Abstract
A newly isolated strain, SJY1, identified as Ochrobactrum sp., utilizes nicotine as a sole source of carbon, nitrogen, and energy. Strain SJY1 could efficiently degrade nicotine via a variant of the pyridine and pyrrolidine pathways (the VPP pathway), which highlights bacterial metabolic diversity in relation to nicotine degradation. A 97-kbp DNA fragment containing six nicotine degradation-related genes was obtained by gap closing from the genome sequence of strain SJY1. Three genes, designated vppB, vppD, and vppE, in the VPP pathway were cloned and heterologously expressed, and the related proteins were characterized. The vppB gene encodes a flavin-containing amine oxidase converting 6-hydroxynicotine to 6-hydroxy-N-methylmyosmine. Although VppB specifically catalyzes the dehydrogenation of 6-hydroxynicotine rather than nicotine, it shares higher amino acid sequence identity with nicotine oxidase (38%) from the pyrrolidine pathway than with its isoenzyme (6-hydroxy-l-nicotine oxidase, 24%) from the pyridine pathway. The vppD gene encodes an NADH-dependent flavin-containing monooxygenase, which catalyzes the hydroxylation of 6-hydroxy-3-succinoylpyridine to 2,5-dihydroxypyridine. VppD shows 62% amino acid sequence identity with the hydroxylase (HspB) from Pseudomonas putida strain S16, whereas the specific activity of VppD is ∼10-fold higher than that of HspB. VppE is responsible for the transformation of 2,5-dihydroxypyridine. Sequence alignment and phylogenetic analysis suggested that the VPP pathway, which evolved independently from nicotinic acid degradation, might have a closer relationship with the pyrrolidine pathway. The proteins and functional pathway identified here provide a sound basis for future studies aimed at a better understanding of molecular principles of nicotine degradation.
INTRODUCTION
Nicotine, which easily passes biological membranes, is significantly harmful to humans and is considered a potentially addictive drug (1, 2). Large quantities of tobacco waste with high concentrations of nicotine are generated annually by cigarette and cigar manufacturing (3). Due to its water solubility, nicotine can easily spread in the environment, emerging as a threat to public health (3). As a result, the U.S. Environmental Protection Agency classified nicotine as a “toxic release inventory” chemical in 1994 (4). Microbes play significant roles in removing nicotine from the environment, and in the meantime, they serve as the catalysts for biotransformation. Nicotine could potentially be used for the production of value-added chemicals. 6-Hydroxy-3-succinoylpyridine (HSP), an important precursor for the synthesis of compounds with biological activities, was efficiently produced by an engineered nicotine-degrading strain (5). Therefore, the study of the mechanism of nicotine degradation provides useful information for both bioremediation and biocatalysis.
Many microorganisms that use various nicotine degradation pathways have been isolated from the environment, including Arthrobacter (6), Pseudomonas (7, 8), Ochrobactrum (9), Agrobacterium (10), Shinella (11), and Aspergillus (12). The pathways of nicotine degradation have been widely investigated by researchers. Pyridine and pyrrolidine pathways are the two best-elucidated nicotine metabolic pathways, and most of the catalytic enzymes in these two pathways have been functionally characterized (6, 13). In the pyrrolidine pathway, nicotine degradation is initiated by dehydrogenation of the pyrrolidine ring to form N-methylmyosmine, which is then converted to 6-hydroxynicotine (6HN), 6-hydroxy-N-methylmyosmine (6HMM), 6-hydroxypseudonicotine (6HPON), 2,6-dihydroxypseudooxynicotine, 2,6-dihydroxypyridine, and 2,3,6-trihydroxypyridine. 2,3,6-Trihydroxypyridine forms a blue pigment spontaneously (6). The pyridine pathway in Pseudomonas putida S16 is nicotine, N-methylmyosmine, pseudoxynicotine, 3-succinoyl-pyridine (SP), HSP, and 2,5-dihydroxypyridine (2,5-DHP). The 2,5-DHP is converted via a ring cleavage reaction to render N-formylmaleamic acid, maleamic acid, maleic acid, and fumaric acid (13). In addition to the pyridine and pyrrolidine pathways, some other nicotine degradation pathways were also proposed, including a variant of the pyridine and pyrrolidine pathways, which is designated the VPP pathway in bacteria (10, 11). In the upper VPP pathway, nicotine is converted to 6HPON through 6HN and 6HMM, just like in the pyridine pathway. Subsequently, 6HPON is oxidized to form HSP. In the lower VPP pathway, HSP is catabolized to fumaric acid as in the pyrrolidine pathway. Two strains, Agrobacterium tumefaciens strain S33 and Shinella sp. strain HZN7, with the VPP pathway, were isolated and characterized recently (10, 11). However, the molecular mechanisms involved were rarely reported. Only the enzyme activities of nicotine dehydrogenase, 6HN oxidase, and HSP hydroxylase in crude cells of strain S33 were detected (10). In Shinella sp. HZN7, a DNA fragment that was considered responsible for the conversion from 6HPON to HSP was identified by transposon mutagenesis (11), whereas the enzyme has not been studied in vitro. Analysis of genes and biochemical characterization of nicotine catabolism enzymes form a basis for rationally improving strain employment in the disposal of nicotine wastes.
In this study, a potential nicotine-degrading strain, Ochrobactrum sp. strain SJY1, was newly isolated and identified. Along with characterizing this new nicotine-degrading strain, we also determined the conditions for cell growth and nicotine degradation. Recently, the genome sequence of strain SJY1 was determined (14), and it helps us to further identify the genomic and metabolic diversity of the species. Strain SJY1 degrades nicotine via the VPP pathway, based on the identification of intermediate metabolites. A DNA fragment of the vpp gene cluster catalyzing nicotine degradation was newly identified using in silico analysis of the genome sequence of strain SJY1. Six genes in the upper and lower VPP pathway were predicted in this DNA fragment. The functions of three genes (vppB, vppD, and vppE) in the VPP pathway were first characterized in detail. Some evolutionary considerations regarding the nicotine degradation genes are also discussed. The objective of this work is to demonstrate the unknown catabolism involved in the nicotine degradation pathway.
MATERIALS AND METHODS
Chemicals and media.
l-Nicotine (99% purity) was purchased from Fluka Chemie GmbH (Buchs Corp., Switzerland) as a standard. 2,5-DHP was purchased from SynChem OHG (Kassel Corp., Kassel, Germany). HSP was prepared as previously described (5). 6HN and 6HPON were obtained from Roderich Brandsch. All other reagents and solvents used in this study were of analytical grade and commercially available. Nicotine medium was prepared by adding filtration-sterilized nicotine to minimal salt medium (MSM) (15). A solid agar plate was prepared by the addition of 1.5% (wt/vol) agar to liquid medium.
Isolation and identification of strain SJY1.
One strain with high nicotine-degrading capacity was isolated and named SJY1. The cell morphology of strain SJY1 was observed with a scanning electron microscope (SEM) (JEOL 7500F; Japan). The 16S rRNA gene was amplified by PCR using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGYTACCTTGTTACGACTT-3′). 16S rRNA genes of other typical strains for sequence analysis were obtained from the LSPN database (16). Sequence alignment and phylogenetic analysis were performed by MEGA6 (17) using a neighbor-joining method with a bootstrap value of 1,000.
