Abstract
Bifidobacterium breve is a common and sometimes very abundant inhabitant of the human gut. Genome sequencing of B. breve JCM 7017 revealed the presence of an extrachromosomal element, designated pMP7017 consisting of >190 kb, thus representing the first reported bifidobacterial megaplasmid. In silico characterization of this element revealed several genomic features supporting a stable establishment of the megaplasmid in its host, illustrated by predicted CRISPR-Cas functions that are known to protect the host against intrusion of foreign DNA. Interestingly, pMP7017 is also predicted to encode a conjugative DNA transfer apparatus and, consistent with this notion, we demonstrate here the conjugal transfer of pMP7017 to representative strains of B. breve and B. longum subsp. longum. We also demonstrate the presence of a megaplasmid with homology to pMP7017 in three B. longum subsp. longum strains.
INTRODUCTION
Bifidobacterium breve is a common inhabitant of the infant gut, in addition to Bifidobacterium longum subsp. longum, Bifidobacterium longum subsp. infantis, Bifidobacterium pseudocatenulatum, Bifidobacterium bifidum (1), and the less frequently encountered Bifidobacterium kashiwanohense (2). Furthermore, representatives of this species have been isolated in other environments, including human milk, the adult gut, and human vagina (3). The presence of bifidobacteria (being members of the Actinobacteria phylum) in the gut has been correlated with particular health-promoting effects, such as enforcement of the intestinal barrier, activation/modulation of the host's immune response and protection against particular infections (4), and for this reason representatives of this genus have been included as active ingredients in functional foods (5).
Since the first genome sequence of B. longum subsp. longum NCC2705 was released in 2002, further sequencing endeavors have produced additional bifidobacterial genome sequences in an effort to elucidate the mode of action of their beneficial effects. In this respect genome sequencing and subsequent functional analyses have revealed that members of this genus possess genetic features that support the process of colonization and adhesion to the intestinal mucosa (6), evasion/stimulation of the host's immune system (7), and a metabolic ability to use host-derived glycans, all of which are believed to contribute to competitive exclusion of pathogens (4) and perhaps other beneficial activities.
The presence of plasmids among representatives of the genus Bifidobacterium has been reported for certain bifidobacterial species (e.g., Bifidobacterium pseudolongum subsp. globosum, Bifidobacterium indicum, Bifidobacterium asteroides, Bifidobacterium bifidum, Bifidobacterium catenulatum, Bifidobacterium pseudocatenulatum, Bifidobacterium kashiwanohense, Bifidobacterium breve, Bifidobacterium longum subsp. longum, and Bifidobacterium longum subsp. infantis) (8–11). Typically, when plasmids are found, just a single plasmid per isolate is present, with some notable exceptions: B. kashiwanohense JCM 15439, B. longum subsp. longum DJO10A, B. longum subsp. infantis 157F, and B. longum subsp. longum KACC 91563, which each harbor two such genetic elements (2, 8). Among the identified bifidobacterial plasmids, most have been isolated from strains of B. longum subsp. longum (8). The bifidobacterial plasmids that have been characterized thus far exhibit a size not exceeding 10.5 kb and the possible presence of extrachromosomal genetic elements larger than 100 kb, which are designated megaplasmids (12), in Bifidobacterium has only been suggested for two porcine cecal isolates (13).
Bacterial conjugation is one of the possible means of horizontal transfer of genetic material and it has been intensively studied in Gram-negative bacteria, in particular in the plant pathogen Agrobacterium tumefaciens due to its pathogenic and biotechnological relevance (e.g., tumor-inducing plasmid Ti) (14–16). Conjugation has been described for Gram-positive bacteria, with the difference that a somewhat simpler conjugative machinery appears to be required due to the fact that DNA traverses just a single membrane layer (17). With regard to bifidobacteria, recent genome analyses for members of this genus has indeed provided evidence of horizontal transfer events (3, 6, 18–20), where prophages are believed to be one of the principal vehicles of such DNA transfer phenomena (21).
The presence of (conjugative) megaplasmids in representatives of the phylum Actinobacteria has thus far only been observed for members of the Gordonia, Rhodococcus, Brevibacterium, Nocardiopsis, and Mycobacterium spp. (with sizes ranging between 100 and 700 kb) (12, 22), of which p1CP from Rhodococcus opacus PD630 (23), pREA400, pREA250, and pREA100 from Rhodococcus erythropolis AN12 (24), and pKB1 from Gordonia westfalica Kb1 (25) have conclusively been shown to be conjugative. With regard to conjugative transfer in bifidobacteria, only a recent study proposed a new shuttle vector pDOJHR-WD2 based on the conjugative machinery of Escherichia coli WM3064(pBB109), which was successfully used for DNA transfer between E. coli and different Bifidobacterium species (26); however, no functional, native bifidobacterial conjugation system has as yet been identified.
In this study we present the sequence and in silico annotation of a megaplasmid present in B. breve JCM 7017. This 190.17-kb plasmid, designated pMP7017, the first and largest megaplasmid to be identified among a member of the genus Bifidobacterium and actinobacterial gut commensals in general, is able to transfer to other bifidobacteria by an apparent conjugative mechanism. Interestingly, and consistent with the horizontal transfer ability of pMP7017, we also detected homologs of this megaplasmid in three representatives of B. longum subsp. longum.
MATERIALS AND METHODS
Sequencing and annotation.
DNA sequencing of pMP7017 was performed using protocols developed by the sequencing provider Macrogen (Seoul, Republic of Korea) and employing a combination of two next-generation sequencing (NGS) platforms (see below). An initial 454 run using a Roche genome sequencer FLX Titanium instrument employing a long-tag, paired-end library (average read length of 400 bp) resulted in the construction of an initial scaffold, which was subsequently improved with an additional Illumina Hiseq 2000 paired-end sequencing run with an average read length of 101 bp.
The resulting sequences were then assembled in a hybrid 454-Illumina approach using a combination of Newbler v2.9 (98.83% of aligned reads and 0.04% inferred read error) (454 Sequencing; Roche) for long reads and Abyss v1.3.4 (http://www.bcgsc.ca/) for short reads, resulting in a high-quality and ungapped final consensus sequence, where the average sequence coverage of each base pair was >200-fold.
Replicon analyses and bioinformatic analysis.
Open reading frame (ORF) prediction was performed with a combined approach of the tool Prodigal v2.0 (http://prodigal.ornl.gov). Functional assignment of the predicted ORFs was first performed on the basis of BLASTP v2.2.26 (27) searches against the nonredundant protein database curated by the National Centre for Biotechnology Information (ftp://ftp.ncbi.nih.gov/blast/db/) and further refined, when necessary, employing BLASTX v2.2.26 searches (27).
