification might be expected to increase and AOA may become proportionately more important toward the estuary mouth. In addition, the estuarine salinity gradient might also tend to favor AOA over AOB as salinity increases down the estuary (6). Our study aimed to examine the activity, abundance, diversity, and distribution of these different groups of ammonia oxidizers (AOA and AOB) in the surface oxic sediments where nitrification can occur. The overall goal was to test whether there is spatial and temporal variation in the relative abundances of AOA and AOB amoA genes in relation to sediment nitrification potentials in this hypernutrified estuary that exhibits gradients of salinity and ammonium concentration.
MATERIALS AND METHODS
Field sampling.
Samples were taken at approximately three monthly intervals to cover temporal variations between June 2009 and January 2010 at three sites along the Colne estuary, United Kingdom (Fig. 1), the upper estuary at the Hythe (51°52.4′N, 0°55.5′E), the mid-estuary at Alresford Creek (51°50.5′ N, 0°58.4′E), and the estuary mouth at Brightlingsea (51°45′N, 1°30′E), as described previously (10, 12). At each site, triplicate surface sediment samples (depth, 0 to 1 cm) were collected using a sterile spatula. (The oxic layers in which nitrification may occur range from a maximum depth of 1.5 mm in winter at the Hythe to approximately 5 mm in winter in the sandier sediment at the estuary mouth at Brightlingsea, and in the summer, oxic layer depths are even shallower [10]). Collected samples were returned to the laboratory on ice within 1 h of sampling. Each replicate was quickly homogenized by mixing, and aliquots (1 g wet weight) of sediment from each replicate were stored at −80°C prior to nucleic acid isolation. Nitrate and ammonium concentrations in the pore water were measured colorimetrically (17) using an autoanalyzer (Skalar Analytical, Netherlands). Sediment water content was determined by oven-drying samples of sediment (5 g wet weight) at 85°C for 48 h to a constant weight.
Nitrification potential measurements.
Nitrification potential is the maximum capacity of a soil's or sediment's population of nitrifying microorganisms to transform NH4+-N to NO3−-N. Changes in nitrification potentials provide quantitative information on how nitrifying communities respond to changes in environmental conditions and reflect potential changes in the in situ nitrification rates. Nitrification potentials were measured with sediment slurries from each site by mixing each of triplicate 10-g (wet weight) samples of sediment from the depth layer of 0 to 1 cm with 100 ml of sterile ESAW medium (18) amended with 300 μM NH4Cl and 60 μM KH2PO4. A further triplicate set of slurries containing allylthiourea (ATU) (172 μM final concentration) were also set up as controls to differentiate autotrophic nitrification from heterotrophic nitrification and examine the relative contributions of AOA and AOB to nitrification activity. All slurries were incubated in the dark at 25°C with gentle shaking (110 rpm) to maintain aeration. Subsamples were removed and analyzed for NH4+ at intervals over 48 h (19). Ammonium concentrations in sediment pore water were analyzed by the indophenol blue spectrophotometric method (20). Rates of ammonium removal were determined by linear regression analysis of the concentrations of ammonium with time.
Real-time quantitative PCR (Q-PCR) of AOB and AOA amoA genes.
Total nucleic acids were extracted from 0.5-g (wet weight) sediment samples (21, 22). Unfortunately, the October (autumn) samples for measurements of gene abundances were lost through equipment malfunction, so only three such temporal samples were available; nonetheless, those samples covered the seasonal extremes of temperature. The number of amoA gene copies per g of sediment (dry weight) was measured using primers amoA-1F and amoA-2R (23) to target the amoA gene from AOB and primers CrenamoA23F and CrenamoA616R (24) to target the amoA gene from AOA. DNA standards were created by PCR amplification of sediment DNA extracts. The resulting amplicons were purified using a QIAquick PCR purification kit (Qiagen) prior to quantification using a Nanodrop ND-1000 spectrophotometer. The target abundance for standards was calculated by assuming a molecular mass of 660 Da for double-stranded DNA using the following formula: gene abundance = 6.023 × 1023 (copies mol−1) × standard concentration (g ml−1)/molecular mass (g mol−1).
