Abstract
Water quality monitoring techniques that target microorganisms in the order Bacteroidales are potential alternatives to conventional methods for detection of fecal indicator bacteria. Bacteroidales and members of the genus Bacteroides have been the focus of microbial source tracking (MST) investigations for discriminating sources of fecal pollution (e.g., human or cattle feces) in environmental waters. For accurate source apportionment to occur, one needs to understand both the abundance of Bacteroides in host feces and the survival of these host-associated microbial markers after deposition in the environment. Studies were undertaken to evaluate the abundance, persistence, and potential for growth of Bacteroidales originating from poultry litter under oxic and anoxic environmental conditions. Bacteroidales abundance, as determined by quantitative PCR (qPCR) with GenBac primers and probe, increased 2 to 5 log gene copies ml−1 and 2 log gene copies g litter−1 under most conditions during incubation of poultry litter in a variety of laboratory microcosm and field mesocosm studies. DNA sequencing of the Bacteroidales organisms in the litter identified taxa with sequences corresponding exactly to the GenBac primer and probe sequences and that were closely related to Bacteroides uniformis, B. ovatus, and B. vulgatus. These results suggest that MST studies using qPCR methods targeting Bacteroidales in watersheds that are affected by poultry litter should be interpreted cautiously. Growth of Bacteroidales originating from poultry litter in environmental waters may occur while Bacteroidales growth from other fecal sources declines, thus confounding the interpretation of MST results.
INTRODUCTION
It has been estimated that current farming practices are responsible for 70% of the pollution in U.S. rivers and streams (1), in large part from animal manure (2). The volume of animal manure produced from concentrated animal feeding operations is three times the volume of human fecal waste in the United States (3). Microbiological water quality in the United States has been based on enumeration of cultured fecal indicator bacteria (FIB), such as fecal coliforms, Escherichia coli, and Enterococcus, for over 100 years (4). Drawbacks to the use of FIB for monitoring recreational water quality include (i) a 24- to 48-h cultivation period, (ii) the potential for extraintestinal growth, and (iii) a lack of correlation with waterborne pathogens in environmental waters. Alternatives to the use of FIB include techniques that target microorganisms such as those in the order Bacteroidales with methods such as quantitative PCR (qPCR). Bacteroidales are attractive alternatives to FIB for water quality monitoring because they are primarily obligate anaerobes that are predominately found in the intestines of warm-blooded animals. In fact, Bacteroidales are found in much greater abundance than E. coli in animal feces, as more than 90% of fecal phylotypes belong to taxa of the Firmicutes and Bacteroidetes divisions (5–7). It has been reported previously that Bacteroidales survival in extraintestinal environments is limited (8, 9) and that decay of fecal Bacteroidales occurs in surface water (10–13).
Several species in the genus Bacteroides have been the focus of microbial source tracking (MST) investigations for diagnosing fecal pollution and discriminating sources (e.g., human or cattle feces) in environmental systems (for recent reviews, see references 14, 15, and 16). It has been suggested that qPCR methods can be used to allocate fecal contributions from various hosts to degraded water quality by comparing the relative abundances of different host-specific Bacteroides markers and the total Bacteroidales abundance (17, 18). For accurate source apportionment to occur, one needs to understand both the abundance of Bacteroides bacteria in host feces and the survival of these host-specific microbial markers after deposition in the environment. As such, many studies were undertaken recently to assess persistence and decay of host-specific Bacteroides markers in freshwater (13, 19–23), marine waters (11, 12, 17, 24, 25), manure-amended soils (26, 27), and animal feces (9), under a variety of conditions. In general, studies have reported a positive relationship between Bacteroidales decay rates and temperature (17, 20), a negative relationship between salinity and decay rates (17, 25), and a variable relationship between decay rates and the presence or absence of light (13, 28). Given that Bacteroidales is a deeply divergent order based on 16S rRNA gene phylogeny, it is not surprising that different Bacteroidales clades may have differing environmental persistences after leaving the intestines of warm-blooded animals in fecal material. Despite these concerns, the U.S. Environmental Protection Agency (USEPA) is considering adopting a Bacteroidales qPCR method for water quality surveys (29).
The literature published to date consistently indicates that Bacteroidales decay after deposition in feces and exposure to extraintestinal environments. The sensitivity of these organisms to oxygen is generally given as the major reason for rapid die-off of these bacteria outside the host (11, 19). Only a few recent publications suggest that some Bacteroidales taxa are not obligately anaerobic. Bacteroides fragilis is capable of growth in nanomolar concentrations of oxygen (30, 31), and Bacteroides thetaiotaomicron maintained viability for up to 20 h after exposure to oxygen (32). Another report evaluated Bacteroidales levels in unchlorinated drinking water originating from groundwater wells, finding mean densities of 3.1, 2.7, and 3.3 log cells/100 ml in groundwater and in wastewater treatment plant influent and effluent, respectively. Another recent report found low densities of Bacteroidales in soils (2.5 log gene copies g−1) and chlorinated tap water (1.4 log gene copies liter−1) (33).