Artemis v.14 (http://www.sanger.ac.uk/resources/software/artemis/) was used to inspect the results of the ORF prediction and used for manual editing, if necessary, of the start codon of a predicted gene. Gene annotations were further integrated with the information retrieved from alternative databases, e.g., protein family (Pfam) (http://pfam.sanger.ac.uk).
TRNA genes were identified using tRNAscan-SE v1.4 (28) and the presence of Restriction-Modification (R-M) systems was investigated using BLASTP (27) alignments against the downloaded REBASE database (http://rebase.neb.com/rebase/rebase.html; cutoff E value of 0.0001 and showing at least 30% similarity across at least 80% of the complete length of the deduced amino acid sequence).
The presence of CRISPR loci was inspected using the CRISPR finder tool (http://crispr.u-psud.fr), while codon usage and amino acid frequency were computed using the GCUA (29) tool and visualized with scatter3D module of the statistical package R (http://www.r-project.org/). Sequence comparisons at the protein level were performed using an bidirectional BLASTP alignment (27) (cutoff E value of 0.0001, with at least 50% identity across at least 50% of either protein sequence).
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. Bifidobacterial strains were routinely cultured in reinforced clostridial medium (RCM [Oxoid, Ltd., Basingstoke, Hampshire, United Kingdom]) or modified de Man, Rogosa, and Sharpe medium (mMRS) prepared from first principles (30). Prior to inoculation, mMRS was supplemented with cysteine-HCl (0.05% [wt/vol]) and lactose (1.0% [wt/vol]) (Sigma). Bifidobacterial cultures were incubated at 37°C under anaerobic conditions, which were maintained using an anaerobic hood (Davidson and Hardy, Belfast, Ireland). E. coli was cultured in Luria-Bertani (LB) broth (31) at 37°C with agitation. Where appropriate, growth media contained tetracycline (10 μg ml−1), ampicillin (100 μg ml−1 for E. coli), erythromycin (100 μg ml−1 for E. coli), or kanamycin (50 μg ml−1 for E. coli). Recombinant E. coli cells containing pORI19 were selected on LB agar containing erythromycin and supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 40 μg ml−1 and 1 mM IPTG (isopropyl-β-d-galactopyranoside).
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant feature(s)b | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| E. coli EC101 | Cloning host, repA+ Kmr | 53 |
| Bifidobacterium breve | ||
| JCM 7017 | Isolate from human feces | JCM |
| JCM 7017-199 | pORI19-tet-0199 insertion mutant of pMP7017 in JCM7017 | This study |
| UCC2003 | Isolate from nursling stool | 19 |
| JCM 7019 | Isolate from infant feces | JCM |
| NCFB 2258 | Isolate from infant intestine | NCFB |
| UCC2003-TC | B. breve UCC2003 transconjugant harboring pMP7017_199 | This study |
| JCM 7019-TC | B. breve JCM7019 transconjugant harboring pMP7017_199 | This study |
| NCFB 2258-TC | B. breve NCFB 2258 transconjugant harboring pMP7017_199 | This study |
| B. longum subsp. longum | ||
| NCIMB 8809 | Isolate from nursling stool | NCIMB |
| 1-6B | Isolate from 6-year-old child | 51 |
| 2-2B | Isolate from 6-year-old child | 51 |
| 44B | Isolate from infant at 1 year | 51 |
| 35B | Isolate from infant at 1 year | 51 |
| NCIMB 8809-TC | B. longum NCIMB 8809-TC transconjugant harboring pMP7017_199 | This study |
| Plasmids | ||
| pAM5 | pBC1-puC19-Tcr | 54 |
| pORI19 | Emr; ΔrepA; ori+, cloning vector | 53 |
| pORI19-tet-0199 | Internal 937-bp fragment of pl_0199 and tetW cloned in pORI19 | This study |
JCM, Japan Collection of Microorganisms; NCFB, National Collection of Food Bacteria; NCIMB, National Collection of Industrial and Marine Bacteria.
Emr, erythromycin resistance.
DNA manipulations.
Chromosomal DNA was isolated from B. breve JCM 7017 as previously described (32). Minipreparation of plasmid DNA from E. coli was achieved using a QIAprep spin plasmid miniprep kit (Qiagen GmBH, Hilden, Germany). Single-stranded oligonucleotide primers (see Table S1 in the supplemental material) used in the present study were synthesized by Eurofins (Ebersberg, Germany). Standard PCRs were performed using TaqPCR mastermix (Qiagen), whereas high-fidelity PCR was achieved using KOD polymerase (Novagen, Darmstadt, Germany). B. breve colony PCRs were performed as described previously (33). PCR fragments were purified using the Qiagen PCR purification kit. Electroporation of plasmid DNA into E. coli was performed as described by Sambrook et al. (31), and into B. breve JCM 7017 as described by Maze et al. (34).
Antibiotic marking of pMP7017 through creation of an insertion mutant of pMP7017_0199.
An internal 937-bp fragment of ORF pMP7017_0199 was amplified by PCR using B. breve JCM 7017 chromosomal DNA as a template and primer pairs IM199F and IM199R (see Table S1 in the supplemental material). The insertion mutation in B. breve JCM 7017 was generated essentially as described previously (35) to produce B. breve JCM 7017-199. Site-specific recombination in tetracyclin-resistant isolates was confirmed by colony PCR using the primer combination tetWFw and tetWRv to verify tetW gene integration and the primer combination 0199-FW (positioned upstream of the selected internal fragment of pMP7017_0199) and pORI19For to confirm integration at the correct genetic location.
Conjugal matings.
Conjugations were performed by inoculating overnight cultures of donor and recipient at 1% into mMRS broth supplemented with cysteine-HCl (0.05% [wt/vol]) and lactose (1.0% [wt/vol]) and growing at 37°C under anaerobic conditions for 7 h to late log phase. Next, 1 ml of each culture (donor and recipient) was mixed and concentrated by centrifugation, and the resultant pellet was resuspended in 200 μl of RCM prior to spread plating on reinforced clostridial agar (RCA) plates. RCA plates were incubated overnight at 37°C under anaerobic conditions. Subsequently, the conjugation mixture was recovered from the plates and resuspended in 2 ml of phosphate-buffered saline and serially diluted. Donor and recipient cells in each conjugation mixture were enumerated based on selective carbohydrate utilization by the donor or recipient strain on mMRS agar, whereas transconjugants were isolated by adopting the appropriate carbohydrate to select for recipient cells in combination with tetracycline (10 μg ml−1) (Table 2).
TABLE 2.
