Abstract
Polyubiquitination by E2 and E3 enzymes is a predominant mechanism regulating protein function. Some RING E3s, including Anaphase Promoting Complex/Cyclosome (APC), catalyze polyubiquitination by sequential reactions with two different E2s. An initiating E2 ligates ubiquitin to an E3-bound substrate. Another E2 grows a polyubiquitin chain on the ubiquitin-primed substrate through poorly defined mechanisms. Here we show that human APC’s RING domain is repurposed for dual functions in polyubiquitination. The canonical RING surface activates an initiating E2~ubiquitin intermediate for substrate modification. However, APC engages and activates its specialized ubiquitin chain elongating E2 UBE2S in ways that differ completely from current paradigms. During chain assembly, a distinct APC11 RING surface helps deliver a substrate-linked ubiquitin to accept another ubiquitin from UBE2S. Our data define mechanisms of APC/UBE2S-mediated polyubiquitination, reveal unexpectedly diverse functions of RING E3s and E2s, and provide a framework for understanding distinctive RING E3 features specifying ubiquitin chain elongation.
Introduction
Regulating protein function often involves precisely coordinated post-translational modification by ubiquitin (Ub). First, an E1 enzyme generates a covalent E2~Ub intermediate, linked by a thioester bond between the catalytic Cys of an E2 enzyme (≈30 in humans) and the C-terminus of the “donor” Ub to be transferred (“~” denotes covalent bond; thioester in E2~Ub and isopeptide in Ub~Ub). An E2~Ub intermediate then functions with an E3 to transfer Ub to a remotely bound protein substrate. The ≈600 human E3s in the RING family are thought to function by their RING domains binding specific E2~Ub intermediates through homologous yet distinctive E3-E2~Ub interfaces (Metzger et al., 2014). RING-dependent stabilization of a particular “closed” E2~Ub conformation immobilizes the thioester bond to spark reactivity toward a lysine nucleophile (Berndsen et al., 2013; Dou et al., 2012, 2013; Plechanovova et al., 2012; Pruneda et al., 2012; Reverter and Lima, 2005; Saha et al., 2011; Scott et al., 2014; Wickliffe et al., 2011). Targeting specificity depends in part on the E2 active site: some E2s react promiscuously with many lysines, whereas others target particular protein lysines or N-termini such as in Ub itself during formation of polyUb chains with specific Ub~Ub linkages (Mattiroli and Sixma, 2014). Although this canonical mechanism has been implicated in activating over a dozen RING E3-E2~Ub intermediates, whether any of the hundreds of other RING E3 and E2 enzymes together promote Ub ligation through other means remains unknown.
A particularly vexing question is whether distinct mechanisms can regulate Ub chain formation. Indeed, polyubiquitination, wherein different chain lengths, sites, and linkage types may be generated, plays a major role in determining fates of modified targets. Some RING E3s use a two-step/two-E2 mechanism to catalyze polyubiquitination (Rodrigo-Brenni and Morgan, 2007; Wu et al., 2010a). First, an “initiating” E2 transfers one or a few Ubs to a substrate. Next, a polyUb chain is assembled with a “chain-elongating” E2 dedicated to producing Ub~Ub (aka di-Ub) linkages. This second E2 generally transfers a “donor Ub” from its catalytic Cys to a specific Lys on a substrate-linked “acceptor Ub”. This mechanism is used by the Anaphase Promoting Complex/Cyclosome (APC) to control passage through mitosis by catalyzing timely polyubiquitination of cell cycle regulators such as Cyclin B (Primorac and Musacchio, 2013).