Bacterial growth and nicotine degradation.
In order to obtain the optimal growth conditions for strain SJY1, culture temperatures, pH values, and nicotine concentrations were investigated. Strain SJY1 was first cultured in 250-ml flasks with 50 ml sterilized MSM with 1.0 mg ml−1 nicotine at 30°C at 200 rpm. After 24 h of cultivation, the culture was inoculated into 50 ml new MSM containing 1.0 mg ml−1 nicotine and inoculated at 30°C, 37°C, and 42°C. Media with different pH values (pH 5.0, 6.0, 7.0, 8.0, and 9.0) adjusted with potassium-phosphate buffer were used to determine the optimal pH value for SJY1 cultivation. Under optimal temperature and pH values, a nicotine toleration test of strain SJY1 was performed with different initial nicotine concentrations (0.5, 1.0, 1.5, 2.0, 3.0, 4.0, and 5.0 mg ml−1). Strain SJY1, for nicotine degradation, was cultivated under optimal conditions with MSM (1.5 mg ml−1 nicotine), LB medium, and LB medium with the addition of 1.5 mg ml−1 nicotine. Cells were collected by centrifugation at 6,000 × g for 5 min and then washed twice with 50 mM phosphate buffer (pH 7.0). The cells were suspended with the same phosphate buffer to 3.6 g liter−1 dry cell weight (as resting cells [18]), and the resting-cell reactions were performed at 30°C at 120 rpm.
Intermediates and genes in nicotine degradation.
Nicotine (5 mg ml−1) and several intermediates, such as 6HN (3 mg ml−1), SP (3 mg ml−1), HSP (3 mg ml−1), and 2,5-DHP (3 mg ml−1), were used as the substrates for resting-cell reactions of strain SJY1. The reaction samples were analyzed by high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS).
Gaps between contigs in the SJY1 draft genome were closed by direct PCR strategies (14, 19). The linkage information for contigs was obtained by sequence alignment with the complete genome sequence of Ochrobactrum anthropi ATCC 49188. The reported nicotine-degrading genes were used for alignment through all the proteins of strain SJY1.
RT-PCR and RT-qPCR experiments.
MSM with 1 mg ml−1 (NH4)2SO4 and 1 mg ml−1 sodium citrate was used as the control medium for reverse transcription-quantitative PCR (RT-qPCR) experiments. Cultures in the presence and absence of nicotine were cultivated at 30°C to mid-exponential phase (optical density at 600 nm [OD600] = 1.0). Total RNAs were extracted from ∼1 × 109 cells of Ochrobactrum sp. SJY1 with an RNAprep pure cell/bacteria kit (TianGen, China). The cDNA was prepared using random-hexamer primers and SuperScript III reverse transcriptase (Invitrogen). The cDNA served as the template for RT-PCR and RT-qPCR analysis. The primers for RT-PCR were 5′-CGCACATCACCAATCCCACA-3′ and 5′-AACTCCGAACTTTGTCCACCA-3′ for region I, 5′-TGCCCGGCATTGTCCGTTCGA-3′ and 5′-ATGGCATCGAGCGAGAAGTCG-3′ for region II, 5′-CGACGGGTTTCACTCCGATGA-3′ and 5′-CAACGGCATCTGGCATCGCG-3′ for region III, and 5′-TCGGCTGGGGCATGAACCCCCA-3′ and 5′-GACCAGACGGCTGCCCTCAA-3′ for region IV. The primers for RT-qPCR were 5′-AAGTCTTGAGTATGGTAG-3′ and 5′-GGTATCTAATCCTGTTTG-3′ for 16S rRNA, 5′-TAAGTTCGGTGCCGCCCTGAT-3′ and 5′-GGTGGTCGGTGAGTCTTGC-3′ for the gene vppB, 5′-GTGGACTACATCCGCAACGA-3′ and 5′-TGAACGCAGGCGGACAGCAC-3′ for the gene vppH, and 5′-GATGAAACATTTCTGTCC-3′ and 5′-ATGTCATCGGAGTGAAAC-3′ for the gene vppF. The RT-qPCR was performed using the CFX96 Real-Time PCR Detection system (Bio-Rad, Hercules, CA) with SYBR green RealmasterMix (TianGen, China) and RT-qPCR primers.
Cloning and expression of the vppB, vppD, and vppE genes.
The vppB gene was amplified from genomic DNA of strain SJY1 by PCR using primers vppB-F (5′-ATACCATGGGGACAGAAAAGATTTATGATGCAA-3′) and vppB-R (5′-CCCAAGCTTAGCGGTCGCCTTCATAAAGTGC-3′). Then, the vppB gene fragment was digested with NcoI and HindIII and ligated into the NcoI-HindIII sites of pET28a to form recombinant plasmid pET28a-vppB. The vppD gene was amplified by PCR using primers vppD-F (5′-ATACCATGGGCAGCGCACATGTCGTTGTCGT-3′) and vppD-R (5′-GTGCTCGAGATATACTGTCCGCATCTGTTC-3′) and ligated into plasmid pET28a with NcoI and XhoI sites to form recombinant plasmid pET28a-vppD. The gene vppE was amplified using primers vppE-F (5′-ATACCATGGACCACACGAGTTTCACCGAAATC-3′) and vppE-R (5′-CCCAAGCTTTTATCGTTCCATTTTCATGTCCG-3′) and ligated into plasmid pET28a with NcoI and HindIII sites to form recombinant plasmid pET28a-vppE without a His tag. The recombinant plasmids were sequenced and transformed into Escherichia coli BL21(DE3). E. coli cells were grown in 50 ml LB medium containing 50 μg ml−1 kanamycin at 37°C at 200 rpm. When the optical density reached to 0.6 to 0.8, a final concentration of 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the culture. The culture was further cultivated for 20 h at 16°C, and the cells were harvested by centrifugation and broken by ultrasonic treatment as previously described (20). His-tagged proteins were purified on a His-trapped column using a liquid chromatography system (Akta; GE Healthcare, Little Chalfont, United Kingdom). The protein concentration was determined by the Bradford method using bovine serum albumin as a standard. SDS-PAGE was carried out according to the method of Laemmli (21).
Enzymatic assays of VppB, VppD, and VppE.
All the enzyme activities were measured at 25°C in 50 mM Tris-HCl buffer, pH 8.0, in a total volume of 1 ml using a UV-2550 spectrophotometer (Shimadzu, Kyoto, Japan). The reaction of VppB was carried out with the addition of VppB and 125 μM 6HN, and enzyme activity was calculated according to the absorbance increase at 300 nm (the molar extinction coefficient change at 300 nm is 6,768 M cm−1). The enzyme assay of 6-hydroxy-l-nicotine oxidase (6HLNO) from Arthrobacter nicotinovorans is the same as that of VppB. The enzyme activity of VppD was determined spectrophotometrically by monitoring the decrease of NADH (or NADPH) at 340 nm as previously described (20). Briefly, 250 μM HSP and 250 μM NADH were added to the reaction system (800 μl total), and the detection started immediately after the addition of appropriate VppD. VppE activity was measured at 320 nm with the addition of VppE, 250 μM 2,5-DHP, and 250 μM Fe2+ (FeCl2) as previously reported (7). The products of enzyme catalytic reactions were further determined by HPLC or LC-MS. Buffers at pH values from 5 to 6.5 (citric acid/sodium citrate), 7 to 9 (Tris-HCl), and 9.5 to 11 (sodium carbonate/sodium bicarbonate) were used in pH-dependent experiments. One unit of activity was defined as the amount of enzyme that catalyzed the conversion of 1 μmol of substrate in 1 min.
