Abstract
The BCL6 gene, which is expressed in certain B- and T-cell human lymphomas, is involved with chromosomal rearrangements and mutations in a number of these neoplasms. Lymphomagenesis is believed to evolve through a multi-step accumulation of genetic alterations in these tumors. We used retroviral insertional mutagenesis in transgenic mice expressing the human BCL6 transgene in order to identify genes that cooperate with BCL6 during lymphomatous transformation. We previously reported PIM1 as the most frequently recurring cooperating gene in this model. We now report three newly identified cooperating genes—GFI1B, EVI5, and MYB—that we identified in the lymphomas of retroviral-injected BCL6 transgenic mice (but not in retroviral-injected non-transgenic controls); mRNA and protein expression of GFI1B and EVI5 were decreased in the murine tumors, whereas MYB mRNA and protein expression were increased or decreased. These findings correlated with protein expression in human lymphomas, both B- and T-cell. Improved therapy of lymphomas may necessitate the development of combinations of drugs that target the alterations specific to each neoplasm.
Keywords: BCL6 transgenic mice, Cooperating genes, Lymphoma development, GFI1B, EVI5, MYB
Introduction
It has long been known that chromosomal rearrangements and/or mutations of the BCL6 gene are associated with diffuse large B-cell lymphomas (DLBLs) in humans [1]. BCL6 expression also is known to occur in certain T-cell lymphomas [2]. Because multiple oncogenic hits are believed to accumulate during lymphoma development, we used retroviral insertional mutagenesis in mice containing the human BCL6 transgene in an effort to identify mutated genes that cooperate with BCL6 during lymphoma development [3].
We previously reported that retroviral insertional mutagenesis in mice transgenic for human BCL6 leads to enhanced lymphoma development, and we described the gene proviral integration site for Moloney murine leukemia virus 1 (PIM1) as the most frequently recurring gene cooperating with BCL6 to promote lymphoma development in this model [3]. We now report additional recurring retroviral integration sites that we identified in the lymphomas which developed in retroviral-injected BCL6 transgenic mice but not in lymphomas from retroviral-injected non-transgenic controls. The mRNA and proteins encoded by two of the genes involved, growth factor independent 1B (GFI1B) and ecotropic viral integration site 5 (EVI5), each of which we identified in the lymphomas from three transgenic animals (three B-cell and three T-cell, respectively), showed decreased expression in the lymphomas from transgenic (BCL6-positive) mice as compared with the lymphomas from non-transgenic retroviral injected controls (BCL6-negative) as well as in BCL6-positive (as compared with BCL6-negative) human lymphomas. The mRNA and protein encoded by a third involved gene, myeloblastosis oncogene (MYB), which we also identified in the lymphomas from three transgenic animals (two B-cell, one T-cell) but not in controls, was variably increased or decreased. Although these three genes have been associated with lymphoma development [4–6], they have not been reported previously to cooperate with BCL6 in lymphomagenesis.
Materials and methods
BCL6 transgenic mice and retroviral insertional mutagenesis
Our transgenic mice (described previously) [7], which express BCL6 constitutively in B- and T-lymphocytes, contain two transgenes: tet-o-BCL6 (human BCL6 cDNA under control of the tetracycline-responsive minimal promoter), and EμSR-tTA (the tetracycline- transactivating protein under control of the Ig heavy-chain enhancer and the SRα promoter). One of our transgenic lines contains four copies of the human BCL6 transgene, the other 20 copies. Under approved institutional protocols, neonatal animals from both transgenic lines and controls (non-transgenic mice of the same background, wild-type or positive for either the EμSR-tTA or tet-o-BCL6 transgenes, but not both) were injected with 105 infectious units of the retrovirus MOL4070LTR [3] intraperitoneally and followed until the need for euthanasia or natural death.
Histology, flow cytometry, inverse PCR, database searches, and real-time quantitative PCR
Processing of tissues, FACS analysis, inverse PCR, cloning, sequencing, database searches, total RNA extraction, and real-time RT-PCR methodologies have been described previously [3]. The classification of murine lymphoid neoplasms was according to the Bethesda proposals [8]. Primers included β-actin as described [3] or glyceraldehyde-3-phosphate dehydrogenase (Real Time Primers, Elkins Park, PA) and primers common to all transcript variants of the murine GFI1B, EVI5, or MYB genes (GFI1B, forward: 5′-CTCTCCAGGCATGGACACTT-3′; reverse: 5′-GACGTGAGTATGCTGCTCCA-3′; EVI5, forward: 5′-CCCATCAAAGTTG AGTCCAG-3′; reverse: 5′-TTCTTCCCCAGAGAATCCAA-3′; MYB, Real Time Primers, forward: 5′-CTGGACAGAAGAGGAGGACA-3′; reverse: 5′-TTGTT CTTCTGGAAGCTCGT-3′).
Immunohistochemistry
Human lymphoma blocks were retrieved from the surgical pathology archives under an Institutional Review-Board approved protocol. Human and murine tissues were stained with anti-BCL6 as described previously [3]. For additional immunohistochemistry, antigen retrieval was performed in pH 6 solution (Epitomics, Burlingame, CA) in a near boiling waterbath for 40 min. Anti-GFI1B (#HPA007012, for human tissues) and anti-EVI5 (#HPA027339, for human and murine tissues) were affinity-isolated Prestige antibodies produced in rabbit (Sigma-Aldrich, Saint Louis, MO). Anti-GFI1B was diluted 1:20 and anti-EVI5 1:35 for incubation overnight at 4 °C. Murine tissues were stained with GFI1B affinity isolated antibody produced in rabbit (Sigma Aldrich, #AV30093), 20 μg/ml, overnight at 4 °C. For immunohistochemistry with MYB antibody, human tissues were incubated overnight at 4 °C with a 1:20 dilution of a rabbit monoclonal antibody (Epitomics, Burlingame, CA, cat. # 3195-1). Murine tissues were incubated with the same MYB antibody diluted 1:1000 for 1 h at room temperature. For all antibodies, antigen-antibody binding was detected with DAB chromogen, and tissues were counterstained with hematoxylin. Images were taken with a BX41 microscope (Olympus), a DP72 digital camera, and cellSens Standard imaging software (Olympus).