Relatively few reports have presented evidence of Bacteroidales densities in chicken and turkey feces (6, 34) and in poultry litter (33, 35). Additionally, we were unable to find peer-reviewed reports regarding the fate of Bacteroidales originating from poultry feces and deposited in poultry litter. Therefore, studies were undertaken to evaluate the abundance and persistence of Bacteroidales organisms originating from poultry litter under various environmental conditions, and the results are reported herein.
MATERIALS AND METHODS
Poultry litter collection and handling.
Poultry litter incubation experiments were conducted at both West Virginia University (WVU) and the University of South Florida (USF). Poultry litter was collected from the WVU Animal Sciences Farm on three separate occasions for the river-deployed field mesocosm and laboratory microcosm studies (litter samples for WVU and USF studies were collected approximately 2 months apart), using the following sampling approach. Random grab samples of 10 scoops (0 to 2.5 cm deep) of wet, soiled poultry litter from three to eight pens containing adult broiler chickens and roosters were collected, composited, and homogenized in the field. Large feathers and clumps of litter were removed in the field. Approximately 500 ml litter was placed in a Whirl-Pak bag and transported to the laboratory on ice. Immediately upon receipt in the laboratory, the litter was homogenized by shaking the litter through a 2-mm (ASTM no. 10) sterile sieve. Litter shipped to the University of South Florida was immediately placed on wet ice and shipped overnight for delivery the next day. Litter used at West Virginia University was collected and homogenized on the same day as micro- or mesocosm preparation.
Laboratory microcosm construction and sampling.
Oxic and anoxic microcosms at WVU were constructed by placing 250 ml of Monongahela River water and 2.5 g of homogenized soiled poultry litter into sterile 1-liter Wheaton bottles (Millville, NJ). The initial density of Bacteroidales in litter was 109 gene copies g−1 (wet weight), and that in the river water was 104.5 gene copies ml−1. In the anoxic microcosms, the river water was sparged with N2 gas (99.9% purity; Air Gas, Morgantown, WV) for 10 min, and immediately after construction, the headspace of the vials was sparged with N2 gas. Bottles were capped with screw caps and incubated at 35°C with shaking at 250 rpm for 2 weeks in the dark. Microcosms were sampled on days 0, 5, and 8 by withdrawing 3.9 ml of solution. Specifically, 1.3 ml was placed into a lysing matrix B tube (MP Biomedical) and centrifuged at 10,000 × g for 5 min at 4°C. Supernatant was removed, and the cell and litter debris pellet was retained. This concentration method was repeated twice more to achieve a total volume of 3.9 ml. DNA was extracted from the concentrated cell and litter debris pellet by use of bead beater tubes, using a manual extraction method as described previously (36). DNA was directly extracted from 0.25 g of homogenized poultry litter (used as the microcosm inoculum) by use of the same manual extraction method. Dissolved oxygen concentrations were not measured in the WVU microcosms.
Microcosm studies at USF were conducted in 2013 (single microcosm for each treatment) and repeated in 2014 (triplicate microcosms for each treatment). Replicate microcosms at USF were constructed as similarly as possible to the WVU microcosms by placing 250 ml of day-old Monongahela River water (shipped overnight from WVU at 4°C on wet ice) and 2.5 g of homogenized soiled poultry litter into two sterile 1-liter Erlenmeyer flasks with screw caps (Fisher Scientific, Pittsburgh, PA). Microcosms were inoculated at USF approximately 24 to 36 h after collection of the litter at WVU. The initial density of Bacteroidales in litter was 106.1 to 109 gene copies g−1 (wet weight), and that in the river water was 104.4 to 104.5 gene copies ml−1. Anoxic conditions were created in one of these flasks by sparging the river water with N2 gas (99.9% purity; Airgas, Tampa, FL) for 15 min before addition of the litter, and the headspace was sparged for 10 min after litter was added. The screw cap on the anoxic flask was tightened and sealed with Parafilm to maintain anoxic conditions. The other flask was loosely capped, without Parafilm. A third, control flask contained river water without litter, and oxic conditions were maintained. All flasks were incubated in the dark for 2 weeks at 35°C with shaking at 250 rpm. Dissolved oxygen concentrations in the second microcosm studies at USF were monitored with a dissolved oxygen probe (YSI, Inc., Yellow Springs, OH) on days 0, 2, 9, 12, and 15. A 10-ml sample for DNA extraction was drawn from each microcosm on days 0, 2, 6, 9, 12, and 15. The headspace of the anoxic treatment flask was sparged with N2 gas for 10 min every time it was opened to retrieve a sample. Samples were filtered through 0.4-μm polycarbonate filters (Fisher Scientific, Pittsburgh, PA), and filters were stored at −80°C until extracted using a MoBio PowerWater DNA isolation kit (MoBio Laboratories, Carlsbad, CA).
River-deployed field mesocosm construction and sampling.
River-deployed field mesocosms were constructed of Spectra/Por 5 molecular porous membrane tubing (molecular weight cutoff [MWCO] of 12,000 to 14,000; Spectrum Laboratories, Inc., Rancho Dominguez, CA) and were deployed in duplicate. Each microcosm contained 50 ml of Monongahela River water and 0.5 g of fresh, homogenized poultry litter. Control mesocosms consisted of 50 ml of Monongahela River water without added poultry litter. Mesocosms were sealed with clamps and placed on metal wire frames (2.65-cm by 2.65-cm holes) attached to polyvinyl chloride (PVC) frames deployed in the Monongahela River near Star City, WV. The mesocosms were submerged beneath 5 to 20 cm of water over the course of 12 days. Half of the mesocosms were exposed to light in the river, and half were covered with a black mat, preventing exposure of the mesocosms to light.