Analytical methods.
Intermediates of nicotine degradation were measured by HPLC with diode array detection, using an Eclipse XDB-C18 reverse-phase column (5 μm; 4.6 by 150 mm; Keystone Scientific, Bellefonte, PA) at 30°C. The mobile phase was 92% (vol/vol) 1 mM sulfuric acid and 8% (vol/vol) methanol at a flow rate of 0.5 ml min−1. The reaction sample of VppD was detected with a mobile phase of 80% (vol/vol) 1 mM sulfuric acid and 20% (vol/vol) methanol on a 250-mm XDB-C18 reverse-phase column. Flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) were determined with a mobile phase of 80% (vol/vol) 5 mM ammonium acetate and 20% (vol/vol) methanol on a 150-mm XDB-C18 reverse-phase column at a flow rate of 0.5 ml min−1. The retention time and absorption spectra were compared with those of standard compounds for each run to identify the substrates or products. LC-MS analysis was performed on an Agilent 6230 time of flight (TOF)-MS equipped with electrospray ionization (ESI) sources in 40% (vol/vol) methanol (0.1% [vol/vol] formic acid) and 60% (vol/vol) deionized water (18 MΩ cm−1; 0.05% formic acid [vol/vol]) at a flow rate of 0.2 ml min−1. All samples were treated with the addition of 2 volumes of methanol at 4°C for 10 min, centrifuged for 2 min at 12,000 × g, and then filtered through a 0.22-μm Millipore filter prior to HPLC and LC-MS analyses.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA gene and vpp cluster from strain SJY1 are available in GenBank under accession numbers KM065744 and KM065745.
RESULTS
Isolation and identification of the nicotine degrader SJY1.
A nicotine-degrading bacterium, strain SJY1, was isolated from nicotine-contaminated water. Strain SJY1 is a rod-shaped Gram-negative bacterium with dimensions of 0.3 to 0.35 by 1.0 to 2.0 μm under the SEM. Based on its 16S rRNA gene sequence, strain SJY1 was found to have the highest sequence identity (99.4%) to Ochrobactrum rhizosphaerae. Thus, the morphological and physiological characterization data supported the idea that strain SJY1 should be classified as an Ochrobactrum strain. Ochrobactrum sp. strain SJY1 has been deposited at the China Center for Type Culture Collection (CCTCC) under accession number AB2014146.
Cultivation conditions and nicotine degradation.
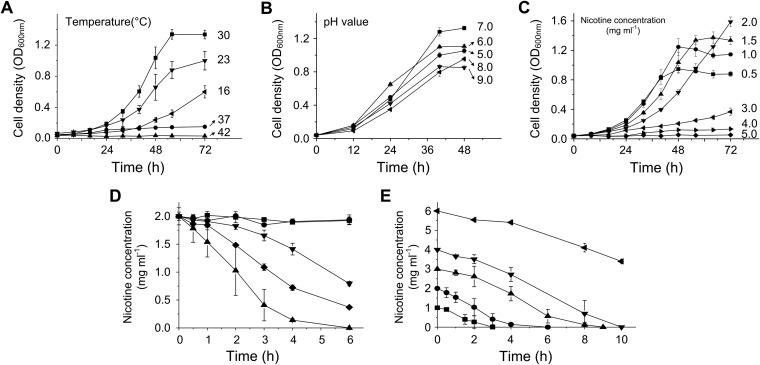
The optimal temperature was observed to be 30°C (Fig. 1A). Strain SJY1 could grow well in the pH range of 5.0 to 9.0, with an optimum at pH 7.0. The growth of strain SJY1 was observed at up to 4.0 mg ml−1 in nicotine medium. The optimal nicotine concentration was 1.5 mg ml−1, whereas growth was poor above a 3.0-mg ml−1 nicotine concentration (Fig. 1C). Nicotine degradation by resting cells of strain SJY1 prepared from nicotine medium (containing 1.5 mg ml−1 nicotine; induced), LB medium (containing 1.5 mg ml−1 nicotine; induced), and LB medium (without nicotine; noninduced) reveals that nicotine-induced resting cells of strain SJY1 have a higher degradation rate (Fig. 1D). The nicotine concentrations in the control samples (cell-free system and heat-killed resting cells) were basically unchanged. The results indicate that the enzyme responsible for the conversion of nicotine is induced by nicotine. Degradation with different initial nicotine concentrations showed that strain SJY1 almost completely degraded 4 mg ml−1 nicotine after 10 h (Fig. 1E). The rate of degradation by the resting cells did not decrease with the increase of the initial nicotine concentration from 1 mg ml−1 to 4 mg ml−1; however, the degradation was inhibited when the nicotine concentration was higher than 4 mg ml−1.
FIG 1.
Optimization of growth conditions and nicotine degradation by resting cells. (A, B, and C) Effects of temperature, pH, and nicotine concentration on growth of Ochrobactrum sp. SJY1. The temperatures, pH values, and nicotine concentrations are indicated at the end of each curve. (D) Nicotine degradation by resting cells of Ochrobactrum sp. SJY1 prepared from MSM with nicotine (▲), LB medium with nicotine (◆), and LB medium (▼). Reactions with a cell-free system (■) and heat-killed cells (●) were performed as controls. (E) Nicotine degradation by resting cells of strain SJY1 with different initial nicotine concentrations: 1 mg ml−1 (■), 2 mg ml−1 (●), 3 mg ml−1 (▲), and 4 mg ml−1 (◆). Each value is the mean of results from three parallel replicates ± the standard deviation (SD).
The nicotine degradation pathway in strain SJY1.