Conjugation between B. breve JCM 7017 and representative B. breve and B. longum subsp. longum strains
| Donor | Carbohydrate selection for donor | Recipient | Carbohydrate selection for recipient | Conjugation efficiencya |
|---|---|---|---|---|
| B. breve JCM 7017_199 | Sorbitol | B. breve UCC2003 | Ribose | 5.1 × 10−9 |
| B. breve JCM 7017_199 | Sorbitol | B. breve NCFB 2258 | Ribose | 1.3 × 10−8 |
| B. breve JCM 7017_199 | Cellobiose | B. breve JCM 7019 | Melezitose | 1.33 × 10−6 |
| B. breve JCM 7017_199 | Sorbitol | B. longum NCIMB 8809 | Arabinose | 4.7 × 10−8 |
Expressed as the average number of transconjugants per recipient from triplicate conjugation experiments.
PFGE plug preparation.
Agarose gel plugs of high-molecular-weight DNA for pulsed-field gel electrophoresis (PFGE) were prepared according to previously described protocols (36, 37), with minor modifications, as outlined below. All bifidobacterial strains were grown in mMRS broth supplemented with 0.05% cysteine and 1% (wt/vol) lactose to early stationary phase. A volume of culture containing ∼109 bacteria (equivalent to an optical density at 600 nm of 2.0) was centrifuged (5,000 × g for 1 min), washed once with 1 ml of NT buffer (1 M NaCl, 10 mM Tris-HCl [pH 7.6]), and repelleted (5,000 × g for 1 min). The cell pellet was resuspended in 300 μl of NT buffer, to which an equal volume of melted 2% (wt/vol) low-melting-point (LMP) agarose, prepared in 0.125 M EDTA (pH 7.6) and maintained at 50°C, was added. The cell suspension and LMP agarose were mixed carefully without introducing bubbles. Gel plugs were formed by pipetting 300-μl volumes into plug molds and allowed to solidify at 4°C for 20 min. Subsequently, two plugs per strain were added to 2 ml of EC buffer (1 M NaCl, 6 mM Tris-HCl, 100 mM EDTA [pH 7.6]) containing 1% (wt/vol) Sarkosyl, 10 mg ml−1 lysozyme, and 40 U ml−1 mutanolysin, followed by incubation at 37°C for 24 h. The lysozyme solution was replaced with 5 ml of 0.5 M EDTA (pH 8.0) containing 1% (wt/vol) Sarkosyl and 0.5 mg/ml proteinase K and then incubated at 37°C for 24 h. Plugs were then washed with 5 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) containing 1 mM PMSF (phenylmethylsulfonyl fluoride; freshly prepared) at 37°C for 1 h to inactivate the proteinase K. This was followed by three 30-min incubations in 5 ml of TE buffer at room temperature to remove the PMSF. Plugs were stored in 10 mM Tris-HCl–100 mM EDTA (pH 8.0) at 4°C.
Treatment with S1 nuclease or restriction endonucleases.
Single slices (2 mm by 2 mm) were soaked in either 200 μl of S1 buffer (50 mM NaCl, 30 mM sodium acetate [pH 4.5], 5 mM ZnSO4) or an appropriate restriction buffer at room temperature for 30 min. The S1 buffer was replaced with another 200 μl of S1 buffer containing 1 unit of Aspergillus oryzae S1 nuclease and then incubated at 37°C for 45 min. The restriction buffer was replaced with fresh restriction buffer containing 15 U of the appropriate restriction enzyme (SpeI and AscI), followed by incubation at 37°C for 16 h. Reactions were stopped by suspending the treated plugs in 200 μl of 0.5 M EDTA (pH 8.0) and held at room temperature for 10 min. The 0.5 M EDTA was replaced with 200 μl of TE and left at room temperature for at least 30 min before loading onto a gel.
PFGE.
Plug slices were loaded directly into the wells of a 1% (wt/vol) PFGE agarose gel melted in 0.5× TBE (89 mM Tris-borate, 2 mM EDTA [pH 8.3]) buffer. The wells were sealed with molten agarose in 0.5 TBE buffer. DNA fragments were resolved using a CHEF-DR III pulsed-field system (Bio-Rad Laboratories, Hercules, CA) at 6 V/cm for 18 h with 0.5× TBE running buffer maintained at 14°C. Linear ramped pulse times were selected depending on the size of DNA fragments to be resolved; for the S1 nuclease-treated samples, a linear ramped pulse time of 3 to 50 s was used; for SpeI- and AscI-digested samples, a linear ramped pulse time of 1 to 15 s was used. A molecular size Chef DNA lambda ladder was included in each gel (number 170-3635; Bio-Rad Laboratories). The gels were stained in distilled water containing ethidium bromide for 120 min under light-limited conditions and destained in distilled water for 60 min. The gels were visualized, and images were captured under UV light.
Nucleotide sequence accession numbers.
The sequences generated in this study have been submitted to GenBank database with the accession number (GenBank accession no. KM406416). All of the sequences used for our analysis have been retrieved from NCBI database with the following accession numbers: B. breve UCC2003 (GenBank accession no. NC_020517), B. breve JCM 7017 (GenBank accession no. CP006712), B. breve NCFB 2258 (GenBank accession no. CP006714)], B. breve JCM 7019 (GenBank accession no. CP006713), B. longum subsp longum 1-6B (GenBank accession no. AJTF00000000), B. longum subsp. longum 2-2B (GenBank accession no. AJTJ00000000), B. longum subsp. longum 35B (GenBank accession no. AJTI00000000), and B. longum subsp. longum 44B (GenBank accession no. AJTM00000000).
RESULTS AND DISCUSSION
Sequence analysis and general features of pMP7017.
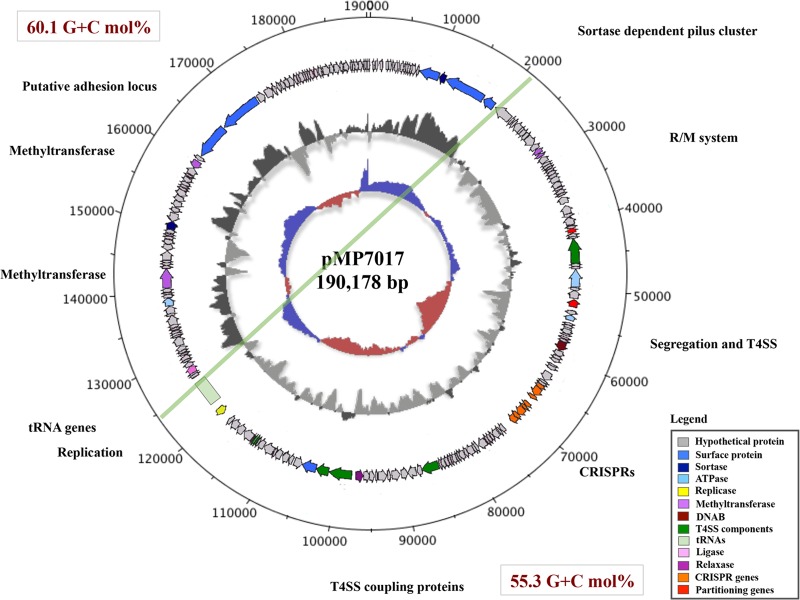
During the genome sequencing and assembly of the Bifidobacterium breve strain JCM 7017 (3), a sequence scaffold of 190,178 bp was detected, corresponding to a DNA element that was not part of the chromosome, thereby providing preliminary evidence for the presence of a megaplasmid in this strain. Sequence analysis of this pMP7017 replicon showed an overall G+C mol% of 57.43, comparable with the G+C content of its host (58.65 G+C mol%) and of bifidobacterial genomes in general (9). However, the fact that a slightly different G+C content was observed for about half of its sequence, roughly corresponding to nucleotide coordinates 22,000 to 125,000 (Fig. 1), suggests that this megaplasmid is the result of cointegration of two originally separate modules (having a G+C mol% of either 60.1 or 55.3) (Fig. 1). The pMP7017 plasmid harbors 228 ORFs, of which 70% do not have significant hits in database searches, thus representing hypothetical genes which is a common genetic feature of plasmids, while the remaining ORFs are associated with predicted functions that are principally involved in DNA replication, DNA methylation, partitioning, DNA transfer, and colonization (Fig. 1).
