Abstract
In Australia, the egg industry is periodically implicated during outbreaks of Salmonella food poisoning. Salmonella enterica serovar Typhimurium and other nontyphoidal Salmonella spp., in particular, are a major concern for Australian public health. Several definitive types of Salmonella Typhimurium strains, but primarily Salmonella Typhimurium definitive type 9 (DT9), have been frequently reported during egg-related food poisoning outbreaks in Australia. The aim of the present study was to generate a pathogenicity profile of nontyphoidal Salmonella isolates obtained from Australian egg farms. To achieve this, we assessed the capacity of Salmonella isolates to cause gastrointestinal disease using both in vitro and in vivo model systems. Data from in vitro experiments demonstrated that the invasion capacity of Salmonella serovars cultured to stationary phase (liquid phase) in LB medium was between 90- and 300-fold higher than bacterial suspensions in normal saline (cultured in solid phase). During the in vivo infection trial, clinical signs of infection and mortality were observed only for mice infected with either 103 or 105 CFU of S. Typhimurium DT9. No mortality was observed for mice infected with Salmonella serovars with medium or low invasive capacity in Caco-2 cells. Pathogenicity gene profiles were also generated for all serovars included in this study. The majority of serovars tested were positive for selected virulence genes. No relationship between the presence or absence of virulence genes by PCR and either in vitro invasive capacity or in vivo pathogenicity was detected. Our data expand the knowledge of strain-to-strain variation in the pathogenicity of Australian egg industry-related Salmonella spp.
INTRODUCTION
Salmonella is one of the most common causes of food-borne infection worldwide. In Australia, the egg industry is periodically implicated in cases of Salmonella food poisoning (1). Uncooked or partially cooked foods containing raw egg as an ingredient accounted for 14% of food-borne outbreaks in 2006, 13% in 2007, and 28% in the first quarter of 2008 (2). It has been shown that some Salmonella serovars, such as Salmonella enterica serovar Enteritidis, have the capacity to infect developing eggs within the oviduct, and therefore contaminated eggs act as an ecological amplifier (3). It is believed that this could then facilitate the dissemination of Salmonella into the food chain and its eventual transmission to humans. These studies, however, for the most part have been focused on Salmonella Enteritidis with limited investigation of S. enterica serovar Typhimurium. The dramatic increase in Salmonella Enteritidis infections occurring in overseas countries has not been observed in Australia (4). Salmonella Typhimurium and other nontyphoidal Salmonella spp., however, have become a major concern for the Australian egg industry.
S. enterica serovars are a diverse group of pathogens that have evolved to survive in a wide range of environments and across multiple hosts (5). There are several definitive types of Salmonella Typhimurium but Salmonella Typhimurium definitive type 9 (DT9) has been frequently reported from egg-related food poisoning outbreaks in Australia (6). Other nontyphoidal serovars such as S. enterica Infantis, S. enterica Oranienburg, and S. enterica Lille are also frequently isolated from egg-layer farms or eggs (6); however, these serovars have not been reported in egg-related food poisoning cases in Australia. In humans, Salmonella Typhimurium can cause self-limiting gastroenteritis and has been found to be the most commonly reported serovar in both North America and Oceania (7). Septicemia associated with nontyphoidal Salmonella is also a growing public health concern, which can affect healthy as well as certain populations of immunodeficient individuals (8).
Complex pathogenesis is characteristic of Salmonella infection. The virulence of Salmonella is encoded by multiple genes which are clustered on Salmonella pathogenicity islands (SPIs) (9). SPIs have the potential to contribute to the overall pathogenesis of the bacterium (9). Genomic variability among bacterial strains arises primarily as a consequence of horizontal gene transfer (10). This inherent variability is likely the source of the various pathogenicities among nontyphoidal Salmonella strains. Consequently, characterization of the virulence gene repertoire by PCR has been used by many groups to profile the virulence of Salmonella (11, 12).
In Australia, there is limited current information regarding the characterization of the overall virulence of nontyphoidal Salmonella strains isolated from eggs or the egg farm environment. Our hypothesis is that Salmonella serovars recovered specifically from layer farm environments or egg shell wash possess various levels of pathogenicity. Other studies that have characterized the pathogenic potential of nontyphoidal Salmonella strains have focused mainly on Salmonella Enteritidis isolates recovered from either human clinical cases or non-poultry-associated environmental sources (13, 14).
Previously, Salmonella pathogenicity has been characterized using an in vitro invasion assay utilizing the human colon tumorigenic cell line Caco-2 (15). Major studies investigating the pathogenicity of Salmonella serovars using a human epithelial cell model system have shown variation in the levels of pathogenicity among different serovars (10). Invasion in cultured epithelial cells is routinely used as a measure of pathogenicity of Salmonella isolates (12). To date, no studies have been conducted to characterize the pathogenicity of field isolates of Salmonella obtained from egg farms in Australia.
The central aim of the present study was to generate an overall pathogenicity profile of multiple nontyphoidal Salmonella isolates sourced from Australian egg farms. To achieve this, the capacity of these isolates to cause gastrointestinal and invasive disease was investigated using both in vitro and in vivo models.
MATERIALS AND METHODS
Salmonella isolates.
All Salmonella serovars were obtained from the Salmonella Reference Laboratory, Institute of Veterinary Medical Science (IMVS), Adelaide, South Australia. The cultures were collected on xylose lysine deoxycholate (XLD) agar (Oxoid, Australia) and incubated overnight at 37°C. Long-term stocks were generated by preparing a bacterial suspension in brain heart infusion broth containing 20% glycerol and storing the stocks at −80°C. The list of cultures is provided in Table 1. The serovars selected in this study are commonly isolated from egg farms or egg shell wash (Table 1).
TABLE 1.
Salmonella serovars isolated from Australian egg shell wash and egg farm environmenta
| S. enterica serovarb | Source of isolate | No. of isolates from egg farms by year |
|||
|---|---|---|---|---|---|
| 2010 | 2011 | 2012 | 2013 | ||
| Typhimurium DT9 | Egg shell wash | 17 | 69 | 17 | 50 |
| Infantis | Egg shell wash | 27 | 122 | 9 | 7 |
| Agona | Egg belt | 18 | 85 | 25 | 4 |
| Mbandaka | Egg belt | 11 | 10 | 1 | 129 |
| Johannesburg | Feces | 0 | 3 | 0 | 0 |
| Livingstone | Feces | 2 | 0 | 0 | 0 |
| Subspecies I 4,12:d:− | Feces | 6 | 8 | 1 | 4 |
| Chester | Feces | 3 | 1 | 0 | 3 |
| Zanzibar | Feces | 0 | 5 | 0 | 0 |
| Kiambu | Feces | 7 | 19 | 0 | 0 |
| Lille | Feces | 3 | 4 | 0 | 0 |
| Ohio | Feces | 17 | 25 | 6 | 0 |
| Anatum | Feces | 16 | 46 | 0 | 13 |
| Havana | Feces | 0 | 26 | 0 | 0 |
| Oranienburg | Feces | 0 | 0 | 12 | 73 |
| Montevideo | Feces | 12 | 49 | 1 | 0 |
| Orion var. 15+,34+ | Dust in layer shed | 10 | 28 | 0 | 0 |
DNA extraction and PCR.