Mesocosms were destructively sampled on days 0, 3, 7, and 12. During sampling, the dialysis bags were removed from the submerged frames, placed in buckets containing river water, and transferred to the laboratory for analysis. Immediately upon receipt of the sample in the laboratory, DNA was extracted from 10 ml of sample according to the following method. Ten milliliters of sample was placed into a 50-ml centrifuge tube (Nalge Nunc International, Rochester, NY) and centrifuged at 20,000 rpm (Avanti J-E centrifuge; Beckman Coulter, Inc., Brea, CA) for 10 min. Supernatant was removed, and the pellet of cells and litter debris was added to lysing matrix B tubes (MP Biomedicals, Santa Ana, CA). DNAs were extracted from centrifuged samples placed in bead beater tubes by using a manual extraction method as described previously (36). Dissolved oxygen concentrations of the river water were determined in the laboratory by use of a dissolved oxygen probe (YSI Inc., Yellow Springs, OH).
Hen house-deployed field microcosm construction and sampling.
Homogenized soiled poultry litter was collected from eight random pens containing juvenile (1 or 2 weeks old) broiler chicks at the WVU Animal Sciences Farm (Morgantown, WV). Immediately prior to the placement of poultry birds on unsoiled wood shavings, the barn was sanitized by use of disinfectant (Virocid; CID Lines N.V., Belgium). Random grab samples of eight scoops (0 to 2.5 cm deep) of wet soiled poultry litter from the 1- and 2-week-old chicks were composited, large clumps and feathers were removed, and the litter was homogenized by hand. A total of 14 ml of homogenized litter was then placed in sterile 15-ml centrifuge tubes (Corning Inc., Corning, NY), capped, and hung in the pens approximately 0.3 m from the ground. A total of 14 replicate tubes were incubated, and 2 tubes were destructively sampled each week for a total of 7 weeks. Duplicate tubes were transported on ice to the laboratory, where they were deconstructed and homogenized. A total of 0.25 g (wet weight) of poultry litter was taken for DNA extraction, which was performed using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany) following the manufacturer's instructions.
Quantitative PCR analysis of Bacteroidales.
Prior to quantification of Bacteroidales in samples, extracted DNA was quantified at 260 nm by using a Nanodrop ND-1000 UV spectrophotometer (Nanodrop Technologies, Wilmington, DE) (at WVU) or a NanoDrop 2000 UV-visible (UV-Vis) spectrophotometer (Thermo Scientific, Wilmington, DE) (at USF). A standard curve was constructed from serial dilutions of a plasmid DNA containing the GenBac target sequence. Plasmid copy numbers were calculated by measuring DNA concentrations by use of a spectrophotometer and converting them from ng DNA μl−1 to plasmid copy numbers. We assumed one gene copy per plasmid and a weight of 4.96 × 10−18 g per plasmid. DNA plasmids ranged in concentration from 106 to 101 gene copies per reaction mixture.
All qPCRs conducted at WVU were performed according to the following methods. The qPCR amplification reaction mixtures (25 μl) contained template DNA (5 μl targeting 25 to 100 ng), 1× TaqMan universal master mix (Applied Biosystems, Foster, City, CA), 1 μM (each) primers (Integrated DNA Technologies, Inc., Coralville, IA), and 80 nM TaqMan probe (Biosearch Technologies, Inc., Novato, CA). The forward primer sequence (GenBacF3) was 5′-GGGGTTCTGAGAGGAAGGT-3′, and the reverse primer sequence (GenBacR4) was 5′-CCGTCATCCTTCACGCTACT-3′. The TaqMan probe sequence was 5′-6-carboxyfluorescein (FAM)-CAATATTCCTCACTGCTGCCTCCCGTA-BHQ-1-3′ (37). All reactions were carried out on a model 7300 Real Time PCR system (Life Technologies, Grand Island, NY), using previously published thermocycler conditions (37). A plasmid containing the target gene sequence (IDT-DNA, Coralville, IA) was used as a positive control in all reaction mixtures. All samples were tested in duplicate with an additional plasmid matrix-spiked sample to determine if the DNA-extracted samples were inhibited in the qPCRs. Control samples for each qPCR run included qPCR negative controls (e.g., DNA-free water instead of template and extraction controls) and qPCR positive controls (i.e., a plasmid containing the GenBac gene).
All qPCRs conducted at USF used the same primers and probe, thermocycler conditions, and other parameters as those described above, but they differed as follows. All reactions were carried out in triplicate in a model 7500 Real Time PCR system (Life Technologies, Grand Island, NY). Standard curves using serially diluted DNA were included in each qPCR run.
Cloning and sequencing of Bacteroidales in poultry litter.