The 1.2 MDa multisubunit APC can be viewed as structurally comprising two conformationally dynamic and functionally linked superdomains: the RING-containing “Platform”, and the substrate-binding “Arc lamp” (Fig. 1A) (Buschhorn et al., 2011; Chang et al., 2014; Dube et al., 2005; Herzog et al., 2009; Schreiber et al., 2011). In the Platform, APC1, APC4, and APC5 anchor a cullin-RING-like APC2-APC11 catalytic core (Fig. 1A). The Arc Lamp provides a substrate-binding site by securing the C-termini of APC10 and a coactivator (CDC20 or CDH1), which co-recruit substrate motifs such as the “D-box” found in the N-terminal domain (NTD) of Cyclin B. APC10 and coactivators also have domains that bind the Platform, and their substrate engagement is thought to propagate conformational changes that enhance APC11 RING domain binding to an initiating E2 that modifies substrate (in humans, typically UBCH10 but also UBCH5 in vitro), and also to the Ub chain elongating E2 in yeast (Chang et al., 2014; Kimata et al., 2008; Van Voorhis and Morgan, 2014). However, a lingering question is how APC from higher eukaryotes functions with the distinctive chain elongating E2, UBE2S, to mediate polyubiquitination.
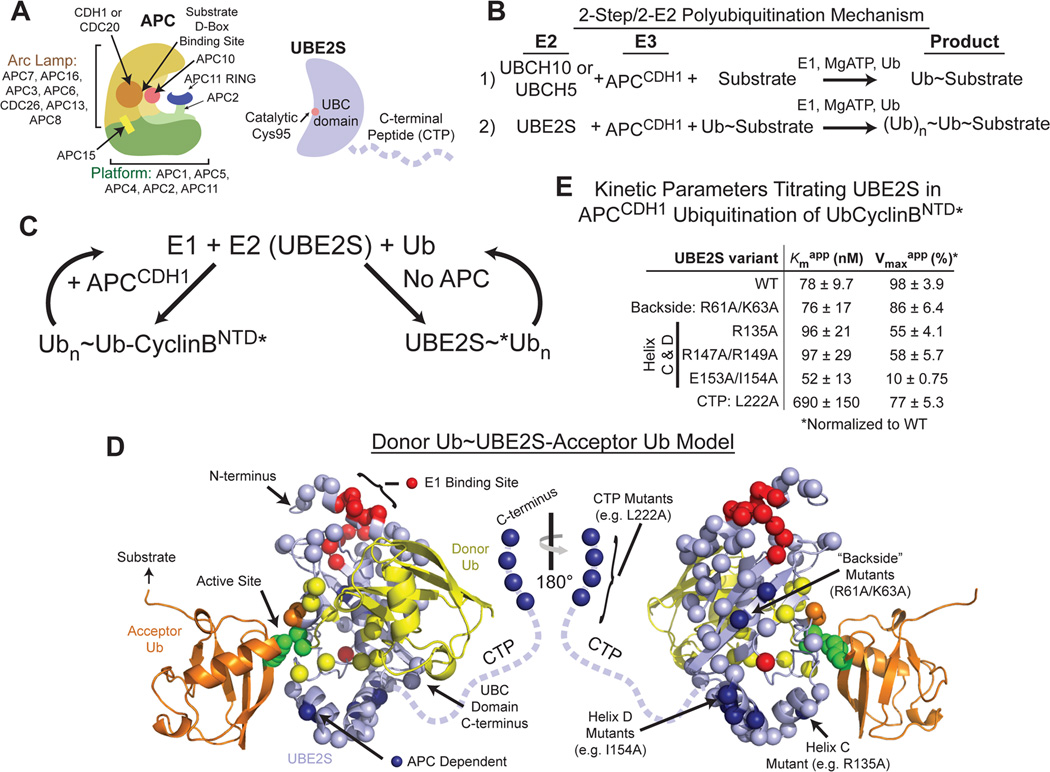
Figure 1. Multiple atypical E2 surfaces dictate UBE2S binding and activation by APC. (relates to Fig. S1).
A, Cartoons of APC and UBE2S highlighting their domains.
B, Reaction scheme for 2-step/2-E2 polyubiquitination by human APC. An “initiating” E2, either UBCH10 or UBCH5, ligates Ub directly to a substrate. The “elongating” E2 UBE2S extends a polyUb chain on the Ub-primed substrate.
