Abstract
Adipose tissue is an endocrine organ that specializes in lipid metabolism and is distributed throughout the body in distinct white adipose tissue (WAT) and brown adipose tissue (BAT) depots. These tissues have opposing roles in lipid metabolism with WAT storing excessive caloric intake in the form of lipid, and BAT burning lipid through non-shivering thermogenesis. As accumulation of lipid in mature adipocytes of WAT leads to obesity and increased risk of comorbidity (Pi-Sunyer et al., 1998), detailed understanding of the mechanisms of BAT activation and WAT accumulation could produce therapeutic strategies for combatting metabolic pathologies. As morphological changes accompany alterations in adipose function, imaging of adipose tissue is one of the most important tools for understanding how adipose tissue mass fluctuates in response to various physiological contexts. Therefore, this chapter details several methods of processing and imaging adipose tissue, including brightfield colorimetric imaging of paraffin sectioned adipose tissue with a detailed protocol for automated adipocyte size analysis; fluorescent imaging of paraffin and frozen sectioned adipose tissue; and confocal fluorescent microscopy of whole mounted adipose tissue. We have also provided many example images showing results produced using each protocol, as well as commentary on the strengths and limitations of each approach.
Keywords: adipose, whole mount, confocal, frozen, paraffin, cell profiler, lineage tracing
1. Introduction
Adipose tissue is distributed throughout the body in distinct “white” and “brown” adipose tissue depots. White adipose tissue (WAT) is largely composed of unilocular lipid-filled adipocytes that specialize in lipid storage, whereas brown adipose tissue (BAT) is largely composed of multilocular adipocytes that specialize in lipid burning. Although adipocytes compose the majority of WAT and BAT volume, both tissue types contain a large number of stromal cells including blood, endothelial, fibroblastic and adipocyte precursor cells which are essential for adipose tissue function. Changes in adipose tissue morphology accompany adipose tissue development (Birsoy et al., 2011), the onset of obesity (Sun, Kusminski, & Scherer, 2011) and response to cold challenge (Seale et al., 2011), making imaging of adipose tissue a powerful tool for understanding the basic biology of adipose tissue development, maintenance, growth and remodelling. Furthermore, imaging of adipose tissue from genetic mouse models allows for study of adipocyte precursor localization (Berry & Rodeheffer, 2013; Gupta et al., 2012; Lee, Petkova, Mottillo, & Granneman, 2012; Tang et al., 2008) and adipocyte lineage derivation (Berry & Rodeheffer, 2013; Tang, et al., 2008; Tran et al., 2012), providing insight into how tissue organization allows WAT to participate in and respond to systemic metabolism. In this chapter, we will provide detailed protocols for preparing and imaging whole mount, paraffin sectioned and frozen sectioned adipose tissue. We will also provide discussion on the benefits and limitations of each approach to guide the application of these imaging approaches to future studies of adipose tissue biology.
2. Imaging of Whole Mounted Adipose Tissue
Adipose tissue that has been stained with fluorescent antibodies/dyes or isolated from fluorescent reporter mice can easily be visualized in whole mount through confocal microscopy. The advantage of imaging adipose tissue in whole mount is that it does not require fixation, processing, embedding, or sectioning. As these steps can decrease antigen recognition, deplete fluorescent signal, and lead to increased auto-fluorescence, imaging of adipose tissue in whole mount generally provides a high signal/noise ratio and allows for clear distinction of fluorescently labelled cells. This approach has recently been used by our group to perform lineage tracing of WAT by clearly differentiating mature adipocytes from stromal cells in situ (Berry & Rodeheffer, 2013). The disadvantage of this technique is that antigen labelling with fluorescently conjugated antibodies can be less robust than what is observed in tissue prepared for IHC as the antibody must permeate the tissue, but this is antibody and antigen dependent.
1. Preparation of Slides
Materials Needed
Microscope slides (Thermo Scientific, MA USA, 4951F-001)
Coverslips (Fischer Scientific, MA USA, 12-545-F)
10 mL syringe (Sigma-Aldrich, MO USA, Z248029)
16 gauge needle (BD Biosciences, CA USA, 305198)
Fluoromount-G (Southern Biotech, AL USA, 0100-01)
Rapid Dry Nail Polish
Sterile PBS (Life Technologies, NY USA, 14190-144)
Vasoline
Prior to Starting
Fill 10mL syringe with vasoline.
Attach 16 gauge needle to filled syringe.
Protocol
-
❖
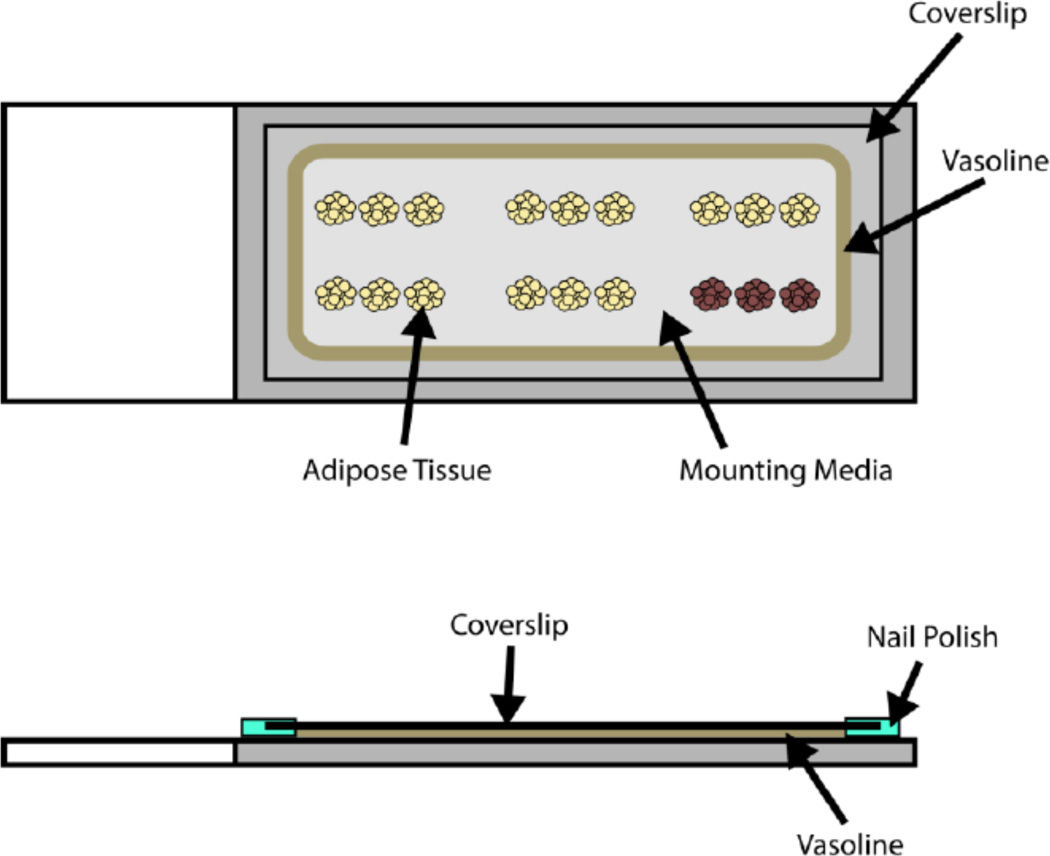
A diagram of a completed slide prepared for imaging of whole mounted adipose tissue is shown in Figure 1.
Figure 1.
A depiction of a slide prepared for imaging of adipose tissue in whole mount.
-
1
Dissect adipose tissue from mouse.
-
2
Cut samples into pieces that are approximately 4 mm × 4 mm × 2 mm.
-
3Stain samples with application specific fluorescent antibodies or dyes.
-
❖A list of commonly used stains, antibodies, and fluorescent reporter proteins along with recommended concentrations and staining times is provided in Table 1.