Statistical analysis
The data from real-time PCR were analyzed by the comparative CT method [9]. Data from the study and control groups were compared by the Wilcoxon rank-sum test. Statistical analyses were performed with Stata software (version 12: StataCorp, College Station, TX).
Results
Lymphoma development in MOL4070LTR-infected mice
As previously reported, 92 of 99 transgenic and 32 of 33 control mice infected with retrovirus were analyzable [3]. Lymphomas developed in 24 of 53 animals (45.3%) in the four copy transgenic line (18 T-cell, six B-cell) and in 10 of 39 (25.6%) mice from the 20 copy line (three T-cell, seven B-cell). Additionally, as MOL4070LTR is known to cause lymphomas and leukemias in mice [10], lymphoid neoplasms also were expected in the control group; 11 of 32 retroviral-injected controls (34.4%) developed lymphomas (5 T-cell, 6 B-cell). The nature of the lymphomas (both B- and T-cell), mouse survival, and murine lymphoma-related mortality rates have been discussed previously [3]. Briefly, the lymphomas usually were aggressive tumors; most of the B-cell lymphomas were mature B-cell, and most T-cell tumors were precursor T-cell lymphoblastic lymphoma/leukemias. The lymphomas from the transgenic mice contained three to 29 insertion sites (average 11), and the lymphomas from the controls contained eight to 20 insertion sites (average 12.7) [3]. Only one of the animals studied here (with an insertion site 5′ to the GFI1B gene) also had an insertion site 3′ to PIM1.
RNA expression in lymphomas containing retroviral insertions in or near the GFI1B, EVI5, and MYB genes
As previously described, PIM1 was the gene most commonly affected by viral insertion sites in this study [3]. Here we have chosen to study the genes that were the next most commonly affected by viral insertions (after PIM1). The types of lymphomas, insert locations, and relative expression levels following real-time quantitative RT-PCR performed on RNA derived from the lymphomas of transgenic mice as compared with RNA from lymphomas of the same type (B or T) in the same organ from multiple randomly selected non-transgenic controls are shown in Table 1.
Table 1.
Location of retroviral inserts in transgenic mice and relative RNA expression levels in lymphomas from BCL6 transgenic mice as compared with lymphomas from retroviral-injected non-transgenic (control) mice.
| Mouse gene (chromosome) | Lymphoma type | Location of insert | Expression level vs. control |
|---|---|---|---|
| GFI1B (Chr 2) | B | 8.692 kb 5′ to GFI1B | 5.7-fold decreased |
| GFI1B | B | 8.686 kb 5′ to GFI1B | 2.6-fold decreased |
| GFI1B | B | 7.877 kb 5′ to GFI1B | 1.8-fold decreased |
| EVI5 (Chr 5) | T | Terminal intron* | 2.0-fold decreased |
| EVI5 | T | 3′ untranslated region** | 1.5-fold decreased |
| EVI5 | T | 3′ untranslated region*** | 1.3-fold decreased |
| MYB (Chr 10) | T | 34.794 kb 3′ to MYB | 2.2-fold increased |
| MYB | B | 73.684 kb 3′ to MYB | 2.0-fold increased |
| MYB | B | 70.687 kb 3′ to MYB | 3.2-fold decreased |
Nucleotide position 108,201,513.
Nucleotide position 108,183,744.
Nucleotide position 108,182,569.
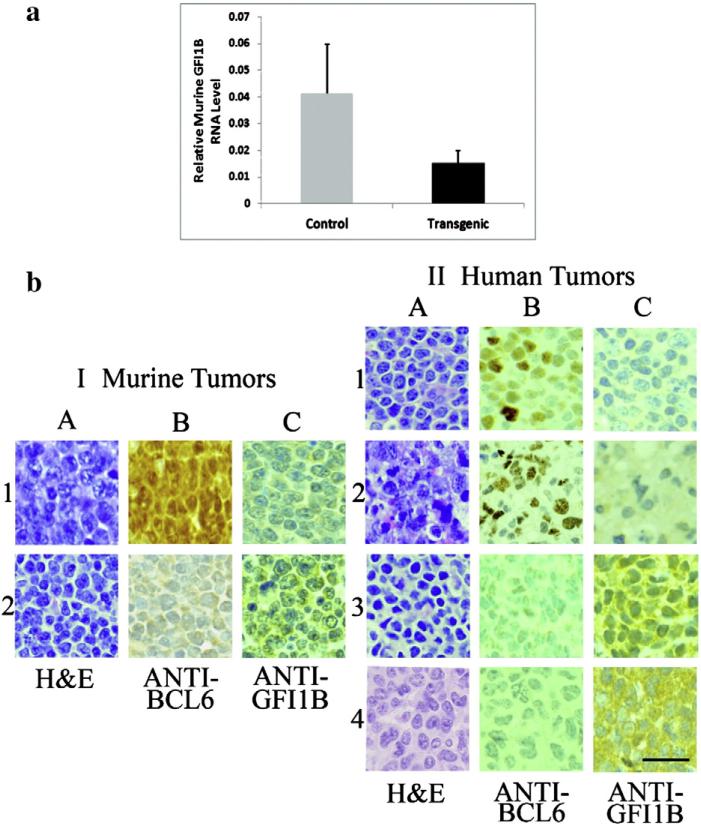
The insertion sites near GFI1B were all within 7.9–8.7 kb 5′ of the GFI1B gene, and these murine tumors were all large B-cell lymphomas. The transgenic animals were females, ranging in age from 4.2 mos to 7.3 mos (average, 6.2 mos), and included both BCL6 transgenic lines. In each case, relative GFI1B RNA expression level in the lymphomas from transgenic mice was decreased as compared with that in randomly selected B-cell lymphomas from retroviral-injected non-transgenic controls (none of which had inserts in or near GFI1B), see Table 1 (overall decrease, 2.71-fold; mean ± SEM = 0.041 ± 0.018 in the controls as compared with 0.015 ± 0.005 in the transgenics; P < 0.05, Fig. 1a).
Fig. 1.