FIG 1.
pMP7017 general features. Genome atlas representing the circular sequence of the magaplasmid pMP7017 with the relative ORFs organization is shown. Highlighted are the four main functional regions (REG1 to -4) with relative genetic features. A green line also divides pMP7017 into two putative cointegrates.
The first (arbitrary) half of the pMP7017 sequence (corresponding to locus tag names pMP7017_0001 through to pMP7017_0100) contains ORFs which, among others, represent a sortase-dependent pilus cluster (cluster I) encoding a predicted sortase (pMP7017_0021) and three putative surface-anchored proteins (pMP7017_0022-24). Interestingly, this cluster shows significant homology (above 50% of similarity in a BLASTP alignment) with predicted pilus clusters of particular B. longum strains, though it is not significantly similar to those identified in other bifidobacteria (19, 38, 39). This portion of pMP7017 also contains two genes encoding a potential type II R/M system (pMP7017_0033-34), followed by genetic functions putatively involved in plasmid segregation and DNA transfer, encompassing a predicted chromosome partitioning genes parA (pMP7017_0053), as well as adjacent genes predicted to encode a mating-pair formation complex (Mpf) for DNA translocation represented by two VirD4-B4 ATPases (pMP7017_0059 and pMP7017_0062, respectively) and a transmembrane pore-forming VirB6 (pMP7017_0056) (Table 3). The VirD4-B4 and VirB6 proteins in the plant pathogen Agrobacterium tumefaciens are known to form the structural scaffold and motor of a type IV secretion system (T4SS) that acts as a DNA secretion channel (40–42). The observed similarity may thus indicate a similar role in the conjugal transmission of pMP7017 between bifidobacterial species (see below). Finally, two adjacent CRISPR/Cas loci (corresponding to locus tags pMP7017_0080-85 and pMP7017_0086-90) are present in this section of the megaplasmid, which are known to provide immunity against invading phages and plasmids in bacteria (43, 44). Further discussion on these CRISPR/Cas systems will follow below.
TABLE 3.
Segregation and conjugal transfer genes in pMP7017
| Cellular process | ORF | Annotation | Pfam no. |
|---|---|---|---|
| Segregation | pMP7017_0053 | Chromosome partitioning protein parA | PF01656 |
| DNA translocation | pMP7017_0062 | VirB4 | PF01656 |
| pMP7017_0056 | VirB6 | PF04610 | |
| pMP7017_0059 | VirD4 | PF10412 | |
| pMP7017_0136 | Protein translocase subunit secE | PF00584 | |
| pMP7017_0138 | Protein translocase subunit secG | PF03840 | |
| Conjugal transfer coupling proteins (CP) | pMP7017_0114 | Ftsk/SpoIIIE family protein | PF01580 |
| pMP7017_0115 | CHAP possible extracellular subunit | ||
| pMP7017_0116 | Hypothetical protein with CHAP domain | PF05257 | |
| pMP7017_0125 | Ftsk/SpoIIIE family protein | PF01580 | |
| pMP7017_0126 | Ftsk/SpoIIIE family protein | PF01580 | |
| pMP7017_0127 | Hypothetical protein with flagellar basal body-associated domain (putative VirB10) | PF03739 | |
| pMP7017_0134 | DivIVA domain containing protein (putative VirC1) | PF05103 | |
| Replication and DNA processing | pMP7017_0124 | Putative relaxase | PF04796 |
| pMP7017_0073 | Replicative DNA helicase dnaB | PF03796 | |
| pMP7017_0108 | Dna polymerase III alpha subunit dnaE | PF00929 | |
| pMP7017_0146 | Predicted repA replicase protein | PF04796 |
The second half of the replicon (locus tags pMP7017_0101 through to pMP7017_0146) encodes the presumed replicative apparatus consisting of a possible relaxase (pMP7017_0124) and a predicted replication protein (pMP7017_0146) (Pfam PF04796.5), as well as several genes that specify coupling proteins (CP) of a type IV secretion system associated with the Mpf complex directing DNA translocation (40–42): FtsK/SpoIIIE family proteins (pMP7017_0114-0116 and pMP7010_0125-26) and a DivIVA domain-containing protein (pMP7017_0134) (Table 3). This second portion of the megaplasmid also specifies biosynthetic functions for a putative adhesion locus encoding two predicted surface proteins (pMP7017_0199-0200), of which the corresponding genes are located in the vicinity of a sortase-encoding gene (pMP7017_0184), as well as two genes that each encode a putative methyltransferase (pMP7017_0174/0197); this region also harbors the largest density of hypothetical genes.
Interestingly, the pMP7017 replicon also specifies three predicted toxin-antitoxin (T-A) systems (pMP7017_0014-15, pMP7017_101-102 and pMP7017_164-165), as well as a putative antidote protein (pMP7017_0019). Such T-A systems are known to be involved in ensuring plasmid maintenance (45).
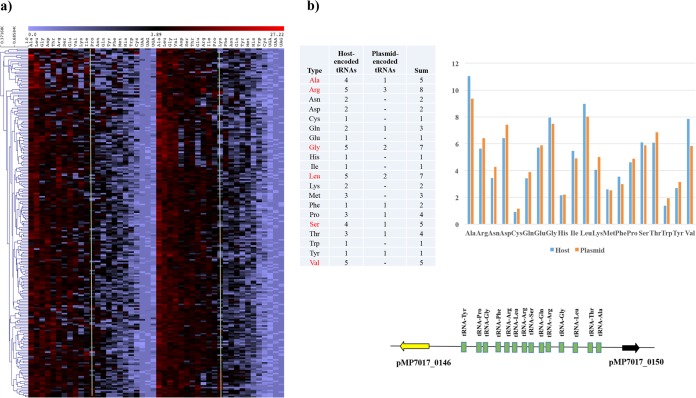
Finally, downstream of the gene encoding the predicted replication protein (pMP7017_0146), a cluster of 14 tRNA genes (base positions 121532 to 125960) is present, and their possible role will be discussed below.