Clone libraries were constructed from 16S rRNA gene PCR products generated from DNAs extracted from the WVU anoxic microcosm on days 5 and 8 and the USF laboratory anoxic microcosms on days 0 and 9. The WVU clone libraries were generated from PCR products amplified using the GenBacF and GenBacR primers and the methods described above, which produced an amplicon of 129 bp. The purpose of the 129-bp clones was to examine the variability in sequences obtained by qPCR using the GenBac primers. The USF clone libraries were generated from PCR products amplified using Bac32F (5′-AACGCTAGCTACAGGCTT-3′) and Bac708R (5′-CAATCGGAGTTCTTCGTG-3′) and previously published methods (38), which generated amplicons of >570 bp. The purpose of the 570-bp clones was to evaluate the phylogeny of the Bacteroidales species present in the poultry litter microcosms. The clone libraries at WVU were constructed using a TOPO cloning reaction kit (Invitrogen, Carlsbad, CA) following the manufacturer's instructions, while the USF libraries were constructed using a CloneJet PCR cloning kit (Thermo Scientific, Wilmington, DE). Four clones from the WVU libraries, including two clones from the day 5 and two from the day 8 sampling event, were randomly selected and were sequenced at the WVU Genomics Core Facility on an ABI model 3130XL genetic analyzer, using an ABI Prism BigDye Terminator cycle sequencing kit with the M13-f and M13-r primers. Six clones from the USF library, three from day 0 and three from day 9, were randomly selected and sequenced by Eurofins Genomics (Huntsville, AL), resulting in five unique sequences.
Clone sequences were assembled and aligned using BioEdit ver. 7.0.5.3 (www.mbio.ncsu.edu/bioedit/bioedit.html) and checked for chimeras using the Ribosomal Database Project II Chimera Check program (http://rdp.cme.msu.edu/) and Bellerophon (39). The 16S rRNA gene sequences with the greatest sequence identities identified by a BLASTn search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were downloaded for inclusion in the phylogenetic analysis. Additionally, sequences of Bacteroidales from various fecal sources, shown by Bernhard and Field to be amplified with Bac32F and Bac708R (38), were included in the phylogenetic analysis. Multiple-sequence alignments were constructed using ClustalW implemented in Mega 6.0 (40). Maximum likelihood phylogenetic analysis was conducted using Mega 6.0 under the Kimura 2-parameter model (41) with gamma-distributed splits. All positions based on a partial deletion treatment with a site coverage cutoff of 95% were included in the phylogenetic analysis. Bootstrap analysis included 500 replicates in order to increase the confidence in the tree topology.
Nucleotide sequence accession numbers.
The sequences of the clones from this study were deposited in the GenBank database under accession numbers KJ958897 to KJ958901.
RESULTS
Extraintestinal growth of Bacteroidales in microcosms and mesocosms.
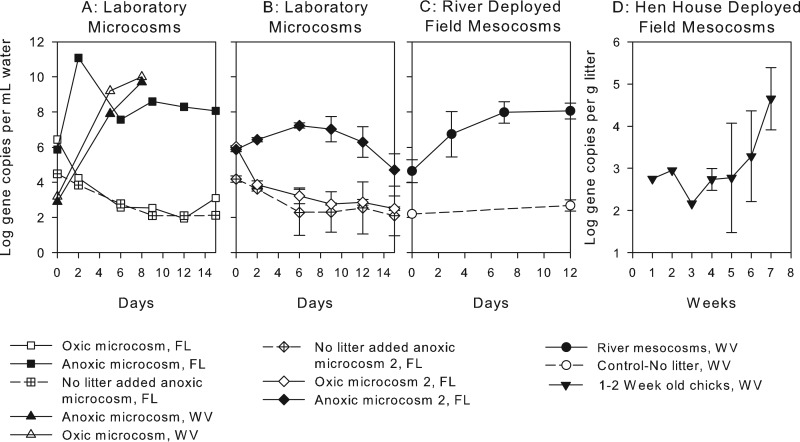
Abundances of Bacteroidales in microcosms and mesocosms over time are shown in Fig. 1. Mean increases in Bacteroidales density in the anoxic laboratory microcosms ranged from 2 to 6 log gene copies ml−1 over 15 days (Fig. 1A and B). The uninoculated control containing only river water had an initial density of ∼104 Bacteroidales gene copies ml−1 in both laboratory studies and decreased approximately 2 log over time. GenBac gene concentrations declined >2 log in control and aerobic microcosms during both studies at USF (Fig. 1A and B) but increased in the single aerobic laboratory microcosm conducted at WVU (Fig. 1A). Based on the initial abundance of Bacteroidales in the litter used to construct the WVU laboratory microcosms, the maximum density of Bacteroidales that could be released from the litter to water without growth was ∼107 gene copies ml−1. Since the maximum density observed in the laboratory microcosms was over 109 gene copies ml litter−1, there was likely both growth of Bacteroidales and release of these organisms from the litter during the course of the study. In the replicate laboratory microcosm studies, the dissolved oxygen concentration in the oxic and control treatments was initially 9.2 mg liter−1 and decreased within 2 days, to 4.4 ± 0.8 mg liter−1, for the rest of the experiment. The dissolved oxygen concentration in the anoxic treatments was 0.3 ± 0.2 mg liter−1 for the entire experiment.
FIG 1.