Standard curves were created using a dilution series of each DNA standard ranging from 102 to 106 target genes ml−1 for AOB amoA and from 101 to 105 target genes ml−1 for AOA amoA. Standards, samples, and no-template controls (NTC) were amplified in triplicate with each primer set. Reactions were performed on a CFX96 real-time system (Bio-Rad) with initial denaturation for 5 min at 95°C, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Each 20-μl reaction mixture contained 20 ng of DNA template, 10 μl of 2× SensiFAST SYBR No-ROX master mix (Bioline), and a 100 nM concentration of each primer. A dissociation curve analysis was performed at the end of each reaction to verify amplification of a single PCR product. The samples were quantified against the corresponding standard curve using CFX Manager version 2.0 software (Bio-Rad).
PCR-DGGE analysis of the AOB 16S rRNA and amoA genes.
PCR amplification of the amoA gene was undertaken (to further define the AOB present) using primers amoA-1F-GC and amoA-2R and PCR cycling conditions as described previously (23). PCR amplification of the AOB 16S rRNA gene used primers CTO189f-GC and CTO654r and cycling conditions as previously described (25). All PCR amplifications were performed in a GeneAmp PCR System 9700 Thermocycler (Applied Biosystems). RNA extraction and reverse transcription-PCR (RT-PCR) reaction experiments were performed as previously described (26). Denaturing gradient gel electrophoresis (DGGE) was performed as previously described (27) except that the gels were silver stained (22). DGGE bands were excised and sequenced using a reverse primer (either amoA-2R or CTO654r) (23, 24) by Geneservice Ltd. (Cambridge, United Kingdom). Phylogenetic analysis of amoA and 16S rRNA gene sequences was performed using PHYLIP 3.4 (28) with the Jukes-Cantor DNA distance and neighbor-joining methods (29, 30). Bootstrap analysis was based on 1,000 replicates. Trees were constructed using Treeview (WIN32 version 1.5.2) (31).
454 pyrosequencing.
In order to determine whether the total microbial community followed spatiotemporal patterns similar to those seen with the AOA and AOB taxa, we used a broad community-screening approach targeting 16S rRNA phylogenic marker genes. Samples were PCR amplified and pyrosequenced by the Research and Testing Laboratory (Lubbock, Texas, USA) using a Roche 454 FLX instrument with Titanium reagents for tag-encoded FLX amplicon pyrosequencing on the basis of their standard PCR methods and protocols. For bacterial 16S rRNA gene libraries, primers Gray28F and Gray519R were applied, producing a fragment of 491 bp (32). For archaeal 16S rRNA gene libraries, primers ARCH349F and ARCH806R were applied, producing a fragment of 457 bp (33).
High-throughput community pyrosequencing results were analyzed using the QIIME pipeline and its associated modules (34). All sequences were checked for the presence of correct pyrosequencing adaptors, 10-bp bar codes, and taxon-specific primers, and those containing errors in these regions were removed. Sequences less than 450 bp in length were removed, and sequences over 550 bp in length were removed. Sequences with low (<25) quality scores and sequences containing homopolymer inserts were also removed. All pyrosequence reads were clustered into operational taxonomic units (OTUs) at the 95% similarity level using the UCLUST algorithm (35), and any chimeras present were removed using ChimeraSlayer. Representative sequences from each OTU were identified using RDP Classifier (36). Finally, all singletons were removed before further analysis was performed (37).
Statistical analysis.
Bacterial and archaeal amplicon libraries were analyzed separately, treating these taxa as two distinct assemblages. Data were analyzed via nonmetric multidimensional scaling (NMDS) based on Jaccard's index as a measure of community dissimilarity. NMDS was supported by permutational multivariate analysis of variance (PERMANOVA), which was also based on Jaccard's index. Species richness was calculated using rarefaction and normalized to the sample with the fewest amplicon reads. All community analyses were conducted using R statistical language version 2.7.2 and the R standard libraries and the community-analysis-specific package “vegan” (38).
A paired-samples t test was used to compare AOB and AOA amoA gene abundances, and two-way ANOVAs and Tukey's honestly significant difference (HSD) post hoc analysis at a 95% confidence interval were used to determine the effect of site and season on AOB and AOA amoA gene abundance (39). Pearson's correlation analyses were performed to determine whether there was a correlation between AOB and AOA amoA gene abundance and nuclear paramagnetic resonance (NPR) (39).
