Abstract
Understanding molecular mechanisms of toxicity is facilitated by experimental manipulations, such as disruption of function by gene targeting, that are especially challenging in non-standard model species with limited genomic resources. While loss-of-function approaches have included gene knock-down using morpholino-modified oligonucleotides and random mutagenesis using mutagens or retroviruses, more recent approaches include targeted mutagenesis using zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technology. These latter methods provide more accessible opportunities to explore gene function in non-traditional model species. To facilitate evaluations of toxic mechanisms for important categories of aryl hydrocarbon pollutants, whose actions are known to be receptor mediated, we used ZFN and CRISPR-Cas9 approaches to generate aryl hydrocarbon receptor 2a (AHR2a) and AHR2b gene mutations in Atlantic killifish (Fundulus heteroclitus) embryos. This killifish is a particularly valuble non-traditional model for this study, with multiple paralogs of AHR whose functions are not well characterized. In addition, some populations of this species have evolved resistance to toxicants such as halogenated aromatic hydrocarbons. AHR-null killifish will be valuable for characterizing the role of the individual AHR paralogs in evolved resistance, as well as in normal development. We first used five-finger ZFNs targeting exons 1 and 3 of AHR2a. Subsequently, CRISPR-Cas9 guide RNAs were designed to target regions in exon 2 and 3 of AHR2a and AHR2b. We successfully induced frameshift mutations in AHR2a exon 3 with ZFN and CRISPR-Cas9 guide RNAs, with mutation frequencies of 10% and 16%, respectively. In AHR2b, mutations were induced using CRISPR-Cas9 guide RNAs targeting sites in both exon 2 (17%) and exon 3 (63%). We screened AHR2b exon 2 CRISPR-Cas9-injected embryos for off-target effects in AHR paralogs. No mutations were observed in closely related AHR genes (AHR1a, AHR1b, AHR2a, AHRR) in the CRISPR-Cas9-injected embryos. Overall, our results demonstrate that targeted genome-editing methods are efficient in inducing mutations at specific loci in embryos of a non-traditional model species, without detectable off-target effects in paralogous genes.
Keywords: non-model organisms, gene knock-outs, mummichog, adaptation, zinc finger nucleases, CRISPR-Cas9
1. INTRODUCTION
The Atlantic killifish (Fundulus heteroclitus) is one of the most ecologically and environmentally important estuarine fish distributed along the East coast of the United States. Their ability to tolerate wide changes in environmental conditions, including temperature, salinity, oxygen and pH, have made them an ideal model species to investigate the biochemical, physiological and evolutionary basis of environmental adaptation (Burnett et al., 2007; Lister et al., 2011; Schulte, 2014; Schulte et al., 2011; Scott and Brix, 2013; Whitehead, 2010; Whitehead et al., 2012). Some populations of killifish are also valuable models for understanding the mechanisms of evolved resistance to toxicants (Hahn, 1998; Van Veld and Nacci, 2008; Wirgin and Waldman, 2004). Populations of killifish inhabiting contaminated coastal waters along the North Atlantic U.S. coast have evolved resistance to some contaminants representing major categories of aryl hydrocarbon pollutants, such as polynuclear aromatic hydrocarbons (PAHs), and halogenated aromatic hydrocarbons such as polychlorinated biphenyls (PCBs), 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, ‘dioxin’) and other dioxin-like compounds (DLCs) (Bello et al., 2001; Elskus et al., 1999; Meyer et al., 2002; Nacci et al., 1999; Nacci et al., 2010; Powell et al., 2000). This evolved resistance involves alterations in signaling through the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor through which some PAHs, PCBs, and TCDD alter gene expression and cause toxicity. Killifish express four AHR paralogs (AHR1a, AHR1b, AHR2a and AHR2b), the products of distinct loci (Hahn et al., 1997; Karchner et al., 1999; Reitzel et al., 2014). Although the respective functions of these AHRs are not well understood, AHR2 proteins appear to play a major role in mediating the developmental effects of PAHs and PCBs in fish (Clark et al., 2010; Jonsson et al., 2007; Prasch et al., 2003).
While there is a great deal of understanding about the physiological and biochemical basis of environmental adaptation in killifish, very little is known about its genetic basis, mainly due to the lack of genetic tools to evaluate gene function. Recently, the antisense morpholino oligonucleotide (MO)-based gene knockdown method was adapted to killifish to determine the role of aryl hydrocarbon receptors and cytochrome P4501A (CYP1A) in developmental toxicity (Clark et al., 2010; Matson et al., 2008), and to study the role of aquaporins in adult killifish (Notch et al., 2011). However, MOs are effective only for a limited period of time and do not completely eliminate gene expression. In order to conclusively demonstrate the function of any protein, gene knockouts are essential.
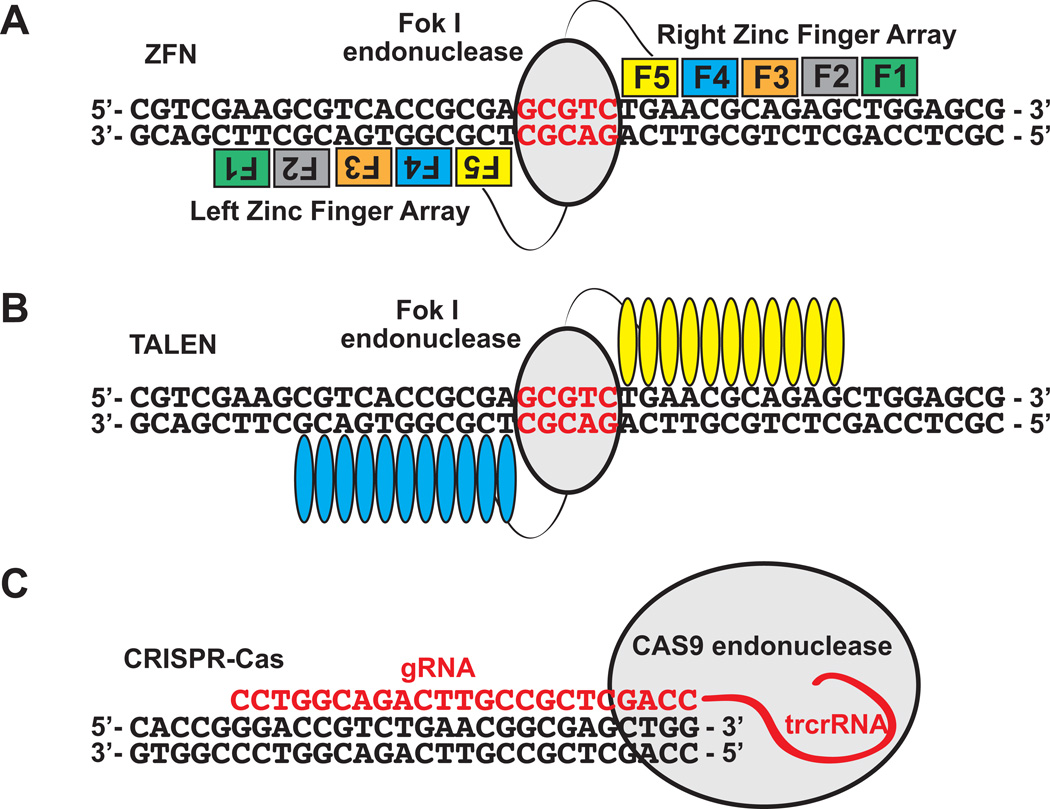
In the past decade, several targeted genome-editing methods have been successfully employed to manipulate genes in a variety of model and non-model organisms. They include zinc-finger nucleases (ZFNs), transactivator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system (Gaj et al., 2013; Kim and Kim, 2014; Peng et al., 2014; Sander and Joung, 2014) (Fig. 1). ZFN and TALENs utilize engineered nucleases consisting of a sequence-specific DNA-binding domain fused to a nonspecific DNA cleavage module. The DNA-binding domains can be customized to recognize any sequence of interest. Distinct from the site-specific nucleases, CRISPR-Cas9 is an RNA-guided DNA nuclease (RGN), where a small guide RNA (sgRNA) complementary to a 20 base target DNA sequence guides Cas9, a bacterially derived nuclease, to induce a double-strand break (DSB) at the target site (Hwang et al., 2013). Because the DSB repair process by non-homologous end-joining (NHEJ) is error prone, it leaves insertions or deletions (indels) at the target site. The relative ease of generating target-specific mutations using these techniques has opened up new avenues for conducting functional studies in any organism of interest. Several studies have demonstrated the efficacy of ZFNs, TALENs and RGNs in producing heritable mutations in non-model organisms such as rainbow trout, medaka, tilapia and yellow catfish (Dong et al., 2011; Li et al., 2013; Yano et al., 2014; Zhang et al., 2014) (Table 1). Mutagenesis rates of up to 95% have been observed with CRISPR-Cas9 (Jao et al., 2013; Li et al., 2014). So far, there are no studies describing the generation of gene knock-outs in killifish or of targeted AHR mutants in any non-mammalian vertebrate. In this study, we utilized ZFN and CRISPR-Cas9 approaches to induce mutations in paralogous killifish AHR2a and AHR2b genes, and we observed indels in the injected embryos. We discuss strategies for the application of this method to knock out closely related paralogs in a species with a long generation time.