Host adaptation: codon usage and amino acid frequency.
The evolutionary process that drives codon usage bias across a genome and indirectly determines the overall tRNA abundance in a given organism is believed to be mediated by natural selection in favor of those codons for which sufficient cognate tRNA is present in order to optimize protein translation efficiency (46, 47).
In order to explore the possible relationship between the B. breve JCM 7017 chromosome and its coexisting megaplasmid pMP7017 a multivariate analysis on codon usage by plasmid and chromosome-encoded proteins was performed (see Fig. S1a in the supplemental material). The resulting tridimensional plot demonstrated that the pMP7017 replicon possesses a codon bias that is compatible with that of the chromosome, while it also revealed that plasmid-borne ORFs with a deviating codon bias mostly specify hypothetical proteins (see Table S2 in the supplemental material). This result indicates that pMP7017 and its host DNA currently coexist following convergent evolution that has led to optimal production of chromosome- and plasmid-encoded functions.
The presence of 14 clustered tRNA genes on pMP7017 suggests a correspondence between codon usage and tRNA abundance. In order to investigate this notion, amino acid frequencies for pMP7017 and B. breve JCM 7017-encoded ORFs were computed and the resulting heatmap, based on hierarchical clustering, showed that concordance exists between frequently used codons in the plasmid and the host and that this is consistent with the tRNA genes present in the plasmid (Fig. 2a and b). For this reason is tempting to argue that pMP7017 followed an evolutionary process which made its codon usage fitting the one of the host allowing efficient protein translation, but also that its maintenance and expression is supported by a selected set of tRNA genes that provides translational backup to the host.
FIG 2.
Amino acid usage comparison and tRNA genes. (a) Heatmap and hierarchical clustering showing the amino acid frequencies computed for the predicted proteome of pMP7017 and the host. For representative reasons, a sample of equal numbers of 227 randomly selected ORFs of B. breve JCM 7017 and pMP7017 are displayed in this heatmap. (b) Bidimensional barplot representing the cumulative amino acid frequency in pMP7017 and the host with locus map indicating the 14 consecutive tRNA genes encountered in pMP7017 sequence. A table with the total number of tRNA in the plasmid and chromosome is also included.
Protection against DNA invasion: CRISPRs.
Plasmid pMP7017 is predicted to encode several methyltransferases (pMP7017_0033-34, pMP7017_0174, and pMP7017_0197) which may be part of R-M systems, while it also encompasses two adjacent CRISPR/Cas systems (pMP7017_0080-85 and pMP7017_0086-90) (Fig. 1). These features may promote the exclusion of (other) invading genetic elements in concert with the chromosomally encoded protection systems (constituted by two R-M systems and a CRISPR/Cas locus) (3), thus providing mutual benefit.
Comparative analysis of the three type CRISPR/Cas systems encoded by B. breve JCM 7017 revealed a subtype I-C system present on the JCM 7017 chromosome and two subtype I-E systems harbored by pMP7017, and named B7017-CRISPR1, pMP7017-CRISPR1, and pMP7017-CRISPR2, respectively. In silico analysis of these three type I CRISPR/Cas systems allowed the identification of 74 nonredundant spacer sequences (Fig. 3a and b), which together are believed to constitute a genetic memory of previous challenges by phages (or other invading DNA elements) mounted against this B. breve strain or against previous hosts of the conjugative pMP7017 megaplasmid.
FIG 3.
CRISPR/Cas systems. (a) Locus map showing the organization of the CRISPR/cas locus in B. breve JCM 7017 chromosome and in the megaplasmid. (b) Heatmap showing the BLASTP analysis of the 74 spacer regions found in the CRISPR/cas systems of the B. breve JCM 7017 chromosome and relative megaplasmid, aligned against NCBI nonredundant (nr) database. A green line also indicates matches within representatives of B. breve species.
Of these three systems, B7017-CRISPR1 located on the B. breve JCM 7017 chromosome (∼18 kb in size with gene coordinates 1632794 to 1650595) possesses a total of 49 nonredundant spacers, of which the distal end (spacers 27 to 49) returned several hits with CRISPR-associated regions of other B. breve representatives (in particular B. breve S27, B. breve ACS-071-V-Sch8b, and B. breve NCFB 2258), while the proximal end (spacers 1 to 26) appears to be more variable, suggesting that this CRISPR/Cas system was originally acquired by certain representatives of B. breve and diversified over time as a result of subsequent phage infections and consequent spacer acquisition events (Fig. 3b).
The presence of at least one cas-1 and cas-2 gene couple in the B7017_CRISPR1 and pMP7017_CRISPR2, being a signature of an active CRISPR/Cas system (48), suggests that they constitute fully functional CRISPR systems with proto-spacer processing/acquisition activity, as a crucial part of the adaptation stage. In contrast, the pMP7017_CRISPR1 locus only appears to encode endonuclease activity and may therefore be active solely in the CRISPR/Cas-mediated immunity phase (49, 50) or rely on the cas-1 and cas-2 activities of the other two systems. Interestingly, BLASTN-based searches of the identified spacer sequences against the National Center for Biotechnology Information (NCBI) nonredundant (nr) database returned a number of significant hits against phage-related sequences (i.e., located in predicted bifidobacterial prophage regions) and other CRISPR-associated spacers, as well as against sequences that are associated with horizontal gene transfer (HGT) (such as portions of putative integrative conjugative elements or integrated plasmids) of various bifidobacteria, mostly those that are commonly found in the infant gut (e.g., B. breve, B. longum subsp. longum, B. longum subsp. infantis, and B. bifidum) (48) (Fig. 3b). These results support the notion that CRISPR/Cas systems not only constitute a barrier to phage infection but also to other DNA invasion events that promote HGT mediated by mobilizable elements (44).
Presence of a megaplasmid in B. breve JCM 7017 and strains of B. longum subsp. longum.