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank under the following accession numbers: JX567314 to JX567343.
RESULTS
Potential nitrification rates.
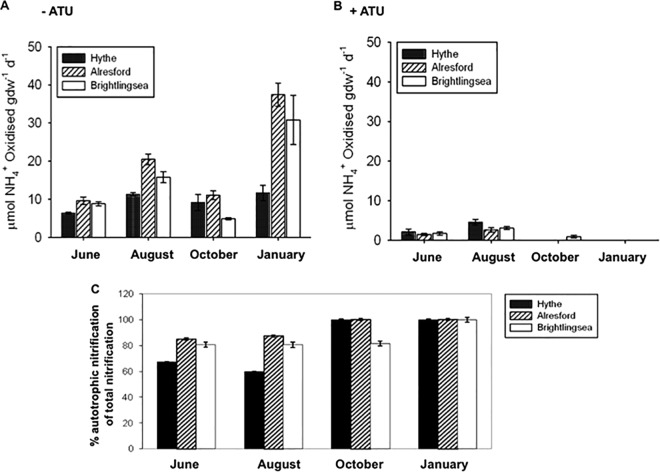
There were significant differences, both spatial and temporal, between nitrification potentials along the estuary (P < 0.001, Tukey HSD test) (Fig. 2). In the absence of ATU, nitrification potentials tended to be greatest in the mid-estuary and lowest in the upper estuary at the Hythe, where oxic layer depths were shallower than at the other sites. Nitrification potentials were significantly (P < 0.009, Tukey HSD test) higher in January than in the warmer summer and autumn months, when the surface oxic layers were shallow (Fig. 2A).
FIG 2.
Mean (± standard error [SE], n = 3) nitrification potential rates along the Colne estuary. (A) Without ATU (allylthiourea); (B) with ATU; (C) percentage autotrophic nitrification to total nitrification. d, day.
In the presence of ATU, nitrification potentials decreased significantly compared to control results (P < 0.001, Tukey HSD test) and were detectable only in the summer months at all sites and also from Brightlingsea in October (Fig. 2B). The nitrification potentials were completely inhibited by ATU in October and January and decreased to about 80% in June and August.
ATU-insensitive nitrification was of greatest significance (at up to 33% of the measured nitrification potential) in the upper estuary and mid-estuary sediments during high-temperature periods (e.g., June to August) (Fig. 2C). Within the oxic layer of all the sediments, nitrification was almost entirely ATU sensitive in October and January (Fig. 2C).
AOA and AOB amoA gene abundances.
AOB amoA gene abundances were significantly higher (by approximately 2 orders of magnitude) than AOA amoA gene abundances across all sites and seasons (Fig. 3) [paired-samples t test; t(25) = 3.92, P = 0.001]. Moreover, AOB amoA gene abundances differed significantly across sites [F(2,25) = 42.28, P < 0.001] and seasons [F(2,25) = 14.86, P < 0.001], and there were significant spatial and temporal interaction effects on AOB amoA gene abundance [F(4,25) = 16.68, P < 0.001]. Post hoc analysis revealed significantly higher AOB amoA gene abundance at the estuary mouth at Brightlingsea than at the Hythe (P < 0.001) and Alresford (P = 0.001). In addition, AOB amoA abundances were significantly higher across sites in June than in August (P < 0.001). In contrast to AOB results, there were no significant spatial or temporal differences in AOA amoA gene abundances. In general, the ratio of AOB/AOA amoA gene copy numbers in surface sediments increased strongly from the upper to lower regions of the estuary throughout the seasons (Table 1). In the present study, there was no significant correlation between potential nitrification rates and AOB amoA gene abundance across the different sites in the Colne estuary (r = 0.044, P = 0.831).
FIG 3.