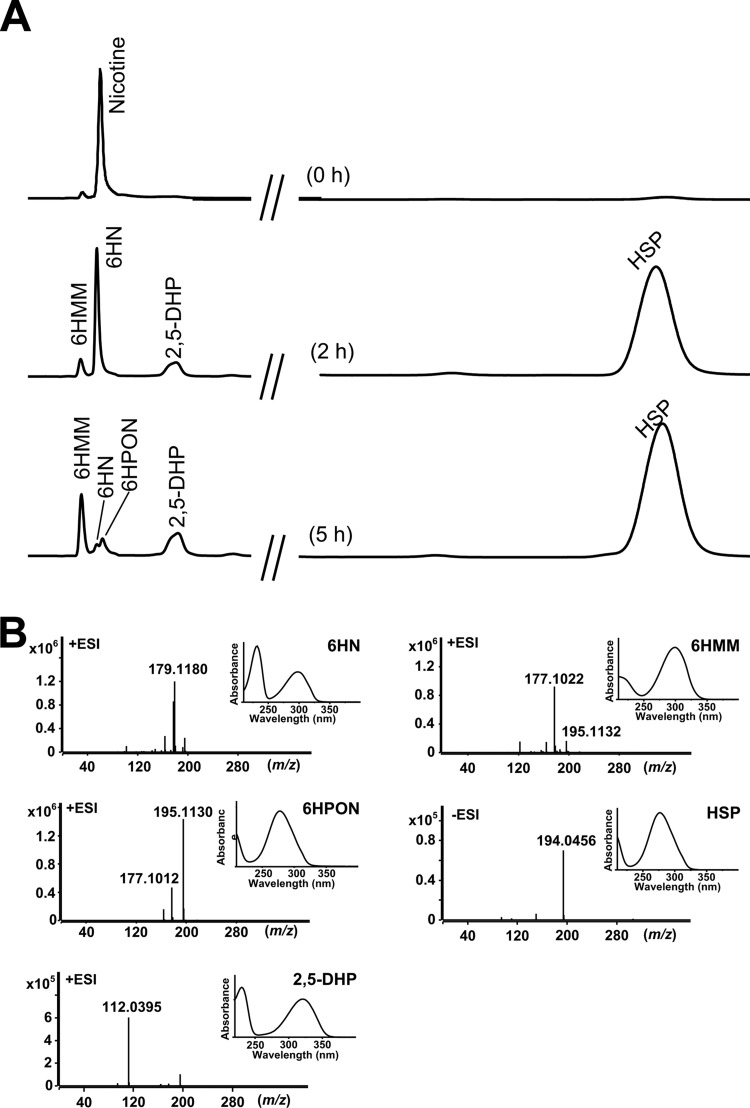
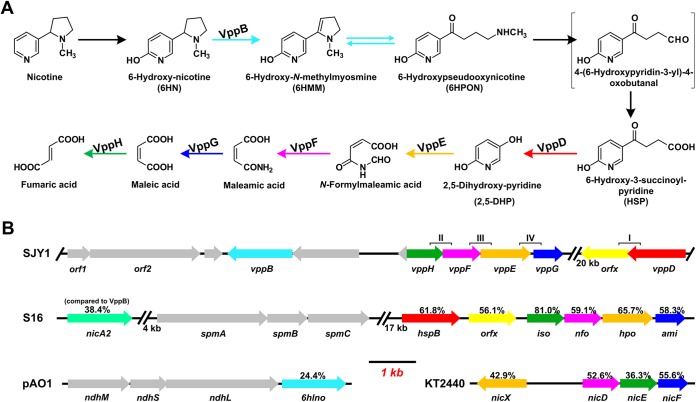
Five subsequent intermediates during nicotine degradation by resting cells of strain SJY1 were analyzed by HPLC and LC-MS. The intermediates were identified as 6HN, 6HMM, 6HPON, HSP, and 2,5-DHP (Fig. 2) by comparing the retention times, spectra, and molecular weights with those of standard compounds. The biotransformation of 6HN to 6HMM and 6HPON was confirmed by using 6HN as the substrate; HSP was transformed to form 2,5-DHP, and 2,5-DHP could be further converted (data not shown). However, SP could not be transformed by resting cells of strain SJY1, which confirmed that HSP was not transformed from SP as in the pyrrolidine pathway (13). No blue pigment was produced during nicotine degradation, indicating that 6HPON was not transformed into 2,3,6-trihydroxypyridine. These results suggested that strain SJY1 possesses a VPP pathway for nicotine degradation (Fig. 3A). Nicotine degradation is initiated by hydroxylation in the pyridine ring to form 6HN. 6HN is catalyzed by a dehydrogenation reaction to 6HMM and then by spontaneous hydrolysis to generate 6HPON. Subsequently, 6HPON is converted to HSP. HSP is transformed directly to 2,5-DHP, which is further degraded. The pathway was confirmed by the enzymatic experiments described below.
FIG 2.
Intermediate identification in nicotine degradation of strain SJY1. (A) HPLC profile of intermediate determination by reaction of resting cells of Ochrobactrum sp. SJY1 at pH 7.0 and 30°C using nicotine as the substrate. (B) Spectrum and LC-MS analysis of intermediates 6HN, 6HMM, 6HPON, HSP, and 2,5-DHP from reactions of resting-cell degradation by strain SJY1.
FIG 3.
The VPP pathway of nicotine catabolism in Ochrobactrum sp. SJY1. (A) The whole predicted nicotine degradation VPP pathway of Ochrobactrum sp. SJY1. (B) Genetic organization around the vpp cluster involved in nicotine degradation in Ochrobactrum sp. SJY1. The genes are functionally annotated following the color code used in panel A. The arrows indicate the size and direction of transcription of each gene. vppB, 6HN oxidase gene; vppH, maleate isomerase gene; vppF, N-formylmaleamic acid deformylase gene; vppE, 2,5-DHP dioxygenase; vppG, maleamate amidase gene; vppD, HSP 3-monooxygenase. The numbers within the arrows indicate the percent amino acid sequence identity with the orthologous gene product from Ochrobactrum sp. SJY1. SJY1, Ochrobactrum sp. SJY1; S16, P. putida S16; pAO1, A. nicotinovorans pAO1; KT2440, P. putida KT2440.
Analysis of a nicotine-degrading gene cluster.
Genome sequencing has the potential to accelerate the study of molecular mechanisms rather than requiring significant production scale searching (13, 22). A 97.6-kbp DNA fragment designated the vpp cluster was assembled by gap closing from the draft genome sequence of strain SJY1. The G+C content of the vpp cluster (59.01%) is higher than that of the genome of strain SJY1 (54.17%). Six genes were found in the vpp cluster that encode proteins showing 38% to 81% amino acid sequence identity with reported nicotine degradation proteins from A. nicotinovorans and P. putida S16 (Fig. 3) (7). VppB shows 24.4% amino acid sequence identity with 6HLNO (6) and 38.4% sequence identity with nicotine oxidase (NicA2) from P. putida S16 (13) and is predicted to catalyze the dehydrogenation step in nicotine degradation. The deduced amino acid sequence of the gene vppD shares 61.8% sequence identity with HspB from P. putida S16, indicating that the gene may be responsible for the conversion of HSP to 2,5-DHP. Proteins from VppH to VppG were supposed to catalyze the conversion of 2,5-DHP to fumaric acid (Fig. 3B).