C, 2-part in vitro assay to identify UBE2S surfaces working with APC: 1) 135 UBE2S mutants were tested for APCCDH1-catalyzed polyubiquitination of fluorescent Ub-CyclinBNTD*; 2) Faulty mutants in Part 1 were filtered out for APC-independent defects by assaying UBE2S autoubiquitination.
D, Results of UBE2S mutant scan depicted on modeledDonorUb~UBE2S-AcceptorUb intermediate, with donor Ub (yellow) from UBCH5~Ub docked on the UBE2S UBC domain (Plechanovova et al., 2012; Sheng et al., 2012) and acceptor Ub (orange) modeled from (Wickliffe et al., 2011). UBE2S CTP is represented with dashed line. Active site - green spheres (UBE2S catalytic Cys, donor Ub G76, acceptor Ub K11). Sites of APC-dependent defects - blue spheres (UBC domain backside and helices C and D, and CTP terminus). Sites of APC-independent defects - red, yellow, and orange spheres for E1, donor Ub, and acceptor Ub binding sites, respectively. Sites mutated with no defects – slate spheres.
E, Kinetic parameters from titrating concentration of UBE2S (WT or mutant) in APCCDH1-mediated ubiquitination of Ub-CyclinBNTD*, to provide insights into how mutations impaired binding to and activation by APC. SEM, n≥3.
UBE2S has many features differing from E2s that are typically activated by RING E3s. First, the available data imply that UBE2S engages APC in a distinct but unknown manner, because the canonical E2 UBCH10 does not compete with UBE2S for binding to APC (Williamson et al., 2009). Second, the UBE2S~Ub intermediate adopts the closed, activated E2~Ub conformation on its own, obviating the need for a RING to stabilize the reactive architecture (Wickliffe et al., 2011). Third, UBE2S’s catalytic ubiquitin conjugating (UBC) domain generates free Lys11-linked polyUb chains by substrate-assisted catalysis, with residues in the acceptor Ub contributing to the active site (Baboshina and Haas, 1996; Bremm et al., 2010; Wickliffe et al., 2011). Fourth, UBE2S has a unique disordered C-terminal peptide-like “CTP” extension (Fig. 1A) that is multifunctional. UBE2S’s CTP binds CDC20 to assemble with APC in a cell-cycle-dependent manner in vivo, binds the APC core to polyubiquitinate substrates, and is subject to autoubiquitination for proteasomal turnover when not engaged in APC-mediated substrate ubiquitination (Garnett et al., 2009; Kelly et al., 2014; Williamson et al., 2009; Wu et al., 2010b). Finally, unlike APC binding to Ub chain initiating E2s, in vitro interactions between APC and UBE2S are not stimulated by CDH1 and a D-box peptide (Chang et al., 2014). Thus, it is unknown how APC coordinates UBE2S activity with the presence of its Ub-primed substrates.
Despite fundamental importance, mechanisms by which E3s and their Ub chain-elongating E2 partners are functionally linked to drive polyubiquitination remain incompletely understood. Here we address this by taking advantage of our recombinant human APC system (Uzunova et al., 2012). Our study reveals that APC engages and stimulates UBE2S and supports Ub chain elongation in a manner that differs completely from known mechanisms by which canonical RING E3s activate E2s. Instead, APC uses an unprecedented RING-dependent mechanism that increases UBE2S’s catalytic efficiency towards forming Ub~Ub linkages, largely by lowering the Kmapp for the acceptor Ub. Our data, and complementary work from the Rape lab (Kelly et al., 2014), provide a framework for understanding catalytic features of APC, UBE2S, and RING-mediated polyubiquitination different from heretofore known E3 elements that activate E2~Ub intermediates, and that are specialized for Ub chain elongation.