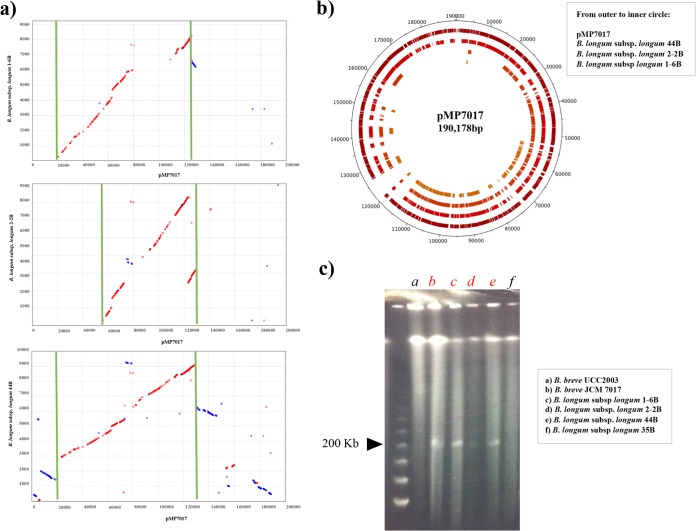
The process of gene annotation, followed by an in silico comparative analysis conducted on the ORFeome of pMP7017, allowed the identification of 90 to 100 kb of homologous sequences in the draft genomes (deposited as a varying number of contigs) of B. longum subsp. longum strains 1-6B, 2-2B, and 44B (51). These homologous sequences (when translated) display an overall conservation of BLASTP similarity between 60 and 100%, whereas at nucleotide level (BLASTN), high levels of identity (80 to 100%) were only observed for portions of these contigs. In order to investigate synteny between the pMP7017 plasmid sequence and the above-mentioned B. longum subsp. longum contigs, a dot plot alignment was performed, showing that a degree of synteny is observed for the pMP7017 region corresponding to the approximate nucleotide coordinates 20,000 to 125,000 (Fig. 4a), which also specifies the predicted conjugative apparatus (Fig. 1) and which supports the above-mentioned notion that this megaplasmid represents a cointegrate of two plasmids.
FIG 4.
Presence of pMP7017 in B. breve and B. longum subsp. longum. (a) Dot plot alignment showing the comparison of pMP7017 megaplasmid with the contigs of B. longum subsp. longum of which ORFs displayed significant match in BLASTP alignment (e.g., B. longum subsp. longum 1-6B, B. longum subsp. longum 2-2B, and B. longum subsp. longum 44B). Delimited by green bars are the regions displaying the most of synteny. (b) BLAST-based genome atlas showing the comparative analysis conducted on pMP7017 against the draft sequence of B. longum subsp longum 1-6B, B. longum subsp. longum 2-2B, and B. longum subsp. longum 44B. From the outer to the inner circle and shaded in gradient from dark red to orange: B. breve JCM 7017 pMP7017, B. longum subsp. longum 44B, B. longum subsp. longum 2-2B, and B. longum subsp. longum 1-6B. (c) PFGE experiment showing the presence of a megaplasmid of a size comparable to that of pMP7017 (about 200 kb) also in B. longum subsp. longum 1-6B, B. longum subsp. longum 2-2B, and B. longum subsp. longum 44B, but absent in B. longum subsp. longum 35B and B. breve UCC2003 as a negative control.
The comparative analysis at protein level showed that ca. 70% of the pMP7017 ORFs returned significant similarities to proteins encoded by B. longum subsp. longum (Fig. 4b). The ORFs that were shown to be similar between pMP7017 and the draft genome sequence of one or more of these three B. longum subsp. longum strains include those that encode sortase-dependent pilus clusters I and II, the ORFs predicted to be involved in plasmid replication and conjugation, and the characteristic 14 clustered tRNA genes and CRISPR/cas genes, the latter showing 80 to 100% similarity at amino acid level (see Fig. S2 in the supplemental material) and identical repeats (see Table S3 in the supplemental material), while the spacers appeared different. However, the draft nature of these B. longum subsp. longum sequences prevented a full-length alignment of these contigs with pMP7017, while it was also not clear whether these sequences were associated with a megaplasmid. For this reason, in order to not only verify the presence of pMP7017 in B. breve JCM 7017 but also evaluate the presence of a megaplasmid in the three aforementioned strains of B. longum subsp. longum, S1 nuclease PFGE was performed (Fig. 4c). This indeed confirmed the presence of pMP7017 in B. breve JCM 7017 and also revealed the presence of a megaplasmid, of equivalent size, in B. longum subsp longum 1-6B, B. longum subsp. longum 2-2B and B. longum subsp. longum 44B. In contrast, the negative controls, B. breve UCC2003 and B. longum subsp longum 35B, did not appear to contain a megaplasmid, concordant with what was expected from the in silico comparative analysis (Fig. 4b).
Conjugal transfer of pMP7017 to representative B. breve and B. longum strains.
In silico characterization of pMP7017 (see above) suggests that this plasmid specifies a DNA conjugation mechanism involving the concerted action of an indigenous partitioning, replication, membrane fusion and DNA translocation system, not previously described for bifidobacteria (Table 3).
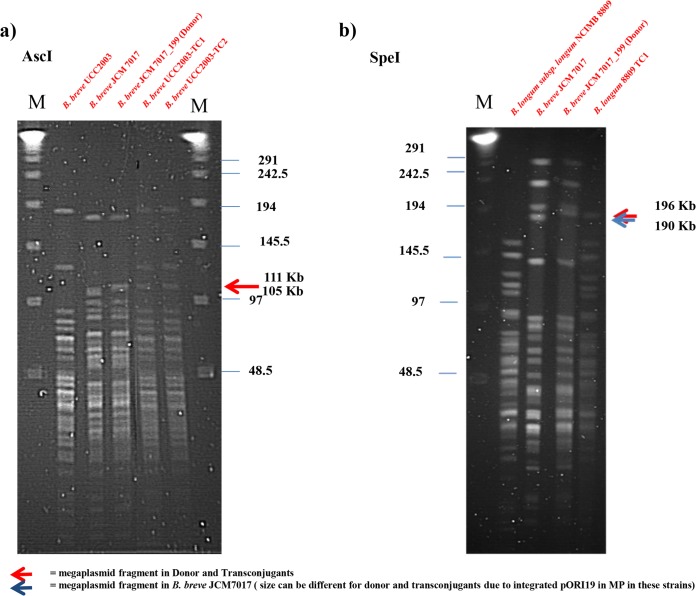
In order to establish if pMP7017 is able to transfer to other strains by conjugation, a derivative strain, designated B. breve JCM 7017-199, was constructed which carries the megaplasmid in which pMP7017_0199, encoding a predicted surface protein (with no predicted function in conjugation), was interrupted by site-specific homologous recombination, with the resultant megaplasmid being marked with a gene that confers tetracycline resistance. Conjugation experiments were performed using B. breve JCM 7017-199 as a donor and strains B. breve UCC2003, B. breve JCM 7019, B. breve NCFB 2258, B. longum subsp. longum NCIMB 8809 as recipients with selection of transconjugants based on selective carbohydrate utilization by each recipient strain and tetracycline resistance (see Materials and Methods and Table 2). Transconjugants were obtained for each recipient strain at frequencies ranging from approximately 10−9 to 10−6. The presence of pMP7017_199 in the transconjugants was confirmed by PCR targeting genes on the megaplasmid and, for each recipient strain, genes for selective carbohydrate utilization. Furthermore, additional primer pairs based on B. breve JCM 7017 chromosomal genes were also included and as expected, these gave no PCR product for the transconjugant strains (see Fig. S3 in the supplemental material). To further verify conjugative transfer of pMP7017_199 to B. breve and B. longum PFGE of restricted total DNA was performed on representative B. breve UCC2003_pMP7017_199 and B. longum NCIMB 8809_pMP7017_199 transconjugants. Adopting the restriction enzymes AscI for B. breve UCC2003 and SpeI for B. longum subsp. longum NCIMB 8809, we clearly demonstrate the presence of expected pMP7017_199 restriction fragments in the presumed transconjugants and also the correct recipient strain restriction profile thereby confirming the conjugal transfer of pMP7017_199 from B. breve JCM 7017_199 to strains of B. breve and B. longum (Fig. 5).