Figure 1.
Outline of the targeted mutagenesis techniques currently used in generating mutants. A. Zinc finger nucleases B. TALENs and C. CRISPR-Cas system.
Table 1.
Published studies employing gene targeting in non-traditional model species.
| Gene-editing approach |
Species | Reference |
|---|---|---|
| ZFN | Monarch butterfly (Danaus plexippus) Two-spotted cricket (Gryllus bimaculatus) Sea urchin (Hemicentrotus pulcherrimus) Ascidian (Ciona intestinalis) Western clawed frog (Xenopus tropicalis) Yellow catfish (Pelteobagrus fulvidraco) Medaka (Oryzias melastigma) Rainbow trout (Oncorhynchus mykiss) |
(Merlin et al., 2013) (Watanabe et al., 2012) (Ochiai et al., 2010) (Kawai et al., 2012) (Young et al., 2011) (Nakajima et al., 2012) (Dong et al., 2011) (Ansai et al., 2012) (Yano et al., 2014) |
| TALEN | Nematode (Pristionchus pacificus) Marine polycheate (Platyneries dumerilii) Culicine mosquito (Aedes aegypti) Silk worm (Bombyx mori) Sea urchin (Hemicentrotus pulcherrimus) Ascidian (Ciona intestinalis) Western clawed frog (Xenopus tropicalis) Iberian ribbed newts (Pleurodeles waltl) Tilapia (Oreochromis niloticus) Medaka (Oryzias melastigma) Yellow catfish (Pelteobagrus fulvidraco) |
(Lo et al., 2013) (Bannister et al., 2014) (Aryan et al., 2013b) (Aryan et al., 2013a) (Ma et al., 2012) (Takasu et al., 2013) (Sajwan et al., 2013) (Wang et al., 2013) (Hosoi et al., 2014) (Treen et al., 2014) (Yoshida et al., 2014) (Ishibashi et al., 2012) (Lei et al., 2012) (Nakajima and Yaoita, 2013) (Liu et al., 2014) (Hayashi et al., 2014) (Li et al., 2013) (Wang and Hong, 2014) (Ansai et al., 2014) (Ansai et al., 2013) (Dong et al., 2011) |
| CRISPR-Cas9 | Western clawed frog (Xenopus tropicalis) Axolotl (Ambystoma mexicanum) Atlantic Salmon (Salmo salar) Tilapia (Oreochromis mossambicus) |
(Nakayama et al., 2013) (Guo et al., 2014) (Flowers et al., 2014) (Ji-Feng et al., 2014) (Edvardsen et al., 2014) (Li et al., 2014) |
2. MATERIALS AND METHODS
2.1. Killifish adults and embryos
Mature male and female Atlantic killifish were collected from Scorton Creek (Sandwich, MA) during full moon and new moon in Spring and Summer of each year (2011–2014) using minnow traps, as described previously (Karchner et al., 1999). Although killifish populations are known to vary dramatically in their sensitivity to DLCs, this source population is known to be relatively sensitive to DLCs, characteristic of other populations of this species resident to similarly uncontaminated sites (Nacci et al., 2010). Fish were maintained in tanks with continuous flow-through seawater (SW) at 18–20°C and 14h:10h light/dark photoperiod conditions. The animal husbandry practices were approved by the Animal Care and Use Committee of the Woods Hole Oceanographic Institution. Female fish were lightly anesthetized with Tricaine (MS222; buffered with sodium bicarbonate, Sigma-Aldrich, St. Louis, MO, USA) and oocytes were obtained for in vitro fertilization (IVF) by gently squeezing the abdomen. Oocytes were collected in glass petri dishes with filtered SW (25 parts per thousand; ppt). Milt was obtained by euthanizing mature males in MS222, dissecting out the gonads, and chopping them with a scalpel blade in seawater. A few drops of milt were added to the oocytes for fertilization. Each IVF experiment included a pool of oocytes stripped from at least 2–3 females and milt from 1 or 2 males. Approximately 20 minutes after the addition of milt, embryos were rinsed with filtered SW to remove any excess sperm. Fertilized embryos were maintained at 23°C until further use.
2.2. Zinc Finger Nuclease design
AHR2a exon 2 and 3 (Fig. 2) were each targeted with a five-finger ZFN pair using the CompoZr® custom ZFN service (Sigma-Aldrich, St. Louis, MO, USA) (Table 2; Fig. 2). Left- and right-ZFN plasmids were in vitro transcribed and polyadenylated using the mMessage mMachine T7 kit (Life Technologies, Carlsbad, CA, USA). ZFN mRNA from each half-site was diluted to a concentration of 100–400 ng/µl and 2.5 nl of each was co-injected into 1 or 2-cell stage killifish embryos, as described below.
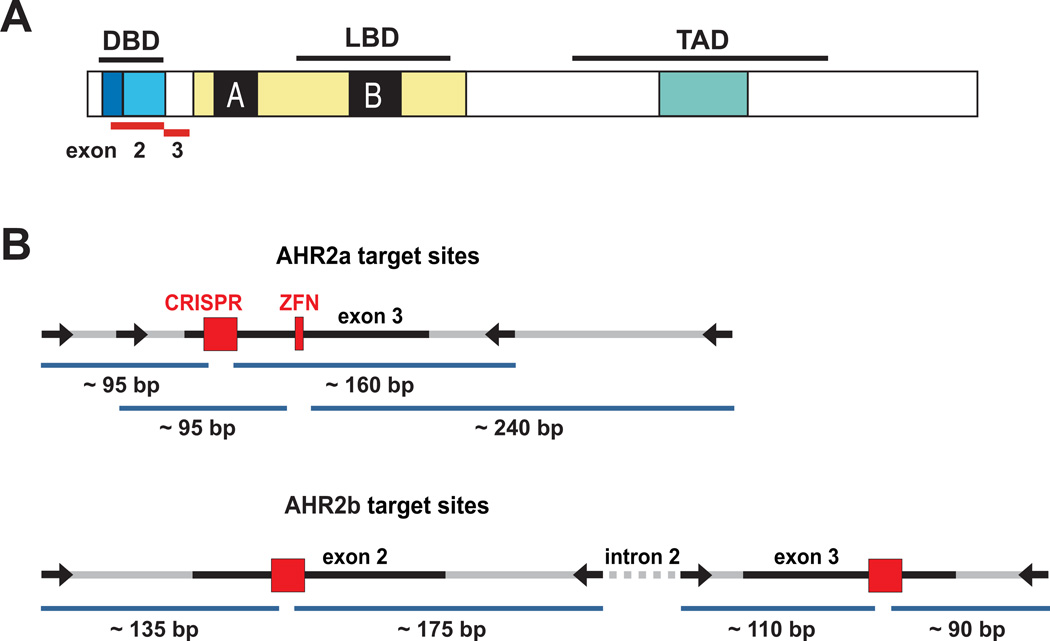
Figure 2.
AHR target sites. A. Functional domain structure of the AHR protein. The location of the targeted exons 2 and 3 in relation to the functional domains is shown as red bars. DBD: DNA-binding domain, LBD: ligand-binding domain, TAD: transcriptional activation domain. B. Schematic representations of the ZFN and CRISPR-Cas target regions in AHR2a and AHR2b loci. Arrows represent the primers used in the PCR amplification of genomic DNA for screening. Surveyor nuclease fragments are shown below the target regions.
Table 2.
AHR2a and AHR2b target regions and oligonucleotides used in ZFN and CRISPR-Cas9-based mutagenesis. The ZFN target site is shown in capital letters. The sequence flanking the target sites are recognized by the ZFN proteins.