Overnight cultures of the Salmonella serovars selected for this study were grown at 37°C in 4 ml of Luria-Bertani (LB) broth. DNA was purified from 1 ml of bacteria using a Promega genomic DNA kit (Promega, USA). Purified DNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific, Australia). Working DNA solutions were prepared by diluting stock solutions to 5 ng/μl. Primers for each gene were designed using the primer design feature in GenBank or obtained from Hughes et al. (16). Primers were obtained from GeneWorks (Adelaide, South Australia) (Table 2). PCR mix was comprised of 1× Taq polymerase buffer (Fisher Biotec, Australia), 1.0 mM MgCl2, 0.5 μM each forward and reverse primer, 0.2 μM each deoxynucleoside triphosphate (dNTP), 0.3 units of Taq polymerase (Fisher Biotec, Australia), and 10 ng of Salmonella DNA.
TABLE 2.
Details of the genes, their role, primer sequence and targeted amplicon size used for Salmonella typing
| Virulence gene | Function | Forward primer (5′–3′) | Reverse primer (5′–3′) | Annealing temp (°C) | Reference or source |
|---|---|---|---|---|---|
| prgH | Invasion protein | GCCCGAGCAGCCTGAGAAGTTAGAAA | TGAAATGAGCGCCCCTTGAGCCAGTC | 55 | 32 |
| sopB | Invasion and intracellular replication | GAAGACTACCAGGCGCACTT | TTGTGGATGTCCACGGTGAG | 55 | 32 |
| invA | Invasion protein | CTGGCGGTGGGTTTTGTTGTCTTCTCTATT | AGTTTCTCCCCCTCTTCATGCGTTACCC | 60 | 32 |
| sitC | Iron transporter | CAGTATATGCTCAACGCGATGTGGGTCTCC | CGGGGCGAAAATAAAGGCTGTGATGAAC | 64 | 32 |
| spiC | Disruption of Golgi apparatus and lysosomes | CCTGGATAATGACTATTGAT | AGTTTATGGTGATTGCGTAT | 56 | 32 |
| sifA | Required for maintenance of SCVa | TTTGCCGAACGCGCCCCCACACG | GTTGCCTTTTCTTGCGCTTTCCACCCATCT | 62 | 32 |
| misL | Adhesin | GTCGGCGAATGCCGCGAATA | GCGCTGTTAACGCTAATAGT | 58 | 32 |
| orfL | Survival within macrophages | GGAGTATCGATAAAGATGTT | GCGCGTAACGTCAGAATCAA | 56 | 32 |
| pipD | Colonization | CGGCGATTCATGACTTTGAT | CGTTATCATTCGGATCGTAA | 58 | 32 |
| iroN | Iron transport | ACTGGCACGGCTCGCTGTCGCTCTAT | CGCTTTACCGCCGTTCTGCCACTGC | 60 | 32 |
| pefA | Plasmid encoded fimbriae | GCGCCGCTCAGCCGAACCAG | CAGCAGAAGCCCAGGAAACAGTG | 58 | 33 |
| spvC | Modulation of host immune response | TCTCTGCATTTCGCCACCAT | TGCACAACCAAATGCGGAAG | 58 | 33 |
| sipA | Invasion protein | TACCCCTGCTGCTACGTAAT | CTCCAGGGCTTTACGTATCA | 60 | 33 |
| sipB | Invasion protein | TGGCAGGCGATGATTGAGTC | CCCATAATGCGGTTCGTTTC | 58 | This study |
| sipC | Invasion protein | TGCCCTGGCAAATAATGTCA | CATCGATTCGGGTCATATCC | 58 | 33 |
| fliC | Flagella protein | TACGCTGAATGTGCAACAAA | TACCGTCATCTGCAGTGTAT | 58 | This study |
| sopA | Induction of proinflammatory response | GCCCACGGTTTCTGAAGGTA | AAAGAGTCCGCTGTGAGTGG | 60 | This study |
| sipD | Invasion protein | TGCGTCAGCGTCTGTAATGT | GGCCTTATTTAGCGCTTCGC | 58 | This study |
| avrA | Modulation of host immune response | ATACTGCTTCCCGCCGC | ACACCGAAGCATTGACCTGT | 58 | This study |
| sptP | Disruption of actin cytoskeleton | TTCACCCTATCCGCCAGGTA | GTGTAGCCCGGTTCTCACAA | 58 | 16 |
| hilA | Activates expression of invasion genes | CACCAACCCGCTTCTCTCTT | ATTGTGGTCCAGCTCTGTCG | 58 | 35 |
| xthA | Survival in egg | CGAAAAACACCAGCCCGATG | CCGGCAGGAAGGAGCATTTA | 55 | 36 |
| yafD | Survival in egg | CGGATCCGTATCCTCGTGTG | ATCGTCAGTGAAACGCACCT | 55 | 36 |
| stn | Enterotoxin | CTTTGGTCGTAAAATAAGGCG | TGCCCAAAGCAGAGAGATTC | 55 | 32 |
SCV, Salmonella-containing vacuoles.
Product sizes for each of the genes were relatively similar; therefore, PCR conditions were nearly universal. The primer annealing temperatures were the only variation during PCR, and these are listed in the Table 2. The general reaction protocol was as follows: 95°C for 5 min (step 1); 30 cycles (step 2) of 95°C for 30 s (melt), 45 s at the annealing temperature specified in Table 2, and 72°C for 1 min (extension); 72°C for 4 min (step 3); and a final hold at 8°C (step 4).
Tissue culture.
A human colonic carcinoma cell line, Caco-2, was obtained from the American Tissue Culture Collection (ATCC). Caco-2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone, Australia) supplemented with 10% fetal bovine serum (FBS) (HyClone, Australia) containing 100 U/ml penicillin and 100 μg/ml streptomycin (HyClone, Australia). Caco-2 cells were expanded and maintained as frozen stocks. Cell line stocks tested negative for mycoplasma. Cells were used between passages 4 and 9 for gentamicin protection invasion assays.
Gentamicin protection in vitro invasion assay.
The gentamicin protection assay was performed using Caco-2 cells to characterize the capacity of the selected Salmonella serotypes for invasion into epithelial tissue. Polarized monolayers of Caco-2 cells were obtained by seeding cells into each well of a 48-well tray. The progression to polarization was monitored by measuring alkaline phosphatase production. When alkaline phosphatase levels plateaued (after 12 to 13 days), cells were considered to be polarized and were used within 24 h. Cell culture medium was changed every other day during polarization. All invasion assays were performed in replicate and repeated five times.