Numbers (averages and standard deviations) of 16S rRNA gene copies for Bacteroidales in various micro- and mesocosms seeded with poultry litter. (A) Laboratory microcosms conducted in 2013 at USF and WVU (n = 1). (B) Replicate laboratory microcosms conducted in 2014 at USF (n = 3). (C) River mesocosm dialysis bags (n = 16) and no-litter-added control dialysis bags (n = 4). (D) Wet litter from 1- and 2-week-old chicks, isolated from other birds and incubated in a hen house.
Average Bacteroidales abundances in the light- and dark-exposed field-deployed mesocosms (n = 16) increased, on average, 4 log gene copies ml water−1 over 12 days. In control treatments without poultry litter (n = 4) in the river-deployed field mesocosms, gene target concentrations did not increase significantly (Fig. 1C). Bacteroidales abundances in wet poultry litter incubated in hen houses but isolated from additional poultry fecal input also increased an average of 2 log gene copies g litter−1 over 7 weeks (Fig. 1D).
Phylogeny of poultry litter Bacteroidales.
Nine unique clones (of 10 total clones) were sequenced from the WVU (4 clones) and USF (5 clones) clone libraries generated from DNAs isolated from the anoxic, in-laboratory microcosms. All the clone sequences clustered in the order Bacteroidales. Multiple-sequence alignments of the Bacteroidales sequences obtained from poultry litter compared to the GenBac qPCR primer and probe sequences are shown in Fig. 2. All WVU sequences corresponded exactly to the GenBac primer and probe sequences used for qPCR. All USF sequences differed from the GenBac forward primer by at least one nucleotide but perfectly matched the probe and reverse primer sequences used for qPCR. A variable region in the DNA sequence (highlighted in gray in Fig. 2) was observed in all clone sequences, between the regions targeted by the qPCR forward primer and probe.
FIG 2.
Multiple-sequence alignments of Bacteroidales 16S rRNA genes from four WVU clones (WVU5-1 to WVU8-2) and five USF clones (USF-A1 to USF-A6). The GenBac row shows, in order, the GenBacF, GenBacP, and GenBacR primer and probe sequences. Dots denote positions identical to primer/probe sequences. The shaded positions represent sequence variability among the clones.
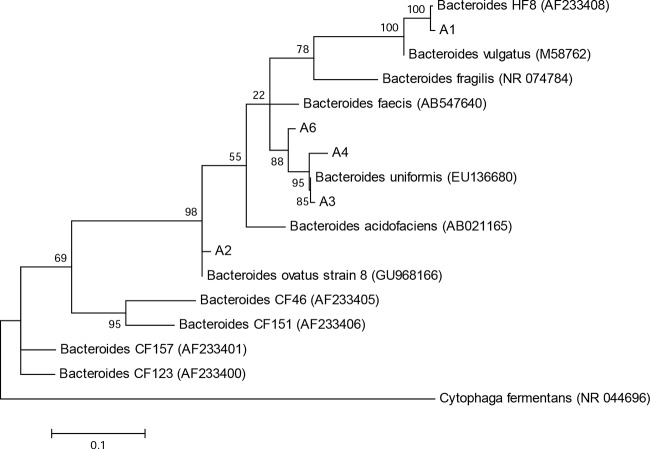
Phylogenetic analysis of the USF sequences compared to Bacteroidales sequences isolated from human and animal feces (38) and to closely related GenBank database sequences is shown in Fig. 3. The USF clones (clone names beginning with “A”) were closely aligned with the Bacteroides species. One clone isolated from the poultry litter, namely, A1, clustered with the previously identified human-associated Bacteroides organism HF8 (accession no. AF233408) (38) and showed 100% sequence identity over the 561-bp sequence analyzed. The clones were not as closely related to previously identified cow fecal (CF) microorganisms. Clones A3, A4, and A6 had 99, 97, and 96% sequence similarity to Bacteroides uniformis (accession no. EU136690) over the 694 bp, 688 bp, and 552 bp analyzed, respectively. Finally, clone A6 had 100% sequence identity to Bacteroides ovatus strain 8 (accession no. GU968166) over the 687 bp analyzed. Phylogenetic alignments of the WVU clones were not constructed due to the short sequence length of 129 bp.
FIG 3.
Molecular phylogenetic analysis of DNA sequences amplified by GenBac PCR. Sequences from this study start with the designation “A” (A1 to A4 and A6).
DISCUSSION
MST methods show great promise for improved water quality monitoring (14). A host of studies have suggested that qPCR methods targeting the order Bacteroidales and the genus Bacteroides may be able to discriminate between fecal contamination of water from a variety of sources and/or be used for detection of these as general fecal pollution indicators. Additionally, there is some evidence that host-specific Bacteroides assays are correlated with pathogens in environmental systems (42) and are positively related to gastrointestinal symptoms in recreational water users (43). As such, the USEPA is evaluating the adoption of qPCR methods for detection of Bacteroidales in support of MST studies (29). Arguments against the use of Bacteroidales-level assays are tied to reports that Bacteroidales markers persist in the environment longer than species-specific markers do or, in some cases, may increase. For example, Dick et al. (20) reported that river water and sediment microcosms spiked with human feces exhibited decay of E. coli and two human-associated Bacteroides markers (HF183 and BacHum) over 11 days, to 1% of the initial densities. However, AllBac, a general marker of Bacteroidales, decayed only 50% over the course of the study. Dick et al. suggested that the observed biphasic decay curve may indicate a reservoir of persistent members of the Bacteroidales (20). Others reported that Bacteroidales fecal bacteria grew for up to 24 h during aerobic incubation of human sewage (44).