Results
Multiple atypical E2 surfaces dictate UBE2S specificity and activation by APC
Polyubiquitination by APC and UBE2S involves: 1) E1 generation of the UBE2S~DonorUb intermediate, 2) APC and UBE2S interacting, and 3) a catalytic architecture with an acceptor Ub properly placed relative to the UBE2S~DonorUb active site (Fig. 1B). We identified mechanisms by which APCCDH1 harnesses UBE2S in a 2- part in vitro assay with 135 purified mutant versions of UBE2S (Fig. 1C, S1A–B). In the first part, the need for an initiating E2 was bypassed by incorporating a priming acceptor Ub into a fluorescent linear Ub-CyclinBNTD* substrate that is readily polyubiquitinated by APCCDH1 and wild-type UBE2S, but not by several of the mutants (Fig. S1A). Next, we filtered out defects in E1 charging and catalytic placement of the donor or acceptor Ubs by examining mutational effects on UBE2S autoubiquitination with fluorescent Ub (*Ub), without APC (Fig. S1B). Mutations decreasing APC-dependent substrate ubiquitination but not autoubiquitination mapped to three UBE2S surfaces, which are remote from the active site and differ from the canonical RING binding site on an E2 UBC domain: 1) the UBC domain “backside” (R61A/K63A) distal from the active site, 2) the “C” and “D” helices at the C-terminus of the UBC domain and 3) the extreme C-terminus of the CTP (Fig. 1D, S1A–B). However, because the E2 backside mutant showed increased APC-independent autoubiquitination and no other obvious major defects, we reasoned that its reduced activity in the presence of APC could indirectly arise from self-targeting. Thus, we focused on roles of the other two UBE2S surfaces with APC.
To gain insights into how the different classes of UBE2S mutations impaired their activation by APC, we measured kinetic parameters for APCCDH1-mediated ubiquitination of Ub-CyclinBNTD* while titrating UBE2S concentration (Fig. 1E, S1C). Importantly, the apparent Km (Kmapp) of ≈78 nM for wild-type UBE2S approximates the ≈200 nM Kd recently reported for UBE2S binding to APC (Chang et al., 2014). The mutants in the UBC domain C and D helices displayed nearly wild-type apparent Km (Kmapp) values for UBE2S but decreased apparent Vmax (Vmaxapp). The Helix D (E153A/I154A) mutant was most impaired, with ≈10-fold decreased Vmaxapp. By contrast, the major effect of the CTP L222A mutation was a ≈9-fold increase in Kmapp for UBE2S. Thus, at least two mechanisms couple UBE2S to APC, and one involves binding via the CTP.
Distinctive E2–E3 interactions: UBE2S CTP is recruited to APC Platform even in the absence of the APC11 RING domain
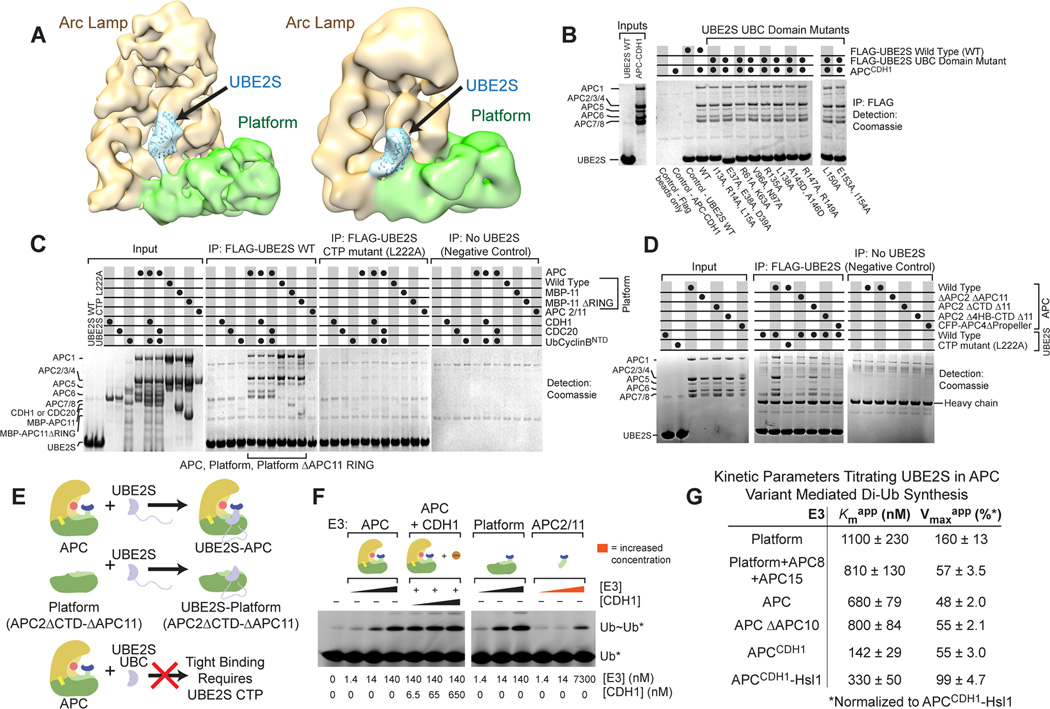