GFI1B: relative RNA levels in lymphomas from BCL6 transgenic and control mice and immunohistochemistry of BCL6-positive and -negative murine and human lymphomas. (a) The graph depicts average relative GFI1B RNA expression from the lymphomas (all large B-cell) of three retroviral-injected BCL6 transgenic mice (black bar) containing inserts 7.9 to 8.7 kb 5′ to the GFI1B gene as compared with three randomly selected B-cell lymphomas from retroviral-injected non-transgenic controls that did not contain inserts in or near GFI1B (gray bar). Expression in each transgenic animal was decreased as compared with the controls [range, 1.8 to 5.7-fold (Table 1), overall decrease 2.71-fold; mean ± SEM = 0.041 ± 0.018 in the controls as compared with 0.015 ± 0.005 in the transgenics; P < 0.05]. (b) I, Representative murine lymphomas from (1) a transgenic mouse (nuclei positive for BCL6, column B) and (2) a non-transgenic animal (nuclei negative for BCL6). The cytoplasmic staining, evident in the anti-GFI1B (C) column, is decreased in the transgenic mouse as compared with the non-transgenic control. II, Representative human lymphomas: (1) BCL6-positive (B-cell), (2) BCL6-positive (T-cell), (3) BCL6-negative (B-cell), (4) BCL6-negative (T-cell). As in the murine lymphomas, the GFI1B cytoplasmic staining (column C) is decreased in the BCL6-positive lymphomas as compared with the stronger expression in the BCL6-negative neoplasms. The bar (lowest right panel) indicates 50 μm.
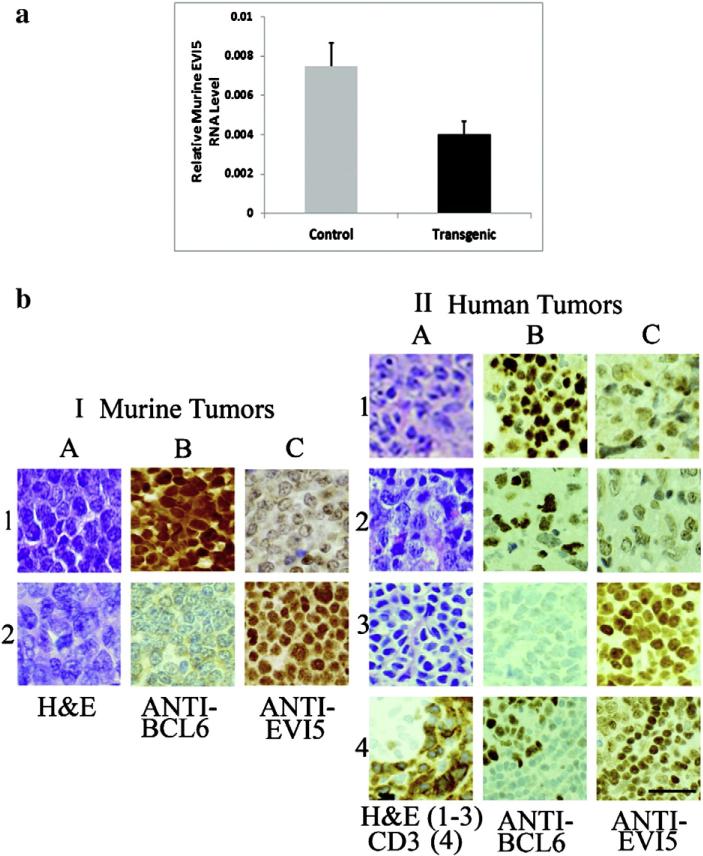
In contrast, the insertion sites involving EVI5 all occurred in T-cell tumors (precursor T-cell lymphoblastic lymphoma/leukemias) from the four copy BCL6 transgenic line. One of these insertions was in the terminal intron, and two were in the 3′ untranslated region of the EVI5 gene. One mouse was female, two were males, and these animals ranged in age from 3.8 mos to 5.9 mos (average, 4.6 mos). Relative EVI5 RNA expression levels were decreased in the T-cell lymphomas from the transgenic mice as compared with T-cell tumors from randomly selected controls, none of which had inserts in or near EVI5 (overall decrease, 1.86-fold; mean ± SEM = 0.007 ± 0.001 in the controls as compared with 0.004 ± 0.0007 in the transgenics; P < 0.05, Fig. 2a).
Fig. 2.
EVI5: relative RNA levels in lymphomas from BCL6 transgenic and control mice and immunohistochemistry of BCL6-positive and -negative murine and human lymphomas. (a) The graph depicts average relative EVI5 RNA expression from the lymphomas (all T-cell) of three retroviral-injected BCL6 transgenic mice (black bar) containing inserts within the terminal intron or 3′-untranslated region of the EVI5 gene as compared with three randomly selected T-cell lymphomas from retroviral-injected non-transgenic controls that did not contain inserts in or near EVI5 (gray bar). Expression in each transgenic animal was decreased as compared with the controls [range, 1.3 to 2-fold (Table 1), overall decrease, 1.86-fold; mean ± SEM = 0.007 ± 0.001 in the controls as compared with 0.004 ± 0.0007 in the transgenics; P < 0.05]. (b) I, Representative murine lymphomas from (1) a transgenic mouse (nuclei positive for BCL6, column B) and (2) a non-transgenic mouse (nuclei negative for BCL6). The nuclear staining evident in the anti-BCL6 (C) column is decreased in the transgenic mouse as compared with the non-transgenic control. II, Representative human lymphomas: (1) BCL6-positive B-cell, (2) BCL6-positive (T-cell), (3) BCL-negative (B-cell), (4) BCL-negative (T-cell); the cytoplasm of the tumor cells stains strongly positive with the T-cell marker CD3 (II, 4A). A germinal center in 4A (upper left corner) does not stain with CD3 and is BCL6-positive (column B), whereas the tumor cells around it are BCL-negative. As in the murine lymphomas, the EVI5 nuclear staining (column C) is decreased in BCL6-positive cells (tumors 1 and 2, or germinal center cells, tumor 4) as compared with the stronger expression in the BCL-negative tumor cells [tumors 3 and 4 (outside the germinal center)]. The length of the bar in the lowest right panel depicts 50 μm.