-
❖We have found that fixation and permeabilization is not required for labelling of antigens with the antibodies listed in Table 1. However, labelling of some antigens may require fixation and/or permeabilization ("Whole mount staining protocols | Abcam, " 2013). In our hands, incubation of small samples in mild fixative (1% paraformaldehyde) or mild detergent (0.5% Tween 20) for 30 minutes prior to labelling does not disrupt antigen labelling or lead to high levels of auto-fluorescence.
-
❖
-
4
Wash samples in sterile PBS.
-
5
Place samples onto the center of a slide.
-
6Make a boundary of vasoline around the samples that is approximately 2 mm high using the vasoline filled syringe.
-
❖The vasoline boundary should be about the same thickness (height off of the slide) as the samples.
-
❖
-
7
Fill the inside of the vasoline boundary with Fluoromount-G.
-
8
Gently place a coverslip onto the slide such that it is resting on top of the vasoline boundary.
-
7Lightly push down on the coverslip until the top of each sample is pressed against the underside of the coverslip.
-
❖For best results, ensure that the coverslip is pressed down evenly on all sides, resulting in a uniform 1 mm gap between the coverslip and the slide.
-
❖
-
8
Remove excess Fluoromount-G that has seeped out from inside the vasoline boundary after pressing down the coverslip.
-
9
Apply nail polish to all edges of the coverslip to adhere the coverslip to the slide.
-
10
Allow nail polish to dry.
Table 1.
Commonly used fluorescent stains, antibodies, and reporter proteins for whole mount confocal imaging of adipose tissue.
| Antibody / Stain | Fluorochrome / Reporter Protein |
Company, Catalog # | Excitation Laser |
Emission Filter |
|---|---|---|---|---|
| N/A | dTomato | N/A | 543 nm | 590–650 nm |
| N/A | eGFP | N/A | 488 nm | 505–540 nm |
| HCS LipidTox1 | “Green” | Invitrogen, H34475 | 488 nm | 505–540 nm |
| HCS LipidTox1 | “Deep Red” | Invitrogen, H34477 | 633 nm | 645–700 nm |
| Isolectin GS-IB41 | Alexa Fluor 488 | Invitrogen, I21411 | 488 nm | 505–540 nm |
| Isolectin GS-IB41 | Alexa Fluor 647 | Invitrogen, I32450 | 633 nm | 645–700 nm |
| Cell Mask2 | “Orange” | Invitrogen, C10045 | 543 nm | 565–585 nm |
| CD453 | Alexa Fluor 647 | Biolegend, 103123 | 633 nm | 645–700 nm |
| F4/803 | Alexa Fluor 647 | Biolegend, 123121 | 633 nm | 645–700 nm |
| CD11b3 | Brilliant Violet 421 | Biolegend, 101235 | 405 nm | 420–470 nm |
| CD243 | Brilliant Violet 421 | Biolegend, 101825 | 405 nm | 420–470 nm |
| DAPI4 | “Blue” | Invitrogen, D1306 | 405 nm | 420–470 nm |
stain at 1:100 in PBS for 1 hour
stain at 1:2000 in PBS for 1 hour
stain at 1:100 for 24 hours
stain at 1:10,000 in PBS for 1 hour
2. Confocal Imaging of Whole Mounted Adipose Tissue
-
❖
For brevity, all fluorescent molecules including fluorochromes conjugated to antibodies, fluorescent stains, and fluorescent reporter proteins are referred to as “fluorochromes” in this section.
Equipment Needed
- Leica TCS SP5 Spectral Confocal Microscope or equivalent
-
❖For excitation, this microscope is equipped with a violet laser (405 nm), an argon laser (split into 4 excitation wavelengths: 458, 476, 488, 514 nm) and a He/Ne laser (provides two excitation wavelengths: 543 and 633 nm).
-
❖For emission detection, this microscope uses a prism spectrophotometer detection system that allows the user to set the desired emission wavelengths to be detected for each excitation laser - fluorochrome combination.
-
❖
Software Needed
Leica AF6000 digital imaging software or equivalent
Materials Needed
- Slide prepared as described in part 1. containing the following samples:
- Sample containing all fluorochromes of interest.
- Sample fluorescent-minus-one (FMO) controls for each fluorochrome.
-
❖A FMO control is a sample that contains all fluorochromes of interest except for one of them. Example: To image adipose tissue that contains eGFP, dTomato, and Alexa Fluor 647, three FMO controls are needed.
- FMO control for Alexa Fluor 647 that contains only eGFP and dTomato.
- FMO control for dTomato that contains only eGFP and Alexa Fluor 647.
- FMO control for eGFP that contains only dTomato and Alexa Fluor 647.
-
❖
Protocol
- In the software, set excitation laser and emission filter parameters as listed in Table 1 for each fluorochrome of interest.
-
❖For best results, images are acquired through “sequential scanning” in which each fluorochrome is excited and detected in a separate scan of the tissue. Therefore a new scan is made for each fluorochrome such that only the laser and filter needed to excite and detect that fluorochrome is active in that scan. When an image is acquired, each scan is performed sequentially and the detected signals from each scan can be over-layed to create a composite image.
-
❖
Apply a small drop of immersion oil onto the coverslip of the slide and mount the slide onto the confocal microscope.
- Set laser voltage and gain settings for each fluorochrome as follows:
- Focus on the first FMO sample using a scan that is designated to excite/detect a fluorochrome present in this FMO sample.
- Switch to the scan that is designated to excite/detect the fluorochrome that is not present in this FMO sample.
- Adjust laser voltage and gain settings for this scan such that the background fluorescence is barely visible.
- Save the laser voltage and gain settings for this scan.
- Repeat steps a. – d. to set the laser voltage and gain settings for each fluorochrome using the appropriate FMO controls.
Focus on a sample that contains all fluorochromes of interest.
Acquire the image at the desired resolution using the laser voltage and gain settings determined in step 4 for each scan. Example images of WAT acquired with the fluorochromes, excitation laser and emission filter settings listed in Table 1 are shown in Figure 2.
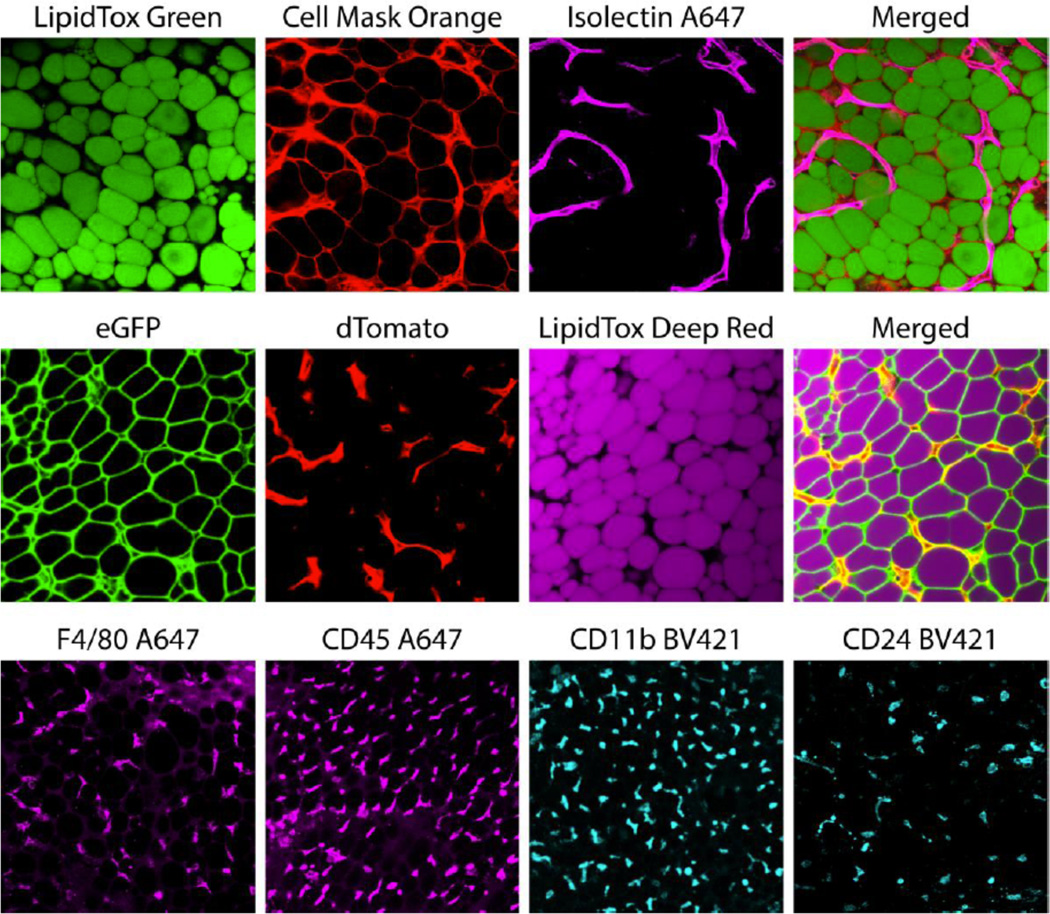
Figure 2.