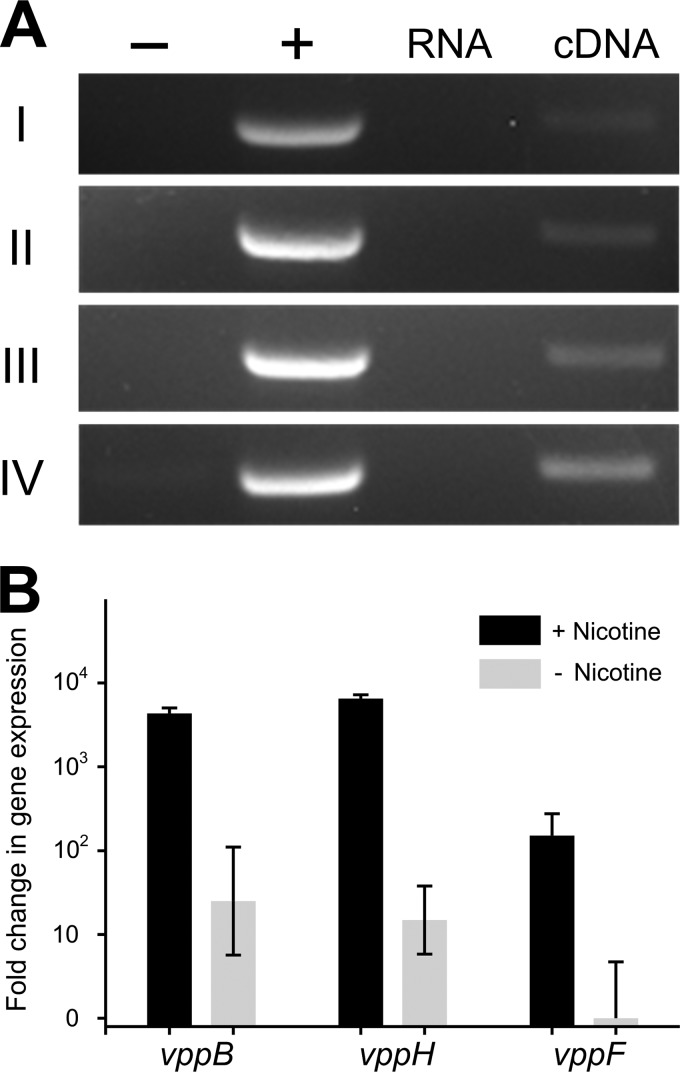
To examine whether the genes in the vpp cluster are transcribed as a single contiguous transcript, the intergenic regions were analyzed by RT-PCR. The vppD-orfx intergenic region (region I) was successfully amplified (Fig. 4A), indicating that the two genes vppD and orfx are cotranscribed. The vppH-vppF (region II), vppF-vppE (region III), and vppE-vppH (region IV) intergenic regions were all detected, suggesting that the four genes are transcribed as a single contiguous transcript. The mRNA levels of three genes (vppB, vppH, and vppF) in strain SJY1 grown in the presence or absence of nicotine were compared using RT-qPCR. The transcript levels were significantly increased in response to nicotine (vppB, 84.6-fold increase; vppH, 346.3-fold increase; and vppF, 100.3-fold increase) (Fig. 4B), indicating that these genes are nicotine inducible in strain SJY1.
FIG 4.
Organization and transcription analysis of the vpp cluster. (A) RT-PCR analysis of regions I, II, III, and IV as shown in Fig. 3B. The genomic DNA of strain SJY1 (lane +) and double-distilled H2O (ddH2O) were used as positive and negative templates for PCR with oligonucleotide pairs for regions I, II, III, and IV. The RNA (lane RNA) and cDNA (lane cDNA) from Ochrobactrum sp. SJY1 cells grown in the presence of nicotine were used as the templates for PCR analysis. (B) Quantitative RT-PCR analysis of the genes in the vpp cluster. The relative expression levels of the vppB, vppH, and vppF genes were measured using RNA extracted from Ochrobactrum sp. SJY1 grown in the presence (black bars) or absence (gray bars) of nicotine. All data were normalized to the 16S rRNA and are expressed as fold change relative to the expression level in cells. Each value is the mean from three parallel replicates ± SD.
The vppB gene encodes the 6-hydroxynicotine oxidase.
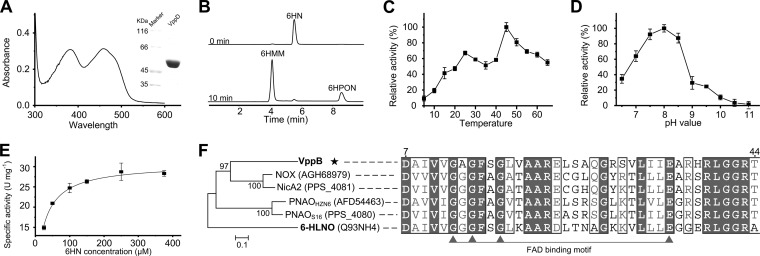
The second enzymatic step in the VPP pathway for nicotine degradation is the dehydrogenation of 6HN to render 6HMM (Fig. 3). This reaction is catalyzed in the pyridine pathway by 6HLNO from A. nicotinovorans (6), which shows only 24.4% amino acid sequence identity with the product of the vppB gene. The purified recombinant VppB turned yellow and exhibited absorption maxima at 382 nm and 459 nm, representing the characteristics of a typical flavoprotein (Fig. 5A). The flavin in VppB was determined to be FAD, and the enzyme activity did not change with an excess of FAD or FMN, indicating that FAD was bound tightly to VppB. The HPLC and LC-MS results showed that 6HN was transformed to 6HMM and 6HPON, which was consistent with the above-mentioned results of resting-cell reactions (Fig. 5B). No change of enzyme activity was detected when excess EDTA was mixed with VppB in the oxidative reaction, indicating that the reaction progressed without the participation of metal ions. The activity of nicotine transformation by VppB was not detected under the same conditions. The optimal pH and temperature values of VppB were observed to be 8.0 and 45°C, respectively (Fig. 5D). The apparent Km and kcat of 6HN in 50 mM Tris-HCl buffer, pH 8.0, at 25°C are 24.6 ± 1.67 μM and 25.8 ± 0.47 s−1, respectively (Fig. 5E). Phylogenetic analysis revealed that VppB has higher amino acid sequence identity with the amine oxidases from the pyrrolidine pathway than its isoenzyme, 6HLNO, from the pyridine pathway. The multiple alignment shows that all these enzymes, including VppB, have a conservative flavin-binding motif in the N termini of proteins (Fig. 5F).
FIG 5.
Characterization of VppB. (A) Spectrum scanning and SDS-PAGE of purified VppB. (B) HPLC analysis of the reaction of VppB at 0 min and 20 min. (C) Temperature-dependent enzyme activity of VppB. (D) pH optimization of VppB. VppB was unstable and participated when the pH value was less than 6.5. (E) Kinetic studies of VppB, fitted to the Michaelis-Menten curve for 6HN. (F) Multiple alignment and phylogenetic analysis of VppB and related amine oxidases. The GenBank accession number for each protein is shown in parentheses. (D and E) The values are the means from three parallel replicates ± SD.
The vppD gene encodes the HSP 3-monooxygenase.
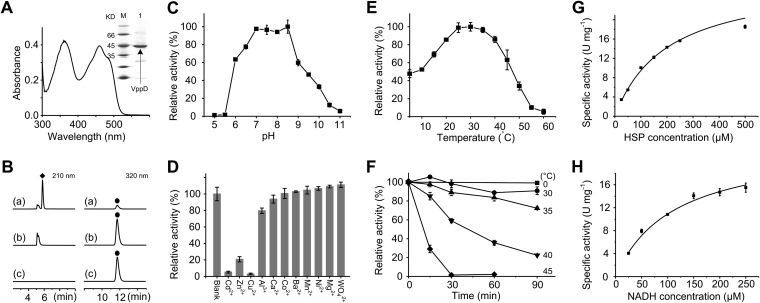
The reaction from HSP to 2,5-DHP is one of the key steps in the pyrrolidine pathway (20). The deduced amino acid sequence of the vppD gene shares 62% sequence identity with HspB from P. putida S16. The spectrum shows that purified recombinant VppD is a flavin-dependent monooxygenase like HspB (Fig. 6A), and the flavin in VppD (FAD) was the same as HspB. The enzyme activity did not change with an excess of FMN; however, it increased by ∼25.2% when an excess of FAD was added to the reaction system, indicating that FAD was partly lost during the purification. VppD could transform HSP into 2,5-DHP only in the presence of NAD(P)H (Fig. 6B), revealing an NAD(P)H-dependent HSP 3-monooxygenase activity. The enzyme activity when using NADPH as an electron donor was 68.2% of that using NADH as the substrate. The optimal pH and temperature of the VppD reaction were around 8.0 and 25°C (Fig. 6C and E), and Cd2+, Zn2+, and Cu2+ strongly inhibited the activity (Fig. 6D). Kinetic analysis revealed that the apparent Km and kcat values for HSP (at 250 μM NADH) at 25°C were 201 ± 21.3 μM and 21.6 ± 1.22 s−1, respectively. The apparent Km and kcat values for NADH at 250 μM HSP were 112 ± 17.5 μM and 17.5 ± 1.48 s−1, respectively (Fig. 6G and H).