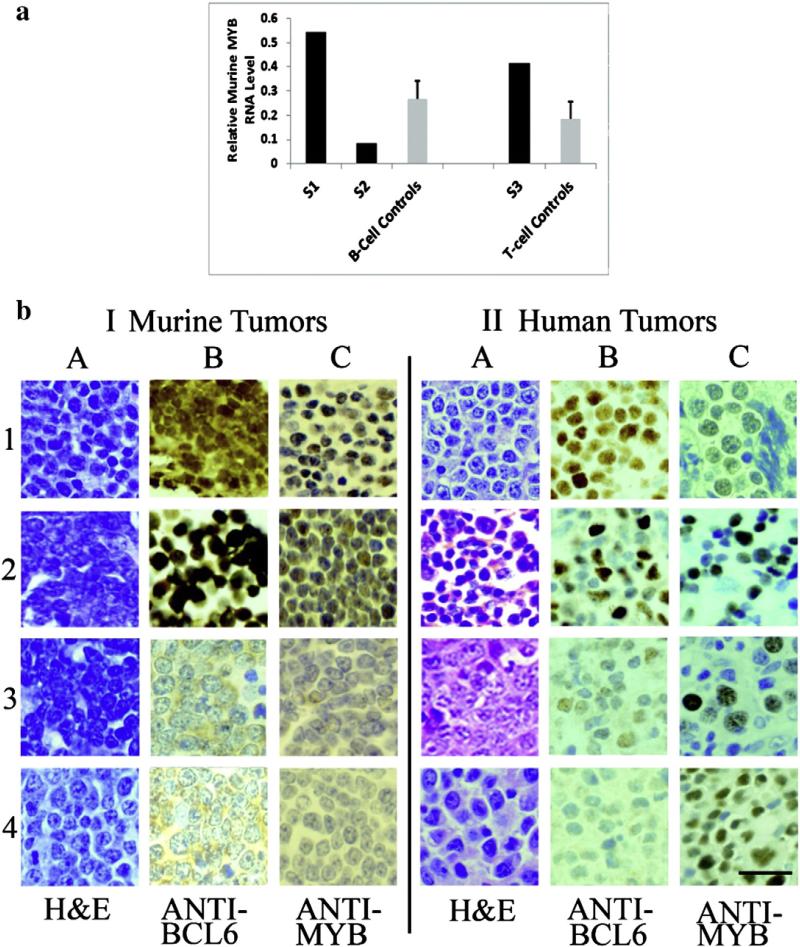
The retroviral inserts near MYB were all 3′ (34.8 to 73.7 kb) and occurred in three females ranging in age from 4.7 mos to 7.8 mos (average, 6.6 mos). Two were large B-cell lymphomas from the 20 copy transgenic line and one was a precursor T-cell lymphoblastic lymphoma/leukemia from the four copy transgenic line. Relative RNA expression in one of the B-cell tumors and in the T-cell lymphoma was increased ≥2-fold as compared with that in the three randomly selected control B-cell lymphomas and two randomly selected control precursor T-cell lymphoblastic lymphoma/leukemias, respectively (which did not contain inserts in or near the MYB gene), but relative RNA expression in the other B-cell lymphoma was 3.2-fold decreased as compared with that in the lymphomas from the three randomly selected B-cell controls. These data are depicted in Fig. 3a.
Fig. 3.
MYB: relative RNA levels in lymphomas from BCL6 transgenic and control mice and immunohistochemistry of BCL6-positive and -negative murine and human lymphomas. (a) The graph depicts average relative MYB RNA expression in the lymphomas of three retroviral-injected BCL6 transgenic mice (black bars, study mice S1, S2, S3) containing inserts ~35 to ~74 kb 3′ to the MYB gene as compared with randomly selected B- or T-cell lymphomas (gray bars) from retroviral-injected control mice that did not contain inserts in or near MYB. S1 is a large B-cell lymphoma from a transgenic mouse whose relative expression is 2-fold higher than the mean of three randomly selected B-cell control tumors (S1 expression = 0.54 vs. control B-cell tumors, mean ± SEM = 0.27 ± 0.07), whereas the relative RNA expression of S2 (0.08), also a large B-cell lymphoma from a transgenic animal, is 3.2-fold decreased as compared with the mean of the three randomly selected control B-cell lymphomas (see above). S3 is a precursor T-cell lymphoblastic lymphoma/ leukemia from a transgenic mouse whose relative RNA expression is 2.2-fold higher than the mean of two randomly selected precursor T-cell lymphoblastic lymphoma/leukemia control tumors; S3 expression = 0.42 vs. control T-cell tumors, mean ± SEM = 0.19 ± 0.07. (b) I, Representative murine lymphomas: tumors (1) and (2) depict the B- and T-cell lymphomas, respectively, from transgenic mice (BCL6-positive nuclear staining, column B) with positive MYB nuclear staining (column C); tumors (3) and (4) show B- and T-cell lymphomas, respectively, from non-transgenic mice (nuclei do not stain with anti-BCL6, column B); staining with anti-MYB is also negative in these animals (column C). (b) II, Representative human lymphomas: tumors (1) and (2) show BCL6-positive (column B) B- and T-cell lymphomas, respectively, which show positive nuclear staining for MYB (column C). Tumor (3) is a T-cell lymphoma that is weakly BCL-6 positive (column B), and tumor (4) is a BCL-negative T-cell neoplasm; in tumors 3 and 4, the cells that are MYB positive (column C) are BCL-negative. The bar (lowest right panel) depicts 50 μm.
GFI1B, EVI5, and MYB protein levels in lymphomas from retroviral-injected BCL6 transgenic mice and controls, and in human BCL6-positive and negative lymphomas
Protein levels in murine lymphomas detected by immunohisto-chemistry correlated with RNA levels from murine tumors quantified by real-time PCR as described above, and human lymphomas had a similar staining pattern as the murine counterparts (Figs. 1b, 2b, 3b). Whereas GFI1B expression was detected in the cytoplasm, EVI5 and MYB had a nuclear localization.