Example images of whole mounted fluorochrome-containing WAT acquired through confocal microscopy. WAT was isolated from fluorescent reporter mice (eGFP and dTomato) and/or incubated in fluorescent stains/antibodies to label lipids (LipidTox), plasma membranes (Cell Mask Orange), endothelial cells (Isolectin GS-IB4), macrophages (F4/80 and CD11b), non-specific blood lineage cells (CD45), B-cells and adipocyte progenitor cells (CD24) as described in Table 1. Images were acquired with the appropriate laser/filter settings listed in Table 1 and laser voltage/gain settings determined by FMO controls.
3. Imaging of Sectioned Adipose Tissue
3.1 Paraffin Sectioned Adipose Tissue
As staining of whole mount tissue requires the antibody/stain to permeate the tissue, more robust staining can be accomplished by sectioning the tissue prior to staining. The most common method of sectioning involves tissue fixation and embedding in paraffin. Although these methods tend to mask antigens, they maintain excellent tissue morphology and therefore allow for tissue analysis following cellular staining. Additionally, antigens can be “unmasked” through an antigen retrieval step to facilitate antibody labelling. The following protocols for paraffin sectioning, H&E staining and immunohistochemical antigen labelling have been adapted from previously published protocols (Cinti, Zingaretti, Cancello, Ceresi, & Ferrara, 2001), but contain several specific modifications.
1. Preparation of Slides
Materials Needed
Microscope slides (Thermo Scientific, MA USA, 4951F-001)
5× Zinc-Formalin Concentrate (Anatech LTD, MI USA, 141).
Sterile PBS (Life Technologies, NY USA, 14190-144)
- Sodium Citrate Dihydrate (Avantor Performance Materials, PA USA, 3646-01)
-
❖Only needed if performing antigen retrieval for IHC
-
❖
Acetone (Avantor Performance Materials, PA USA, 9006-01)
CitriSolv (Fischer Scientific, MA USA, 22-143-975), or xylene
100% Ethanol (Decon Labs, PA USA, 2701)
95% Ethanol (Decon Labs, PA USA, 2801)
Biobond (Electron Microscopy Services, PA USA, 71304)
Paraffin (Leica Microsystems, Germany, EM-400)
Embedding cassettes (Sakura Finetek USA, CA USA, 4135)
Equipment Needed
- Embedding station (containing heating and cooling plates)
-
❖Individual heating and cooling plates can be used in place of an embedding station.
-
❖
Metal embedding molds (Sakura Finetek USA, CA USA, 4124)
Microtome
Heating water bath
- 2100 Retriever (Electron Microscopy Services, PA USA, 62706)
-
❖Only needed if performing antigen retrieval for IHC
-
❖There are several alternative protocols for antigen retrieval, including some that do not require specialized equipment ("D. Antigen retrieval (IHC-P guide) | Abcam," 2013).
-
❖
Prior to Starting
- Prepare 75% and 70% ethanol:
- Mix together 750 mL 100% EtOH and 250 mL dH2O (75% EtOH).
- Mix together 500 mL 75% EtOH and 35 mL dH2O (70% EtOH).
- Prepare 10 mM citrate buffer for antigen retrieval:
-
aTo make 1 L, add 2.94g Sodium Citrate Dihydrate to 1 L dH2O.
-
bpH to 6.0 using hydrochloric acid.
-
❖Tris/EDTA pH 9.0 buffer can be used in place of 10 mM citrate buffer and may be better suited for retrieval of some antigens ("D. Antigen retrieval (IHC-P guide) | Abcam," 2013).
-
a
Prepare 1× Zinc-formalin by adding 10 mL 5× Zinc-formalin to 40 mL dH2O.
Protocol
-
1
Dissect adipose tissue from mouse.
-
2Incubate samples in 1× Zinc-formalin overnight at 4°C.
-
❖For downstream labelling of phosphorylated antigens, animals must be perfused with fixative to preserve phosphorylation events.
-
❖
-
3
Wash samples 3 times in sterile PBS.
-
4
Place samples into labelled embedding cassette.
-
5Incubate cassette in 70% ethanol until tissue processing is performed.
-
❖Samples can be stored in 70% ethanol at 4°C until tissue processing.
-
❖
Tissue Processing
-
❖
Tissue processing can be performed manually as described below or by using a histology tissue processing machine.
-
6
Incubate cassette in 75% ethanol for 30 minutes.
-
7
Incubate cassette in 95% ethanol for 75 minutes. Repeat this step a 2nd time with fresh 95% ethanol.
-
8
Incubate cassette in 100% ethanol for 60 minutes. Repeat this step a 2nd and 3rd time with fresh 100% ethanol.
-
9
Incubate cassette in CitriSolv for 60 minutes. Repeat this step a 2nd time with fresh CitriSolv.
-
10
Incubate cassette in melted paraffin for 60 minutes at 60°C.
-
11
Transfer cassette to fresh melted paraffin and incubate overnight at 60°C.
-
12
Transfer cassette to fresh melted paraffin and incubate for 60 minutes at 60°C.
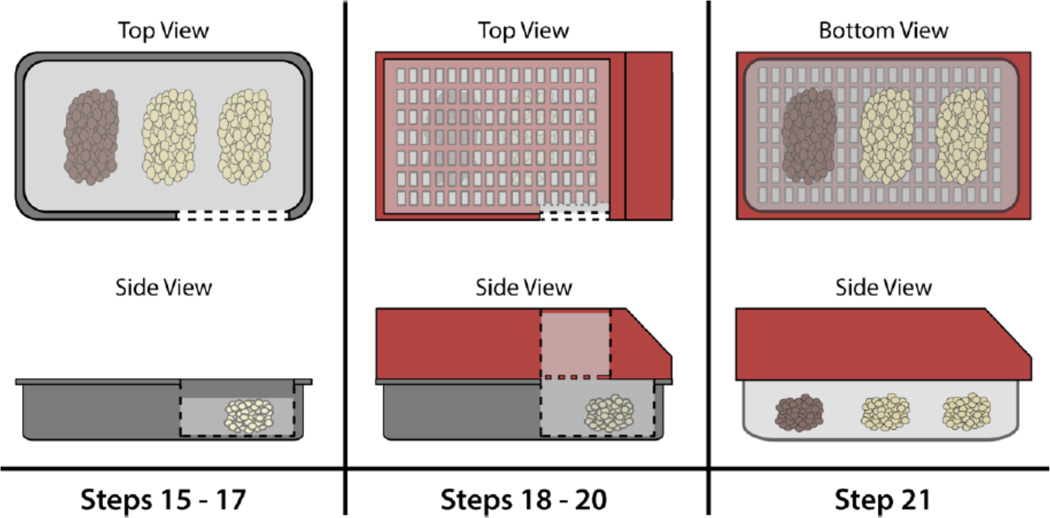
Paraffin Embedding
-
13
Remove cassette from paraffin and place on a heating plate set to 60°C.
-
14
Place metal embedding mold on a heating plate set to 60°C.
-
15
Fill metal embedding mold 75% full with melted paraffin.
-
16
Remove samples from cassette and place in melted paraffin within the metal embedding mold in the desired orientation.