The gentamicin MIC was determined for all serovars included in this study using the Clinical and Laboratory Standards Institute (CLSI) protocol (17). The gentamicin MIC for all serovars was less than 0.25 μg/ml. For invasion assays, 400 μg/ml gentamicin was used to eliminate bacteria that had not invaded (18).
For the invasion assay experiments, bacteria were plated fresh 24 h prior to the experiments. Suspensions were prepared in normal saline by selecting individual colonies from agar plates. The optical densities at 600 nm (OD600) of the suspensions for each serovar were measured and adjusted to between 0.150 and 0.200, corresponding to 108 CFU/ml. An inoculum check was performed by doing a dilution series of the stock on nutrient agar plates.
Invasion assays were also performed using bacteria prepared in LB suspensions. These were prepared by selecting a single colony and placing it into 3 ml of LB broth. Tubes were incubated at 37°C for 6 h with shaking at 50 rpm. Ten microliters of this suspension was then added to 4 ml of LB medium and incubated overnight at 37°C with shaking at 50 rpm. This method yielded bacterial cultures that were in a stationary (liquid) phase of growth.
To investigate the difference in invasion abilities of stationary and non-stationary-phase cells (overnight cultures in solid phase versus liquid phase) of Salmonella spp., the invasion assays were performed using bacteria suspended in normal saline and LB medium. On the day of the invasion assay, Caco-2 cells were lifted from a single well from each of two 48-well trays and counted using a hemocytometer. An average number of cells was obtained and used to calculate the multiplicity of infection (MOI). An MOI of 100 was selected to statistically distinguish between low-invasive, moderately invasive, and highly invasive Salmonella serovars.
Prior to the addition of bacteria, Caco-2 cells were washed three times with DMEM containing no supplements to remove any residual culture medium antibiotics or FBS. The final DMEM wash was replaced with 500 μl of fresh DMEM. Bacteria were added to individual wells. Experiments were performed in replicate. Following the addition of Salmonella, Caco-2 cells were incubated with bacteria for 2 h at 37°C and 5% CO2. Cells were then washed three times with DMEM with no supplements. To kill any adherent bacteria, 400 μg/ml gentamicin diluted in DMEM was added to the Caco-2 cells. Cells were incubated in gentamicin for 15 min at 37°C and 5% CO2. Cells were then washed three times with calcium- and magnesium-free phosphate-buffered saline (PBS). Finally, 1 ml of sterile 1% Triton X-100 was added to each well to lyse the cells and collect the intracellular bacteria. A serial 10-fold dilution series was prepared from the 1 ml of bacteria/cell lysate. One hundred microliters of each dilution was plated onto nutrient agar plates. Plates were incubated at 37°C overnight, and colonies were counted for each dilution of each serovar.
Mouse infection trial with selected isolates.
The BALB/c mouse strain was selected for these studies. Mouse challenge studies were performed according to animal ethics protocol approved by the University of Adelaide (UA) Animal Care and Use Committee. Six- to 8-week-old, specific-pathogen-free, female mice weighing between 10 and 14 g were obtained from Laboratory Animal Services (University of Adelaide). Mice were raised in isolator cages and fed on a Salmonella-free commercial diet. Food and water were supplied ad libitum until the end of the experiment.
Salmonella serovars with different cell invasion potentials (as determined from data obtained from in vitro experiments) were used for in vivo experiments (Table 3). Along with Salmonella Typhimurium DT9, other nontyphoidal Salmonella spp., such as Salmonella Infantis, S. enterica serovar Montevideo, and Salmonella Oranienburg, were also isolated from human cases.
TABLE 3.
Treatment groups for Salmonella infection triala
| Salmonella serovar | Invasiveness level | Invasiveness group |
|---|---|---|
| Typhimurium DT 9 | High | C |
| Infantis | Low | A |
| Lille | Low | A |
| Oranienburg | Moderate | B |
| Montevideo | Moderate | B |
| Typhimurium ATCC 14028 (positive control) | High | C |
Animals received doses of either 103 CFU or 105 CFU of Salmonella (n = 7 animals/treatment for each serovar).
Bacterial isolates were grown overnight in Luria-Bertani (LB) broth and serially diluted in PBS to obtain desired cell counts. Mice from each treatment group were inoculated by oral gavage with either 103 or 105 CFU/ml of LB containing Salmonella culture. Control mice were inoculated with sterile LB broth. Each treatment group contained seven animals. To exclude observer bias in the interpretation of results, all clinical and bacteriological assessments were conducted by personnel blinded to the identity of the challenge isolates.
Challenged mice were observed at least twice daily for mortality and clinical parameters of disease, including ruffled fur, hunching behavior, and lethargy, for up to 21 days postinfection (p.i.). Mice showing all of the above symptoms were considered moribund and were euthanized by carbon dioxide asphyxiation. An experiment was terminated at day 21 p.i. when all the surviving mice were euthanized. The ATCC Salmonella Typhimurium strain 14028 was included as a positive control. The negative-control was LB broth.
Salmonella isolation from fecal samples by culture method.
Fecal pellets were collected from each mouse at 3, 6, 9, 12, 15, and 18 days postinfection (p.i.) and processed for isolation of challenge isolates by culture (fecal enrichment method) according to a previously described method (4, 19).
DNA extraction from fecal samples for real-time PCR.
DNA from feces was extracted using a QIAamp DNA stool minikit (Qiagen, Australia) as per the manufacturer's guidelines. Fecal samples (0.2 g) collected from 0 to 18 days postinfection (p.i.) from each treatment group were weighed and treated with 2 ml of QIAamp stool lysis buffer. The samples were centrifuged at 4,800 × g for 10 min, and 120 μl of the supernatant was treated with Inhibitex (tablet; Qiagen, Australia). The samples were then centrifuged, and the supernatant was treated with proteinase K and lysis buffer. Washing and elution were performed using a spin column according to the manufacturer's instructions. Extracted DNA was quantified using a spectrophotometer (NanoDrop ND 1000) and stored at −70°C until used for real-time PCR (RT-PCR). Five nanograms of fecal DNA was used for the real-time PCR.
Quantitative PCR (qPCR).
Salmonella shedding in fecal material was quantified using real-time PCR (RT-PCR). RT-PCR was performed using a Rotor Gene 3000 real-time PCR machine (Qiagen, Australia) and a TaqMan Salmonella enterica detection kit (Applied Biosystems, Australia). Each reaction mixture contained 9 μl of qPCR supermix and 6 μl of DNA template (12 ng) in a total reaction volume of 15 μl. The cycling parameters were 95°C for 10 min and 40 cycles at 95°C for 15 s, followed by 60°C for 60 s. All real-time PCR runs included negative and positive controls. The data were analyzed by two-way analysis of variance (ANOVA).