| Name | Description | Sequence |
|---|---|---|
| AHR2a exon 2 | ZFN target site | ctgcgcctcagcgtgGGATAcctgagggtcaagag |
| AHR2a exon 3 | ZFN target site | gactcattcagcttcTCTGAaggagagctgctgct |
| AHR2a exon 2 | CRISPR-Cas9 target site | GGAGCGAATCTCATCAGGGA |
| AHR2a exon 3 | CRISPR-Cas9 target site | GGGTGGCTCCTGGAGTCAAC |
| AHR2b exon2 | CRISPR-Cas9 target site | GGACCGTCTGAACGGCGAGC |
| AHR2b exon3 | CRISPR-Cas9 target site | GGGCGAAGGTCTGAGCGGTC |
| 2-ex3Fwd | AHR2a ZFN pair3 screening | TGTTTCACCTCCTGCACAGCTTCC |
| 1XRR1 | AHR2a ZFN pair3 screening | TACTAGCTAAACGTGACCTGTCG |
| 2a-ex3CrF | AHR2a CRISPR-Cas exon3 screening | CTGTTCTATCAGGTGGAGAGTCC |
| 2a-ex3CrR | AHR2a CRISPR-Cas exon3 screening | GTTGTTGGATGAGTATGCAGGTGG |
| AHR2b-CrF | AHR2b CRISPR-Cas9 exon2 screening | AATCTTTGATCCGGTCTGATCTGG |
| AHR2b-CrR | AHR2b CRISPR-Cas9 exon2 screening | AGACACAACCAAGCAGCAACACG |
| 2bex3-Fwd | AHR2b CRISPR-Cas9 exon3 screening | ATCGCCCAGCCTTAGCTTCCAGTCG |
| 2bex3-Rev | AHR2b CRISPR-Cas9 exon3 screening | GCGTTTTTGCTTTGTCTGCTGTCGG |
| Fh2A-ex3-CR1 | AHR2a exon3 gRNA oligo1 | TAGGGTGGCTCCTGGAGTCAAC |
| Fh2A-ex3-CR2 | AHR2a exon3 gRNA oligo2 | AAACGTTGACTCCAGGAGCCAC |
| FhAHR2b-CR1 | AHR2b exon2 gRNA oligo1 | TAGGACCGTCTGAACGGCGAGC |
| FhAHR2b-CR2 | AHR2b exon2 gRNA oligo2 | AAACGCTCGCCGTTCAGACGGT |
| Fh2B-ex3-CR1 | AHR2b exon3 gRNA oligo1 | TAGGGCGAAGGTCTGAGCGGTC |
| Fh2B-ex3-CR2 | AHR2b exon3 gRNA oligo2 | AAACGACCGCTCAGACCTTCGC |
| 2b-1aF | AHR1a off-target screening | GCTTAATCCTGCTTTCGTCTCTGC |
| Fh1a-ex2rev | AHR1a off-target screening | AAGAAGTGTTTGGCTCGTAGGTAGC |
| 2b-2aF | AHR2a off-target screening | TTTTACCCCTCACGGAAGAAGATGG |
| Fh2a-ex2rev | AHR2a off-target screening | GTAGCTCTTGACCCTCAGGTATCC |
| 2b-1bF | AHR1b off-target screening | TTTCCTCCTCTCCAGAGCCAAACC |
| Fh1b-ex2rev | AHR1b off-target screening | AAGAAGCTCTTGGTGCGCAGGTAGC |
| RR-ex2fwd | AHRR off-target screening | CTCATCCCTTCTGCCGTCTCGTCC |
| RR-ex2rev | AHRR off-target screening | AAGAAGCTTTTGACGCGGAGGTAGG |
2.3. CRISPR-Cas guide RNA design and construction
Guide RNAs (gRNA) targeting AHR2a and AHR2b were designed using the ZiFiT Targeter website (http://zifit.partners.org/ZiFiT_Cas9). Exons 1, 2 and 3 of AHR2a and AHR2b were searched for target sites in order to obtain the shortest truncated mutant protein. A single target site was found in each exon. The AHR2a exon 1 target site did not begin with GG residues, which prevented it from being cloned into the gRNA expression vector. The AHR2b exon 1 target site was not optimal due to repetitive sequences. Therefore, the target sites for exons 2 and 3 of AHR2a and AHR2b were selected for gRNA synthesis (Table 2). PAGE-purified complementary oligonucleotides (Eurofins Genomics, Huntsville, AL, USA) were annealed and cloned into an expression vector following a published protocol (Hwang et al., 2013). The gRNA oligonucleotide sequences are provided in Table 2. Expression vectors for guide RNA (pDR274) and Cas9 endonuclease (MLM3613) were obtained from Addgene (https://www.addgene.org/). The gRNA was transcribed using the MAXIscript T7 kit, and the Cas9 mRNA was transcribed and poly(A)-tailed with the mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies, Carlsbad, CA, USA). gRNA and Cas9 mRNA were mixed at a ratio of 1:12 for the microinjections.
2.4. Microinjection of ZFN mRNA and CRISPR-Cas9 gRNA
Microinjection of killifish embryos was performed following a previously established protocol (Matson et al., 2008) with some modifications. Briefly, prior to microinjection, 1-cell stage embryos were rolled on wet paper towels to prevent the filaments on the chorion from adhering to the microinjection needle. They were subsequently placed on custom-made agarose microinjection embryo trays and oriented so that the cell directly faces the microinjection needle. The microinjection setup consisted of a Zeiss Stemi 2000-C stereomicroscope, Narishige IM-300 microinjector and a micromanipulator (Narishige International, East Meadow, NY, USA). Aluminosilicate glass tubes (1.0 mm outer diameter, 0.64 mm inner diameter) were used to prepare microinjection needles. This type of needle was found to be superior to borosilicate needles for microinjection of killifish embryo, which have a thick chorion. A P-30 vertical micropipette puller (Sutter Instruments, Novato, CA, USA) at a heater output setting of 97 was used to pull aluminosilicate glass tubing into microinjection needles. The injection volume was calibrated by injecting the solution into mineral oil and measuring the diameter of the droplet using a stage micrometer. The microinjection volume was approximately 5 nL per embryo. To compensate for variations in needle opening size, injection time and pressure were altered during microinjection. Injected embryos were maintained in filtered SW at 23°C. Some of the injected embryos were transferred to the fish-rearing facility at the National Health and Environmental Effects Research Laboratory, Atlantic Ecology Division, Environmental Protection Agency, Narragansett, RI, to be raised to adulthood. The numbers of injected fish that are being raised for identifying founders are provided in Supplementary Table 1.
2.5. ZFN protein expression
ZFN-injected and uninjected embryos were sampled at 3, 6, 9 and 24 hours post-injection to determine the temporal profile of ZFN protein expression. At each time point, one pool of 10 embryos was sampled. Embryos were homogenized in 1× sample treatment buffer with β-mercaptoethanol and boiled for 5 minutes to denature the protein. Samples were separated on an 8% polyacrylamide gel using the discontinuous buffer system (Laemmli, 1970). The proteins were transferred onto a nitrocellulose membrane with a semi-dry transfer unit (BioRad, Waltham, MA, USA), and the membrane was blocked with 5% milk in TBST (20 mM Tris pH 7.5, 300 mM NaCl and 0.1% (v/v) Tween 20) overnight. ZFN protein was detected using a mouse monoclonal anti-Flag antibody (Sigma, St. Louis, MO, USA) at 1:5000 dilution. The secondary antibody used was horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Santa Cruz Biotechnology, Dallas, TX, USA) at 1:5000 dilution. The membranes were incubated in the primary antibody for 60 min at room temperature, washed with TBST twice (10 min each), incubated with secondary antibody for 60 min, and finally washed with TBST (15 min). Detection was done using the ECL Prime Western blotting reagent (GE Healthcare, Pittsburgh, PA, USA).
2.6. Surveyor endonuclease assay
Somatic mutations were determined using the Surveyor® mutation detection kit (Transgenomic Inc., Omaha, NE, USA) following the manufacturer’s instructions (Qiu et al., 2004). Embryos (pool of 5 embryos per replicate) were sampled at 48 hours post-injection and genomic DNA was isolated using the proteinase K digestion method (Goldenberger et al., 1995). The AHR2a and AHR2b target regions were amplified by polymerase chain reaction (PCR) using Advantage polymerase (Clontech, Mountain View, CA, USA). The primer sequences are given in Table 2. PCR cycling conditions were 94°C for 1 min, followed by 35 cycles of [94°C for 5 s, 68°C for 1 min], followed by 72°C for 7 min. The PCR products were melted and re-annealed to form hetero-duplex DNA and digested with Surveyor nuclease for 20 min at 42°C. The resulting products were electrophoresed on a 15% TBE gel and visualized by ethidium bromide staining.
2.7. DNA sequencing
PCR products from samples that were positive in the Surveyor endonuclease assay were cloned into the pGEM-T easy vector (Promega, Madison, WI, USA) and sequenced in 96-well plates (Functional Biosciences, Madison, WI, USA). Insertions and deletions were identified by aligning the sequences with the wildtype AHR sequence (Sequencher 5.1, Gene Codes Corp., Ann Arbor, MI, USA). Off-target screening for AHR2b CRISPR-Cas9 exon2 was done by direct-sequencing of PCR products. The amplification reactions were purified using the GeneClean kit (MP Biomedicals, Santa Ana, CA, USA). Direct-sequencing of the amplicons for AHR1a, AHR2a, and AHRR was done with primers Fh2a-ex2rev, 2b-1aF, and RR-ex2fwd, respectively (Table 2). The chromatograms were visually inspected for regions of double peaks indicating insertions or deletions.
3. RESULTS
3.1. ZFN-induced AHR2a mutations
We tested different concentrations of ZFN mRNA on killifish embryos by injecting 500, 1000 and 2000 pg per embryo. No overt toxic effects or phenotypic abnormalities were observed at any of the concentrations as determined by microscopic observation of embryos during development. Hence, we selected 2000 pg to inject in subsequent experiments. ZFN protein expression was confirmed in the embryos injected with 2000 pg ZFN mRNA. Embryos were sampled at 3, 6, 9 and 24 hours post-microinjection. The maximum ZFN protein expression was observed at 6 hours post-microinjection (Fig. 3A). At 3 and 9 hours, ZFN protein expression was much lower, and at 24 hours, no expression was detected. These results suggest that the maximum efficiency of ZFNs in inducing DSBs in killifish is around 6 hours post-injection.
Figure 3.