-
17
Place metal embedding mold on a cold plate set to 0°C for a few seconds to harden the paraffin just enough to maintain the sample orientation.
-
18
Place embedding mold back on the heating plate set to 60°C and place the embedding cassette on top of embedding mold.
-
19Quickly pour melted paraffin into the embedding cassette to fill the remainder of the embedding mold and the embedding cassette.
-
❖The melted paraffin will seep through the holes in the bottom of the cassette to fill the mold before filling the cassette.
-
❖
-
20Immediately transfer the mold with cassette to the cold plate and leave the mold/cassette on the cold plate until the paraffin has fully hardened.
-
❖The paraffin will harden through the holes in the cassette to attach the cassette to the molded paraffin block.
-
❖
-
21Carefully remove the paraffin block from the mold by pulling the attached cassette away from the mold.
-
❖If the paraffin block is not completely hardened, the molded block may detach from the cassette, necessitating re-embedding of the samples. Therefore, if the paraffin block does not easily detach from the mold, incubate the mold at -20°C for 30 minutes before attempting to detach the block.
-
❖
Sectioning
-
22Prior to sectioning, coat slides with Biobond as follows:
- Dilute Biobond 1:50 in acetone.
- Incubate slides in diluted Biobond for 5 minutes.
- Incubate slides in dH2O for 5 minutes.
- Allow slides to dry before sectioning.
-
23
Cut 5 µm thick paraffin sections using microtome.
-
24Carefully transfer paraffin section(s) to a water-bath set to 37°C.
-
❖Paraffin section(s) should float.
-
❖
-
25
Transfer each paraffin section to a slide by submerging the slide into the water bath, placing the slide under the floating section, and raising the slide out of the water such that the section attaches to the slide as it is removed from the water.
-
26Incubate the slides with sectioned adipose tissue at 40°C overnight to dry and adhere the tissue sections to the slides.
-
❖For H&E staining, proceed to part 2. Hematoxylin and Eosin (H&E) Staining.
-
❖For Immunohistochemistry applications, proceed to step 27.
-
❖
De-paraffinization and Re-hydration
-
27
Incubate the dried slides in CitriSolv for 5 minutes to dissolve the paraffin. Repeat this step a 2nd time with fresh CitriSolv.
-
28Re-hydrate the sectioned tissue through sequential incubations in ethanol as described below:
- Incubate slides in 100% ethanol for 10 minutes. Repeat this step a 2nd time with fresh 100% ethanol.
- Incubate slides in 95% ethanol for 10 minutes. Repeat this step a 2nd time with fresh 95% ethanol.
- Incubate slides in 70% ethanol for 5 minutes. Repeat this step a 2nd time with fresh 70% ethanol.
-
29Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
-
❖If you do not want to perform antigen retrieval, skip to step 33.
-
❖
-
30Perform antigen retrieval by running slides submerged in 10 mM citrate buffer through the 2100 Retriever.
-
❖This process takes 35–40 minutes
-
❖Several methods exist for antigen retrieval ("D. Antigen retrieval (IHC-P guide) | Abcam," 2013).
-
❖
-
31
Remove slides from 2100 Retriever and allow slides to cool at room temperature for 15 minutes in 10 mM citrate buffer.
-
32
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
-
33
Proceed to part 4. Immunohistochemistry (IHC) using Horseradish Peroxidase Substrates.
2. Hematoxylin and Eosin (H&E) Staining
Hematoxylin and Eosin (H&E) staining is a useful tool for the study of adipose tissue as it can display changes in tissue composition and morphology such as the formation of crown-like structures, “browning” of WAT, “whitening” of BAT, or changes in adipocyte cell size as described in part 3. Automated Adipocyte Size Analysis using Cell Profiler.
Materials Needed
Slides containing paraffin sectioned adipose tissue dried overnight at 40°C.
Eosin Y Alcoholic (Leica Microsystems, Germany, 3801600)
Shandon Gill 3 Hematoxylin (Thermo Scientific, MA USA, 6765010)
Hydrochloric Acid (Avantor Performance Materials, PA USA, 9535-00)
Lithium Carbonate (Sigma-Aldrich, MO USA, L4283)
Xylene (Avantor Performance Materials, PA USA, 8668-16)
100% Ethanol (Decon Labs, PA USA, 2701)
95% Ethanol (Decon Labs, PA USA, 2801)
Mounting Media (Thermo Scientific, MA USA, 8310-16)
Coverslips (Fischer Scientific, MA USA, 12-545-F)
Prior to Starting
Prepare a 1% lithium carbonate (saturated) solution by adding 5 g lithium carbonate to 500 mL dH2O and mixing well.
Prepare 70% ethanol by mixing 70 mL 100% ethanol and 30 mL dH2O.
Prepare a HCl-ethanol solution (0.125% HCl in 96.25% ethanol) by adding 625 µL hydrochloric acid to 62.5 mL 70% ethanol in a 500 mL flask. Add 437 mL 100% ethanol and gently mix the solution.
Protocol
Incubate slides in xylene for 20 seconds. Repeat this step 3 more times using fresh xylene each time.
Incubate slides in 100% ethanol for 20 seconds. Repeat this step a 2nd time with fresh 100% ethanol.
Incubate slides in 95% ethanol for 15 seconds.
Incubate slides in dH2O for 15 seconds.
Incubate slides in Shandon Gill 3 Hematoxylin for 15 seconds. Repeat this steps 4 more times using fresh hematoxylin each time.
Incubate slides in dH2O for 45 seconds.
Incubate slides in HCl-ethanol solution for 15 seconds.
Incubate slides in dH2O for 15 seconds.
Incubate slides in 1% lithium carbonate solution for 15 seconds.
Incubate slides in dH2O for 15 seconds.
Incubate slides in Eosin Y Alcoholic for 15 seconds.
Incubate slides in 95% ethanol for 15 seconds. Repeat this step a 2nd and 3rd time using fresh 95% ethanol.
Incubate slides in 100% ethanol for 15 seconds.
Incubate slides in fresh 100% ethanol for 45 seconds.
Incubate slides in xylene for 45 seconds.
Allow slides to air dry and coverslip the slides using mounting media.
H&E stained sections can be imaged using bright-field microscopy to produce images as shown in Figure 4A. For automated adipocyte size analysis using H&E stained sections in Cell Profiler, proceed to part 3. Automated Adipocyte Size Analysis using Cell Profiler.
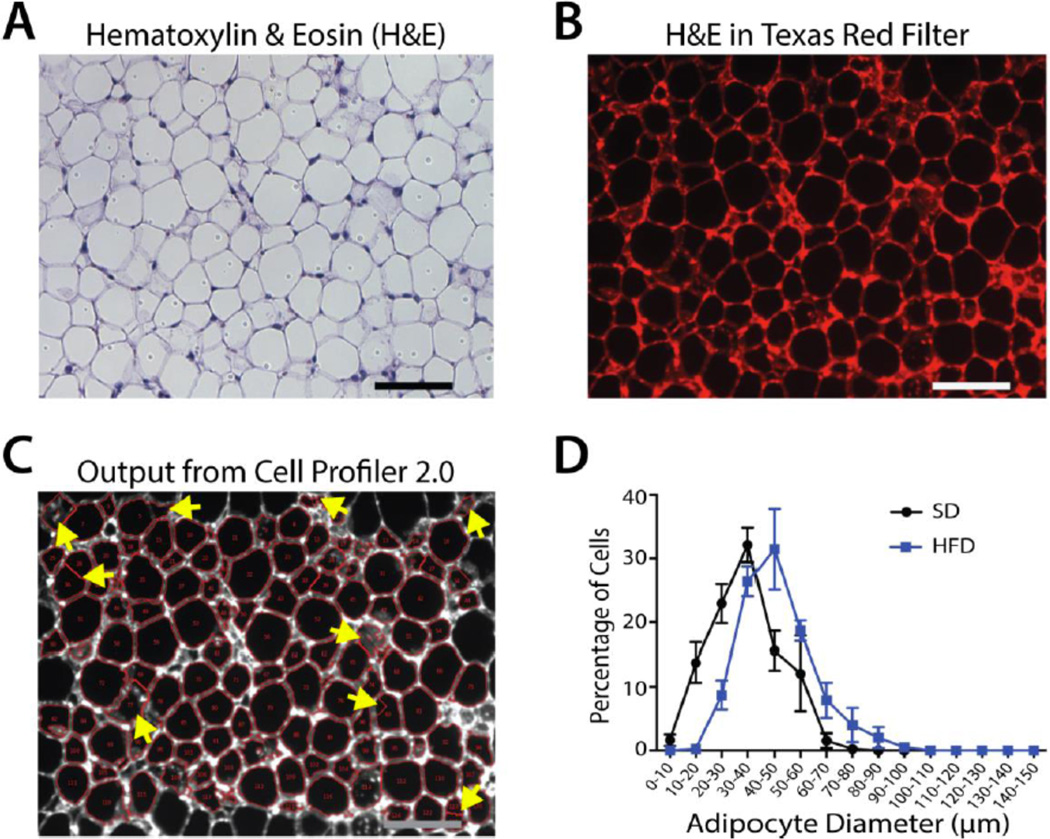
Figure 4.