A standard curve was generated by preparing a serial dilution of a Salmonella strain. Bacteria were resuscitated on XLD agar overnight at 37°C. The individual isolated colonies were then suspended in 2 ml of PBS and matched with a 0.5 McFarland standard (bioMérieux, Australia). Serial dilutions were performed to achieve 108 CFU/ml. The CFU counts were confirmed by spreading serial dilutions on XLD agar plates. In order to determine the limit of detection of qPCR, fecal samples were spiked with various concentrations (108 CFU/ml to 100 CFU/ml) of Salmonella. qPCR was performed on serial dilutions (108 to 100) of genomic DNA, and a proportionality relationship was produced by plotting the threshold cycle (CT) value against the logarithm of the CFU number. Salmonella copies were calculated using a standard curve prepared by serial 10-fold dilution of a cultured Salmonella sp.
Statistical analysis.
The data were normally distributed. One-way analysis of variance (ANOVA) was used with Tukey's correction for multiple comparisons. Kaplan-Meier survival curves were generated for the in vivo mouse infection experiment. A log rank Mantel-Cox test was used to compare survival curves. A k-means cluster analysis was performed to identify invasive group types. All tests were run using either SPSS, version 21, or GraphPad Prism, version 6.0.
RESULTS AND DISCUSSION
The Salmonella genus is comprised of two species, S. enterica and S. bongori. Members of both species possess the ability to infect host cells while members of S. enterica are able to cause systemic infection (20). S. enterica is highly diverse, containing over 2,500 different serovars. Representative serovars from this species are the most commonly isolated serovars during outbreaks of food-borne salmonellosis, including S. Enteritidis, S. Typhimurium, S. enterica serovar Virchow, and S. Infantis (21). Many other S. enterica serotypes, however, are commonly isolated from eggs and egg-related products as well as from the layer farm environment, yet their degree of pathogenicity remains largely uncharacterized. In the present study, we have examined three parameters, in vitro cell invasion, virulence gene profile, and in vivo pathogenicity, of 17 Salmonella serovars isolated directly from egg shell wash or the egg farm environment.
In vitro intestinal epithelial invasion.
Surface contamination of an egg shell with Salmonella has been reported to be as high as 106 CFU (19). Thus, improper egg handling or storage conditions can lead to kitchen contamination or direct exposure of an individual to bacteria. Bacteria on the surface of the egg shell would be in a nutrient-poor environment and as such would not be expressing fimbriae and other virulence genes commonly associated with invasion.
The process of host cell invasion by Salmonella spp. has been widely studied using the intestinal epithelial cell line Caco-2 (22). To determine whether egg-associated Salmonella serovars have equal invasive potentials following direct exposure from eggshell, bacterial suspensions were prepared in normal saline and subsequently coincubated with cell monolayers for 2 h. Salmonella grown on nutrient agar and suspended in normal saline (solid phase) would not have upregulated genes in Salmonella pathogenicity island 1, providing a model for direct exposure. Data obtained are represented as percent recovery, determined by the ratio of the amount of bacteria recovered to the initial MOI. Invasive assays were performed in duplicate and repeated five times.
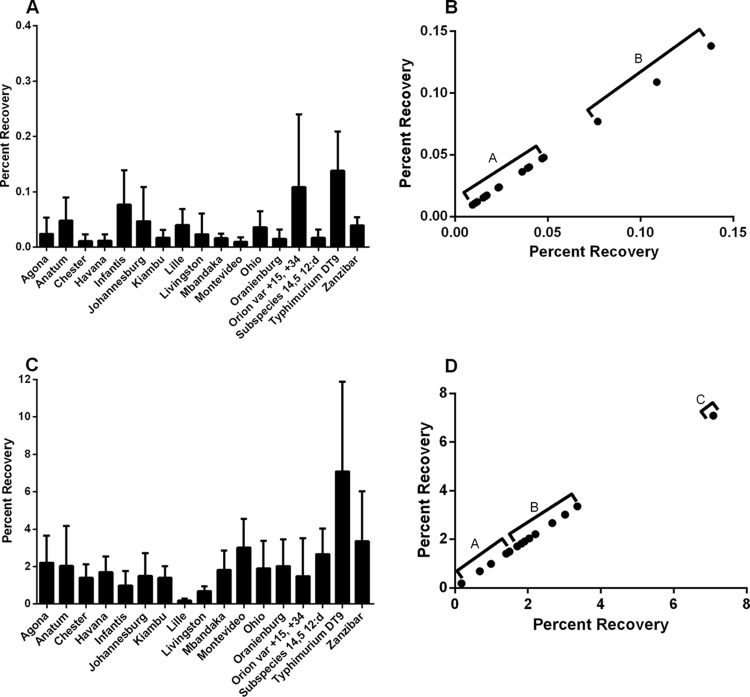
Overall, the invasiveness of the serovars included in this study was low. Despite limited invasion, a significant difference of mean percent recovery between serovars was observed (P < 0.0066) (Fig. 1A). A k-means cluster analysis revealed two distinct invasive groups (Fig. 1B). The S. enterica serovars Agona, Anatum, Chester, Havana, Johannesburg, Kiambu, Lille, Livingston, Mbandaka, Montevideo, Ohio, Oranienburg, subspecies 4,[5],12:d, and Zanzibar had mean percent recoveries of between 0.0095% and 0.047% and clustered in group A (cluster center, 0.03; change about cluster center, 0.016) (Fig. 1A and B). S. Typhimurium DT9, S. Infantis, and S. enterica serovar Orion var. 15+,34+ had mean percent recoveries of greater than 0.1% and clustered in group B (cluster center, 0.11; change about cluster mean, 0.03) (Fig. 1A and B).
FIG 1.
The in vitro invasive capacity of 17 nontyphoidal Salmonella serovars was assessed using the gentamicin protection assay with the human intestinal epithelial cell line Caco-2. Bacteria were either suspended in normal saline (A) or grown to stationary (liquid) phase in LB broth (C) and then added to cell monolayers at an MOI of 100. Data are presented as mean percent recovery. Assays were repeated five times. Statistical analysis was performed by Kruskal-Wallis ANOVA with post hoc analysis utilizing Dunn's multiple-comparison test. A significant effect of serotype for both treatment groups (P < 0.006, saline; P < 0.0002, LB broth) was detected, but no significant differences were observed between individual serotypes. Cluster analysis (k-means) was performed to identify invasion groups. Two invasive types were identified for serovars suspended in normal saline and are identified as group A (low) and group B (moderate) (B). Following growth in LB broth, substantial increases in percent recoveries were observed (C). Three invasion types were identified by cluster analysis and were classified as low (group A), moderate (group B), and high (group C) (D).