FIG 6.
Characterization of VppD. (A) Absorbance spectrum and SDS-PAGE of VppD. M, molecular size markers; lane 1, purified His-tagged VppD. (B) HPLC profiles of biochemical assay of VppD for the conversion of HSP (●) into 2,5-DHP (◆). (a) One millimolar HSP plus 1.2 mM NADH plus VppD; (b) 1 mM HSP plus 1.2 mM NADH; (c) 1 mM HSP plus VppD. The reactions were performed for 20 min at 25°C. (C) pH-dependent enzyme activity of VppD. (D) Effects of metal ions on VppD activity. The final concentration of metal ions was 2 mM. Blank, without metal ion. (E) Temperature-dependent enzyme activity of VppD. (F) Stability of VppD at different temperatures. (G and H) Kinetic studies of VppD for HSP and NADH, fitted to the Michaelis-Menten curve for HSP and NADH. (C to H) The values are the means of results from three parallel replicates ± SD.
Conversion of 2,5-dihydroxypyridine to fumaric acid.
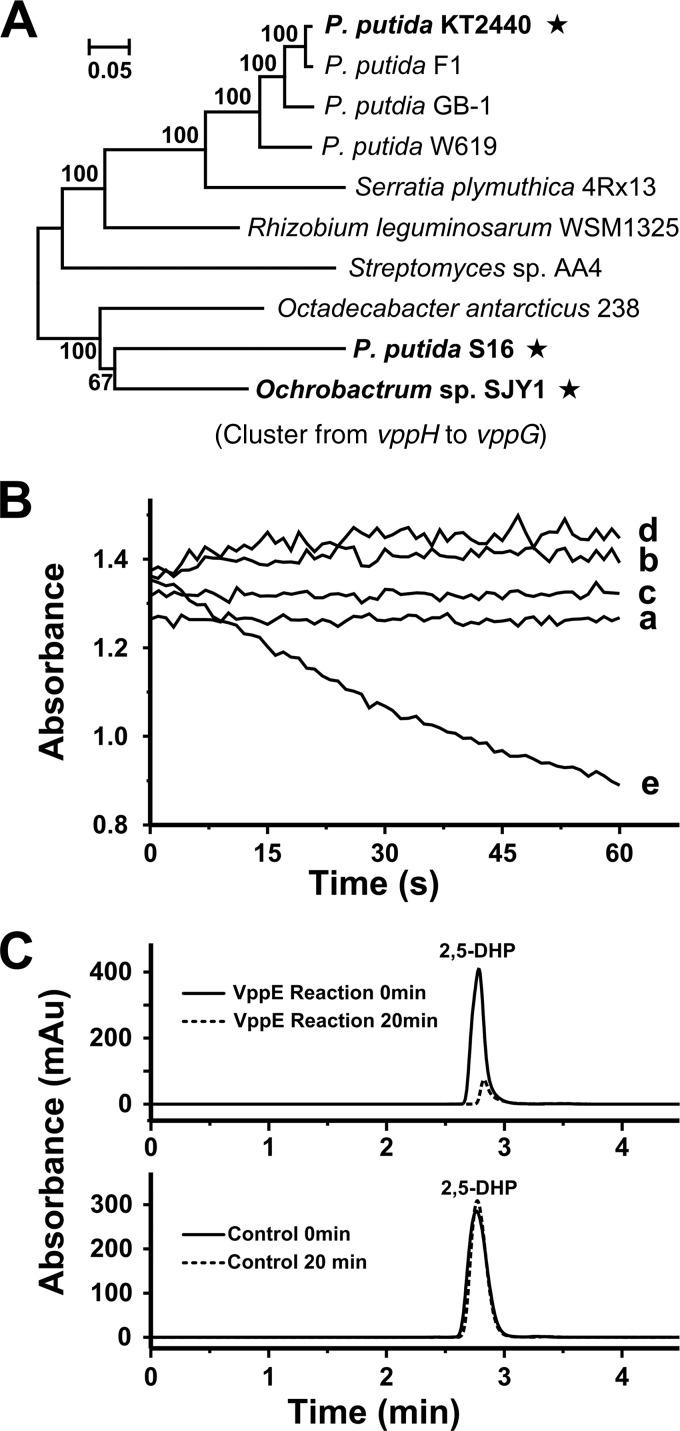
Four genes (vppE, vppF, vppG, and vppH), which are transcribed as a single contiguous transcript, are predicted for the conversion from 2,5-DHP to fumaric acid (Fig. 3B). Phylogenetic analysis of the four-gene cluster reveals that the gene cluster from SJY1 shares homology with sequences from P. putida S16 and Octadecabacter antarcticus 238 (Fig. 7A) (7). Many pyridine derivatives are degraded by aerobic microorganisms to generate 2,5-DHP as an intermediate, which makes it important to study the catabolism of 2,5-DHP (23). Oxidative activity was detected in cell extracts containing VppE (with the addition of 2,5-DHP and Fe2+) according to the absorbance decrease at 320 nm (Fig. 7B) (7), indicating that VppE is responsible for the conversion of 2,5-DHP to form N-formylmaleamic acid. HPLC analysis of reactions with the VppB cell extract and with the control [cell extract of E. coli BL21(DE3) with pET28a] confirmed that 2,5-DHP was transformed only by VppB (Fig. 7C).
FIG 7.
Phylogenetic analysis of genes from the lower VPP pathway and characterization of VppE. (A) Phylogenetic tree of nucleotide sequences of the gene cluster from vppH to vppG constructed using MEGA6. (B) Enzymatic activity of VppE. The enzyme activity was measured at 320 nm with the addition of 0.34 mg ml−1 cell extract, 250 μM 2,5-DHP, and 250 μM Fe2+ (FeCl2). a, negative control, 2,5-DHP; b, negative control, 2,5-DHP plus Fe2+; c, negative control, cell extract containing VppE plus 2,5-DHP; d, negative control, control cell extract plus 2,5-DHP plus Fe2+; e, reactions of VppE (cell extract containing VppE plus 2,5-DHP plus Fe2+). The concentrations of 2,5-DHP and Fe2+ were 250 μM. The control cell extract was from E. coli BL21(DE3) cells with pET28a. The cell extract containing VppE was prepared from E. coli BL21(DE3) cells with pET28a-vppE. (C) HPLC analysis of 2,5-DHP conversion reactions at 0 min and 20 min with cell extract containing VppE (top) and control cell extract (bottom). All the reactions were performed in 50 mM Tris-HCl, pH 8.0, at 25°C.