Example Images and Graph of Cell Profiler Adipocyte Size Analysis. (A) Brightfield image of H&E stained paraffin sectioned WAT. (B) Fluorescent image of H&E stained paraffin sectioned WAT using the Texas Red filter cube. (C) Image from (B) outputted from Cell Profiler 2.0 following adipocyte pixel area analysis. Yellow arrows indicate improperly gated cells that must be manually excluded from the dataset. (D) Adipocyte cell size distribution in WAT depots isolated from standard- (SD) and high fat-diet (HFD) fed mice following Cell Profiler 2.0 analysis and conversion of measured adipocyte pixel area to adipocyte diameter as described in section 3.3.
3. Automated Adipocyte Size Analysis using Cell Profiler
Adipocyte size has been shown to correlate with metabolic phenotypes such as insulin resistance (de Souza et al., 2001; Lundgren et al., 2007). Additionally, adipocyte hypertrophy is an important component of WAT accumulation in development (Birsoy, et al., 2011) and obesity (Hirsch & Batchelor, 1976; Johnson, Zucker, Cruce, & Hirsch, 1971). As the vast majority of WAT volume is accounted for by lipid within mature adipocytes, H&E staining of adipocyte plasma membranes allows for adipocyte size measurements in paraffin sectioned WAT to determine the contribution of adipocyte hypertrophy to WAT mass accumulation. The following protocol details automated adipocyte size measurements using H&E stained WAT sections and Cell Profiler software (Carpenter et al., 2006; "CellProfiler cell image analysis software, " 2013).
Materials Needed
Slide containing paraffin sectioned and H&E stained adipose tissue.
Equipment Needed
- Zeiss Axioplan 2 fluorescent microscope with a Texas Red filter cube (560/55 excitation filter (533–588nm); 645/75 emission filter (608–683nm).
-
❖Any fluorescent microscope with a green laser (514–568 nm) and a detection filter for red wavelengths (575–700 nm) can be used.
-
❖
Software Needed
Zeiss Axiovision 4.2 or equivalent.
Cell Profiler 2.0 (Carpenter, et al., 2006)
- Adipocyte membrane specific pipeline for Cell Profiler 2.0
-
❖The pipeline that our lab uses to automate adipocyte size measurement in Cell Profiler 2.0 can be downloaded from the Cell Profiler Forum ("CellProfiler cell image analysis software," 2013) (http://cellprofiler.org/forum/) under CellProfiler 2.0 Help ----> Adipocyte H&E Cell Profiler Pipeline. The pipeline file is named “Adipocyte Pipeline numbered Rodeheffer Lab.cp” and can be directly downloaded from the following page: http://cellprofiler.org/forum/viewtopic.php?f=14&t=1687&hilit=adipocyte&start=15. This pipeline is a series of commands that allows the Cell Profiler 2.0 software to recognize the plasma membrane of adipocytes in images of H&E stained sections that are acquired as described below; number each recognized adipocyte; measure the pixel area of each adipocyte; export an image that shows the software determined adipocyte boundaries for each adipocyte; and export a .CSV spread-sheet containing the pixel area of each adipocyte. Cell Profiler 2.0 software and the listed pipeline are designed to sequentially analyze large numbers of images that are uploaded at one time, allowing for measurement of hundreds to thousands of adipocytes for comparison of cell size distribution in different adipose depots or the same depot under varying experimental conditions.
-
❖
Protocol
- Acquire low magnification (~20×), high resolution (~1300 ×1030 pixels) RGB images of H&E stained adipose tissue sections using the Texas Red filter cube.
-
❖As the tissue sections are not labelled with fluorescent molecules, this imaging approach utilizes the auto-fluorescence of the H&E stained plasma membranes in the Texas Red emission filter upon exposure to filtered light of 533–588 nm wavelengths to acquire an image in which the adipocyte membranes are easily distinguishable from the cell interior. Example images acquired under bright-field or as-described fluorescent conditions are shown in Figure 4 A and B, respectively.
-
❖
Export images acquired under fluorescent settings as .TIFF files.
Place all .TIFF images from the same depot or experimental condition in an “input” folder on your computer hard-drive.
Make an “output” folder for each depot or experimental condition.
Load “Adipocyte Pipeline numbered Rodeheffer Lab.cp” in Cell Profiler.
- Select experimental “input” folder as the default input folder at the bottom of the Cell Profiler 2.0 window.
-
❖Image files from the input folder are shown in a window in the bottom left of the software window.
-
❖
Select a previously made output folder as the default output folder at the bottom of the Cell Profiler 2.0 window.
Name the output file (.CSV file containing acquired data) at the bottom of the Cell Profiler 2.0 window.
- Click on the “Analyze Images” button.
-
❖The final analysis step for each image produces a .TIFF file as shown in Figure 4C in which the software designated adipocyte number and software determined cell boundaries are shown.
-
❖After Cell Profiler 2.0 has analyzed all of the images in the input folder, a spread-sheet in .CSV format is saved in the output folder that contains the pixel area of every adipocyte from every image in the input folder.
-
❖
- Identify structures that have been improperly gated during the analysis by manually checking each .TIFF image showing the software determined adipocyte boundaries and adipocyte number and deleting the software calculated pixel area from the .CSV file for improperly gated structures.
-
❖This pipeline does not allow for correction of improperly gated structures, so manual exclusion of these structures from the dataset is necessary for obtaining an accurate cell size distribution.
-
❖
In the spread-sheet, convert pixel area to pixel diameter for all of the properly gated adipocytes using the formula: Diameter=4(Area)π
Determine the pixel to µm scaling factor for the imaging settings used to acquire the initial images in step 1. This is usually accomplished through the imaging software or by using a stage micrometer.
In the spread-sheet, convert the diameter in pixels to diameter in µm by multiplying the diameter in pixels by the determined scaling factor.
For each dataset, group the cell diameters into user determined “bins”. For adipose tissue, we recommend 10 µm bins such that each dataset can be displayed in graphical format as the percentage of adipocytes with cell diameters that fit in each bin (i.e. 0–10 µm, 10–20 µm, etc.). This allows for representation of adipocyte size data as a distribution of sizes that can be compared between depots or experimental conditions. A graph comparing the “binned” adipocyte size distribution in a WAT depot dissected from mice that have been fed either a standard diet (SD) or a high-fat diet (HFD) is shown in Figure 4D.
4. Immunohistochemistry (IHC) using Horseradish Peroxidase Substrates
Detection of a specific antigen in paraffin-sectioned tissue is routinely performed through immunohistochemistry, in which an antigen-specific antibody is directly or indirectly coupled to a reporter molecule, allowing microscopic visualization of antibody labelled antigens. The simplest IHC protocol involves antigen labelling with a fluorochrome-conjugated monoclonal antibody. This technique often produces poor results as fixation and processing of tissue can modify antigen structure and reduce antibody recognition. Additionally, this technique relies on a 1/1 ratio of antigen/reporter molecule, limiting detection of sparsely expressed antigens. When tissue preparation only requires light fixation, such as in the frozen sectioning protocol provided in section 3.2, these limitations can be overcome by amplifying the fluorescent reporter through the “sandwich method”, in which the antigen is labelled with a non-fluorescent primary antibody and the primary antibody is labelled with a fluorochrome-conjugated secondary antibody. This method amplifies the reporter signal as multiple secondary antibodies can bind one primary antibody.