ZFN-mediated mutagenesis of killifish AHR2a. A. ZFN protein expression in killifish embryos. Uninjected (−) or ZFN-injected (+) embryos were sampled at 3, 6, 9, and 24 hours after micro-injection. Homogenates were resolved on SDS-PAGE and probed with a Flag-tag antibody. Lysate from COS-7 cells transfected with the ZFN expression plasmid was run as a positive control. B. Surveyor nuclease detection of mutations in the ZFN target region of AHR2a. Each lane represents a pool of 5 embryos from which a 335 bp genomic DNA fragment was amplified. U: uninjected control, ZFN: injected embryos. Lanes 1 and 2 are two representative samples that were positive in the mutation detection assay. Approximate sizes of the Surveyor nuclease-digested fragments containing the deletions are shown (240 and 95 bp). The full-length PCR product from sample 2 was cloned and sequenced. C. AHR2a exon 3 sequence surrounding the ZFN target site (in red). Four types of deletion mutants were observed among the sequenced clones (deletions of 2, 4, 5, and 28 nucleotides).
Approximately 400 embryos were injected with the ZFN pair targeting exon 2 of AHR2a, and all were screened for mutations using the Surveyor mutation detection assay. We did not detect any insertions or deletions in these embryos; therefore, we next tested the ZFN pair targeting exon 3. Using the Surveyor assay, we analyzed 14 pools of ZFN-injected embryos (5 embryos per pool), and detected the presence of mutations in 4 pools (Fig. 3B). Endonuclease digestion resulted in fragments of approximately 240 bp and 95 bp (Fig. 2B, 3B). The PCR product from one of the embryo pools that was positive in the Surveyor assay was cloned into the pGEM-T Easy vector. Sequencing of multiple clones revealed 6 mutant sequences out of 59 (10%), represented by four different types of deletions at the target site (2, 4, 5 and 28-base pair deletions) (Fig. 3C). We did not observe any cuts with the Surveyor endonuclease in the 2 pools of uninjected embryos. The remaining fish have been raised to adulthood, and their progeny are being screened to identify potential founders (Supplemental Table 1).
3.2. CRISPR-Cas9-induced AHR2a mutations
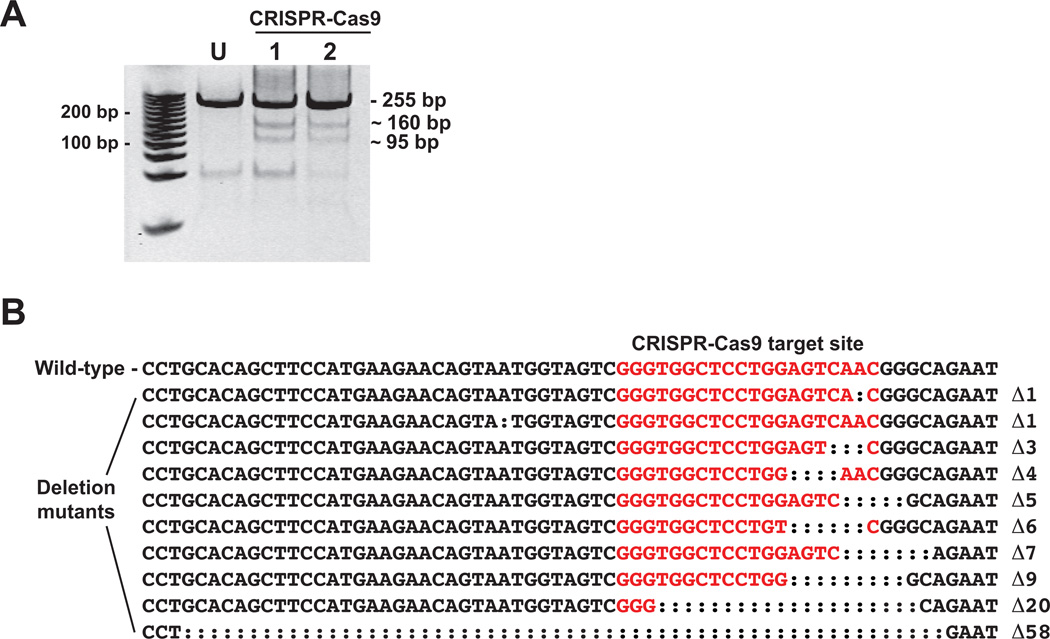
Microinjection of the CRISPR-Cas9 guide RNA targeting exon 2 of AHR2a was not successful in generating mutations (20 pools of 5 embryos/pool were analyzed). However, for the guide RNA targeting exon 3, Surveyor mutation detection assay results were positive in 2 out of 6 pools of injected embryos analyzed (30 embryos total; 5 embryos per pool) (Fig. 4A). The mutated target sites resulted in fragments of approximately 160 bp and 95 bp (Fig. 2B, 4A). The PCR products from the two positive samples (10 embryos total) were combined, cloned into the pGEM-T Easy vector, and sequenced. We observed 13 deletions in 81 sequences (16%). The 13 deletions varied in length from 1 to 58 bp, representing 10 distinct deletions (Fig. 4B).
Figure 4.
CRISPR-Cas9-mediated mutagenesis of killifish AHR2a targeting exon 3. A. Surveyor nuclease detection of mutations in the CRISPR-Cas9 target region of AHR2a exon 3. Each lane represents a pool of 5 embryos from which a 255 bp genomic DNA fragment was amplified. U: uninjected, lanes 1 and 2: CRISPR-Cas9-injected embryos. Approximate sizes of the digested fragments containing the deletions and insertions are shown (160 and 95 bp). The full-length PCR products from samples 1 and 2 were pooled, cloned, and sequenced. B. AHR2a exon 3 sequence surrounding the CRISPR-Cas9 target site (in red). Ten different deletion mutants were observed among the sequenced clones.
3.3. CRISPR-Cas9-induced AHR2b mutations
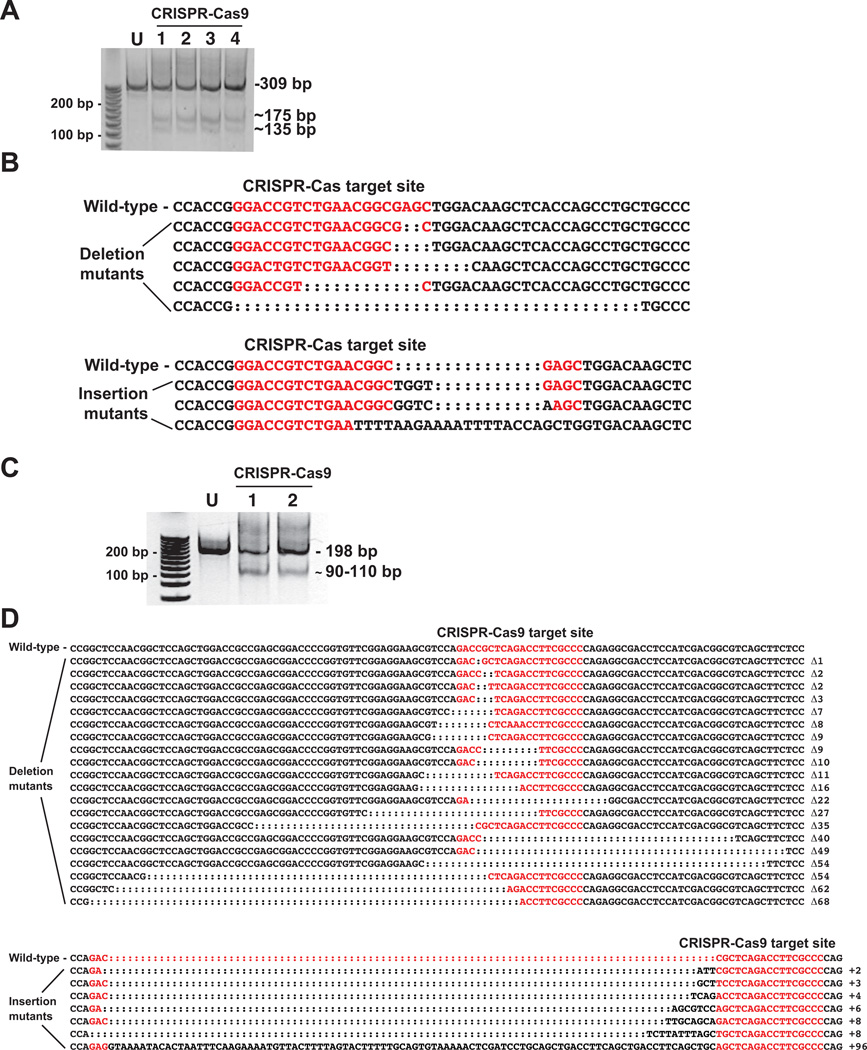
CRISPR-Cas9 guide RNAs targeting exon 2 and 3 of the AHR2b locus were both efficient at generating mutations in the microinjected embryos. Surveyor mutation detection assay showed the presence of mutations in all 4 pools of AHR2b exon 2 CRISPR-Cas-injected embryos analyzed (20 embryos total; 5 embryos per pool) (Fig. 5A). The PCR products from these 4 pools were combined and cloned into the pGEM-T Easy vector. Sequencing results revealed that 16 sequences out of 70 (23%) had indels at the target site, represented by 8 distinct mutations (5 deletions and 3 insertions) (Fig. 5B).