To gain insights into interactions, we examined the structure of APC in complex with UBE2S by cryo-electron microscopy (EM) (Fig. 2A, S2). Compared to the structure of human APC alone (Chang et al., 2014), extra density was readily visible, which we attribute to UBE2S. Two divergent APC-UBE2S conformers were refined at 13 and 23 Å resolution, which showed that 1) the APC Platform is flexible, with several orientations of APC4/APC2/APC11 relative to APC1/APC5, and 2) UBE2S occupies various locations, albeit with common features. Fitting UBE2S’s UBC domain structure (Sheng et al., 2012) into the maps localized UBE2S as protruding from the Platform region comprising the APC2, APC11, and APC4 subunits and extending toward and contacting APC10.
Figure 2. APC Platform and UBE2S CTP mediate high-affinity interactions. (relates to Fig. S2).
A, EM structures showing two conformations of APC-UBE2S at 13 Å (left) and 23 Å (right) resolution. Platform - green, Arc Lamp – tan, density attributed to UBE2S - cyan with modeled UBC domain.
B, Coomassie-stained SDS-PAGE showing binding of recombinant APCCDH1 with FLAG-UBE2S (WT and UBC domain mutants) after anti-FLAG IP.
C, As in B showing FLAG-UBE2S, but not CTP L222A mutant, co-IPs recombinant APC with or without coactivator, Platform with or without His6-MBP-APC11 RING (MBP-11 and MBP-11ΔRING), but not APC2-APC11 (APC2/11). APC subcomplexes are described in Fig. S2.
D, As in B showing FLAG-UBE2S co-IP with APC lacking APC2 C-terminal domain and APC11 (APC2ΔCTD Δ11), but not with further deletions of APC2 or a mutant disrupting APC4 structure (CFP-APC4ΔPropeller).
E, Model summarizing results from A–D. UBE2S CTP is required and APC Platform is sufficient for high-affinity APC-UBE2S interaction. From the Platform, APC4 and APC2 4-helix bundle are required for co-IP with UBE2S, but APC2 CTD and APC11 are not.
F, Phosphorimager scans examining effects of indicated APC variants on UBE2S-mediated synthesis of unanchored Ub~Ub* chains between unlabeled donor Ub and fluorescein-labeled acceptor Ub*. Concentrations of APC variants and CDH1 are indicated.
G, Kinetic parameters comparing APCCDH1 and subcomplexes for recruiting and activating of UBE2S, upon titrating UBE2S concentration in assays monitoring unanchored di-Ub chain synthesis. SEM, n≥3.
To localize high-affinity interactions, we tested coimmunoprecipitation of various purified APC assemblies and subcomplexes with FLAG-tagged wild-type and mutant versions of UBE2S (Fig. 2B–D, S2). Mutations in the UBE2S UBC domain, including the backside (R61A/K63A) and C-terminal helix (E153A/I154A), did not affect high-affinity binding to APC (Fig. 2B). However, the UBE2S CTP L222A mutant failed to bind recombinant APC (Fig. 2C), in agreement with prior studies (Chang et al., 2014; Garnett et al., 2009; Williamson et al., 2009; Wu et al., 2010b). This binding defect explains the increased Kmapp value for the UBE2S CTP L222A mutant.