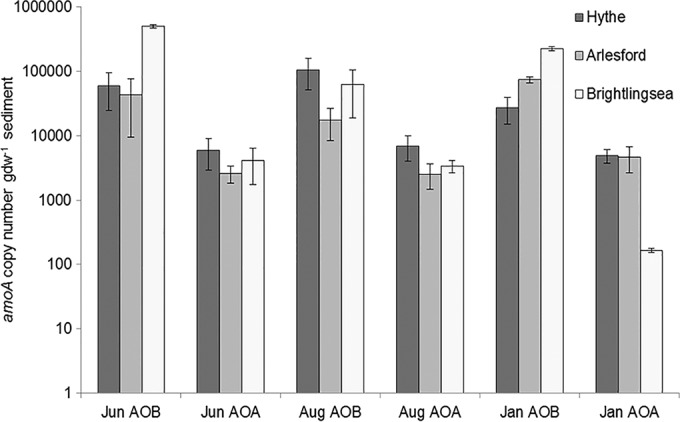
AOB and AOA amoA gene abundance (mean ± SE, n = 3) at Hythe, Alresford, and Brightlingsea in June (Jun), August (Aug), and January (Jan).
TABLE 1.
Ratio of AOB/AOA amoA gene abundance and physicochemical characteristics of the estuarine sites
| Site (mo) | AOB/AOA ratio | Salinity | Water content (%) | NH4+-Na (μM) | NO2−-N + NO3−-Na (μM) |
|---|---|---|---|---|---|
| Hythe (June) | 10.0 | 3.0 | 64.3 | 14.13 | 20.11 |
| Alresford (June) | 16.5 | 20.0 | 51.2 | 4.46 | 3.52 |
| Brightlingsea (June) | 121.0 | 33.5 | 70.0 | 2.10 | 0.94 |
| Hythe (August) | 15.0 | 3.0 | 66.6 | 6.69 | 6.56 |
| Alresford (August) | 6.9 | 20.0 | 53.2 | 6.31 | 2.99 |
| Brightlingsea (August) | 18.2 | 33.5 | 61.4 | 3.21 | 1.30 |
| Hythe (January) | 5.6 | 3.0 | 70.3 | 4.48 | 4.98 |
| Alresford (January) | 15.8 | 20.0 | 57.5 | 5.01 | 2.53 |
| Brightlingsea (January) | 1340.6 | 33.5 | 34.9 | 3.68 | 1.39 |
Sediment pore water was used to measure nutrient concentrations.
AOA and AOB community structures.
Since AOB were both temporally and spatially dominant over AOA in the Colne estuary, AOB communities were further analyzed by DGGE band sequencing of the amoA and 16S rRNA genes from extracted DNA and reverse-transcribed RNA from ammonia-oxidizing bacteria. In total, 30 bands with distinct positions in the DGGE fingerprints (amoA gene,14 bands; 16S rRNA gene, 16 bands) from across sites and sample time points were obtained (see Fig. S1 and S2 in the supplemental material). Based on both DNA and RNA profiles from amoA and 16S rRNA genes, AOB communities from the Hythe were more distinct than those from Alresford and Brightlingsea (see Fig. S1 and S2 in the supplemental material). In general, the DNA profiles of the amoA gene showed that there were a greater number of bands at the Hythe and that the number of bands decreased downstream and with time (see Fig. S1). In the RNA profiles for both the amoA and 16S rRNA genes, there were a number of unique fragments which were absent in the DNA profiles that had high sequence identity to Nitrosomonas spp. (bands 5 to 7 [see Fig. S1A]; bands 4 to 5 [Fig. S1B]; band 4 [Fig. S1C]; bands 5 to 6 [Fig. S2B]; and band 6 [Fig. S2C]), suggesting that there was a less complete picture of the AOB community in the DNA fingerprints.
Phylogenetic analysis of amoA gene sequences revealed that several DGGE bands retrieved from the Hythe and Brightlingsea in June and August grouped with 100% bootstrap confidence to a Nitrosomonas clade (see Fig. S3A in the supplemental material). In addition, amoA gene sequences recovered from the Hythe (in June and October) and Brightlingsea (in August and October) formed two discrete clades which grouped with Nitrosomonas cryotolerans (see Fig. S3A). Phylogenetic analysis of the 16S rRNA gene sequences from DGGE bands corroborated the amoA gene sequence data showing a clustering with Nitrosomonas spp. (with the exception of one DGGE band recovered from Brightlingsea in August which clustered within a Nitrosospira clade) (see Fig. S3B).