Figure 5.
CRISPR-Cas9-mediated mutagenesis of killifish AHR2b targeting exon 2 and exon 3. A. Surveyor nuclease detection of mutations in the AHR2b CRISPR-Cas9 target region. Each lane represents a pool of 5 embryos from which a 309 bp genomic DNA fragment was amplified. U: uninjected control, lanes 1–4: CRISPR-Cas9-injected embryos. Approximate sizes of the digested fragments containing the deletions and insertions are shown (175 and 135 bp). The full-length PCR products from samples 1–4 were pooled, cloned, and sequenced. B. AHR2b exon 2 sequence surrounding the CRISPR-Cas9 target site (in red). Five types of deletion mutants and 3 types of insertions were observed among the sequenced clones. C. Surveyor nuclease detection of mutations in the CRISPR-Cas9 target region of AHR2b exon 3. Each lane represents a pool of 5 embryos from which a 198 bp genomic DNA fragment was amplified. U: uninjected, lanes 1 and 2: CRISPR-Cas9-injected embryos. Approximate sizes of the digested fragments containing the deletions and insertions are shown (110 and 90 bp). The full-length PCR products from samples 1 and 2 were pooled, cloned, and sequenced. D. AHR2b exon 3 sequence surrounding the CRISPR-Cas9 target site (in red). Twenty different types of deletion mutants and seven types of insertion mutants were observed among the sequenced clones.
Similarly, injection of guide RNA targeting exon 3 of AHR2b induced mutations as observed with Surveyor mutation detection assay (Fig. 5C). All 4 pools of injected embryos showed the presence of mutations (5 embryos per pool). PCR products from two of the samples were combined, cloned into the pGEM-T easy vector and sequenced. We observed 55 sequences with indels out of 87 (63%). There were 20 different deletions and 7 different insertion mutants (Fig. 5D).
3.4. Screening for CRISPR-Cas9 off-target effects
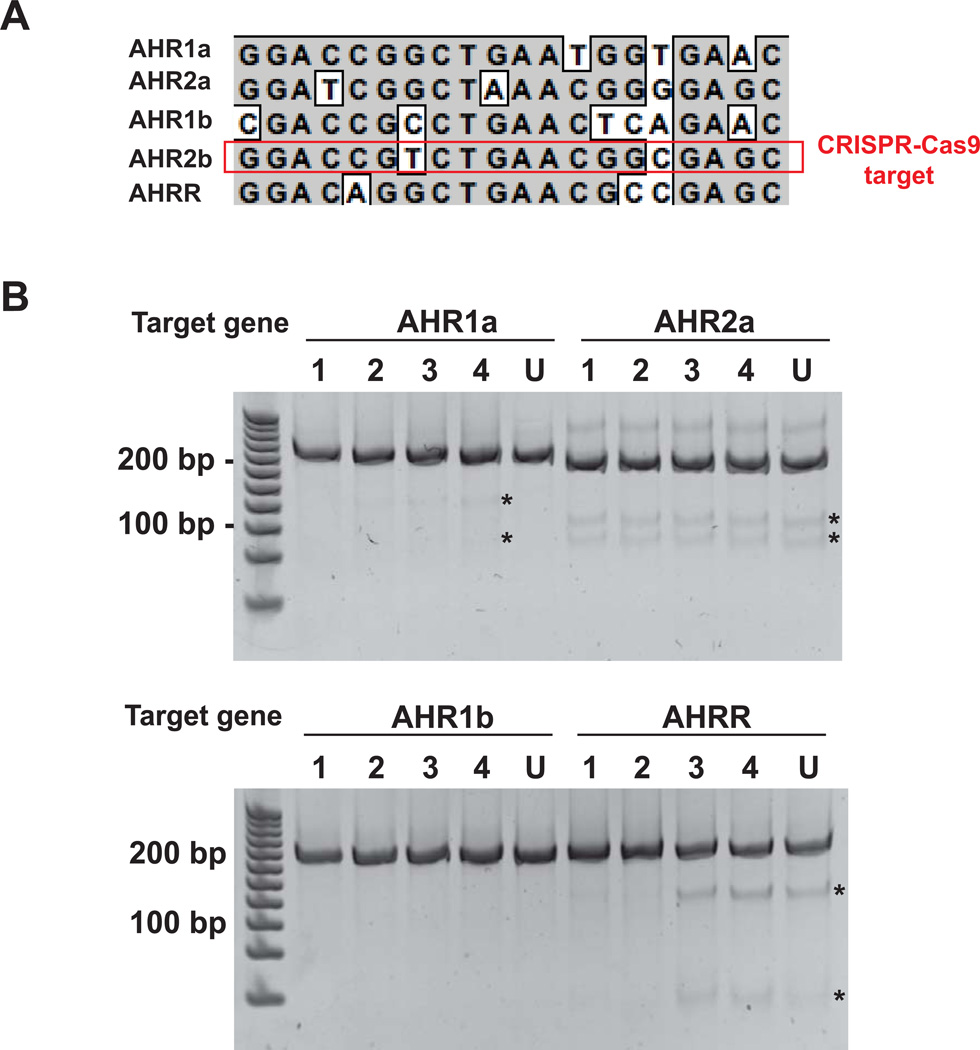
In order to assess potential off-target effects of the CRISPR-Cas9 experiments, we chose one of the guide RNAs that gave positive results (exon 2 of AHR2b) to screen for possible mutations in other AHR paralogs and AHRR. The multiple AHR paralogs and the AHRR are closely related genes and share substantial sequence identity, especially in the N-terminus. Alignment of the AHR2b CRISPR-Cas9 target sequence with the other genes is shown in Fig. 6A. The numbers of mismatches between the 20 bp AHR2b target site and AHR1a, AHR2a, AHR1b, and AHRR are 4, 4, 6, and 3, respectively. These corresponding regions were amplified from the uninjected and AHR2b CRISPR-Cas9-injected embryo genomic DNA samples in which AHR2b mutations were observed. The PCR products were analyzed for mutations using the Surveyor endonuclease assay. The AHR1a, AHR2a, and AHRR samples contained small fragments indicating endonuclease digestion of the PCR products (Fig. 6B). These fragments were also present in the AHR2a and AHRR reactions from the uninjected embryos. These amplification reactions were purified and analyzed by direct-sequencing. The chromatograms of the AHR1a and AHR2a sequencing reactions contained double peaks at previously characterized SNP locations (#141 and #159 for AHR1a, and #156 for AHR2a) (Reitzel et al., 2014). Surveyor endonuclease is sensitive to even a single nucleotide mismatch between the two DNA strands, therefore it will cleave DNA at the SNP sites. The sizes of the observed Surveyor endonuclease fragments in these samples (Fig. 6B) can be explained by these SNPs, thereby suggesting that they do not represent off-target mutations. On the other hand, we did not detect a SNP in the AHRR sequences that would explain the fragments observed in the Surveyor assay. However the presence of these fragments in both uninjected and injected samples suggest that they are rare SNPs that may not be detected by direct sequencing.
Figure 6.
AHR2b CRISPR-Cas9 off-target analysis. A. Nucleotide sequence alignment of the AHR2b CRISPR-Cas9 target sequence with the corresponding regions of closely related genes. AHR1a, AHR2a, AHR1b, and AHRR have 4, 4, 6, and 3 mismatches to AHR2b in this region, respectively. B. Surveyor nuclease detection of mutations in AHR1a, AHR2a, AHR1b, and AHRR in the AHR2b CRISPR-Cas9 target region. Digested fragments are indicated by a *. Each lane represents a pool of 5 embryos from which genomic DNA fragments were amplified. U: uninjected control, 1–4: CRISPR-Cas9 injected embryos. AHR1a, AHR2a, and AHRR PCR products from uninjected and injected embryos were sequenced.
4. DISCUSSION
In this study, we demonstrated the feasibility of targeted mutagenesis approaches in inducing mutations in an environmental model species with limited genomic and genetic resources. The ease with which ZFN and CRISPR-Cas9 methods can be utilized to cleave specific genomic DNA sequences has led to the generation of gene knockouts in species not traditionally used for conducting genetic and functional studies (Table 1). Using these methods, we produced mutations in AHR2a and AHR2b loci in killifish embryos. These embryos are currently being raised to adulthood, and their progeny will be screened to identify potential founders. As killifish have a long generation time (2 years minimum) and lack established breeding techniques, the time it takes to generate homozygous mutants is relatively long. Here, we propose some strategies to expedite the process of generating homozygous mutants in killifish. AHR-null killifish will be valuable for characterizing the role of the individual AHR paralogs in evolved resistance, as well as in normal development.
We initially adopted the ZFN approach for generating mutations, as their design and activity are well characterized (Urnov et al., 2010). In addition, ZFNs were the first targeted mutagenesis approach that was successfully used in generating mutations in a number of model and non-model organisms (Bibikova et al., 2003; Doyon et al., 2008; Scott, 2005). ZFNs are assembled using zinc finger motifs, each of which recognizes a unique 3-bp DNA sequence (Kim et al., 2010; Urnov et al., 2010). Initial studies with ZFNs used 3-finger zinc-finger proteins (ZFPs), but recent studies have used up to six fingers per ZFP to increase specificity and target rare cleavage sites (Kim and Kim, 2014; Kim et al., 2010; Sander et al., 2011; Segal, 2011). We used five-finger zinc finger proteins that enabled ZFN dimers to specifically target a 30-bp region in exon 2 and exon 3 of AHR2a (Table 2).