Some of the Gram-negative organisms in the genus Bacteroides, such as Bacteroides ovatus, are known to be nutritionally versatile and capable of degrading complex polysaccharides, such as starches (45) and hemicelluloses or xylans found in plant materials (46). The use of selective enrichment media has been proposed for Bacteroides fragilis, to capitalize on this anaerobe's ability to utilize ammonium sulfate (47). Soiled poultry litter, which is a mixture of feces, bedding material (wood shavings or straw), waste feed, dead birds, and feathers, may be a suitable growth medium for Bacteroidales. Specifically, it may supply suitable nitrogen and carbon sources, as poultry litter has been shown to contain up to 1.8% dry weight of ammonia nitrogen (48) and up to 35% hemicellulose (i.e., pine shavings) (46). Additionally, poultry litter can be saturated with water just a few centimeters from the litter surface, particularly near water nozzles in barns. Therefore, with a high water content and elevated carbon and nitrogen levels in litter, anoxic and microaerophilic conditions can be generated quickly in poultry litter within a poultry house. These conditions likely support the extraintestinal environments necessary for support of Bacteroidales growth or persistence in poultry litter. In fact, a recent study reported the occurrence of Bacteroides species in every wet litter sample obtained from poultry houses in the Delmarva Peninsula (35).
In the present studies, Bacteroidales organisms were shown to increase in extraintestinal environments under both anoxic (laboratory microcosms) and oxic or microaerobic (river mesocosms and hen houses) conditions. In most cases, the interlaboratory microcosm incubation results were in good agreement. However, Bacteroidales gene concentrations increased in the aerobic laboratory microcosms at WVU but not at USF, where they decreased 2 log over time. While utmost care was taken to replicate the experimental conditions in the laboratory microcosms at USF and WVU, the litter used in the USF experiments had an additional 24-h transit time. The longer storage time may have caused physiological stress to the bacteria, which may have been reflected in the differing results, as the level of Bacteroidales increased under oxic conditions in the WVU experiments but decreased under oxic conditions in both rounds of USF experiments. Bacteroidales increases in the anoxic laboratory microcosm data were consistent between laboratories, but these conditions would be expected to be more permissive for Bacteroidales than oxic conditions, so physiological stress would not play as great a role.
While oxygen concentrations were measured only in the river-deployed microcosms and the replicate USF laboratory studies, there were likely low concentrations of oxygen present in the laboratory microcosms at WVU and the initial studies at USF, as evidenced by the negligible measured concentrations in the replicate USF experiment. Recent evidence suggests that several members of the Bacteroides class can grow in the presence of 70 μM dissolved oxygen. In fact, B. fragilis bacteria were shown to contain an O2-dependent respiratory chain that may give them an evolutionary advantage over strict anaerobes in colonizing surfaces (30). The ability to tolerate and proliferate in low-oxygen environments fits with observations of Bacteroides species in larger numbers on the mucosal surfaces of intestines than in the intestinal lumen, where the is lower. Other evidence for the survival of Bacteroidales in various oxygen environments includes detection of these organisms by qPCR in shallow soils (<10 cm) not directly contaminated with fecal material (33, 49), in domestic tap water samples, and in aerobic groundwater (33, 50).
The results of these studies suggest that caution should be used in interpreting the results of source tracking studies using qPCR methods targeting Bacteroidales in watersheds likely affected by poultry litter. If significant growth of Bacteroidales originating from poultry litter occurs after release into the environment, then the results of MST studies may be confounded. The findings here indicate that studies should be carried out using fecal matter from other host species in which Bacteroidales are important indicators, such as cattle. Other questions arising from this research that would benefit from additional study include the following. (i) Do Bacteroidales originating from chicken feces grow in bedding material besides wood shavings, such as the commonly used rice or peanut hulls, straw, and corn stalks? (ii) What are Bacteroidales densities in composted or aged litter, and how do they vary over time? (iii) Are there poultry litter-specific Bacteroides sp., and what is their persistence compared to that of Bacteroidales? Finally, an evaluation of Bacteroidales densities in litter applied to agricultural fields over time, as well as temporal variations in Bacteroidales in these soils, should be conducted. These proposed studies are required to aid in our understanding of the variability in poultry litter-associated Bacteroidales survival, persistence, and growth under environmental conditions so that accurate MST studies and remedial efforts can be conducted.
ACKNOWLEDGMENTS
Partial funding for this project was obtained from a summer undergraduate research fellowship from the WVU Department of Civil and Environmental Engineering and from National Science Foundation (NSF) Environmental Engineering Program funding, to Jennifer Weidhaas (grant 1234366) and Valerie J. Harwood (grant 1234237).
We thank the Morgantown Utility Board for allowing us unrestricted use of their property for the river studies. We appreciate the insightful comments and constructive criticism of the independent peer reviewers and editorial board, which strengthened the manuscript.