Tissue sections from the lymphomas of the three transgenic mice containing retroviral inserts near GFI1B (all large B-cell) and from B-cell lymphomas of three randomly selected retroviral-injected nontransgenic controls (no control tumors contained inserts near GFI1B) were studied with anti-GFI1B. Representative sections, shown in Fig. 1b, panel I, indicate that GFI1B staining is decreased in transgenic tumors (nuclear staining is positive for BCL6) as compared with the tumors in non-transgenic retroviral-injected controls (nuclear staining is negative for BCL6). Additionally, sixteen human lymphomas were studied by immunohistochemistry for GFI1B. Nine were B-cell tumors, seven were T-cell neoplasms; eight were BCL6-positive (six B-cell, two T-cell), and eight were BCL6-negative (three B cell, five T-cell). GFI1B expression was decreased in the BCL6-positive lymphomas as compared with the BCL6-negative tumors, which showed strong cytoplasmic expression of GFI1B (Fig. 1b, panel II). Thus, the findings in human lymphomas paralleled those in mice. Additionally, whereas in mice inserts near GFI1B were noted only in BCL6-positive B-cell tumors (which demonstrated decreased expression of GFI1B RNA and protein levels as compared with BCL-negative murine lymphomas), in humans, BCL6-positive T-cell as well as B-cell neoplasms demonstrated decreased expression of GFI1B protein as compared with BCL-negative lymphomas of each cell type (Fig. 1b, panel II).
Representative immunohistochemical staining of the murine lymphomas containing retroviral inserts in the EVI5 gene (all precursor T-cell lymphoblastic lymphoma/leukemias) is shown in Fig. 2b, panel I and reveals that EVI5 staining (nuclear) is decreased in the BCL6-positive T-cell neoplasms as compared with the non-transgenic retroviral-injected BCL-negative T-cell controls, none of which contained inserts in or near EVI5. Twenty-seven human lymphomas were studied with anti-BCL6. Eight were negative for BCL6 (3 B-cell, 5 T-cell), 17 were BCL6-positive (13 B, 4 T), and two (T-cell) stained weakly with anti-BCL6. Representative sections (Fig. 2b, panel II) indicate that BCL6-positive lymphomas contain less EVI5 than do BCL-negative neoplasms. As in the case of GFI1B, findings for EVI5 in human lymphomas paralleled the findings in murine lymphomas, and, additionally, whereas in the mouse tumors inserts in the EVI5 gene were noted only in BCL6-positive T-cell tumors (which had decreased expression of EVI5 RNA and protein as compared with BCL-negative T-cell lymphomas), in human lymphomas, decreased levels of EVI5 protein were present in BCL6-positive B- as well as T-cell tumors as compared with BCL-negative controls (Fig. 2b, panel II). Non-neoplastic BCL6-positive germinal center cells also contained less EVI5 than the surrounding BCL-negative tumor cells (Fig. 2b, panel II, 4C).
Representative immunohistochemistry of MYB staining (Fig. 3b, I) of two murine lymphomas from transgenic mice (a B-cell tumor, panel 1 and a T-cell tumor, panel 2), reveals that these BCL6-positive neoplasms also show nuclear staining for MYB, whereas two murine lymphomas from non-transgenic animals (B-cell, panel 3, and T-cell, panel 4) are BCL-negative (column B) as well as MYB-negative (column C). A third transgenic mouse with a large B-cell lymphoma (not shown) showed little to no staining for MYB. Murine lymphomas stained readily with anti-MYB, which was used in a dilution of 1:1000 for 1 h at room temperature. Human lymphomas were more difficult to stain, requiring a 1:20 dilution and overnight incubation, and even then, positive staining was often focal and noted in only six (24%) of the 25 lymphomas studied. Of the 25 human lymphomas (12 B-cell, 13 T-cell) studied, 17 were BCL6-positive (10 B-cell, seven T-cell), five were BCL-negative (one B-cell, four T-cell), and three stained weakly for BCL6 (one B-cell, two T-cell). In four BCL6-positive lymphomas (two B-cell, two T-cell), the MYB-positive cells were also BCL6-positive (Fig. 3b, II, panels 1 and 2); however, in two T-cell lymphomas, one BCL6-positive, but with variably weak BCL6 staining, and one BCL-negative (Fig. 3b, panels 3 and 4, respectively), the MYB-positive cells were BCL-negative. Our observations in human lymphomas paralleled those in murine tumors in the respect that MYB levels could be increased or decreased in BCL6-positive neoplasms, T- as well as B-cell, and in human tumors, MYB positivity could be demonstrated in BCL6-positive as well as BCL-negative neoplastic cells (Fig. 3b, panel II).
Recurring sites in BCL6-positive transgenic mice not found in nontransgenic controls
Table 2 lists the genes nearest to recurring sites found in at least two transgenic animals but not in non-transgenic controls. Additional genes that may cooperate with BCL6 occurred in multiple retroviral-injected transgenic mice and also in at least one retroviral-injected non-transgenic (control) animal (e.g., MYC), and thus may have transforming properties on their own. These are not reported here.
Table 2.
Recurring retroviral integration sites in BCL6 transgenic mice not found in non-transgenic controls.*
| Murine genes containing inserts or, if insert not within a gene, the nearest characterized gene (unless indicated) | Mouse chromosome | Number of mice | Lymphoma type |
|---|---|---|---|
| PIM1 (proviral integration site for Moloney murine leukemia virus 1) | 17 | 7 | 6T, 1B |
| GFI1B (growth factor independent 1B) | 2 | 3 | B |
| EVI5 (ecotropic viral integration site 5) | 5 | 3 | T |
| MYB (transcriptional activator MYB isoform 1) | 10 | 3 | 2B, 1T |
| LMO2 (LIM domain only 2) | 2 | 2 | T |
| MN1 (probable tumor suppressor protein MN1) | 5 | 2 | T |
| AHI1 (jouberin isoform 1; Abelson helper integration site-1) | 10 | 2 | T |
| BCOR (BCL6 corepressor isoform d) | 10 | 2 | B |
| IKZF1 (DNA-binding protein Ikaros isoform a and b) | 11 | 2 | T |
| SYKb (tyrosine protein kinase SYK) | 13 | 2** | T |
| MPPE (metallophosphoesterase 1); IMPA2 (inositol monophosphatase 2) | 18 | 2*** | B |
| DNTT (DNA nucleotidylexotransferase) | 19 | 2 | T |
| GATA1 (erythroid transcription factor) | X | 2 | B |
Sequences have ≥98% identity to the murine databases described in the text.