Environmental conditions are known to influence the expression of Salmonella virulence genes (23), in particular, thee genes that regulate the function of flagella and those that are directly involved in cell invasion. To determine how growth conditions affected the invasive capacities, Salmonella serovars were cultured to stationary phase (or liquid phase) in LB broth. Consistent with previous reports, we observed substantial increases in invasion. Mean percent recoveries increased from 0.002 to 0.13% (normal saline) (Fig. 1A) to 0.18 to 7.08% (LB broth) (Fig. 1C). S. Montevideo was found to have the greatest increase in invasive capacity. Mean percent recovery for S. Montevideo suspended in normal saline was 0.009%, which increased over 300-fold to 3.02% following growth in LB broth. S. enterica serovars Agona, Chester, Havana, Mbandaka, and Oranienburg as well as subspecies 4,[5],12:d also exhibited substantial increases in invasive capacity, increasing between 90- and 158-fold.
Three distinct invasive groups were identified by k-means cluster analysis and were defined as low (group A), moderate (group B), and high (group C) (Fig. 1D). The low-invasive group (cluster center, 1.17; change about cluster mean, 0.99) consisted of S. enterica serovars Chester, Havana, Infantis, Johannesburg, Kiambu, Lille, Livingston, and Orion var. 15+,34+. Mean percent recoveries obtained for the low-invasive group ranged from 0.18% to 1.7%. S. enterica serovars Agona, Anatum, Mbandaka, Montevideo, Ohio, Oranienburg, subspecies 4,[5],12:d, and Zanzibar all clustered into the moderately invasive group (group B) (cluster center, 2.38; change about cluster mean, 0.97) and had mean percent recoveries ranging from 1.83% to 3.36%. S. Typhimurium DT9 exhibited the greatest mean percent recovery (7.08%) of the serovars tested. S. Typhimurium DT9 clustered by itself in group C (cluster center, 7.08; change about cluster mean, 0.00) and was classified as possessing high invasion capacity.
Invasion experiments using bacteria grown to log phase in LB broth were also performed. No significant difference in invasive capacities was observed for log phase growth compared with bacteria grown to stationary phase in LB broth (data not shown).
Patterson et al. (24) reported upregulation of all genes in Salmonella pathogenicity island 1 (SPI-1) of S. Typhimurium upon growth in LB broth. Based upon findings of this current work, it was hypothesized that a similar pattern would be observed for other Salmonella serovars; further gene expression investigations are, however, necessary. It is important to note that the invasive ability of Salmonella Typhimurium DT9 increased from medium to high while other serovars retained their low-invasive status. These results indicate that not all isolates of Salmonella recovered from poultry may be equally invasive even after growth to stationary phase (or in liquid phase).
The findings from these intestinal epithelial invasion assays could have significant implications for egg handling as they suggest that some strains of Salmonella require favorable growth conditions and environments to stimulate pathogenicity. Moreover, these results also indicate that many Salmonella serovars, in particular, the S. Typhimurium definitive types, may have a constitutively active virulence gene(s) that enables invasion under any conditions. This may provide a selective advantage for these strains. Future work will be aimed at determining the molecular and cellular mechanisms responsible for these differences.
PCR typing for virulence genes.
The Salmonella genome possesses many genes whose products are involved in the processes of cellular adhesion and invasion. The 23 genes analyzed in this study were included because they are known to be involved in adhesion, invasion, and survival of Salmonella spp. Furthermore, PCR detection of these genes has been widely used as a predictive measure for Salmonella virulence (11, 25). Results for virulence gene PCR experiments are summarized in Table 4. Most serovars tested were positive for the majority of the selected virulence genes. S. enterica serovars Kiambu, Lille, Livingston, Montevideo, Ohio, and Oranienburg were negative for avrA. No amplification of sopB was detected for Salmonella Lille. S. enterica serovars Chester, Havana, Infantis, Johannesburg, Kiambu, Lille, Mbandaka, Montevideo, Oranienburg, and Zanzibar were negative for sptP. No PCR amplification was detected for fliC except for S. Typhimurium DT9. This was also observed for the genes pefA and spvC. As such, it is likely that PCR results do not indicate that the Salmonella serovars tested were negative for these genes but more likely that they possess sufficient genetic variability preventing primer annealing and subsequent amplification. Full-genome sequence analysis studies are warranted to investigate genetic variability.
TABLE 4.
Virulence gene profile of Salmonella serovars
| Virulence gene |
S. enterica serovar |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Agona | Anatum | Chester | Havana | Infantis | Johannesburg | Kiambu | Lille | Livingston | Mbandaka | Montevideo | Ohio | Oranienburg | Orion var. 15+,34+ | Subspecies I 4,[5],12:d | Typhimurium DT9 | Zanzibar | |
| avrA | + | + | + | + | + | + | − | − | − | + | − | − | − | + | + | + | + |
| hilA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| invA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| prgH | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sipA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sipB | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sipD | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sopA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sopB | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| sptP | + | + | − | − | − | − | − | − | + | − | − | + | − | + | + | + | − |
| sitC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| spiC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sifA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| misL | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| orfL | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| pipD | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| iroN | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| pefA | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| spvC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| fliC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| xthA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| yafD | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| stn | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Comparison of the pathogenicity profiles generated in this study with in vitro cell invasion data did not reveal any correlative association between the presence or absence of a gene or genes with virulence. The Salmonella genome possesses multiple pathogenicity islands (PIs), which are genetic elements within the bacterial genome that harbor genes associated with virulence. In the species S. enterica, 23 Salmonella pathogenicity islands (SPIs) have been identified (26) which are considered to play a vital role in the evolution of Salmonella (27). Entry of Salmonella into intestinal epithelial cells is dependent upon invasion genes that are localized to SPI-1 (28). Bacteria utilize SPI-1 during the gastrointestinal (GI) stage of disease to invade cells and to invoke the inflammatory response (29). For our experiments, different virulence genes for PCR amplification from the Salmonella serovars were selected. Our results showed that the majority of the Salmonella serovars tested were positive for the virulence genes. Primer sets were designed to the published genome of Salmonella Typhimurium LT2. It is known, however, that there is significant genetic variability of virulence gene sequences between Salmonella serovars (10). This variability in genomic sequence may translate into variation of function, leading to differences in pathogenicities. However, it is essential to note that possession of a single or a few virulence genes does not endow a strain with pathogenic status unless that strain has acquired the appropriate virulence gene combination to cause disease in a specific host species (30). It should also be noted that there are several other genes which could play important roles in Salmonella invasion, and the current study detected the limited set of genes by PCR. Future studies could be directed to investigate virulence gene expression of Salmonella serovars at different stages of pathogenesis.
Mouse infection trial with selected isolates.