REFERENCES
- 1.Horrigan L, Lawrence R, Walker P. 2002. How sustainable agriculture can address the environmental and human health harms of industrial agriculture. Environ Health Perspect 110:445–456. doi: 10.1289/ehp.02110445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jongbloed A, Lenis B. 1998. Environmental concerns about animal manure. J Anim Sci 76:2641–2648. [DOI] [PubMed] [Google Scholar]
- 3.USEPA. 2003. National pollutant discharge elimination system permit regulation and effluent limitation guidelines and standards for concentrated animal feeding operations (CAFOs). Fed Regist 68:7177–7274. [Google Scholar]
- 4.Leclerc H, Mossel D, Edberg S, Struijk C. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu Rev Microbiol 55:201–234. doi: 10.1146/annurev.micro.55.1.201. [DOI] [PubMed] [Google Scholar]
- 5.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia GD, Carvalho MAR, Diniz CG, Marques JL, Nicoli JR, Farias LM. 2012. Isolation, identification and antimicrobial susceptibility of Bacteroides fragilis group strains recovered from broiler faeces. Br Poult Sci 53:71–76. doi: 10.1080/00071668.2012.662272. [DOI] [PubMed] [Google Scholar]
- 7.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allsop K, Stickler DJ. 1985. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J Appl Bacteriol 58:95–99. doi: 10.1111/j.1365-2672.1985.tb01433.x. [DOI] [PubMed] [Google Scholar]
- 9.Oladeinde A, Bohrmann T, Wong K, Purucker ST, Bradshaw K, Brown R, Snyder B, Molina M. 2014. Decay of fecal indicator bacterial populations and bovine-associated source-tracking markers in freshly deposited cow pats. Appl Environ Microbiol 80:110–118. doi: 10.1128/AEM.02203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed W, Gyawali P, Sidhu JP, Toze S. 2014. Relative inactivation of faecal indicator bacteria and sewage markers in freshwater and seawater microcosms. Lett Appl Microbiol 59:348–354. doi: 10.1111/lam.12285. [DOI] [PubMed] [Google Scholar]
- 11.Bae S, Wuertz S. 2009. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res 43:4850–4859. doi: 10.1016/j.watres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 12.Green HC, Shanks OC, Sivaganesan M, Haugland RA, Field KG. 2011. Differential decay of human faecal Bacteroides in marine and freshwater. Environ Microbiol 13:3235–3249. doi: 10.1111/j.1462-2920.2011.02549.x. [DOI] [PubMed] [Google Scholar]
- 13.Walters SP, Field KG. 2009. Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ Microbiol 11:1410–1421. doi: 10.1111/j.1462-2920.2009.01868.x. [DOI] [PubMed] [Google Scholar]
- 14.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 15.Field K, Samadpour M. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res 41:3517–3538. doi: 10.1016/j.watres.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 16.Stoeckel D, Harwood V. 2007. Performance, design and analysis in microbial source tracking studies. Appl Environ Microbiol 73:2405–2415. doi: 10.1128/AEM.02473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz CJ, Childers GW. 2011. Fecal Bacteroidales diversity and decay in response to variations in temperature and salinity. Appl Environ Microbiol 77:2563–2572. doi: 10.1128/AEM.01473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silkie SS, Nelson KL. 2009. Concentrations of host-specific and generic fecal markers measured by quantitative PCR in raw sewage and fresh animal feces. Water Res 43:4860–4871. doi: 10.1016/j.watres.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Ballesté E, Blanch AR. 2010. Persistence of Bacteroides species populations in a river as measured by molecular and culture techniques. Appl Environ Microbiol 76:7608–7616. doi: 10.1128/AEM.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM. 2010. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl Environ Microbiol 76:3255–3262. doi: 10.1128/AEM.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Z, He Z, Zhou X, Powell CA, Yang Y, Roberts MG, Stoffella PJ. 2012. High diversity and differential persistence of fecal Bacteroidales population spiked into freshwater microcosm. Water Res 46:247–257. doi: 10.1016/j.watres.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Sokolova E, Åström J, Pettersson TJ, Bergstedt O, Hermansson M. 2012. Decay of Bacteroidales genetic markers in relation to traditional fecal indicators for water quality modeling of drinking water sources. Environ Sci Technol 46:892–900. doi: 10.1021/es2024498. [DOI] [PubMed] [Google Scholar]
- 23.Tambalo DD, Fremaux B, Boa T, Yost CK. 2012. Persistence of host-associated Bacteroidales gene markers and their quantitative detection in an urban and agricultural mixed prairie watershed. Water Res 46:2891–2904. doi: 10.1016/j.watres.2012.02.048. [DOI] [PubMed] [Google Scholar]
- 24.Jeanneau L, Solecki O, Wéry N, Jardé E, Gourmelon M, Communal PY, Jadas-Hécart A, Caprais MP, Gruau G, Pourcher AM. 2012. Relative decay of fecal indicator bacteria and human-associated markers: a microcosm study simulating wastewater input into seawater and freshwater. Environ Sci Technol 46:2375–2382. doi: 10.1021/es203019y. [DOI] [PubMed] [Google Scholar]