DISCUSSION
Microbial catabolism plays a significant role in the dissipation of nicotine residues in the environment. In the present study, an efficient nicotine-degrading strain, SJY1, was isolated and identified as Ochrobactrum sp. (14), which degrades nicotine through the VPP pathway (Fig. 3). Many Ochrobactrum strains were studied for the potential capacity to degrade chemical pollutants and for heavy metal detoxification under a wide range of environmental conditions (24–26). Another Ochrobactrum strain, DN2, could metabolize nicotine; however, the degradation pathway of strain DN2 is still unknown (9). The elucidation of the nicotine degradation pathway for Ochrobactrum sp. SJY1 has potential application in bioremediation of nicotine-contaminated environments. A lack of information about the factors controlling the metabolism of microorganisms in polluted environments often limits its implementation (27). Two strains, A. tumefaciens S33 and Shinella sp. HZN7, possessing the VPP pathway, have been previously isolated; however, information on the genes and enzymes involved is still scarce. Here, with the genome sequencing of strain SJY1, six genes showing various levels of sequence identity with the reported nicotine degradation genes were found in a 97.6-kbp DNA fragment. The functions of three genes, vppB, vppD, and vppE, have been investigated. VppB is responsible for the dehydrogenation of 6HN to 6HMM. The turnover number of VppB is calculated to be 25.8 ± 0.47 s−1, which was less than that of 6HLNO from A. nicotinovorans (293 ± 3 s−1) (28). VppD, catalyzing the conversion of HSP to 2,5-DHP, has potential application in biocatalysis for the production of 2,5-DHP, a precursor for the synthesis of 5-aminolevulinic acid (29). VppD shows catalytic properties in response to different pH values, temperatures, and metal ions similar those of a previously reported HSP 3-monoxygenase (HspB) from P. putida S16 (20). However, the specific activity (kcat) (HSP) of VppD (21.6 s−1) is ∼10-fold higher than that of HspB (2.0 s−1). VppE is a 2,5-DHP dioxygenase that transformed 2,5-DHP to N-formylmaleamic acid. 2,5-DHP is a central metabolic intermediate in the catabolism of many pyridine derivatives (23, 30).
Genome sequencing promotes the study of molecular mechanisms of xenobiotic degradation (13, 22), because genes can be identified through sequence alignment with isoenzymes or enzymes with similar functions. Genes responsible for the conversion of nicotine to 6HN and for the conversion from 6HPON to HSP have still not been found in the genome of strain SJY1. It seems that the genes in the upper VPP pathway show less sequence identity with the isoenzymes (Fig. 3B); therefore, the genes responsible for the conversion of nicotine and 6HPON may have lower sequence identity with reported proteins. The orf1 and orf2 genes in Fig. 3B were predicted to be the subunits of isoquinoline 1-oxidoreductase and show 36.4% and 13.7% sequence identity with the small and the large subunits of NDH, respectively. However, the homology gene of the middle subunit of NDH was not found in the vpp cluster or other parts of the SJY1 genome. Genes for the degradation of xenobiotics are usually clustered, like the genes of the nicotine pyridine pathway in A. nicotinovorans (6) and the genes of the nicotine pyrrolidine pathway in P. putida S16 (13). Further study should be performed to identify genes, especially genes in the vpp cluster, for the above two steps.
Only two Ochrobactrum strains with nicotine-degrading capacity have been reported to date; therefore, determining the diversification and adaptation of the two strains is of great interest. Horizontal gene transfer plays a central role in bacterial evolution to bring new catabolic pathways (31), and the information gathered over past years from the genetic study of nicotine degradation pathways suggests that many genes involved in nicotine degradation are also obtained in this way through plasmids or genomic islands (7, 32–34). The VPP pathway was identified in three strains from three different genera, and the G+C content of the vpp cluster is different from that of the whole genome of strain SJY1, indicating that the genes of the VPP degradation pathway may also have been obtained through horizontal gene transfer. Evolutionary relationships among genes may provide significant information for elucidation of the molecular mechanism of nicotine degradation. Interestingly, VppB shows higher amino acid sequence identity with the enzymes (NicA2, with a different catalytic function) from the pyrrolidine pathway than with its isoenzyme in the pyridine pathway (Fig. 5F). Besides, the proteins in strain SJY1 catalyzing the reactions from HSP to fumaric acid, like those in the pyrrolidine pathway, share more than 56% (from 56.1% to 81.0%) amino acid sequence identity with related proteins from strain S16. These results suggest that the VPP pathway might have a closer evolutionary relationship with the pyrrolidine pathway. Although, homology was observed between the genes from the VPP pathway and those from the pyrrolidine pathway, the gene locations in the two pathways are not the same. In strain S16, the genes from hspB to ami are located in one gene cluster in the same transcriptional direction (Fig. 3B) and in one operon (35). However, in strain SJY1, the vppD and orfx genes are located far from the genes vppH to vppG in the reverse transcriptional direction (Fig. 3B), obviously representing at least two operons. Therefore, the genes of the VPP pathway are not a simple combination of genes from the pyrrolidine and pyridine pathways. Many pyridine derivatives, such as nicotinic acid and nicotine, are degraded by aerobic microorganisms to generate 2,5-DHP as an intermediate and, further, follow the same pathway (23). The VPP nicotine degradation pathway in strain SJY1, the pyrrolidine pathway of P. putida S16, and the nicotinic acid maleamate degradation pathway of P. putida KT2440 all generate 2,5-DHP and then transform 2,5-DHP through N-formylmaleamic acid, maleic acid, and fumaric acid (Fig. 3A), catalyzed by four enzymes, 2,5-DHP dioxygenase, NFM amine oxidase, aminase, and isomerase (7, 30). The gene organizations between the nicotinic acid degradation pathway and the nicotine degradation pathways are different, whereas the gene organizations of the two nicotine degradation pathways are the same. Moreover, each gene in SJY1 shows higher sequence identity with the related gene from P. putida S16 than with the gene from P. putida KT2440 (Fig. 3B). The results imply that the nicotine degradation gene cluster evolved independently from the nicotinic acid degradation gene cluster.
The newly isolated strain Ochrobactrum sp. SJY1 utilizes nicotine through the VPP pathway with an efficient degrading capacity. Three genes in this pathway have been functionally studied for the first time at the molecular level. The study provides new insights into the microbial metabolism of nicotine. The genes responsible for the conversion from nicotine to 6HN and the enzyme catalyzing the conversion from 6HPON to HSP remain to be elucidated.
ACKNOWLEDGMENTS
We thank Roderich Brandsch (Institute of Biochemistry and Molecular biology, University of Freiburg) for providing the compound and enzyme used in this study.
This work was supported in part by grants from the Chinese National Natural Science Foundation (31422004, 31270154, 31230002, and 31121064) and from the National Basic Research Program of China (2013CB733901). We also acknowledge the Shanghai Rising-Star Program (13QA1401700) and the Chen Xing project of Shanghai Jiaotong University.