Because antigen labelling is generally less robust in paraffin-sectioned tissue, protocols for paraffin IHC rely on the sandwich method and replacement of the conjugated fluorochrome with an enzyme capable of repeatedly converting a substrate molecule into a fluorescent or colored reporter to result in high reporter signal amplification. The most common antibody-conjugated enzymes are alkaline phosphatase (AP) and horseradish peroxidase (HRP), both of which can be used in combination with several reporter substrates. Below we provide protocols for antigen labelling with colored (3,3'-Diaminobenzidine (DAB)) and fluorescent (tyramide signal amplification (TSA)) HRP substrates, as these substrates produce high signal amplification and low background staining in paraffin sectioned WAT.
a. 3,3'-Diaminobenzidine (DAB)
When fluorescent antigen labelling is not feasible or desired, labelling of antigens with DAB provides a good alternative. DAB is a soluble organic compound that in the presence of hydrogen peroxide is oxidized by HRP to yield an insoluble compound that is brown in color. Therefore, DAB labelled sections can be visualized though brightfield microscopy.
Materials Needed
Slides containing paraffin-sectioned adipose tissue dried overnight at 40°C
30% hydrogen peroxide (Avantor Performance Materials, PA USA, 2186-01)
Sterile PBS (Life Technologies, NY USA, 14190-144)
Unconjugated IHC grade primary antibody
- VECTASTAIN Peroxidase ABC kit (Vector Labs, CA USA, PK-4000 – PK-4007)
-
❖ABC kits are species specific. Choose the kit that is designed for the species from which the primary antibody is derived (i.e. PK-4001 is an ABC Peroxidase Rabbit IgG kit and should be used when the primary antibody is derived from rabbit).
-
❖
Avidin/Biotin Blocking kit (Vector Labs, CA USA, SP-2001)
DAB Peroxidase Substrate kit (Vector Labs, CA USA, SK-4100)
Pap Pen (Life Technologies, NY USA, 00-8899)
Mounting Media (Thermo Scientific, MA USA, 8310-16)
Coverslips (Fischer Scientific, MA USA, 12-545-F)
Prior to Starting
Prepare 0.3% H2O2 in PBS by mixing 1 mL 30% H2O2 in 100 mL PBS.
- Prepare Normal Blocking Serum by adding three drops of the supplied normal serum concentrate from the VECTASTAIN Peroxidase ABC kit (yellow bottle) in 10 mL sterile PBS.
-
❖The blocking serum used is from the species from which the secondary antibody is derived. For VECTASTAIN Peroxidase ABC kits, the secondary antibody is derived from horse, and therefore the blocking serum is normal horse serum.
-
❖
Protocol
Make a hydrophobic barrier around the tissue sections with the Pap Pen.
Incubate sections in 0.3% H2O2 in PBS for 15 minutes at room temperature (RT) to block endogenous peroxidase activity.
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
Incubate sections in normal blocking serum for 30 minutes at RT to prevent non-specific antibody binding.
Blot away normal blocking serum and wash sections in sterile PBS.
Incubate sections in avidin serum from the Avidin/Biotin blocking kit for 15 minutes at RT to prevent non-specific binding of the ABC complex.
Blot away avidin serum and wash sections with sterile PBS.
Incubate sections in biotin serum from the Avidin/Biotin blocking kit for 15 minutes at RT to prevent non-specific binding of the ABC complex.
Blot away biotin serum and wash sections with sterile PBS.
- Incubate sections in primary antibody diluted in normal blocking serum overnight at 4°C.
-
❖Antibody concentration is user determined.
-
❖
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
Prepare biotinylated secondary antibody by adding one drop of biotinylated secondary antibody stock concentrate from the VECTASTAIN Peroxidase ABC kit (blue bottle) to 10 mL sterile PBS.
- Incubate sections in biotinylated secondary antibody for 30 minutes at RT.
-
❖Immediately after incubation begins, prepare ABC reagent by adding 2 drops of Reagent A from the VECTASTAIN ABC kit to 10 mL sterile PBS. Mix solution and add 2 drops of Reagent B. Mix solution again and allow ABC reagent to stand at RT until step 14.
-
❖
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
Incubate sections in ABC reagent for 30 minutes at RT.
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
- Prepare DAB reagent from the DAB Peroxidase Substrate kit as follows:
-
❖Add 2 drops of Buffer Stock solution to 5 mL dH2O and mix.
-
❖Add 4 drops of DAB Stock solution and mix.
-
❖Add 2 drops of Hydrogen Peroxide solution and mix.
-
❖Add 2 drops of Nickel solution and mix.
-
❖
- Incubate section in DAB reagent for 15 minutes at RT.
-
❖Incubation time needed for effective antigen staining with low background staining is antigen dependent and user determined, but usually varies from 1 to 30 minutes.
-
❖
- Incubate slides in sterile PBS for 45 minutes at RT.
-
❖Several colored HRP substrates are commercially available. Therefore, this protocol can be repeated for labelling of a second or third antigen in the same tissue section.
-
❖
- Coverslip slides using mounting media and visualize staining through brightfield microscopy.
-
❖An example image of DAB labelled PdgfRα in WAT is provided in Figure 5A.
-
❖
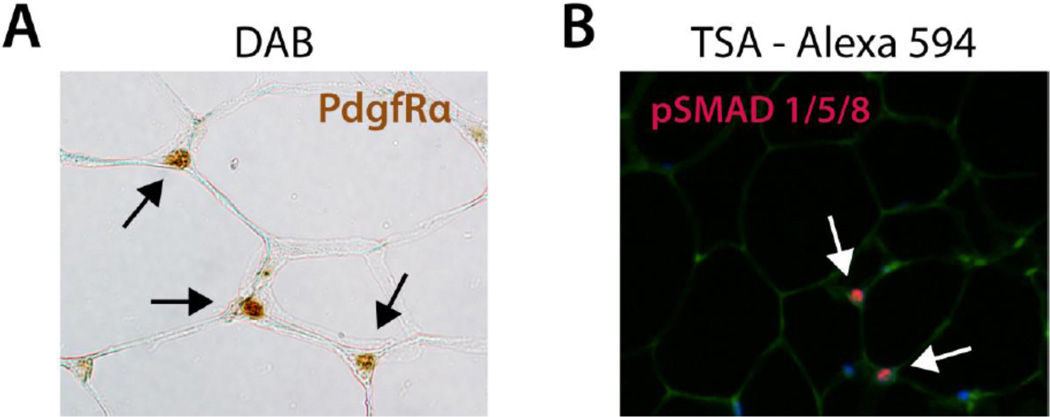
Figure 5.
Immunohistochemistry on Paraffin Sectioned WAT. (A) HRP-DAB mediated immunolabelling of PdgfRα in paraffin sectioned WAT. Black arrows indicate PdgfRα+ cells marked by deposition of oxidized DAB, which is brown in color. (B) HRP-TSA mediated immunolabelling of phospho-SMADs 1,5,8 in paraffin sectioned WAT. White arrows indicate phospho-SMAD+ cells marked by fluorescence of oxidized TSA-A594.
b. Tyramide Signal Amplification (TSA)
When fluorescent antigen labelling is desired, tyramide signal amplification provides high sensitivity to overcome the normal limitations of fluorescent labelling of paraffin sectioned tissue. In TSA, inactive fluorochrome conjugated tyramide derivatives are oxidized by HRP in the presence of hydrogen peroxide to yield highly fluorescent reporter molecules that can be visualized through fluorescent microscopy. There are a large number of fluorochrome labelled tyramide derivatives available, and TSA labelled sections can be counterstained with other fluorescent markers such as Isolectin GS-IB4, Cell Mask and DAPI (Table 1) for detailed fluorescent analysis of adipose tissue.