FIG 5.
Conjugal transfer of pMP7017. PFGE experiments for B. breve UCC2003 or B. longum NCIMB 8809 transconjugants conducted with AscI (a) and SpeI (b) enzymes, respectively. The presence of megaplasmid in the parent strain, B. breve JCM 7017, and the donor strain, B. breve JCM 7017_199, is depicted in each panel. The presence of pMP7017 in B. breve UCC2003 trasconjugants is evidenced by a 111-kb fragment in total DNA digests with AscI, while the megaplasmid presence in B. longum NCIMB 8809 transconjugants is verified by the presence of a 196-kb fragment.
Conclusions.
The presence of plasmids does not represent a typical feature of B. breve and B. longum genomes and all currently available bifidobacterial plasmids are considered cryptic as their encoded functions (besides, in some cases, those involved in replication) are unknown. In this respect the pMP7017 megaplasmid, identified in B. breve JCM 7017, represents an exceptional bifidobacterial plasmid, which not only represents the largest reported conjugative plasmid identified thus far in the genus Bifidobacterium and actinobacterial gut commensals (at >190 kb) but also possesses a number of genomic features indicating the presence of parasite-benefactor balance reached over time through convergent evolution with the host. In addition, it also provides two CRISPR/Cas systems to counter phage infections and other invading DNA elements. Plasmid pMP7017 also possesses a number of genes whose products are similar to proteins that are known to be part of a conjugative machinery present in both Gram-positive and Gram-negative bacteria (40–42).
The capacity of this element of being horizontally transferable between different species was first suspected due to the finding that elements similar to pMP7017 of B. breve JCM 7017 were also present in several members of the B. longum subsp. longum taxon.
Plasmid-mediated conjugation in other bacteria is known to involve the transfer of a protein-single-strand DNA complex (called the relaxosome) pumped through an aqueous pore of a type IV secretion system from the donor to the recipient cell (40–42). In bifidobacteria, a gene specifying a protein similar to the type IV secretion protein has been observed in the chromosome of B. bifidum MIMBb75 (52). The structural scaffold of such DNA transfer system in pMP7017 is believed to be represented by a VirB6 structural scaffold, which is coupled to aVirD4-B4 ATPases, to provide the energy for DNA translocation. The functionality of the predicted conjugative machinery in pMP7017 megaplasmid was conclusively demonstrated by conjugation experiments showing that this element is capable to spread and persist in different bifidobacterial strains and species.
One of the conditions for initiating a conjugative transfer is the establishment of cell-to-cell contact, which in Gram-negative bacteria is accomplished by proteinaceous filaments called sex pili, while in Gram-positive bacteria such structures have not yet been clearly identified. The fact that pMP7017 is predicted to encode two different sortase-dependent pili (pMP7017_0021 to pMP7017_0024 and pMP7017_0184-pMP7017_0199-pMP7017_0200) which have been shown to promote bifidobacterial coaggregation (39) suggests a role in bacterial conjugation.
B. breve and B. longum subsp. longum both represent common inhabitants of the infant gut (1), which supports the hypothesis that the cooccurrence of apparent homologs of this conjugative element in both of these species may be a result of previous DNA exchanges, followed by diversification and stable maintenance of this element over generations. The fact that pMP7017 represents the first native plasmid capable of conjugal transmission between bifidobacterial species is highly relevant as it may confer this replicon a relevant biotechnological potential for improving the future genetic manipulation of members of this genus.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a Science Foundation Ireland grant (SFI/12/RC/2273) and an HRB postdoctoral fellowship (grant PDTM/2011/9) awarded to M.O.M.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02871-14.
REFERENCES
- 1.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahata M, Toh H, Nakano A, Takagi M, Murakami M, Ishii Y, Takizawa T, Tanabe S, Morita H. 2014. Complete sequence analysis of two cryptic plasmids from Bifidobacterium kashiwanohense JCM 15439 (type strain) isolated from healthy infant feces. Anim Sci J 85:158–163. doi: 10.1111/asj.12095. [DOI] [PubMed] [Google Scholar]
- 3.Bottacini F, O'Connell Motherway M, Kuczynski J, O'Connell KJ, Serafini F, Duranti S, Milani C, Turroni F, Lugli GA, Zomer A, Zhurina D, Riedel C, Ventura M, van Sinderen D. 2014. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventura M, Turroni F, Lugli GA, van Sinderen D. 2014. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J Sci Food Agric 94:163–168. doi: 10.1002/jsfa.6356. [DOI] [PubMed] [Google Scholar]
- 5.Stanton C, Ross RP, Fitzgerald GF, Van Sinderen D. 2005. Fermented functional foods based on probiotics and their biogenic metabolites. Curr Opin Biotechnol 16:198–203. doi: 10.1016/j.copbio.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanning S, Hall LJ, van Sinderen D. 2012. Bifidobacterium breve UCC2003 surface exopolysaccharide production is a beneficial trait mediating commensal-host interaction through immune modulation and pathogen protection. Gut Microbes 3:420–425. doi: 10.4161/gmic.20630. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, O'Sullivan DJ. 2010. Genomic insights into bifidobacteria. Microbiol Mol Biol Rev 74:378–416. doi: 10.1128/MMBR.00004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cronin M, Knobel M, O'Connell-Motherway M, Fitzgerald GF, van Sinderen D. 2007. Molecular dissection of a bifidobacterial replicon. Appl Environ Microbiol 73:7858–7866. doi: 10.1128/AEM.01630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayo B, van Sinderen D. 2010. Bifidobacteria: genomics and molecular aspects. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 12.Schwartz E. 2009. Microbial megaplasmids. Springer, Berlin, Germany. [Google Scholar]
- 13.Simpson PJ, Stanton C, Fitzgerald GF, Ross RP. 2003. Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J Bacteriol 185:2571–2581. doi: 10.1128/JB.185.8.2571-2581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piper KR, Beck von Bodman S, Farrand SK. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 15.Lang S, Gruber K, Mihajlovic S, Arnold R, Gruber CJ, Steinlechner S, Jehl MA, Rattei T, Frohlich KU, Zechner EL. 2010. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol Microbiol 78:1539–1555. doi: 10.1111/j.1365-2958.2010.07423.x. [DOI] [PubMed] [Google Scholar]