In one lymphoma, the insert is within SYKb; in the other, SYKb is 61.3 kb 3′ to the insert, but there is a gene closer to the insert at the 5′ side [40.3 kb away, D/RAS2 (GTP-binding protein DI-RAS2)].
MPPE (5′) is closer to the insert in one lymphoma (17.8 kb vs. 50.2 kb), but IMPA2 (3′) is closer to the insert in the other lymphoma (1 kb vs. 33.8 kb).
We previously reported that the most frequent retroviral insertions in our BCL6 transgenic mice occurred in or near the PIM1 gene, and we showed overall higher levels of PIM1 RNA and protein (by immunohistochemistry) in lymphomas (B- or T-cell) containing these insertion sites [3]. Here we have studied the genes involved by the next most common insertion sites (in or near GFI1B, EVI5, and MYB, which were observed in lymphomas from three transgenic mice in each case).
Discussion
Although the BCL6 gene is best known for its association with DLBLs in humans (~40% of these neoplasms are associated with chromosomal rearrangements involving BCL6 and ~16% contain mutations disrupting autoregulation of the BCL6 gene) [1], the role of BCL6 in T-cell development and function has gained increased attention [11,12]. Through the use of retroviral insertional mutagenesis in BCL6 transgenic mice, we have identified three genes that have not been reported previously as cooperating with BCL6 to promote lymphomagenesis. All of these have been recognized as proto-oncogenes [13–15]. Two (GFI1B and MYB) are transcription factors [16,17] and the third (EVI5) is considered an essential regulator of cell membrane trafficking [18,19]. Additionally, immunohistochemistry performed on B- and T-cell human lymphomas (16 in the case of GFI1B, 27 for EVI5, 25 for MYB), both BCL6-positive and negative, confirmed our observations in murine lymphomas at the RNA and protein level: GFI1B and EVI5 are downregulated in BCL6-positive lymphomas as compared with BCL-negative tumors, and MYB can be either up- or down-regulated in BCL6-positive lymphomas as compared with BCL-negative controls.
The GFI1B gene, located on human chromosome 9q34.13, is not a common target in retroviral insertional mutagenesis [20]. It encodes a transcriptional repressor with an N-terminal SNAG (Snail/Gfi1) domain and six C-terminal zinc fingers that are believed to have an important role in hematopoiesis. Of two transcripts in humans, the most frequent is a 330 amino acid protein which binds DNA and suppresses gene expression through recruitment of histone modifying enzymes at target promoters [16]. It is expressed in myeloid progenitor cells as well as in B- and T-cell subsets [21], is known to control expression levels of genes critical for B-cell development [22], and is essential for erythroid and megakaryocytic development [21]. It is not expressed in mature thymocytes [23].
The EVI5 gene, located on human chromosome 1p22, is a common viral integration site in T-cell lymphomas derived from AKXD mice [5]. It encodes a coiled-coil protein [5] that is considered essential for regulation of membrane trafficking [18,19] and has been believed to be involved in T-cell disease [5]. As we noted in our murine lymphomas, a common viral integration site is in a 3′ intron, and integrations are opposite to the transcriptional orientation of the gene [5]. Although real-time RT-PCR revealed decreased RNA expression in all of the lymphomas from the transgenic mice as compared with the nontransgenic controls, the differences were not large; however, it is known that even small perturbations in the expression level of a protein may lead to significant biological effects [24,25]. Alternatively, as it is known that gene expression can be affected over hundreds of kilobases by retroviral integrations [26], it is possible that viral insertions in the EVI5 gene could affect another gene, e.g., growth factor independent 1 (GFI1), which is located 18 kb downstream from EVI5 and is known to be a transcription factor that is crucial for normal hematopoietic development. Thus, Schmidt et al. [27], who could not detect a signal with an EVI5 probe on Northern blots prepared from total RNA of lymphomas bearing proviral EVI integrations in MYC/PIM bitransgenic mice, found that the tumors had enhanced GFI1 RNA expression. However, these authors indicate that other targets which are located several hundred kilobases apart also could be activated by EVI5 integrations.
MYB, a site of recurrent retroviral insertional mutagenesis in a number of murine hematopoietic malignancies [28], is a leucine zipper DNA-binding transcription factor [29] with a short half-life that undergoes post-translational modifications, including ubiquitylation, phosphorylation, acetylation, and sumoylation [30]. Human MYB, located on chromosome 6q23.3 [17], contains 15 exons and encodes a family of related proteins. Alternative splicing leads to splice variants that are predicted to encode proteins with differing transcriptional activities and specificity domains [31]. The MYB protein is expressed in all proliferating hematopoietic cells, is involved in the regulation of proliferation and differentiation of bone marrow progenitor cells (also of colon and adult brain) [30,32], and is required for normal hematopoiesis, T-cell development, pro-B to pre-B transition, and survival of spleen B cells [30,33]. In humans, expression of MYB is known to be relevant for the lymph node germinal center phenotype, and that expression is sustained by BCL6 repression of microRNA (miR)-155 [34]. Although, traditionally, MYB has been touted as a transcriptional activator, several corepressor molecules that interact through various MYB domains imply that MYB could act also as a repressor [28]. In one study, about half of the genes found to be regulated by MYB were repressed [35]. Its target genes have functions in cell cycle progression, cell differentiation, and survival [30,32]. The gene is known to be translocated in T-cell acute lymphocytic leukemia, and its protein is overexpressed in that disease, in part through gene duplication [29].
In addition to the three genes described above, additional genes of potential interest are listed in Table 2. Almost all of these are known to be involved in hematopoiesis or hematologic malignancies, lending validity to the notion that they may well cooperate with BCL6 in lymphomagenesis. For example, LMO2 is up-regulated in non-Hodgkin lymphomas [36] and has been noted to be translocated in certain human T-cell acute lymphoblastic leukemias [37]; MN1 is deregulated in human acute myeloid leukemia [38]; AHI1 is an oncoprotein that interacts with BCR-ABL and Janus kinase 2 (JAK2) in chronic myelogenous leukemia cells [39] and is expressed in cutaneous T-cell lymphomas with intermediate to poor prognosis [40]; BCOR is part of a BCL6 repression complex that facilitates survival and proliferation of lymphomas [41]; IKZF1 encodes the IKAROS transcription factor, which drives lymphoid development [42], and IKZF1 deletions have been noted in childhood B-cell precursor acute lymphoblastic leukemia [43]; SYK was identified as a target in acute myeloid leukemia [44] and, when involved in translocations with the interleukin 2-inducible T cell kinase (ITK) gene, it induces a T-cell lymphoproliferative disease in mice mimicking human disease [45]; DNTT encodes a lymphoid regulator [46]; and, finally, GATA1 encodes an erythroid transcription factor [47].