Observed differences between AOB and AOA communities may also be accounted for by their respective proportions of the total bacterial and archaeal communities. While DGGE analysis provided putative identification of potential key AOB species, it did not provide a robust analysis of total bacterial and archaeal communities. In order to examine whether AOB and AOA populations follow general trends of bacterial and archaeal communities, we examined total bacterial and archaeal communities using 454 pyrosequencing of 16S rRNA genes. NMDS analysis of total bacterial and archaeal pyrosequencing libraries (6,904 bacterial operational taxonomic units [OTUs] and 234 archaeal OTUs from 118,800 and 3,381 amplicon reads, respectively, following quality control checks) showed that both total bacterial and archaeal communities changed in composition along the estuary (see Fig. S4A and C in the supplemental material; PERMANOVA) [for bacteria, F(2,9) = 1.31, P = 0.054; for archaea, F(2,8) = 1.19, P = 0.084] but not across seasons (see Fig. S4A and C; PERMANOVA) (P = >0.1 in all cases). In general, the total bacterial communities were twice as rich as those of the archaea (see Fig. S4B and D), even when normalized (rarefied) for differences in sequencing intensity across kingdoms.
OTUs that were assigned to known AOB species represented only a very small fraction of the total 16S rRNA bacterial libraries. Nitrosospira-like sequences assigned to a single OTU were detected across all samples and comprised <0.07% of the bacterial libraries. A further OTU assigned to the family Nitrosomonadaceae was found in most libraries but was more abundantly detected in August in Arlesford (0.11%) and the Hythe (0.05%). Nitrite-oxidizing bacteria were generally more abundant than AOB, with seven OTUs assigned to the genus Nitrospira and six to the genus Nitrospina which together represented up to a maximum of 0.66% and 0.85% of 16S rRNA sequences in the January Hythe and Brightlingsea samples, respectively. AOA sequences were undetected in the archaeal 16S rRNA libraries, suggesting that they represent only a very small proportion of the total archaeal communities at each site, and this supports the idea of a lower measured abundance of AOA than AOB amoA genes.
DISCUSSION
Nitrification potentials measured in the Colne estuary sediments in the absence of ATU increased in winter in a manner commensurate with increases in sediment oxic zone depth stimulating nitrification. Indeed, previous work (10) has reported sediment-water export of nitrate from sandy sediments at the mouth of the estuary during winter, when, despite the low temperature, the oxic layer depth is maximal. Addition of 172 μM ATU resulted in drastic reductions of nitrification potentials at all sites and times: virtually complete inhibition in October and January and 60% to 80% inhibition in warmer months. AOB are reportedly more sensitive to ATU than AOA, while heterotrophic nitrification is also not inhibited by ATU (40). This would suggest that the largest part of the nitrification potentials in the sediments of the Colne is due to AOB, at virtually 100% during winter, with the small residual potentials in the presence of ATU due to AOA and heterotrophic nitrifiers which are not inhibited by the concentration of ATU used. In the more highly organic sediments in the upper estuary, ATU-insensitive nitrification was 30% to 40% of the total during June and August. AOB seem to be more sensitive to ATU than AOA, with AOA being able to maintain the ability to oxidize ammonium in the presence of 100 μM ATU (41), while the AOA “Candidatus Nitrososphaera viennensis” required 500 μM ATU to stop nitrification (42). This suggests that nitrification in the Colne estuary was largely attributable to ATU-sensitive AOB rather than AOA.
In the Colne estuary, AOB were generally both spatially and temporally dominant over AOA, with the ratio of AOB/AOA amoA gene abundances increasing from the upper (freshwater) to lower (marine) regions. Within the AOB, several sequences relating to Nitrosomonas spp. were recovered from all sites and months, suggesting that members of this genus were dominant in the AOB community. In a study of the Schelde estuary, Nitrosomonas spp. were also found to be dominant over Nitrosospiras (43). A selection for Nitrosospira spp. with increasing salinity in an estuary system has also been shown previously (9). In another study, Nitrosospira spp. were also found at the marine sites of an estuary (7). In the Colne estuary, one DGGE band of a sample which was recovered from the marine site at Brightlingsea in August showed high 16S rRNA gene sequence identity (99%) to Nitrosospira sp. The lack of recovered sequences relating to Nitrosospira spp. in the Colne estuary was supported by the pyrosequencing analysis, whereby Nitrosospira spp. represented <0.07% of the bacterial libraries.