REFERENCES
- 1.Henningfield JE, Miyasato K, Jasinski DR. 1985. Abuse liability and pharmacodynamic characteristics of intravenous and inhaled nicotine. J Pharmacol Exp Ther 234:1–12. [PubMed] [Google Scholar]
- 2.Benowitz NL, Hukkanen J, Jacob P III. 2009. Nicotine chemistry, metabolism, kinetics and biomarkers, p 29–60. In Henningfield JE, Calvento E, Pogun S (ed), Nicotine psychopharmacology. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Civilini M, Domenis C, Sebastianutto N, de Bertoldi M. 1997. Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manag Res 15:349–358. [Google Scholar]
- 4.Novotny TE, Zhao F. 1999. Consumption and production waste: another externality of tobacco use. Tob Control 8:75–80. doi: 10.1136/tc.8.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Tang H, Xu P. 2014. Green strategy from waste to value-added-chemical production: efficient biosynthesis of 6-hydroxy-3-succinoyl-pyridine by an engineered biocatalyst. Sci Rep 4:5397. doi: 10.1038/srep05397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandsch R. 2006. Microbiology and biochemistry of nicotine degradation. Appl Microbiol Biotechnol 69:493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- 7.Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P. 2012. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2:377. doi: 10.1038/srep00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu J, Ma Y, Zhang J, Wen Y, Liu W. 2013. Cloning of a novel nicotine oxidase gene from Pseudomonas sp. strain HZN6 whose product nonenantioselectively degrades nicotine to pseudooxynicotine. Appl Environ Microbiol 79:2164–2171. doi: 10.1128/AEM.03824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan Y, Lu Z, Wu N, Huang L, Lü F, Bie X. 2005. Isolation and preliminary characterization of a novel nicotine-degrading bacterium, Ochrobactrum intermedium DN2. Int Biodeter Biodegr 56:45–50. doi: 10.1016/j.ibiod.2005.04.002. [DOI] [Google Scholar]
- 10.Wang SN, Huang HY, Xie KB, Xu P. 2012. Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl Microbiol Biotechnol 95:1567–1578. doi: 10.1007/s00253-012-4007-2. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, Wei Y, Qiu JG, Wen RT, Hong J, Liu WP. 2014. Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl Microbiol Biotechnol 98:2625–2636. doi: 10.1007/s00253-013-5207-0. [DOI] [PubMed] [Google Scholar]
- 12.Meng XJ, Lu LL, Gu GF, Xiao M. 2010. A novel pathway for nicotine degradation by Aspergillus oryzae 112822 isolated from tobacco leaves. Res Microbiol 161:626–633. doi: 10.1016/j.resmic.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Tang HZ, Wang LJ, Wang WW, Yu H, Zhang KZ, Yao YX, Xu P. 2013. Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet 9:e1003923. doi: 10.1371/journal.pgen.1003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, Li YY, Tang HZ, Xu P. 2014. Genome sequence of a newly isolated nicotine-degrading bacterium Ochrobactrum sp. SJY1. Genome Announc 2:e00720-14. doi: 10.1128/genomeA.00720-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang SN, Xu P, Tang HZ, Meng J, Liu XL, Huang J, Chen H, Du Y, Blankespoor HD. 2004. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol Lett 26:1493–1496. doi: 10.1023/B:BILE.0000044450.16235.65. [DOI] [PubMed] [Google Scholar]
- 16.Parte AC. 2014. LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang HZ, Wang SN, Ma LY, Meng XZ, Deng ZX, Zhang DK, Ma CQ, Xu P. 2008. A novel gene, encoding 6-hydroxy-3-succinoylpyridine hydroxylase, involved in nicotine degradation by Pseudomonas putida strain S16. Appl Environ Microbiol 74:1567–1574. doi: 10.1128/AEM.02529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Hijum SA, Zomer AL, Kuipers OP, Kok J. 2005. Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res 33:W560–W566. doi: 10.1093/nar/gki356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang HZ, Yao YX, Zhang DK, Meng XZ, Wang LJ, Yu H, Ma LY, Xu P. 2011. A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J Biol Chem 286:39179–39187. doi: 10.1074/jbc.M111.283929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Yu H, Tang HZ, Wang LJ, Yao YX, Wu G, Xu P. 2011. Complete genome sequence of the nicotine-degrading Pseudomonas putida strain S16. J Bacteriol 193:5541–5542. doi: 10.1128/JB.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao YX, Tang HZ, Ren HX, Yu H, Wang LJ, Zhang W, Behrman EJ, Xu P. 2013. Iron(II)-dependent dioxygenase and N-formylamide deformylase catalyze the reactions from 5-hydroxy-2-pyridone to maleamate. Sci Rep 3:3235. doi: 10.1038/srep03235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Sayed WS, Ibrahim MK, Abu-Shady M, El-Beih F, Ohmura N, Saiki H, Ando A. 2003. Isolation and identification of a novel strain of the genus Ochrobactrum with phenol-degrading activity. J Biosci Bioeng 96:310–312. doi: 10.1016/S1389-1723(03)80200-1. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XH, Zhang GS, Zhang ZH, Xu JH, Li SP. 2006. Isolation and characterization of a dichlorvos-degrading strain DDV-1 of Ochrobactrum sp. Pedosphere 16:64–71. doi: 10.1016/S1002-0160(06)60027-1. [DOI] [Google Scholar]
- 26.Sultan S, Hasnain S. 2007. Reduction of toxic hexavalent chromium by Ochrobactrum intermedium strain SDCr-5 stimulated by heavy metals. Bioresour Technol 98:340–344. doi: 10.1016/j.biortech.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 27.Lovley DR. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol 1:35–44. doi: 10.1038/nrmicro731. [DOI] [PubMed] [Google Scholar]
- 28.Dai VD, Decker K, Sund H. 1968. Purification and properties of L-6-hydroxynicotine oxidase. Eur J Biochem 4:95–102. doi: 10.1111/j.1432-1033.1968.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakano H, Wieser M, Hurh B, Kawai T, Yoshida T, Yamane T, Nagasawa T. 1999. Purification, characterization and gene cloning of 6-hydroxynicotinate 3-monooxygenase from Pseudomonas fluorescens TN5. Eur J Biochem 260:120–126. doi: 10.1046/j.1432-1327.1999.00124.x. [DOI] [PubMed] [Google Scholar]
- 30.Jimenez JI, Canales A, Jimenez-Barbero J, Ginalski K, Rychlewski L, Garcia JL, Diaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105:11329–11334. doi: 10.1073/pnas.0802273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igloi GL, Brandsch R. 2003. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J Bacteriol 185:1976–1986. doi: 10.1128/JB.185.6.1976-1986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thacker R, Rorvig O, Kahlon P, Gunsalus IC. 1978. NIC, a conjugative nicotine-nicotinate degradative plasmid in Pseudomonas convexa. J Bacteriol 135:289–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang MZ, Yang GQ, Min H, Lv ZM. 2009. A novel nicotine catabolic plasmid pMH1 in Pseudomonas sp. strain HF-1. Can J Microbiol 55:228–233. doi: 10.1139/W08-135. [DOI] [PubMed] [Google Scholar]
- 35.Wang LJ, Tang HZ, Yu H, Yao YX, Xu P. 2014. An unusual repressor controls the expression of a crucial nicotine-degrading gene cluster in Pseudomonas putida S16. Mol Microbiol 91:1252–1269. doi: 10.1111/mmi.12533. [DOI] [PubMed] [Google Scholar]