Materials Needed
Slides containing paraffin sectioned adipose tissue dried overnight at 40°C
30% hydrogen peroxide (Avantor Performance Materials, PA USA, 2186-01)
Sterile PBS (Life Technologies, NY USA, 14190-144)
- Tyramide Signal Amplification (TSA) kit (Life Technologies, NY USA, T-20911 –T-30955)
-
❖TSA kits are species specific. Choose the kit that is designed for the species from which the primary antibody is derived (i.e. T-20925 is a TSA kit with HRP-Goat Anti-Rabbit IgG and Alexa Fluor 594 and can be used when the primary antibody is derived from rabbit). Invitrogen makes HRP-goat anti-rabbit IgG kits, HRP-goat anti-mouse IgG kits, and HRP-streptavidin kits. If the primary antibody is derived from non-rabbit or mouse species, the HRP-streptavidin kit can be used in combination with a biotin conjugated primary antibody.
-
❖
Unconjugated or biotinylated IHC grade primary antibody
- Streptavidin/Biotin Blocking kit (Vector Labs, CA USA, SP-2002)
-
❖Only needed if using a TSA HRP-streptavidin kit.
-
❖
Pap Pen (Life Technologies, NY USA, 00-8899)
Mounting Media (Thermo Scientific, MA USA, 8310-16)
Coverslips (Fischer Scientific, MA USA, 12-545-F)
Prior to Starting
Prepare 0.3% H2O2 in PBS by mixing 1 mL 30% H2O2 in 100 mL PBS.
Prepare tyramide stock solution, 1% blocking reagent, HRP conjugate stock solution and amplification buffer per TSA kit protocols.
Protocol
Make a hydrophobic barrier around the tissue sections with the Pap Pen.
Incubate sections in 0.3% H2O2 in PBS for 15 minutes at RT to block endogenous peroxidase activity.
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
Incubate the sections in prepared 1% blocking reagent for 60 minutes at RT.
- Blot away 1% blocking reagent and wash sections with sterile PBS.
-
❖If using HRP-streptavidin kit, proceed to step 6. If not, proceed to step 10.
-
❖
Incubate sections in streptavidin serum from the Streptavidin/Biotin blocking kit for 15 minutes at RT.
Blot away streptavidin serum and wash sections with sterile PBS.
Incubate sections in biotin serum from the Streptavidin/Biotin blocking kit for 15 minutes at RT.
Blot away biotin serum and wash sections with sterile PBS.
- Incubate sections in primary antibody diluted in 1% blocking reagent overnight at 4°C.
-
❖Antibody concentration is user determined.
-
❖
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
Prepare a HRP-conjugate working solution by diluting the HRP conjugate stock solution 1:100 in 1% blocking reagent.
Incubate sections in 100 µL of HRP-conjugate working solution for 30–60 minutes at RT.
Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
Prepare a tyramide working solution by diluting the tyramide stock solution 1:100 in amplification buffer.
Incubate the sections in 100 µL tyramide working solution for 5–10 minutes at RT.
- Incubate slides in PBS for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS.
-
❖Tissue sections can now be counterstained with additional fluorescent markers such as those listed in Table 1.
-
❖
Coverslip slides using mounting media and visualize staining through fluorescent microscopy using appropriate lasers and filters for each fluorescent stain.
3.2. Frozen Sectioning
To circumvent complex labelling strategies needed to amplify reporter signal in paraffin sectioned adipose tissue, less harsh processing and sectioning protocols have been developed in which adipose tissue is lightly fixed and embedded in gelatin before hardening through freezing. Frozen sectioning maintains antigen integrity and allows for simple immunofluorescent labelling of a single, or multiple, antigen(s). The frozen sectioning and immunofluorescent labelling protocols below have been adapted from previously published protocols (Crisan et al., 2008), with several specific modifications.
1. Preparation of Slides
Materials Needed
Microscope slides (Thermo Scientific, MA USA, 4951F-001)
5× Zinc-Formalin Concentrate (Anatech LTD, MI USA, 141).
Sterile PBS (Life Technologies, NY USA, 14190-144)
Sucrose (Avantor Performance Materials, PA USA, 4072-01)
Gelatin (MP Biomedicals, 960317)
12 well polystyrene plate (BD Biosciences, CA USA, 351143)
Scalpel
Small spatula
Equipment Needed
Cryostat
Prior to Starting
- Prepare PBS-sucrose buffer.
- To make 1 L, add 150 g sucrose to 1 L PBS.
- Store at 4°C.
- Prepare sucrose-gelatin solution.
- To make 100 mL, add 15 g sucrose and 7.5 g gelatin to 100 mL PBS.
- Incubate at 60°C until the gelatin melts.
Prepare 1× Zinc-formalin by adding 10 mL 5× Zinc-formalin to 40 mL dH2O.
Protocol
Dissect adipose depot(s) from mouse.
- Incubate samples in 1× Zinc-formalin at 4°C.
-
❖Incubation times needed for proper fixation varies based on sample size. We recommend:
-
▪For small samples (~10 mg), incubate for 1–2 hours.
-
▪For medium-sized samples (<1 g), incubate overnight.
-
▪For large samples (>1 g), incubate for 36 – 48 hours.
-
▪
-
❖For downstream labelling of phosphorylated antigens, animals must be perfused with fixative to preserve phosphorylation events.
-
❖
Wash samples 3 times in PBS.
Incubate samples in fresh PBS overnight at 4°C.
- Incubate samples in PBS-sucrose buffer overnight at 4°C.
-
❖Samples can be stored in PBS-sucrose buffer at 4°C for up to 2 weeks.
-
❖
Incubate samples in sucrose-gelatin solution for 1–2 hours at 37°C.
Cut samples to 1 cm × 1 cm using a scalpel.
Place each sample in the center of a well in a 12 well plate.
- Fill the well half full with warm sucrose gelatin solution from step 6.
-
❖Adipose tissue sample will float to the top.
-
❖
Incubate the plate for 10–20 minutes at 4°C to solidify the sucrose-gelatin solution.
Fill the well with sucrose-gelatin solution ensuring that the sample remains submerged.
Incubate the plate for 10–20 minutes at 4°C to solidify the sucrose-gelatin solution.
Carefully detach the sucrose-gelatin block from the well using a small spatula ensuring that the sample remains entirely within the block.
Trim the block with a scalpel to ~ 2 cm × 2 cm × 2 cm ensuring that the sample remains entirely within the block.
Place the trimmed block back into the well and incubate the plate containing the block at −80°C for at least 24 hours.
- Section the frozen sucrose-gelatin block at −45°C using a cryostat.
-
❖We recommend a section thickness of 12–20 µm.
-
❖The frozen sucrose-gelatin block must be entirely frozen when sectioning. Therefore, it is essential to adhere to the following sectioning protocol:
- Quickly transfer the sucrose-gelatin block from −80°C to the cryostat pre-set to −45 °C.
- Allow the block to re-freeze at −45°C prior to sectioning.
- Trim the frozen block through sequential 100 µm sections until the tissue is exposed.
- Allow the block to re-freeze at −45°C for 15–30 minutes.
- Section the block at a thickness of 12–20 µm.
-
❖
- Gently transfer each section to a slide by slowing approaching the section with the slide until the section adheres to the slide.
-
❖Care must be taken to prevent mechanical alteration of the section during this transfer process.
-
❖
Allow the section to air dry for 5 minutes at room temperature.
Proceed to part 2. Immunohistochemistry (IHC) using Fluorochrome-Conjugated Antibodies.
2. Immunohistochemistry (IHC) using Fluorochrome-Conjugated Antibodies
As mentioned, frozen sectioning results in less antigen masking and allows for effective antigen labelling with fluorochrome-conjugated antibodies. In our hands, HRP-based signal amplification produces significantly higher background staining in frozen sections as compared to paraffin sections. Because of this, immunofluorescent labelling is our preferred method of staining frozen sections. Additionally, although the provided frozen sectioning protocol produces sections with excellent tissue morphology, paraffin embedded tissue can be sectioned thinner, producing sections more suitable for H&E staining. The following protocol details immunofluorescent labelling of frozen sectioned adipose tissue and provides example images of immunofluorescent labelled WAT in Figure 6.