To determine whether in vitro cell invasiveness was a correlative measure of in vivo pathogenicity, representative Salmonella isolates with low (S. Infantis and S. Lille), medium (S. Montevideo and S. Oranienburg), and high (S. Typhimurium DT9 and ATCC 14028) invasiveness were tested using an in vivo mouse challenge model. While the dose rates (103 and 105 CFU) selected for this study are lower than those used in other reported work, they represent bacterial counts commonly recovered from the egg shell surface (19).
For both the 103 and 105 doses, mice inoculated with Salmonella isolates with high in vitro invasiveness (group C) (Table 3) exhibited a higher rate of mortality than those inoculated with isolates with medium (group B) (Table 3) or low (group A) (Table 3) invasiveness (Mantel-Cox log rank test, P < 0.001) (Fig. 2A and B.). At the 105 dose, the mean survival time for mice inoculated with group C isolates ranged from 5 to 9 days. The majority of mice inoculated with group C isolates died or were euthanized by 9 days p.i.; two animals inoculated with S. Typhimurium DT9 and one in the ATCC 14028 inoculation group survived until day 21. No clinical signs of infection or mortality were recorded in mice challenged with group A or B isolates. Similar results were obtained in mice inoculated with 103 CFU of Salmonella (Fig. 2B). No statistically significant difference was detected in the survival curves between the dose groups (P > 0.1). Based on the results of in vitro findings, the virulence of several Salmonella serovars was tested in a mouse infection model. The clinical signs of disease and mortality were observed in mice infected with both high and low doses of S. Typhimurium DT9 and S. Typhimurium ATCC 14028. Mortality was not recorded for infection with medium- or low-invasive Salmonella serovars. Lack of morbidity and mortality of S. Montevideo-infected mice has been previously reported (31). Similar results were observed for Salmonella serovars Infantis, Oranienburg, and Lille, but the mechanisms driving this lack of in vivo pathogenicity are not known. These serovars have been previously isolated during food poisoning cases. A single isolate of each serovar was used in this study, and lack of pathogenicity in these serovars could possibly be due to within-serotype genetic variation. Translating the in vitro findings into the context of an animal model and subsequently to human disease remains a difficult challenge for any disease process as this introduces a plethora of variables (32).
FIG 2.
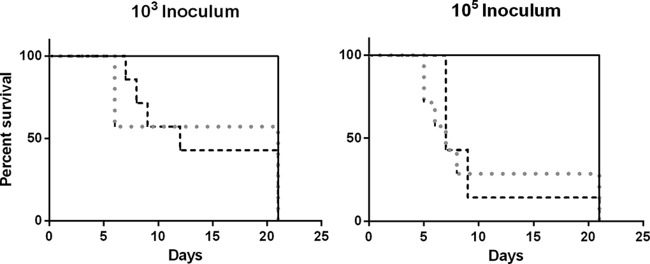
The in vivo pathogenicity of Salmonella serovars with low (S. Infantis and S. Lille), moderate (S. Montevideo and S. Oranienburg), and high (S. Typhimurium DT9 and ATCC 14028) in vitro invasiveness was investigated using BALB/c mice. Mice were inoculated with either 103 or 105 CFU. A Kaplan-Meier survival curve illustrates the percent survival rates of mice challenged with nontyphoidal Salmonella isolates. A Mantel-Cox log rank analysis was performed comparing percent survival. Percent survival of mice infected with both S. Typhimurium DT9 (gray dotted line) and ATCC 14028 (black dashed line) was significantly less than that of all other treatment groups (black line) at both 103 (P < 0.001) and 105 (P < 0.0001) CFU. No significant difference in survival rates was observed between S. Typhimurium DT9 and ATCC 14028 for either the low or high dose (n = 7 mice per group).
The TaqMan Salmonella enterica detection system does not provide quantification of positive fecal samples. Therefore, to determine the limit of detection of the assay, a standard curve prepared from a known concentration of Salmonella spp. (108 to 100 CFU) was used. The standard curve produced a slope of −3.2, a y intercept of 39.4, and an R2 value of 0.91. A cutoff CT of 33.7 was used to exclude detection of false positives. A CT of 33.7 corresponded to 50 CFU of Salmonella. Amplification was not recorded in negative-control (LB) samples or any of the treatment groups at day 0 of infection.
In Salmonella invasive group A (Salmonella serovars Infantis and Lille) infected with a low dose (103 CFU), both qPCR and culture tests indicated that the shedding was detected until days 9 and 15 p.i., respectively. Highest shedding (4.5 log CFU) was recorded on day 6 p.i. in the S. Infantis-infected group (Table 5). Salmonella was isolated up to 18 days p.i. after a higher dose (105 CFU) of infection with S. Infantis and S. Lille. On other hand, Salmonella shedding was detected by qPCR up to 15 and 9 days p.i. after the higher dose (105 CFU) of infection with S. Infantis and S. Lille, respectively. For the S. Infantis and S. Lille infection group, there was significant difference in Salmonella shedding between time points (days p.i.) (P < 0.0001). There was significant interaction between dose and days p.i. (P < 0.0001). There was a significant difference in Salmonella shedding between the high and low doses for the S. Infantis group (P < 0.0001). qPCR results showed that in Salmonella invasion group B (Salmonella serovars Oranienburg and Montevideo), Salmonella shedding was recorded until day 18 p.i. in mice infected with both high and low doses of S. Montevideo. However, S. Oranienburg was isolated from feces until 15 days p.i. in mice infected with a low dose (Table 5). For the S. Oranienburg and S. Montevideo groups, there was a significant difference in Salmonella shedding between doses and time points (days p.i.) (for both parameters, P < 0.0001). There was significant interaction between dose and days p.i. (P < 0.0001). In Salmonella invasion group C infected with a low dose (S. Typhimurium DT9), Salmonella shedding was recorded until day 18 pi. by both qPCR and the culture method. In the high-dose infected group, Salmonella was detected until day 18 p.i. by the qPCR method while Salmonella could be isolated until day 9 p.i. by the culture method. There was no significant difference in shedding results between infection doses; however, a significant difference was recorded for days p.i. (P < 0.0001).