In order to determine the translational efficiency of the injected ZFN mRNA, we measured ZFN protein expression in embryos using an anti-FLAG antibody. Our results demonstrated that maximal ZFN protein expression is observed at 6 hours post-injection, suggesting that ZFN protein binding to the target site and creating DSBs occurs around this time. In comparison, in zebrafish embryos injected with a ZFN targeting the aryl hydrocarbon receptor repressor (ahrra) gene, we observed maximal ZFN protein expression at 3 hours post-injection (Aluru et al., 2013). These species-specific differences in ZFN protein expression could be linked to the developmental rate in the two species. The developmental period for zebrafish from fertilization to hatching is 2 days, whereas killifish hatch at 15 days post-fertilization (Armstrong and Child, 1965). Another factor that could contribute to the differences in protein expression is the water temperature at which embryos are incubated. Zebrafish embryos are reared at a higher temperature (28°C) than killifish (23°C), which may affect the rate of transcription and translation.
We were successful in obtaining mutations in the AHR2a gene with a ZFN targeting exon 3. Sequencing results revealed the presence of 4 different deletions at the target site in 10% of the sequenced clones. These results are comparable to the frequencies observed in other species (McCammon et al., 2011; Ochiai et al., 2010; Young et al., 2011; Zhang et al., 2014). The CRISPR-Cas9 method has been reported to result in higher mutation frequencies in comparison to ZFNs or TALENs (Flowers et al., 2014). Therefore, we used the CRISPR-Cas9 approach to target AHR2a and observed indels in 16% of the sequences from embryos microinjected with a gRNA targeting exon 3. We detected mutations in the CRISPR-Cas9 injected embryos in the very first trial compared to several ZFN trials, even though the proportion of indels showed only a modest increase with the CRISPR-Cas9 method. Similarly, microinjection of gRNAs targeting exon 2 and exon 3 of AHR2b produced indels in 17% and 63% of the sequences. We are currently raising the ZFN-AHR2a, CRISPR-Cas9-AHR2a and -AHR2b fish to reproductive maturity so that we can screen for founders (Supplemental Table 1). To our knowledge, this is the first study comparing the effectiveness of two different targeted genome-editing methods (protein and gRNA based) in a species with limited genomic resources. Additional comparisons with other target loci will be necessary to confirm our observations.
Genome-editing methods have become a powerful resource for generating mutants in non-model species, but there are some concerns with regard to their off-target effects (Gupta et al., 2011; Lin et al., 2014; Porteus, 2006; Porteus and Baltimore, 2003). ZFNs, as well as CRISPR-Cas9, have been associated with cytotoxicity, presumably due to binding and cleavage at non-target sites (Bibikova et al., 2002; Porteus, 2006; Porteus and Baltimore, 2003; Pruett-Miller et al., 2008), although toxic effects could also result from the target gene disruption. Screening the genome for off-target effects a priori by blast searches with the target sequence is not feasible for species that do not have sequenced genomes. However, paralogs and closely related genes that have been characterized can be screened for mutations, as these represent likely targets. We checked for off-target effects of CRISPR-Cas9 in all AHR paralogs, as well as AHRR, a gene closely related to AHR (Fig. 6). Based on the Surveyor assay and sequencing results, no off-target effects were detected, even though the number of sequence mismatches among these genes was as low as 3 residues. If genomic resources are available, several tools exist for in silico identification of potential off-target sites (Fine et al., 2014; Xie et al., 2014). Alternatively, potential off-target sites can be searched using the fuzznuc algorithm (http://embossgui.sourceforge.net/demo/manual/fuzznuc.html) in RNAseq data, if available.
In ZFN and CRISPR-Cas9 injected embryos we did not observe any developmental toxicity, including the highest concentration of ZFN (2000 pg of ZFN mRNA/embryo). These results are in contrast to studies in other fish species, in which overt developmental phenotypes were observed at these concentrations (McCammon and Amacher, 2010; McCammon et al., 2011; Zhang et al., 2014). One reason for the lack of toxicity could be the relative size of the killifish embryos in comparison to zebrafish embryos, in which most of these methods have been optimized. Freshly fertilized killifish embryos are 2 mm in diameter, and are larger than zebrafish (0.7 mm), where ZFNs above 100 pg per embryo have been shown to cause embryo toxicity (McCammon and Amacher, 2010). Similarly, in medaka with an egg diameter (1.2 mm), 160 pg of ZFN caused embryo toxicity (Zhang et al., 2014). In addition to the size of the embryo, rapid turnover of the ZFN protein might be responsible for the lack of embryo toxicity in killifish. Our results suggest that the ZFN protein is expressed transiently around 6 hours post-injection and is degraded by 9 hours (Fig. 3A), suggesting faster degradation of the injected mRNA in killifish. We cannot rule out the possibility of additional factors, yet to be determined, contributing to the low toxicity of injected mRNA in killifish.
Our results demonstrate the utility of the ZFN and CRISPR-Cas9 approaches in generating mutations in an ecologically relevant fish species that have successfully adapted to environments contaminated with PCBs and PAHs. Generating AHR knockout mutants will provide a powerful tool to functionally characterize the role of these proteins in evolved resistance to PCBs and PAHs. We are currently raising the ZFN- and CRISPR-Cas9-injected fish to adults. The numbers of injected animals that are being raised and the breeding strategy that will be followed are provided in the supplemental information (Supp. Table 1 and Supp. Fig. 1). Killifish mature in 1 year; however, larger fish, producing sufficient offspring per pair for testing, require > 2 years of laboratory rearing. Most organisms in which these approaches are extensively used to generate mutants have short life history cycles (e.g. 2–3 months). However, in the past few years targeted mutagenesis approaches have been successfully used in species with longer life cycles such as yellow catfish (Dong et al., 2011), rainbow trout (Yano et al., 2014) and Atlantic salmon (Edvardsen et al., 2014). In order to expedite the process of obtaining homozygous mutants, several strategies can be followed. During certain steps of the homozygous mutant breeding, fin-clips from juvenile fish can be screened by direct-sequencing to identify mutants (Supp. Fig. 1). Direct-sequencing allows screening a single DNA sample per fish, as opposed to cloning the PCR product and screening multiple clones. Another possible approach to accelerate the identification of founders is to screen gametes stripped from juveniles. Once the founder animals are identified, it is possible to artificially induce precocious maturation of gametes by treating the founders with gonadotropins, a common practice in breeding aquaculture species (Chaube et al., 2014; Gwo et al., 1993; Watson et al., 2009).
Other strategies to consider while generating mutants in species with long life history cycles is to generate at least two mutant lines targeting different sites in each gene. As it is time consuming to raise embryos to reproductive maturity, targeting multiple regions of the same gene confirms the specificity of the observed phenotypes associated with the gene knockout. In addition, if a frameshift mutation is not produced in one of the targets, the other target will serve as an alternative. We followed these approaches and designed CRISPR-Cas9 guide RNAs against AHR2a, in addition to the ZFN method. Similarly, we designed two different gRNAs against AHR2b, and observed mutations at both target sites. Another important consideration in target site selection in species such as the Atlantic killifish is to avoid regions with known single nucleotide polymorphisms (SNPs) (Reitzel et al., 2014). Killifish have high genetic diversity (McMillan et al., 2006), and if the target site has multiple mismatches due to SNPs, the efficacy of the ZFN proteins or guide RNAs will be reduced. In addition, SNPs interfere with the mutation detection assays such as Surveyor endonuclease by creating cuts at polymorphic sites. An additional strategy to consider is optimizing injection conditions to maximize the chances for mutating both alleles at the target locus simultaneously. Such bi-allelic mutations in CRISPR-Cas9-injected embryos have been demonstrated previously (Blitz et al., 2013; Edvardsen et al., 2014; Flowers et al., 2014; Jao et al., 2013; Li et al., 2014). The presence of bi-allelic mutations in the germline will expedite the process of generating homozygous individuals.
Overall, targeted mutagenesis approaches have provided new opportunities for conducting functional studies in non-traditional model organisms. As a proof of concept, we and others (Table 1) have demonstrated that it is feasible to generate mutants in non-traditional model species. The relative ease of these methods, as well as the open-source sharing of reagents, have made these methods accessible to a wide range of laboratories.
5. CONCLUSIONS
In this study we demonstrated the utility of targeted gene-editing methods for generating ahr mutants in Atlantic killifish. We observed higher frequency of mutations with CRISPR-Cas9 compared to ZFN mRNA injected embryos. We are currently raising the injected embryos to identify potential germ line mutants. Some of the challenges in generating homozygous mutants in killifish include long generation time and lack of established breeding methods. We have proposed some strategies to overcome these challenges and expedite the process of identifying the founders and generating ahr-null mutants. The strategies proposed here are applicable to any fish species with limited aquaculture resources, providing an opportunity to conduct functional studies.
Supplementary Material
Highlights.
AHR2 genes were mutated without detectable off-target effects in paralogs.