It has been suggested that the AOA and AOB niches are defined by ammonium concentrations (13). Indeed, differences with respect to niche and response to ammonia concentration may also exist between different lineages of AOB and AOA. For example, the thaumarchaeon Nitrosopumilus maritimus SCM1 appears to have a much higher affinity than other AOA for ammonium (13). The available data suggest that thaumarchaeal AOA have extremely low Km values (in the nM range) for ammonium (13) and that higher ammonium concentrations may also inhibit them (41). While low Km values may make AOA highly competitive at low ammonium concentrations, the AOA may be outcompeted by the AOB at higher concentrations. Such nM ammonium concentrations are greatly exceeded (at μM levels) in both water column and sediment pore water (references 10 and 11 and this study) in the Colne estuary, conforming with the predominance of AOB found in this study. Similarly, it has been previously shown that AOA grew at all concentrations of ammonium added to soil microcosms but that AOB grew only at the highest ammonium concentration added (44). AOA have also been shown to dominate over AOB in low-ammonium environments such as some soils (3), in the open ocean (15), and in other estuaries (6). In the present study, if the AOA niche were strongly influenced by lower ammonium concentrations, as previously proposed (13), we might hypothesize a trend in favor of AOA down the estuary as ammonium in the water column declines due to dilution with low-nutrient seawater. However, this was not observed; in general, the ratio of AOB/AOA amoA gene copy numbers in surface sediments increased strongly from the upper to the lower regions of the estuary throughout the seasons, possibly because, even at Brightlingsea, the ambient ammonium concentrations in the water column exceeded that favoring AOA. Furthermore, ammonium concentrations in the sediment pore waters are even greater than in the water column due to ammonification from breakdown of organic matter in the sediments under anoxic conditions (45). In the present study, there were no significant spatial or temporal correlations among the combined AOA and AOB amoA gene abundances, nitrification potentials, and ammonium concentrations. The differences in the relationships between AOA and AOB abundances and nitrification potential rates may also be explained by the presence of ammonia-oxidizing microorganisms not targeted by the amoA assays used.
Salinity is also considered important in controlling the community structure (16) and abundance (46) of ammonia oxidizers and nitrification rates (46, 47). Good correlations between AOB amoA gene abundance and nitrification rates in marine and saltmarsh sediments (48) and between AOA amoA gene abundance and potential nitrification rates in the open ocean and some estuaries (6, 9) have been found. In the Colne estuary, AOB amoA gene abundances generally increased in the lower reaches of the estuary (Table 1). In contrast to the results of the present study, AOA amoA gene abundance has been shown to be greater than AOB gene abundance along a different estuarine salinity gradient (8). Although the potential rates did not correlate significantly with salinity, AOB amoA gene abundance was generally significantly higher at the marine end of the estuary (at Brightlingsea), where salinities are typically between 28 and 32 compared to the levels seen in brackish water at the top of the estuary (the Hythe), with salinities typically between 2 and 17 (12). Salinity variations also play a major role in ammonium adsorption/desorption in sediments (47, 49), and ammonium efflux to the oxic layer from deeper, high-ammonium estuarine sediments is enhanced by tidal changes in salinity (47, 49), with salinity variations being greater at the estuary head than at the estuary mouth. This again tends to favor AOB over AOA. However, other environmental variables such as trace metal and pH levels could also be significant in shaping AOB and AOA communities and potential nitrification rates.
In conclusion, differences in nitrification potential rates occurred both spatially and temporally in the Colne estuary, with the greatest potential autotrophic nitrification rates occurring mid-estuary in January. Although several factors (such as levels of trace metals, pH, and salinity) might selectively promote autotrophic activity by AOB or AOA in an estuarine environment, the sensitivity of AOA to high ammonium concentrations (13, 44) might explain the dominance of nitrification by AOB in this highly nutrified estuary. Furthermore, the greater temporal and spatial abundance of AOB amoA genes suggests that AOB (possibly Nitrosomonas spp.), rather than AOA, were of major significance in nitrogen cycling in the Colne estuary.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Chinese Scholarship Council Fund, Natural Environment Research Council (NERC) (NE/H525289/1), and the University of Essex.