TABLE 5.
qPCR and culture detection of Salmonella from mice inoculated with 103 and 105 CFU
| Salmonella serovar | Dose | Detection by day p.i. and methoda |
P value for qPCR testb |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 |
3 |
6 |
9 |
12 |
15 |
18 |
||||||||||||
| qPCR | Culture | qPCR | Culture | qPCR | Culture | qPCR | Culture | qPCR | Culture | qPCR | Culture | qPCR | Culture | Dose | Day p.i. | Dose × days p.i. | ||
| Typhimurium DT 9 | 103 | ND | − | 3.8 ± 0.13 | − | 3.6 ± 0.09 | + | ND | − | 3.1 ± 0.18 | + | 3.0 ± 0.1 | + | 3.0 ± 0.1 | + | NS | <0.0001 | <0.0001 |
| 105 | ND | − | 3.7 ± 0.01 | − | 2.8 ± 0.02 | + | 4.0 ± 0.01 | + | 2.3 ± 0.2 | − | ND | − | 3.6 ± 0.09 | − | ||||
| Infantis | 103 | ND | − | 3.0 ± 0.02 | + | 4.6 ± 0.02 | + | 3.1 ± 0.07 | + | ND | − | ND | − | ND | − | <0.0001 | <0.0001 | <0.0001 |
| 105 | ND | − | 0 | + | 0 | + | 3.0 ± 0.13 | + | ND | + | 2.7 ± 0.04 | + | ND | + | ||||
| Lille | 103 | ND | − | 3.0 ± 0.05 | + | 1.5 ± 0.2 | + | ND | + | ND | − | 2.9 ± 0.08 | + | ND | − | NS | <0.0001 | <0.0001 |
| 105 | ND | − | 3.1 ± 0.07 | + | 2.9 ± 0.11 | + | 3.0 ± 0.01 | − | ND | − | ND | + | ND | + | ||||
| Oranienburg | 103 | ND | − | 3.0 ± 0.33 | + | ND | − | ND | + | ND | − | ND | + | ND | − | <0.0001 | <0.0001 | <0.0001 |
| 105 | ND | − | 3.7 ± 0.04 | + | 3.1 ± 0.16 | + | 3.0 ± 0.15 | − | 2.8 ± 0.07 | + | 3.2 ± 0.01 | + | ND | + | ||||
| Montevideo | 103 | ND | − | 3.6 ± 0.06 | + | 2.9 ± 0.01 | + | 2.8 ± 0.2 | + | 2.7 ± 0.03 | − | 2.8 ± 0.04 | + | 3.0 ± 0.06 | + | <0.0001 | <0.0001 | 0.0004 |
| 105 | ND | − | 3.2 ± 0.11 | + | 3.5 ± 0.06 | + | 3.4 ± 0.03 | + | 3.5 ± 0.33 | + | 3.6 ± 0.01 | + | 4.1 ± 0.02 | + | ||||
| Typhimurium ATCC 14028 | 103 | ND | − | 3.3 ± 0.08 | + | 3.5 ± 0.18 | + | 4.1 ± 0.23 | + | 4.1 ± 0.94 | + | 3.9 ± 0.25 | + | 3.1 ± 0.72 | + | NS | 0.0001 | NS |
| 105 | ND | − | 4.1 ± 0.08 | + | 3.1 ± 0.32 | + | 1.6 ± 0.6 | + | 2.9 ± 0.45 | + | 3.2 ± 0.24 | + | 3.7 ± 0.06 | + | ||||
Cultures were either positive (+) or negative (−). All qPCR values presented are in CFU/g of feces ± standard error of the mean. ND, none detected; values below the qPCR limit of detection.
The P values indicate the significance of the indicated parameter. NS, not significant; Dose × days p.i., interaction of dose and days p.i.
Some disparity was observed between Salmonella culture and qPCR results. Using the culture method, Salmonella was isolated from qPCR-negative samples. The culture method used in this study enabled the isolation of Salmonella at levels below the detection limit established for the qPCR. Some culture-negative samples, however, tested positive by qPCR (S. Typhimurium DT9, S. Infantis, and S. Montevideo). It is essential to note that qPCR methods detect both viable and nonviable bacteria while culture methods detect only viable bacteria. Such disparity in culture versus qPCR results has also been observed during Salmonella detection/isolation from swine feces (33).
Persistent shedding of Salmonella in feces contributes to the transmission of the bacteria to naive hosts. Variable shedding patterns were observed among the six serovars included in this study. Similar patterns have been shown for other nontyphoidal Salmonella serovars (31). Genes carried on SPI-2 are known to contribute toward the maintenance of long-term infection (34). It is likely, therefore, that genetic variation within these genes may contribute to the ability of some serovars to establish a persistent infection.
Conclusion.
In conclusion, this study has shown that although eggs or egg products are frequently implicated in Salmonella infection in humans, not all Salmonella isolates obtained from egg or egg-associated products are equally invasive and/or pathogenic, as measured by invasion assays using human intestinal epithelial cells (Caco-2) and live BALB/c mice. Further investigations are necessary to study the comparatively high- and low-virulence Salmonella genomes and their detailed molecular mechanisms in order to understand the pathogenesis.
It is also important that only one isolate per serovar was used in this study. Within-serotype variation in Salmonella virulence has been demonstrated for several serotypes (17). Although the current study has characterized the pathogenicity of an individual serovar, future studies need to be directed toward investigation of within-serotype variation. Collectively, our data demonstrated a presence of virulence genes from SPI-1 to SPI-4 in high-, medium-, and low-invasive Salmonella serovars. Our data also greatly expand on the known significant strain-to-strain variation in in vitro and in vivo pathogenicity and highlight the need for further comparative genomic and phenotypic studies.
ACKNOWLEDGMENTS
This research was supported by the Australian Egg Corporation, Ltd.
We acknowledge technical support from Geraldine Laven-Law, Amanda Kidsley, and Vivek Pande, Nikita Nevrekar. We also acknowledge the support of Lab Animal Services at the University of Adelaide.