- 16.Wetzel ME, Kim KS, Miller M, Olsen GJ, Farrand SK. 2014. Quorum-dependent mannopine-inducible conjugative transfer of an Agrobacterium opine-catabolic plasmid. J Bacteriol 196:1031–1044. doi: 10.1128/JB.01365-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, Canchaya C, Claesson MJ, He F, Mantzourani M, Mulas L, Ferrarini A, Gao B, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Gupta RS, Zhang Z, Beighton D, Fitzgerald GF, O'Toole PW, van Sinderen D. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet 5:e1000785. doi: 10.1371/journal.pgen.1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventura M, Lee JH, Canchaya C, Zink R, Leahy S, Moreno-Munoz JA, O'Connell-Motherway M, Higgins D, Fitzgerald GF, O'Sullivan DJ, van Sinderen D. 2005. Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl Environ Microbiol 71:8692–8705. doi: 10.1128/AEM.71.12.8692-8705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dib JR, Wagenknecht M, Hill RT, Farias ME, Meinhardt F. 2010. Novel linear megaplasmid from Brevibacterium sp. isolated from extreme environment. J Basic Microbiol 50:280–284. doi: 10.1002/jobm.200900332. [DOI] [PubMed] [Google Scholar]
- 23.Konig C, Eulberg D, Groning J, Lakner S, Seibert V, Kaschabek SR, Schlomann M. 2004. A linear megaplasmid, p1CP, carrying the genes for chlorocatechol catabolism of Rhodococcus opacus 1CP. Microbiology 150:3075–3087. doi: 10.1099/mic.0.27217-0. [DOI] [PubMed] [Google Scholar]
- 24.Yang JC, Lessard PA, Sengupta N, Windsor SD, O'Brien XM, Bramucci M, Tomb JF, Nagarajan V, Sinskey AJ. 2007. TraA is required for megaplasmid conjugation in Rhodococcus erythropolis AN12. Plasmid 57:55–70. doi: 10.1016/j.plasmid.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Broker D, Arenskotter M, Steinbuchel A. 2008. Transfer of megaplasmid pKB1 from the rubber-degrading bacterium Gordonia westfalica strain Kb1 to related bacteria and its modification. Appl Microbiol Biotechnol 77:1317–1327. doi: 10.1007/s00253-007-1262-8. [DOI] [PubMed] [Google Scholar]
- 26.Dominguez W, O'Sullivan DJ. 2013. Developing an efficient and reproducible conjugation-based gene transfer system for bifidobacteria. Microbiology 159:328–338. doi: 10.1099/mic.0.061408-0. [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2, doi:. [DOI] [PubMed] [Google Scholar]
- 28.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McInerney JO. 1998. GCUA: general codon usage analysis. Bioinformatics 14:372–373. doi: 10.1093/bioinformatics/14.4.372. [DOI] [PubMed] [Google Scholar]
- 30.De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J Appl Bacteriol 23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 31.Sambrook J, Russell DW, Fritsch EF, Maniatis T. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.O'Riordan K, Fitzgerald GF. 1998. Evaluation of bifidobacteria for the production of antimicrobial compounds and assessment of performance in cottage cheese at refrigeration temperature. J Appl Microbiol 85:103–114. doi: 10.1046/j.1365-2672.1998.00474.x. [DOI] [PubMed] [Google Scholar]
- 33.Motherway MO, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol 74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maze A, O'Connell-Motherway M, Fitzgerald GF, Deutscher J, van Sinderen D. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol 73:545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motherway MO, O'Driscoll J, Fitzgerald GF, van Sinderen D. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol 2:321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Canchaya C, Fang F, Raftis E, Ryan KA, van Pijkeren JP, van Sinderen D, O'Toole PW. 2007. Distribution of megaplasmids in Lactobacillus salivarius and other lactobacilli. J Bacteriol 189:6128–6139. doi: 10.1128/JB.00447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wall R, Fitzgerald G, Hussey S, Ryan T, Murphy B, Ross P, Stanton C. 2007. Genomic diversity of cultivable Lactobacillus populations residing in the neonatal and adult gastrointestinal tract. FEMS Microbiol Ecol 59:127–137. doi: 10.1111/j.1574-6941.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 38.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O'Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M. 2011. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Fact 10(Suppl 1):S16. doi: 10.1186/1475-2859-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turroni F, Serafini F, Foroni E, Duranti S, Motherway MO, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol 341:961–977. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christie PJ, Cascales E. 2005. Structural and dynamic properties of bacterial type IV secretion systems. Mol Membr Biol 22:51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahrstrom CT. 2014. Structural biology: solving the T4SS structural mystery. Nat Rev Microbiol 12:312. doi: 10.1038/nrmicro3254. [DOI] [PubMed] [Google Scholar]
- 43.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 44.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Goeders N, Van Melderen L. 2014. Toxin-antitoxin systems as multilevel interaction systems. Toxins 6:304–324. doi: 10.3390/toxins6010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikemura T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34. [DOI] [PubMed] [Google Scholar]
- 47.Akashi H. 1994. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong HJ, Nilsson L, Kurland CG. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 50.Novoa EM, de Pouplana LR. 2012. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet 28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Shkoporov AN, Efimov BA, Khokhlova EV, Chaplin AV, Kafarskaya LI, Durkin AS, McCorrison J, Torralba M, Gillis M, Sutton G, Weibel DB, Nelson KE, Smeianov VV. 2013. Draft genome sequences of two pairs of human intestinal Bifidobacterium longum subsp. longum strains, 44B and 1-6B and 35B and 2-2B, consecutively isolated from two children after a 5-year time period. Genome Announce 1:e00234-13. doi: 10.1128/genomeA.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guglielmetti S, Balzaretti S, Taverniti V, Miriani M, Milani C, Scarafoni A, Corona S, Ciranna A, Arioli S, Santala V, Iametti S, Bonomi F, Ventura M, Mora D, Karp M. 2014. TgaA, a VirB1-like component belonging to a putative type IV secretion system of Bifidobacterium bifidum MIMBb75. Appl Environ Microbiol 80:5161–5169. doi: 10.1128/AEM.01413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177:7011–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alvarez-Martín P, O'Connell-Motherway M, Van Sinderen D, Mayo B. 2007. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl Microbiol Biotechnol 76:1395–1402. doi: 10.1007/s00253-007-1115-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.