We thank John Green and Farid Benyahid for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02654-14.
REFERENCES
- 1.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W. 2004. Environmental shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 2.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 4.Mosier AC, Francis CA. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol 10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- 5.De Corte D, Yokokawa T, Varela MA, Agogué H, Herndl GJ. 2009. Spatial distribution of Bacteria and Archaea and amoA gene copy numbers throughout the water column of the Eastern Mediterranean Sea. ISME J 3:147–158. doi: 10.1038/ismej.2008.94. [DOI] [PubMed] [Google Scholar]
- 6.Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J 1:660–662. doi: 10.1038/ismej.2007.79. [DOI] [PubMed] [Google Scholar]
- 7.Sahan E, Muyzer G. 2008. Diversity and spatio-temporal distribution of ammonia-oxidizing Archaea and Bacteria in sediments of the Westerschelde estuary. FEMS Microbiol Ecol 64:175–186. doi: 10.1111/j.1574-6941.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 8.Bernhard AE, Landry ZC, Blevins A, de la Torre JR, Giblin AE, Stahl DA. 2010. Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl Environ Microbiol 76:1285–1289. doi: 10.1128/AEM.02018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernhard AE, Tucker J, Giblin AE, Stahl DA. 2007. Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ Microbiol 9:1439–1447. doi: 10.1111/j.1462-2920.2007.01260.x. [DOI] [PubMed] [Google Scholar]
- 10.Ogilvie B, Nedwell DB, Harrison RM, Robinson A, Sage A. 1997. High nitrate, muddy estuaries as nitrogen sinks: the nitrogen budget of the River Colne estuary (United Kingdom). Mar Ecol Progr Ser 150:217–228. doi: 10.3354/meps150217. [DOI] [Google Scholar]
- 11.Papaspyrou S, Smith CJ, Dong LF, Whitby C, Dumbrell AJ, Nedwell DB. 2014. Nitrate reduction functional genes and nitrate reduction potentials persist in deeper estuarine sediments. Why? PLoS One 9:e94111. doi: 10.1371/journal.pone.0094111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong LF, Thornton DCO, Nedwell DB, Underwood GJC. 2000. Denitrification in sediments of the River Colne estuary, England. Mar Ecol Progr Ser 203:109–122. doi: 10.3354/meps203109. [DOI] [Google Scholar]
- 13.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 14.Di HJ, Cameron KC, Shen J-P, Winefield CS, O'Callaghan M, Bowatte S, He J-Z. 2010. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol 72:386–394. doi: 10.1111/j.1574-6941.2010.00861.x. [DOI] [PubMed] [Google Scholar]
- 15.Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific subtropical gyre. Environ Microbiol 9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 16.Bernhard AE, Donn T, Giblin AE, Stahl DA. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ Microbiol 7:1289–1297. doi: 10.1111/j.1462-2920.2005.00808.x. [DOI] [PubMed] [Google Scholar]
- 17.Strickland JDH, Parsons TR. 1972. A practical handbook of seawater analysis, 2nd ed. Fisheries Research Board of Canada, Ottawa, Ontario, Canada. [Google Scholar]
- 18.Berges JA, Franklin DJ, Harrison PJ. 2004. Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades (Corrigendum). J Phycol 40:619–619. doi: 10.1111/j.1529-8817.2004.40301.x. [DOI] [Google Scholar]
- 19.Dollhopf SL, Hyun JH, Smith AC, Adams HJ, O'Brien S, Kostka JE. 2005. Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl Environ Microbiol 71:240–246. doi: 10.1128/AEM.71.1.240-246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pai SC, Tsau YJ, Yang TI. 2001. pH and buffering capacity problems involved in the determination of ammonia in saline water using the indophenol blue spectrophotometric method. Anal Chim Acta 434:209–216. doi: 10.1016/S0003-2670(01)00851-0. [DOI] [Google Scholar]
- 21.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for co-extraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicol GW, Tscherko D, Embley TM, Prosser JI. 2005. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol 7:337–347. doi: 10.1111/j.1462-2920.2005.00698.x. [DOI] [PubMed] [Google Scholar]