REFERENCES
- 1.Chousalkar KK, Flynn P, Sutherland M, Roberts JR, Cheetham BF. 2010. Recovery of Salmonella and Escherichia coli from commercial egg shells and effect of translucency on bacterial penetration in eggs. Int J Food Microbiol 142:207–213. doi: 10.1016/j.ijfoodmicro.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 2.OzFoodNet Working Group. 2010. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network, 2009. Commun Dis Intell 34:396–426. [PubMed] [Google Scholar]
- 3.Gast RK, Guraya R, Guard J, Holt PS. 2011. The relationship between the numbers of Salmonella Enteritidis, Salmonella Heidelberg, or Salmonella Hadar colonizing reproductive tissues of experimentally infected laying hens and deposition inside eggs. Avian Dis 55:243–247. doi: 10.1637/9540-092810-Reg.1. [DOI] [PubMed] [Google Scholar]
- 4.Cox JM, Woolcock JB, Sartor AL. 2002. The significance of Salmonella, particularly S. Infantis, to the Australian egg industry. Rural Industries Research and Development Corporation, Barton, Australian Capital Territory, Australia. [Google Scholar]
- 5.Foley SL, Johnson TJ, Ricke SC, Nayak R, Danzeisen J. 2013. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol Mol Biol Rev 77:582–607. doi: 10.1128/MMBR.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Australian Salmonella Reference Centre. 2014. Australian Salmonella Reference Centre laboratory report, January-March 2014. SA Pathology, Adelaide, South Australia, Australia. [Google Scholar]
- 7.Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, Aarestrup FM. 2011. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 8:887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 8.Thornbrough JM, Worley MJ. 2012. A naturally occurring single nucleotide polymorphism in the Salmonella SPI-2 type III effector srfH/sseI controls early extraintestinal dissemination. PLoS One 7:e45245. doi: 10.1371/journal.pone.0045245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hensel M. 2004. Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol 294:95–102. doi: 10.1016/j.ijmm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, Agmon V, McClelland M, Rahav G, Gal-Mor O. 2013. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS One 8:e58449. doi: 10.1371/journal.pone.0058449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziemer CJ, Steadham SR. 2003. Evaluation of the specificity of Salmonella PCR primers using various intestinal bacterial species. Lett Appl Microbiol 37:463–469. doi: 10.1046/j.1472-765X.2003.01430.x. [DOI] [PubMed] [Google Scholar]
- 12.Shah DH, Zhou X, Addwebi T, Davis MA, Orfe L, Call DR, Guard J, Besser TE. 2011. Cell invasion of poultry-associated Salmonella enterica serovar Enteritidis isolates is associated with pathogenicity, motility and proteins secreted by the type III secretion system. Microbiology 157:1428–1445. doi: 10.1099/mic.0.044461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betancor LL, Yim M, Fookes A, Martinez NR, Thomson A, Ivens S, Peters C, Bryant G, Algorta S, Kariuki F, Schelotto D, Dougan MG, Chabalgoity JA. 2009. Genomic and phenotypic variation in epidemic-spanning Salmonella enterica serovar Enteritidis isolates. BMC Microbiol 9:237. doi: 10.1186/1471-2180-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yim L, Betancor L, Martinez A, Giossa G, Bryant C, Maskell D, Chabalgoity JA. 2010. Differential phenotypic diversity among epidemic-spanning Salmonella enterica serovar Enteritidis isolates from humans or animals. Appl Environ Microbiol 76:6812–6820. doi: 10.1128/AEM.00497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cencič A, Langerholc T. 2010. Functional cell models of the gut and their applications in food microbiology: a review. Int J Food Microbiol 141:S4–S14. doi: 10.1016/j.ijfoodmicro.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes LA, Shopland S, Wigley P, Bradon H, Leatherbarrow AH, Williams NJ, Bennett M, Pinna ED, Lawson B, Cunningham AA, Chantrey J. 2008. Characterisation of Salmonella enterica serotype Typhimurium isolates from wild birds in northern England from 2005–2006. BMC Vet Res 4:4–14. doi: 10.1186/1746-6148-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. CLSI document M100-23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Mickael CS, Lam P-KS, Berberov EM, Allan B, Potter AA, Köster W. 2010. Salmonella enterica serovar Enteritidis tatB and tatC mutants are impaired in Caco-2 cell invasion in vitro and show reduced systemic spread in chickens. Infect Immun 78:3493–3505. doi: 10.1128/IAI.00090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gole VC, Torok V, Sexton M, Caraguel C, Chousalkar KK. 2014. Association between the indoor environmental contamination by Salmonella enterica and contamination of eggs on layer farms. J Clin Microbiol 52:3250–3258. doi: 10.1128/JCM.00816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hensel M. 2000. Salmonella pathogenicity island 2. Mol Microbiol 36:1015–1023. doi: 10.1046/j.1365-2958.2000.01935.x. [DOI] [PubMed] [Google Scholar]
- 21.Jones FT, et al. . 2008. Salmonellosis outcomes differ substantially by serotype. J Infect Dis 198:109–114. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- 22.McCormick BA. 2003. The use of transepithelial models to examine host-pathogen interactions. Curr Opin Microbiol 6:77–81. doi: 10.1016/S1369-5274(02)00003-6. [DOI] [PubMed] [Google Scholar]
- 23.Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, Porcella SF, Steele-Mortimer O. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156:1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson SK, Borewicz K, Johnson T, Xu W, Isaacson RE. 2012. Characterization and differential gene expression between two phenotypic phase variants in Salmonella enterica serovar Typhimurium. PLoS One 7:e43592. doi: 10.1371/journal.pone.0043592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu C, Ou JT. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol 34:2619–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett 305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 27.Groisman EA, Ochman H. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791–794. doi: 10.1016/S0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 28.Klein JR, Fahlen TF, Jones BD. 2000. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect Immun 68:3368–3376. doi: 10.1128/IAI.68.6.3368-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galyov EE, Wood MW, Rosqvist R, Mullan PB, Watson PR, Hedges S, Wallis TS. 1997. A secreted effector protein of Salmonella Dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol 25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 30.Gilmore MS, Ferretti JJ. 2003. The thin line between gut commensal and pathogen. Science 299:1999–2002. doi: 10.1126/science.1083534. [DOI] [PubMed] [Google Scholar]
- 31.Swearingen MC, Porwollik S, Desai PT, McClelland M, Ahmer BMM. 2012. Virulence of 32 Salmonella strains in mice. PLoS One 7:e36043. doi: 10.1371/journal.pone.0036043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurley BP, McCormick BA. 2003. Translating tissue culture results into animal models: the case of Salmonella Typhimurium. Trends Microbiol 11:562–569. doi: 10.1016/j.tim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Wilkins W, Waldner Rajić CA, McFall M, Muckle A, Mainar-Jaime RC. 2010. Comparison of bacterial culture and real-time PCR for the detection of Salmonella in grow-finish pigs in Western Canada using a Bayesian approach. Zoonoses Public Health 57:115–120. doi: 10.1111/j.1863-2378.2010.01365.x. [DOI] [PubMed] [Google Scholar]
- 34.Monack DM. 2012. Salmonella persistence and transmission strategies. Curr Opin Microbiol 15:100–107. doi: 10.1016/j.mib.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Guo X, Chen J, Beuchat LR, Brackett RE. 2000. PCR detection of Salmonella enterica serotype Montevideo in and on raw tomatoes using primers derived from hilA. Appl Environ Microbiol 66:5248–5252. doi: 10.1128/AEM.66.12.5248-5252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clavijo R, Loui C, Andersen GL, Riley Lu LWS. 2006. Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl Environ Microbiol 72:1055–1064. doi: 10.1128/AEM.72.2.1055-1064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Australian Salmonella Reference Centre. 2010. Quarterly report. Australian Salmonella Reference Centre, Adelaide, South Australia, Australia. [Google Scholar]
- 38.Australian Salmonella Reference Centre. 2011. Quarterly report. Australian Salmonella Reference Centre, Adelaide, South Australia, Australia. [Google Scholar]
- 39.Australian Salmonella Reference Centre. 2012. Quarterly report. Australian Salmonella Reference Centre, Adelaide, South Australia, Australia. [Google Scholar]
- 40.Australian Salmonella Reference Centre. 2013. Quarterly report. Australian Salmonella Reference Centre, Adelaide, South Australia, Australia. [Google Scholar]