CRISPR-Cas9 method was more efficient in inducing mutations compared to ZFN.
We suggest strategies for generating knockouts in non-traditional fish models.
AHR knockouts will be valuable for understanding evolved resistance to toxicants.
ACKNOWLEDGEMENTS
This work was supported by National Institute of Environmental Health Sciences (NIEHS) grant P42ES007381 (Superfund Research Program at Boston University) and by an Andrew W. Mellon Foundation Award for Innovative Research to MEH. Design of guide RNAs and primers was aided by reference to a preliminary draft of the F. heteroclitus genome sequence, which was supported by funding from the National Science Foundation (collaborative research grants DEB-1120512, DEB-1265282, DEB-1120013, DEB-1120263, DEB-1120333, DEB-1120398). We also acknowledge the help provided by Ms. Kathryn Crawford in fish collection and embryo rearing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aluru N, Karchner SI, Jenny MJ, Franks D, Hahn ME. Understanding the physiological role of aryl hydrocarbon receptor repressor (AHRR) using gene knock-down and targeted mutagenesis in zebrafish. Annual Meeting of Society of Toxicology; San Antonio, Texas. 2013. [Google Scholar]
- Ansai S, Inohaya K, Yoshiura Y, Schartl M, Uemura N, Takahashi R, Kinoshita M. Design, evaluation, and screening methods for efficient targeted mutagenesis with transcription activator-like effector nucleases in medaka. Dev Growth Differ. 2014;56:98–107. doi: 10.1111/dgd.12104. [DOI] [PubMed] [Google Scholar]
- Ansai S, Ochiai H, Kanie Y, Kamei Y, Gou Y, Kitano T, Yamamoto T, Kinoshita M. Targeted disruption of exogenous EGFP gene in medaka using zinc-finger nucleases. Dev Growth Differ. 2012;54:546–556. doi: 10.1111/j.1440-169X.2012.01357.x. [DOI] [PubMed] [Google Scholar]
- Ansai S, Sakuma T, Yamamoto T, Ariga H, Uemura N, Takahashi R, Kinoshita M. Efficient targeted mutagenesis in medaka using custom-designed transcription activatorlike effector nucleases. Genetics. 2013;193:739–749. doi: 10.1534/genetics.112.147645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong PB, Child JS. Stages in Normal Development of Fundulus heteroclitus. Biol Bull. 1965;128:143–168. [Google Scholar]
- Aryan A, Anderson MA, Myles KM, Adelman ZN. Germline excision of transgenes in Aedes aegypti by homing endonucleases. Sci Rep. 2013a;3:1603. doi: 10.1038/srep01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryan A, Anderson MA, Myles KM, Adelman ZN. TALEN-based gene disruption in the dengue vector Aedes aegypti. PLoS One. 2013b;8:e60082. doi: 10.1371/journal.pone.0060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister S, Antonova O, Polo A, Lohs C, Hallay N, Valinciute A, Raible F, Tessmar-Raible K. TALENs mediate efficient and heritable mutation of endogenous genes in the marine annelid Platynereis dumerilii. Genetics. 2014;197:77–89. doi: 10.1534/genetics.113.161091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: In vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicol Sci. 2001;60:77–91. doi: 10.1093/toxsci/60.1.77. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, Cho KW. Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis. 2013;51:827–834. doi: 10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gomez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, Maclatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Rees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL. Fundulus as the premier teleost model in environmental biology: opportunities for new insights using genomics. Comp Biochem Physiol Part D Genomics Proteomics. 2007;2:257–286. doi: 10.1016/j.cbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaube R, Singh RK, Joy KP. Effects of ovaprim, a commercial spawning inducer, on vasotocin and steroid hormone profiles in the catfish Heteropneustes fossilis: in vivo and in vitro studies. Gen Comp Endocrinol. 2014;195:190–200. doi: 10.1016/j.ygcen.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Clark BW, Matson CW, Jung D, Di Giulio RT. AHR2 mediates cardiac teratogenesis of polycyclic aromatic hydrocarbons and PCB-126 in Atlantic killifish (Fundulus heteroclitus) Aquat Toxicol. 2010;99:232–240. doi: 10.1016/j.aquatox.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Ge J, Li K, Xu Z, Liang D, Li J, Jia W, Li Y, Dong X, Cao S, Wang X, Pan J, Zhao Q. Heritable targeted inactivation of myostatin gene in yellow catfish (Pelteobagrus fulvidraco) using engineered zinc finger nucleases. PLoS One. 2011;6:e28897. doi: 10.1371/journal.pone.0028897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardsen RB, Leininger S, Kleppe L, Skaftnesmo KO, Wargelius A. Targeted Mutagenesis in Atlantic Salmon (Salmo salar L.) Using the CRISPR/Cas9 System Induces Complete Knockout Individuals in the F0 Generation. PLoS One. 2014;9:e108622. doi: 10.1371/journal.pone.0108622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elskus AA, Monosson E, McElroy AE, Stegeman JJ, Woltering DS. Altered CYP1A expression in Fundulus heteroclitus adults and larvae: a sign of pollutant resistance? Aquat Toxicol. 1999;45:99–113. [Google Scholar]
- Fine EJ, Cradick TJ, Zhao CL, Lin Y, Bao G. An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res. 2014;42:e42. doi: 10.1093/nar/gkt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers GP, Timberlake AT, McLean KC, Monaghan JR, Crews CM. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development. 2014;141:2165–2171. doi: 10.1242/dev.105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberger D, Perschil I, Ritzler M, Altwegg M. A simple "universal" DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. PCR Methods Appl. 1995;4:368–370. doi: 10.1101/gr.4.6.368. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwo JC, Strawn K, Arnold CR. Induced ovulation in Atlantic croaker (Sciaenidae) using hCG and an LHRH analog: A preliminary study. Theriogenology. 1993;39:353–361. doi: 10.1016/0093-691x(93)90378-i. [DOI] [PubMed] [Google Scholar]
- Hahn ME. Mecahnisms of innate and acquired resistance to dioxin-like compounds. Reviews in Toxicology. 1998;2:395–443. [Google Scholar]
- Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Natl Acad Sci U S A. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Sakamoto K, Sakuma T, Yokotani N, Inoue T, Kawaguchi E, Agata K, Yamamoto T, Takeuchi T. Transcription activator-like effector nucleases efficiently disrupt the target gene in Iberian ribbed newts (Pleurodeles waltl), an experimental model animal for regeneration. Dev Growth Differ. 2014;56:115–121. doi: 10.1111/dgd.12103. [DOI] [PubMed] [Google Scholar]
- Hosoi S, Sakuma T, Sakamoto N, Yamamoto T. Targeted mutagenesis in sea urchin embryos using TALENs. Dev Growth Differ. 2014;56:92–97. doi: 10.1111/dgd.12099. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S, Kroll KL, Amaya E. Generating transgenic frog embryos by restriction enzyme mediated integration (REMI) Methods Mol Biol. 2012;917:185–203. doi: 10.1007/978-1-61779-992-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson ME, Jenny MJ, Woodin BR, Hahn ME, Stegeman JJ. Role of AHR2 in the expression of novel cytochrome P450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3,3',4,4',5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2007;100:180–193. doi: 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Powell WH, Hahn ME. Identification and functional characterization of two highly divergent aryl hydrocarbon receptors (AHR1 and AHR2) in the teleost Fundulus heteroclitus Evidence for a novel subfamily of ligand-binding basic helix loop helix-Per-ARNT-Sim (bHLH-PAS) factors. J Biol Chem. 1999;274:33814–33824. doi: 10.1074/jbc.274.47.33814. [DOI] [PubMed] [Google Scholar]
- Kawai N, Ochiai H, Sakuma T, Yamada L, Sawada H, Yamamoto T, Sasakura Y. Efficient targeted mutagenesis of the chordate Ciona intestinalis genome with zinc-finger nucleases. Dev Growth Differ. 2012;54:535–545. doi: 10.1111/j.1440-169X.2012.01355.x. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kim JS, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods. 2010;7:91. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CH, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci U S A. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yang H, Zhao J, Fang L, Shi H, Li M, Sun Y, Zhang X, Jiang D, Zhou L, Wang D. Efficient and Heritable Gene Targeting in Tilapia by CRISPR/Cas9. Genetics. 2014;197:591–599. doi: 10.1534/genetics.114.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MH, Yang HH, Li MR, Sun YL, Jiang XL, Xie QP, Wang TR, Shi HJ, Sun LN, Zhou LY, Wang DS. Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology. 2013;154:4814–4825. doi: 10.1210/en.2013-1451. [DOI] [PubMed] [Google Scholar]
- Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister AL, Van Der Kraak GJ, Rutherford R, MacLatchy D. Fundulus heteroclitus: ovarian reproductive physiology and the impact of environmental contaminants. Comp Biochem Physiol C Toxicol Pharmacol. 2011;154:278–287. doi: 10.1016/j.cbpc.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Luo D, Lei Y, Hu W, Zhao H, Cheng CH. A highly effective TALEN-mediated approach for targeted gene disruption in Xenopus tropicalis and zebrafish. Methods. 2014;69:58–66. doi: 10.1016/j.ymeth.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, Bian Q, Doudna JA, Meyer BJ. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Zhang S, Wang F, Liu Y, Xu H, Liu C, Lin Y, Zhao P, Xia Q. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One. 2012;7:e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CW, Clark BW, Jenny MJ, Fleming CR, Hahn ME, Di Giulio RT. Development of the morpholino gene knockdown technique in Fundulus heteroclitus: a tool for studying molecular mechanisms in an established environmental model. Aquat Toxicol. 2008;87:289–295. doi: 10.1016/j.aquatox.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon JM, Amacher SL. Using zinc finger nucleases for efficient and heritable gene disruption in zebrafish. Methods Mol Biol. 2010;649:281–298. doi: 10.1007/978-1-60761-753-2_18. [DOI] [PubMed] [Google Scholar]
- McCammon JM, Doyon Y, Amacher SL. Inducing high rates of targeted mutagenesis in zebrafish using zinc finger nucleases (ZFNs) Methods Mol Biol. 2011;770:505–527. doi: 10.1007/978-1-61779-210-6_20. [DOI] [PubMed] [Google Scholar]
- McMillan AM, Bagley MJ, Jackson SA, Nacci DE. Genetic diversity and structure of an estuarine fish (Fundulus heteroclitus) indigenous to sites associated with a highly contaminated urban harbor. Ecotoxicology. 2006;15:539–548. doi: 10.1007/s10646-006-0090-4. [DOI] [PubMed] [Google Scholar]
- Merlin C, Beaver LE, Taylor OR, Wolfe SA, Reppert SM. Efficient targeted mutagenesis in the monarch butterfly using zinc-finger nucleases. Genome Res. 2013;23:159–168. doi: 10.1101/gr.145599.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Nacci DE, Di Giulio RT. Cytochrome P4501A (CYP1A) in killifish (Fundulus heteroclitus): Heritability of altered expression and relationship to survival in contaminated sediments. Toxicol Sci. 2002;68:69–81. doi: 10.1093/toxsci/68.1.69. [DOI] [PubMed] [Google Scholar]
- Nacci D, Coiro L, Champlin D, Jayaraman S, McKinney R, Gleason TR, Munns WR, Specker JL, Cooper KR. Adaptations of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Mar Biol. 1999;134:9–17. [Google Scholar]
- Nacci DE, Champlin D, Jayaraman S. Adaptation of the Estuarine Fish Fundulus heteroclitus (Atlantic Killifish) to Polychlorinated Biphenyls (PCBs) Estuar Coast. 2010;33:853–864. [Google Scholar]
- Nakajima K, Nakajima T, Takase M, Yaoita Y. Generation of albino Xenopus tropicalis using zinc-finger nucleases. Dev Growth Differ. 2012;54:777–784. doi: 10.1111/dgd.12006. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Comparison of TALEN scaffolds in Xenopus tropicalis. Biol Open. 2013;2:1364–1370. doi: 10.1242/bio.20136676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notch EG, Shaw JR, Coutermarsh BA, Dzioba M, Stanton BA. Morpholino gene knockdown in adult Fundulus heteroclitus: role of SGK1 in seawater acclimation. PLoS One. 2011;6:e29462. doi: 10.1371/journal.pone.0029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H, Fujita K, Suzuki K, Nishikawa M, Shibata T, Sakamoto N, Yamamoto T. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells. 2010;15:875–885. doi: 10.1111/j.1365-2443.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- Peng Y, Clark KJ, Campbell JM, Panetta MR, Guo Y, Ekker SC. Making designer mutants in model organisms. Development. 2014;141:4042–4054. doi: 10.1242/dev.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Powell WH, Bright R, Bello SM, Hahn ME. Developmental and tissue-specific expression of AHR1, AHR2, and ARNT2 in dioxin-sensitive and -resistant populations of the marine fish Fundulus heteroclitus. Toxicol Sci. 2000;57:229–239. doi: 10.1093/toxsci/57.2.229. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol Sci. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- Qiu P, Shandilya H, D'Alessio JM, O'Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. Biotechniques. 2004;36:702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Karchner SI, Franks DG, Evans BR, Nacci D, Champlin D, Vieira VM, Hahn ME. Genetic variation at aryl hydrocarbon receptor (AHR) loci in populations of Atlantic killifish (Fundulus heteroclitus) inhabiting polluted and reference habitats. BMC Evol Biol. 2014;14:6. doi: 10.1186/1471-2148-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajwan S, Takasu Y, Tamura T, Uchino K, Sezutsu H, Zurovec M. Efficient disruption of endogenous Bombyx gene by TAL effector nucleases. Insect Biochem Mol Biol. 2013;43:17–23. doi: 10.1016/j.ibmb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte PM. What is environmental stress? Insights from fish living in a variable environment. J Exp Biol. 2014;217:23–34. doi: 10.1242/jeb.089722. [DOI] [PubMed] [Google Scholar]
- Schulte PM, Healy TM, Fangue NA. Thermal performance curves, phenotypic plasticity, and the time scales of temperature exposure. Integr Comp Biol. 2011;51:691–702. doi: 10.1093/icb/icr097. [DOI] [PubMed] [Google Scholar]
- Scott CT. The zinc finger nuclease monopoly. Nat Biotechnol. 2005;23:915–918. doi: 10.1038/nbt0805-915. [DOI] [PubMed] [Google Scholar]
- Scott GR, Brix KV. Evolution of salinity tolerance from transcriptome to physiological system. Mol Ecol. 2013;22:3656–3658. doi: 10.1111/mec.12372. [DOI] [PubMed] [Google Scholar]
- Segal DJ. Zinc-finger nucleases transition to the CoDA. Nat Methods. 2011;8:53–55. doi: 10.1038/nmeth0111-53. [DOI] [PubMed] [Google Scholar]
- Takasu Y, Sajwan S, Daimon T, Osanai-Futahashi M, Uchino K, Sezutsu H, Tamura T, Zurovec M. Efficient TALEN construction for Bombyx mori gene targeting. PLoS One. 2013;8:e73458. doi: 10.1371/journal.pone.0073458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treen N, Yoshida K, Sakuma T, Sasaki H, Kawai N, Yamamoto T, Sasakura Y. Tissue-specific and ubiquitous gene knockouts by TALEN electroporation provide new approaches to investigating gene function in Ciona. Development. 2014;141:481–487. doi: 10.1242/dev.099572. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Van Veld PA, Nacci D. Chemical Tolerance: Acclimation and adaptations to chemical stress. In: Di Giulio RT, Hinton DE, editors. The Toxicology of Fishes. Washington, DC: Taylor and Francis; 2008. pp. 597–644. [Google Scholar]
- Wang F, Ma S, Xu H, Duan J, Wang Y, Ding H, Liu Y, Wang X, Zhao P, Xia Q. High efficiency system for construction and evaluation of customized TALENs for silkworm genome editing. Mol Genet Genomics. 2013;288:683–690. doi: 10.1007/s00438-013-0782-4. [DOI] [PubMed] [Google Scholar]
- Wang T, Hong Y. Direct gene disruption by TALENs in medaka embryos. Gene. 2014;543:28–33. doi: 10.1016/j.gene.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N, Nakamura T, Bando T, Ohuchi H, Yamamoto T, Noji S, Mito T. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun. 2012;3:1017. doi: 10.1038/ncomms2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CA, Hill JE, Graves JS, Wood AL, Kilgore KH. Use of a novel induced spawning technique for the first reported captive spawning of Tetraodon nigroviridis. Mar Genomics. 2009;2:143–146. doi: 10.1016/j.margen.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Whitehead A. The evolutionary radiation of diverse osmotolerant physiologies in killifish (Fundulus sp.) Evolution. 2010;64:2070–2085. doi: 10.1111/j.1558-5646.2010.00957.x. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Roach JL, Zhang S, Galvez F. Salinity- and population-dependent genome regulatory response during osmotic acclimation in the killifish (Fundulus heteroclitus) gill. J Exp Biol. 2012;215:1293–1305. doi: 10.1242/jeb.062075. [DOI] [PubMed] [Google Scholar]
- Wirgin I, Waldman JR. Resistance to contaminants in North American fish populations. Mutat Res. 2004;552:73–100. doi: 10.1016/j.mrfmmm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014;9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A, Nicol B, Jouanno E, Guiguen Y. Heritable targeted inactivation of the rainbow trout (Oncorhynchus mykiss) master sex-determining gene using zinc-finger nucleases. Mar Biotechnol (NY) 2014;16:243–250. doi: 10.1007/s10126-013-9546-8. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Treen N, Hozumi A, Sakuma T, Yamamoto T, Sasakura Y. Germ cell mutations of the ascidian Ciona intestinalis with TALE nucleases. Genesis. 2014;52:431–439. doi: 10.1002/dvg.22770. [DOI] [PubMed] [Google Scholar]
- Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Harland RM, Zeitler B. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad of Sci U S A. 2011;108:7052–7057. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guan G, Chen J, Naruse K, Hong Y. Parameters and efficiency of direct gene disruption by zinc finger nucleases in medaka embryos. Mar Biotechnol (NY) 2014;16:125–134. doi: 10.1007/s10126-013-9556-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.