- 23.Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of national ammonia-oxidizing population. Appl Environ Microbiol 63:4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidising archaea and bacteria in soil microcosms. Environ Microbiol 10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- 25.Kowalchuk GA, Stephen JR, Deboer W, Prosser JI, Embley TM, Woldendorp JW. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol 63:1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKew BA, Coulon F, Osborn AM, Timmis KN, McGenity TJ. 2007. Determining the identity and roles of oil-metabolizing marine bacteria from the Thames estuary, UK. Environ Microbiol 9:165–176. doi: 10.1111/j.1462-2920.2006.01125.x. [DOI] [PubMed] [Google Scholar]
- 27.Muyzer G, De Wall EC, Uitterlinden AG. 1993. Profiling of complex microbial population by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA. [Google Scholar]
- 29.Jukes TH, Cantor CR. 1969. Mammalian protein metabolism. Academic Press, New York, NY. [Google Scholar]
- 30.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 31.Page R. 1996. Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358. [DOI] [PubMed] [Google Scholar]
- 32.Ishak HD. 2011. Bacterial diversity in Solenopis invicta and Solenopsis geminate ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61:821–831. doi: 10.1007/s00248-010-9793-4. [DOI] [PubMed] [Google Scholar]
- 33.Takai K, Horikoshi K. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072. doi: 10.1128/AEM.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pentilde AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickie IA. 2010. Insidious effects of sequencing errors on perceived diversity in molecular surveys. New Phytol 188:916–918. doi: 10.1111/j.1469-8137.2010.03473.x. [DOI] [PubMed] [Google Scholar]
- 38.Development Core Team R. 2007. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 39.Norusis MJ. 2010. PASW statistics 18 guide to data analysis. Prentice-Hall, New York, NY. [Google Scholar]
- 40.Kuenen JG, Robertson LA. 1987. Ecology of nitrification and denitrification, p 162–218. In Cole JA, Ferguson SJ (ed), The nitrogen and sulphur cycles. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 41.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeota from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen T, Stieglmeier M, Dai J, Urich T, Schleper C. 2013. Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett 344:121–129. doi: 10.1111/1574-6968.12164. [DOI] [PubMed] [Google Scholar]
- 43.de Bie MJM, Speksnijder AGCL, Kowalchuk GA, Schuurman T, Zwart G, Stephen JR, Diekmann OE, Laanbroek HJ. 2001. Shifts in the dominant populations of ammonia-oxidizing β-subclass Proteobacteria along the eutrophic Schelde estuary. Aquat Microb Ecol 23:225–236. doi: 10.3354/ame023225. [DOI] [Google Scholar]
- 44.Verhamme DT, Prosser JI, Nicol GW. 2011. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5:1067–1071. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thornton DCO, Dong LF, Underwood GCU, Nedwell DB. 2002. Factors affecting microphytobenthos biomass, species composition and production in the Colne Estuary (UK). Aquat Microb Ecol 27:285–300. doi: 10.3354/ame027285. [DOI] [Google Scholar]
- 46.Cébron A, Berthe T, Garnier J. 2003. Nitrification and nitrifying bacteria in the lower Seine River and estuary (France). Appl Environ Microbiol 69:7091–7100. doi: 10.1128/AEM.69.12.7091-7100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rysgaard S, Thastum P, Dalsgaard T, Christensen PB, Sloth NP. 1999. Effects of salinity on NH4+ adsorption capacity, nitrification, and denitrification in Danish estuarine sediments. Estuaries 22:21–30. doi: 10.2307/1352923. [DOI] [Google Scholar]
- 48.Risgaard-Petersen N, Nicolaisen MH, Revsbech NP, Lomstein BA. 2004. Competition between ammonia-oxidizing bacteria and benthic microalgae. Appl Environ Microbiol 70:5528–5537. doi: 10.1128/AEM.70.9.5528-5537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seitzinger SP, Gardner WS, Spratt AK. 1991. The effect of salinity on ammonium sorption in aquatic sediments—implications for benthic nutrient recycling. Estuaries 14:167–174. doi: 10.2307/1351690. [DOI] [Google Scholar]
- 50.Smith CJ, Nedwell DB, Dong LF, Osborn AM. 2007. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol 73:3612–3622. doi: 10.1128/AEM.02894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.