- 25.Okabe S, Shimazu Y. 2007. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl Microbiol Biotechnol 76:935–944. doi: 10.1007/s00253-007-1048-z. [DOI] [PubMed] [Google Scholar]
- 26.Piorkowski GS, Bezanson GS, Jamieson RC, Hansen LT, Yost CK. 2014. Effect of hillslope position and manure application rates on the persistence of fecal source tracking indicators in an agricultural soil. J Environ Qual 43:450–458. doi: 10.2134/jeq2013.07.0274. [DOI] [PubMed] [Google Scholar]
- 27.Rogers SW, Donnelly M, Peed L, Kelty CA, Mondal S, Zhong Z, Shanks OC. 2011. Decay of bacterial pathogens, fecal indicators, and real-time quantitative PCR genetic markers in manure-amended soils. Appl Environ Microbiol 77:4839–4848. doi: 10.1128/AEM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae S, Wuertz S. 2009. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with propidium monoazide. Appl Environ Microbiol 75:2940–2944. doi: 10.1128/AEM.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.USEPA. June 2010. Method B: Bacteroidales in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay. EPA-822-R-10-003 US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 30.Baughn AD, Malamy MH. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 31.Meehan BM, Baughn AD, Gallegos R, Malamy MH. 2012. Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc Natl Acad Sci U S A 109:12153–12158. doi: 10.1073/pnas.1203796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra S, Imlay JA. 2013. An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol Microbiol 90:1356–1371. doi: 10.1111/mmi.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weidhaas J, Garner E, Basden T, Harwood VJ. 2014. Runoff studies demonstrate parallel transport behavior for a marker of poultry fecal contamination and Staphylococcus aureus. J Appl Microbiol 117:417–429. doi: 10.1111/jam.12543. [DOI] [PubMed] [Google Scholar]
- 34.Kelty CA, Varma M, Sivaganesan M, Haugland RA, Shanks OC. 2012. Distribution of genetic marker concentrations for fecal indicator bacteria in sewage and animal feces. Appl Environ Microbiol 78:4225–4232. doi: 10.1128/AEM.07819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumas MD, Polson SW, Ritter D, Ravel J, Gelb J Jr, Morgan R, Wommack KE. 2011. Impacts of poultry house environment on poultry litter bacterial community composition. PLoS One 6:e24785. doi: 10.1371/journal.pone.0024785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths R, Whiteley A, O'Donnell A, Bailey M. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siefring S, Varma M, Atikovic E, Haugland R. 2008. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J Water Health 6:225–237. doi: 10.2166/wh.2008.022. [DOI] [PubMed] [Google Scholar]
- 38.Bernhard A, Field K. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. doi: 10.1128/AEM.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huber I, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 42.Bae S, Wuertz S. 2012. Survival of host-associated Bacteroidales cells and their relationship with Enterococcus spp., Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus in freshwater microcosms as measured by propidium monoazide-quantitative PCR. Appl Environ Microbiol 78:922–932. doi: 10.1128/AEM.05157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade T, Sams E, Brenner K, Haugland RA, Chern E, Beach M, Wymer L, Rankin C, Love D, Li Q, Noble R, Dufour A. 2010. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ Health 9:66. doi: 10.1186/1476-069X-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walters SP, Field KG. 2006. Persistence and growth of fecal Bacteroidales assessed by bromodeoxyuridine immunocapture. Appl Environ Microbiol 72:4532–4539. doi: 10.1128/AEM.00038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degnan BA, Macfarlane S, Quigley ME, Macfarlane GT. 1997. Starch utilization by Bacteroides ovatus isolated from the human large intestine. Curr Microbiol 34:290–296. doi: 10.1007/s002849900184. [DOI] [PubMed] [Google Scholar]
- 46.Hespell RB, Whitehead TR. 1990. Physiology and genetics of xylan degradation by gastrointestinal tract bacteria. J Dairy Sci 73:3013–3022. doi: 10.3168/jds.S0022-0302(90)78988-6. [DOI] [PubMed] [Google Scholar]
- 47.Ushijima T, Takahashi M, Tatewaki K, Ozaki Y. 1983. A selective medium for isolation and presumptive identification of the Bacteroides fragilis group. Microbiol Immunol 27:985–993. doi: 10.1111/j.1348-0421.1983.tb02929.x. [DOI] [PubMed] [Google Scholar]
- 48.Kelleher BP, Leahy JJ, Henihan AM, O'Dwyer TF, Sutton D, Leahy MJ. 2002. Advances in poultry litter disposal technology—a review. Bioresour Technol 83:27–36. doi: 10.1016/S0960-8524(01)00133-X. [DOI] [PubMed] [Google Scholar]
- 49.Vierheilig J, Farnleitner AH, Kollanur D, Blöschl G, Reischer GH. 2012. High abundance of genetic Bacteroidetes markers for total fecal pollution in pristine alpine soils suggests lack in specificity for feces. J Microbiol Methods 88:433–435. doi: 10.1016/j.mimet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Wielen PWJ, Medema G. 2010. Unsuitability of quantitative Bacteroidales 16S rRNA gene assays for discerning fecal contamination of drinking water. Appl Environ Microbiol 76:4876–4881. doi: 10.1128/AEM.03090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]