Figure 6.
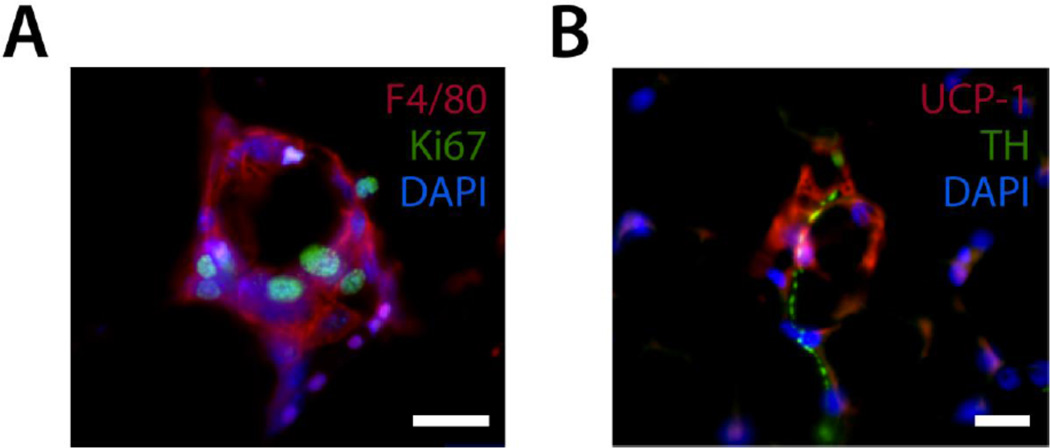
Immunofluorescent staining of frozen sectioned WAT. (A) A classic crown-like structure stained with antibodies for F4/80 and Ki67 and counterstained with DAPI. (B) Sympathetic innervation of a beige adipocyte as shown through staining with antibodies for UCP-1 and the sympathetic nerve fiber marker tyrosine hydroxylase (TH) and counterstained with DAPI. Scale bar is 20 µm.
Materials Needed
Slide containing frozen sectioned adipose tissue
Unconjugated IHC grade primary antibody
Fluorochrome –conjugated secondary antibody
Bovine Serum Albumin, BSA (Sigma-Aldrich, MO USA, A9647)
Sterile PBS (Life Technologies, NY USA, 14190-144)
Triton X-100 (Sigma-Aldrich, MO USA, X100)
Acetone (Avantor Performance Materials, PA USA, 9006-01)
Methanol (Avantor Performance Materials, PA USA, 9070-01)
Pap Pen (Life Technologies, NY USA, 00-8899)
Slide staining Jar (Electron Microscopy Services, PA USA, 71405-01)
Mounting Media (Thermo Scientific, MA USA, 8310-16)
Coverslips (Fischer Scientific, MA USA, 12-545-F)
Prior to Starting
Prepare 0.3% PBS-T by dissolving 600 µL Triton X-100 in 200 mL PBS.
Prepare 1% BSA in PBS-T by dissolving 1 g BSA in 100 mL 0.3% PBS-T.
Prepare 1:1 acetone;methanol by adding 50 mL acetone to 50 mL methanol. Incubate at −20°C until ice-cold.
Protocol
Incubate slides containing frozen sectioned adipose tissue in ice-cold 1:1 acetone:methanol for 10 minutes in a slide staining jar.
Make a hydrophobic barrier around the tissue sections with the Pap Pen.
Incubate sections in 0.3% PBS-T for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS-T.
Incubate sections in 1% BSA in PBS-T for 1 hour at RT to block non-specific antibody binding.
- Incubate sections in primary antibody diluted in 1% BSA in PBS-T overnight at 4°C.
-
❖Antibody concentration is user determined.
-
❖
Incubate sections in 0.3% PBS-T for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS-T.
- Incubate sections in species specific fluorochrome-conjugated secondary antibody diluted in 1% BSA in PBS-T for 60 minutes at RT.
-
❖Antibody concentration is user determined. A 1:200 dilution is generally a good secondary antibody concentration to start with.
-
❖
Incubate slides in 0.3% PBS-T for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS-T.
Tissue can be counterstained with DAPI or other fluorescent stains. Additionally, steps 4–8 can be repeated to label other antigens, using primary antibodies derived from different species for each antigen.
Incubate slides in 0.3% PBS-T for 5 minutes. Repeat this step a 2nd and 3rd time with fresh PBS-T.
Coverslip slides using mounting media and visualize staining through fluorescent microscopy using appropriate lasers and filters for each fluorescent stain.
Figure 3.
A depiction of paraffin embedding of adipose tissue.
Acknowledgements
We thank Michael Schadt, the Section of Comparative Medicine Histology Manager at Yale University for aiding in the development of adipose tissue sectioning and H&E staining protocols. We also thank Dr. Mark Bray from the Carpenter lab at the Broad Institute of MIT and Harvard for assistance in developing and refining a Cell Profiler pipeline suitable for adipocyte cell size analysis. This work was supported by American Diabetes Association Award 7-12-JF-46, DERC pilot project grant DK045735, and NIDDK grant DK090489 to M.S.R; EMBO Long-term postdoctoral fellowship ALTF 132-2011 to C.C; Deutsche Forschungsgemeinschaft DFG-SFB 1052/1: “Obesity mechanisms” (project A04), Helmholtz alliance “Imaging and Curing Environmental Metabolic Disease”, and Research grant from the Medical Faculty, Leipzig University to M.T.G.
References
- Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Berry R, Wang T, Ceyhan O, Tavazoie S, Friedman JM, et al. Analysis of gene networks in white adipose tissue development reveals a role for ETS2 in adipogenesis. Development. 2011;138(21):4709–4719. doi: 10.1242/dev.067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CellProfiler cell image analysis software. 2013 from http://www.cellprofiler.org/ [Google Scholar]
- Cinti S, Zingaretti MC, Cancello R, Ceresi E, Ferrara P. Morphologic techniques for the study of brown adipose tissue and white adipose tissue. Methods Mol Biol. 2001;155:21–51. doi: 10.1385/1-59259-231-7:021. [DOI] [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- D. Antigen retrieval (IHC-P guide) | Abcam. 2013 from http://www.abcam.com/?pageconfig=resource&rid=11488. [Google Scholar]
- de Souza CJ, Eckhardt M, Gagen K, Dong M, Chen W, Laurent D, et al. Effects of pioglitazone on adipose tissue remodeling within the setting of obesity and insulin resistance. Diabetes. 2001;50(8):1863–1871. doi: 10.2337/diabetes.50.8.1863. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Batchelor B. Adipose tissue cellularity in human obesity. Clin Endocrinol Metab. 1976;5:299–311. doi: 10.1016/s0300-595x(76)80023-0. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Zucker LM, Cruce JA, Hirsch J. Cellularity of adipose depots in the genetically obese Zucker rat. J Lipid Res. 1971;12:706–714. [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M, Svensson M, Lindmark S, Renstrom F, Ruge T, Eriksson JW. Fat cell enlargement is an independent marker of insulin resistance and 'hyperleptinaemia'. Diabetologia. 2007;50(3):625–633. doi: 10.1007/s00125-006-0572-1. [DOI] [PubMed] [Google Scholar]
- Pi-Sunyer FX, Becker DM, Bouchard C, Carleton RA, Colditz GA, Dietz WH, et al. Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults. 1998 [Text]. [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121(6):2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KV, Gealekman O, Frontini A, Zingaretti MC, Morroni M, Giordano A, et al. The vascular endothelium of the adipose tissue gives rise to both white and brown fat cells. Cell Metab. 2012;15(2):222–229. doi: 10.1016/j.cmet.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whole mount staining protocols | Abcam. 2013 from http://www.abcam.com/index.html?pageconfig=resource&rid=11330. [Google Scholar]