BCL6 is believed to exert its function by repressing hundreds of proteins [34]. B- as well as T-cell lymphomas are aggressive hemato-logic neoplasms resulting from the malignant transformation of B- or T-cell progenitors, respectively. These transformation processes are believed to be multi-step events in which a series of heterogeneous genetic alterations cooperate to induce perturbations in normal lymphocyte growth and differentiation. DLBLs [48] and T-cell acute lymphoblastic leukemias [29], for example, have been reported to harbor multiple genetic abnormalities (in the case of DLBLs, more than 30 clon-ally represented gene alterations per neoplasm) [48] which differ from tumor to tumor, making approaches to therapeutic intervention diffi-cult. Nonetheless, therapies directed against MYB [30] and the proteins encoded by a number of other genes known to be involved in lymphomatous transformation are currently under investigation. Our studies have added GFI1B, EVI5, and MYB to the list of genes cooperating with BCL6 during lymphomagenesis, thus expanding the arsenal of potential therapeutic targets for DLBL. The finding in this study of several down-regulated cooperating genes suggests that efforts to upregulate them, e.g., by target mRNA manipulation [49], may have some therapeutic benefit. Identification of the alterations in individual lymphomas, with selection of the appropriate combinations of agents targeting the specific lesions, may hold promise for therapy of these neoplasms.
Acknowledgments
We thank Dr. B. Gladstone for technical assistance, Dr. I. Aifantis for antibodies for flow cytometry, D. Lane for expertise in immunohisto-chemistry, and R. Duggan for assistance with FACS. This work was supported by the Department of Pathology at The University of Chicago (to BWB), University of Chicago Cancer Center Support Grant P30 CA014599 (to BWB), and Hematology Research Funds at The University of Chicago donated by S. Samsky and E. Lanzl (to JMB).
Footnotes
Authorship
Contribution: B.W.B, L.J., and M.J.T. designed research; B.W.B., P.L.R., and J.M.B. performed research; J.B. and L.W. contributed new reagents/ analytic tools; B.W.B., J.A., M.J.T., K.W., and J.M.B. analyzed data; and B.W.B. and J.M.B. wrote the paper.
All authors have approved the final article.
Conflict-of-interest disclosure
All authors declare no conflict of interests.
Declaration
The work has not been published previously and is not under consideration for publication elsewhere. The work is approved by all authors. The work, if accepted, will not be published elsewhere, including electronically in the same form, in English or in any other language, without the written consent of the copyright-holder.
References
- 1.Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 2.Hyjek E, Chadburn A, Liu YF, Cesarman E, Knowles DM. BCL6-protein is expressed in precursor T-cell lymphoblastic lymphoma and in prenatal and post-natal thymus. Blood. 2001;97:270–276. doi: 10.1182/blood.v97.1.270. [DOI] [PubMed] [Google Scholar]
- 3.Baron BW, Anastasi J, Hyjek EM, Bies J, Reddy PL, Dong J, et al. PIM1 gene cooperates with human BCL6 gene to promote the development of lymphomas. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5735–5739. doi: 10.1073/pnas.1201168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu W, Kee BL. Growth factor independent 1B (Gfi1b) is an E2A target gene that modulates Gata3 in T-cell lymphomas. Blood. 2007;109:4406–4414. doi: 10.1182/blood-2006-08-043331. [DOI] [PubMed] [Google Scholar]
- 5.Liao X, Du Y, Morse HC, III, Jenkins NA, Copeland NG. Proviral integrations at the Evi5 locus disrupt a novel 90 kDa protein with homology to the Tre2 oncogene and cell-cycle regulatory proteins. Oncogene. 1997;14:1023–1029. doi: 10.1038/sj.onc.1200929. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Wang Z, Dai X, Pan J, Ge J, Han JX, et al. Re-expression of miR-150 induces EBV-positive Burkitt lymphoma differentiation by modulating c-Myb in vitro. Cancer Sci. 2013 Mar 25; doi: 10.1111/cas.12156. (Epub ahead of print, in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baron BW, Anastasi J, Montag A, Huo D, Baron RM, Karrison T, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14198–14203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morse HC, III, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 9.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 10.Wolff L, Koller R, Anver MR. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J. Virol. 2003;77:4965–4971. doi: 10.1128/JVI.77.8.4965-4971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang CC, Vlad G, D'Agati VD, Liu Z, Zhang QY, Witkowski P, et al. BCL6 is required for differentiation of Ig-like transcript 3-Fc-induced CD8+ T suppressor cells. J. Immunol. 2010;185:5714–5722. doi: 10.4049/jimmunol.1001732. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 2012;209:1841–1852. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleque S, Cameron S, Orkin SH. The zinc finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 2002;16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JD, Jackson PK. The Evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor Emi1. Cell. 2006;124:367–380. doi: 10.1016/j.cell.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 15.Rosson D, Dugan D, Reddy EP. Aberrant splicing events that are induced by proviral integration: implications for myb oncogene activation. Proc. Natl. Acad. Sci. U. S. A. 1987;84:3171–3175. doi: 10.1073/pnas.84.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooney JD, Hildick-Smith GJ, Shafizadeh E, McBride PR, Carroll KJ, Anderson H. Teleost growth factor independence (gfi) genes differentially regulate successive waves of hematopoiesis. Dev. Biol. 2013;373:431–441. doi: 10.1016/j.ydbio.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela J-M, Dik WA, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 18.Laflamme C, Assaker G, Ramel D, Dorn JF, She D, Maddox PS, et al. Evi5 promotes cell migration through its Rab-GAP activity. J. Cell Biol. 2012;198:57–67. doi: 10.1083/jcb.201112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westlake CJ, Junutula JR, Simon GC, Pilli M, Prekeris R, Scheller RH, et al. Identification of Rab11 as a small GTPase binding protein for the Evi5 oncogene. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1236–1241. doi: 10.1073/pnas.0610500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vassen L, Fiolka K, Mahlmann S, Moroy T. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 2005;33:987–998. doi: 10.1093/nar/gki243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 2010;24:1834–1843. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 22.Schulz D, Vassen L, Chow KT, McWhirter SM, Amin RH, Moroy T, et al. Gfi1b negatively regulates Rag expression directly and via the repression of FoxO1. J. Exp. Med. 2011;109:187–199. doi: 10.1084/jem.20110645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doan LL, Kitay MK, Yu Q, Singer A, Herblot S, Hoang T, et al. Growth factor independence-1B expression leads to defects in T cell activation, IL-7 receptor α expression, and T cell lineage commitment. J. Immunol. 2003;170:2356–2366. doi: 10.4049/jimmunol.170.5.2356. [DOI] [PubMed] [Google Scholar]
- 24.Oleksiak MF, Roach JL, Crawford DL. Natural variation in cardiac metabolism and gene expression in Fundulus heteroclitus. Nat. Genet. 2005;37:67–72. doi: 10.1038/ng1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senkel S, Waldner C, Ryffel GU, Thomas H. Improved conditional expression systems resulting in physiological level of HN4α expression confirm HN4α induced apoptosis in the pancreatic beta-cell line INS-1. B.M.C. Res. Notes. 2009;2:210–216. doi: 10.1186/1756-0500-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazo PA, Lee JS, Tsichlis PN. Long-distance activation of the Myc protooncogene by provirus insertion in Mlvi-1or Mlvi-4 in rat T-cell lymphomas. Proc. Natl. Acad. Sci. U. S. A. 1990;87:170–173. doi: 10.1073/pnas.87.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt T, Zornig M, Beneke R, Moroy T. MoMuLV proviral integrations identified by Sup-F selection in tumours from infected myc/pim bitransgenic mice correlate with activation of the gfi-1 gene. Nucleic Acids Res. 1996;24:2528–2534. doi: 10.1093/nar/24.13.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pattabiraman DR, Gonda TJ. Role and potential for therapeutic targeting of MYB in leukemia. Leukemia. 2013;27:269–277. doi: 10.1038/leu.2012.225. [DOI] [PubMed] [Google Scholar]
- 29.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Invest. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat. Rev. Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 31.O'Rourke JP, Ness SA. Alternative RNA splicing produces multiple forms of c-Myb with unique transcriptional activities. Mol. Cell. Biol. 2008;28:2091–2101. doi: 10.1128/MCB.01870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Streubel G, Bouchard C, Berberich H, Zeller MS, Teichmann S, Adamkiewicz J, et al. PRMT4 is a novel coactivator of c-Myb-dependent transcription in haematopoietic cell lines. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003343. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, et al. BCL6 positively regulates AID and germinal center expression via repression of miR-155. J. Exp. Med. 2012;209:2455–2465. doi: 10.1084/jem.20121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L, Glazov EA, Pattabiraman DR, Al-Owaidi F, Xhang P, Brown MA, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cubedo E, Gentles AJ, Huang C, Natkunam Y, Bhatt S, Lu X, et al. Identification of LMO2 transcriptome and interactome in diffuse large B-cell lymphoma. Blood. 2012;119:5478–5491. doi: 10.1182/blood-2012-01-403154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homminga I, Vuerhard MJ, Langerak AW, Buijs-Gladdines J, Pieters R, Meijerink JP. Characterization of a pediatric T-cell acute lymphoblastic leukemia patient with simultaneous LYL1 and LMO2 rearrangements. Haematologica. 2012;97:258–261. doi: 10.3324/haematol.2011.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandilci A, Surtel J, Janke L, Neale G, Terranova S, Grosveld GC. Mapping of MN1 sequences necessary for myeloid transformation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061706. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M, Gallipoli P, DeGeer D, Sloma I, Forrest DL, Chan M, et al. Targeting primitive chronic myeloid leukemia cells by effective inhibition of a new AHI-1-BCR-ABL-JAK2 complex. J. Natl. Cancer Inst. 2013;105:405–423. doi: 10.1093/jnci/djt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litvinov IV, Kupper TS, Sasseville D. The role of AHI1 and CDKN1C in cutaneous T-cell lymphoma progression. Exp. Dermatol. 2012;21:964–966. doi: 10.1111/exd.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parekh S, Polo JM, Shaknovich R, Juszczynski P, Lev P, Ranuncolo SM, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer C, Zur Stadt U, Escherich G, Hofmann J, Binato R, Barbosa TC, et al. Refinement of IKZF1 recombination hotspots in pediatric BCP-ALL patients. Am. J. Blood Res. 2013;3:165–173. [PMC free article] [PubMed] [Google Scholar]
- 43.Palmi C, Valsecchi MG, Longinotti G, Silvestri D, Carrino V, Conter V, et al. What is the relevance of Ikaros gene deletions as prognostic marker in pediatric Philadelphia negative B-cell precursor acute lymphoblastic leukemia? Haematologica. 2013 Apr 12; doi: 10.3324/haematol.2012.075432. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hahn CK, Berchuck JE, Ross KN, Kakoza RM, Clauser K, Schinzel AC, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16:281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dierks C, Adrian F, Fisch P, Ma H, Maurer H, Herchenback D, et al. The ITK-SYK fusion oncogene induces a T-cell lymphoproliferative disease in mice mimicking human disease. Cancer Res. 2010;70:6193–6204. doi: 10.1158/0008-5472.CAN-08-3719. [DOI] [PubMed] [Google Scholar]
- 46.Greif PA, Konstandin NP, Metzeler KH, Herold T, Pasalic Z, Ksienzyk B, et al. RUNX1 mutations in cytogenetically normal acute myeloid leukemia are associated with a poor prognosis and up-regulation of lymphoid genes. Haematologica. 2012;97:1909–1915. doi: 10.3324/haematol.2012.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Freudenberg J, Cui K, Dale R, Song SH, Dean A, et al. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood. 2013;121:4575–4585. doi: 10.1182/blood-2013-01-479451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA. 2012;3:311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]