In terms of APC, similar levels of the APC1-APC5-APC4-APC2-APC11 “Platform” and holo-APC coimmunoprecipitated with FLAG-UBE2S (Fig. 2C, S2). This did not require CDC20, CDH1, or substrate. Although the Platform contains APC2-APC11 that might be expected to bind E2s like other cullin-RING ligases, our data suggest distinctive interactions because the isolated APC2–APC11 subcomplex is not sufficient for high-affinity binding, and significant binding was retained upon deleting the APC11 RING domain from the Platform (Fig. 2C) and upon co-deleting the APC2 C-terminal domain and entire APC11 subunit from the full APC (Fig. 2D). However, UBE2S binding was eliminated by co-deleting APC11 either with the full APC2 subunit or with APC2’s 4-helix bundle and C-terminal domain, or by disrupting APC4’s β-propeller (Fig. 2D, S2). Thus, the APC2–APC4 region of the Platform, near but excluding the APC11 RING, is required for high affinity binding to UBE2S (Fig. 2E).
UBE2S-mediated Ub chain elongation is minimally activated by the cullin-RING-like APC2–APC11 and progressively more enhanced by Platform and APCCDH1
Overall, our data indicated that UBE2S’s CTP and the APC Platform confer noncanonical E2–E3 interactions. Thus, we tested if the APC Platform, like APCCDH1, also influences UBE2S activity. Because the isolated Platform lacks ability to recruit a substrate D-box, we could not compare it with APCCDH1 for ubiquitinating the Ub-CyclinBNTD* substrate. Instead, we examined effects on UBE2S-mediated transfer of an unlabeled donor Ub to a free fluorescein-labeled acceptor Ub, producing unanchored Ub~Ub* chains. Di-Ub chain synthesis was efficiently stimulated by adding APC, either alone or with CDH1, or by adding the Platform subcomplex (Fig. 2F). However, the isolated APC2–APC11 complex was only able to stimulate UBE2S activity at very high concentration (~70-fold molar excess), suggesting that other Platform subunits contribute either directly or indirectly to catalysis (Fig. 2F).
It was possible to quantitatively compare APC subcomplexes for ability to recruit and activate UBE2S by titrating UBE2S concentration in kinetic assays monitoring di-Ub synthesis (Fig. 2G, S1–3). The Platform subcomplex displayed a Kmapp for UBE2S of 1100 nM and Vmaxapp of 160% (normalized to APCCDH1-Hsl1) in synthesizing di-Ub chains. Increasing the number of APC subunits associated with the Platform, such as in a complex containing the Platform, APC8 and APC15, or in the entire APC, led to progressively lower Kmapp values for UBE2S. Saturating APC with CDH1 decreased the Kmapp for UBE2S to 142 nM during synthesis of free di-Ub chains. This is close to the value of 78 nM for polyubiquitination of Ub-CyclinBNTD (Fig. 1E), and is consistent with similar APC-UBE2S interactions in the presence or absence of a D-box substrate. The improved Kmapp values for the larger complexes may reflect APC8, APC15, and/or CDH1 influencing the conformation of the Platform, as was observed in a recent EM study (Chang et al., 2014). Also, some subunits in the larger complexes may make direct but weak contacts to UBE2S (Kelly et al., 2014; Williamson et al., 2009), or could indirectly stabilize key Platform elements involved in catalysis.
Interestingly, the positive effect of CDH1 on di-Ub synthesis is slightly offset by Hsl1 (Fig. 2G), whose D-box tightly engages CDH1 and APC10 but which lacks a priming Ub and thus is not a direct substrate of UBE2S (Burton et al., 2005; Buschhorn et al., 2011; da Fonseca et al., 2011). Furthermore, deleting APC10 did not substantially influence APC activation of UBE2S mediated di-Ub synthesis (Fig. 2G, S3). These results highlight differences between mechanisms by which APC activates UBE2S versus initiating E2s. As binding to initiating E2s was reportedly stimulated by CDH1, APC10, and D-box substrates (Chang et al., 2014; Van Voorhis and Morgan, 2014), the data led us to consider whether distinct mechanisms establish synergy between APC, UBE2S, and the acceptor Ub.
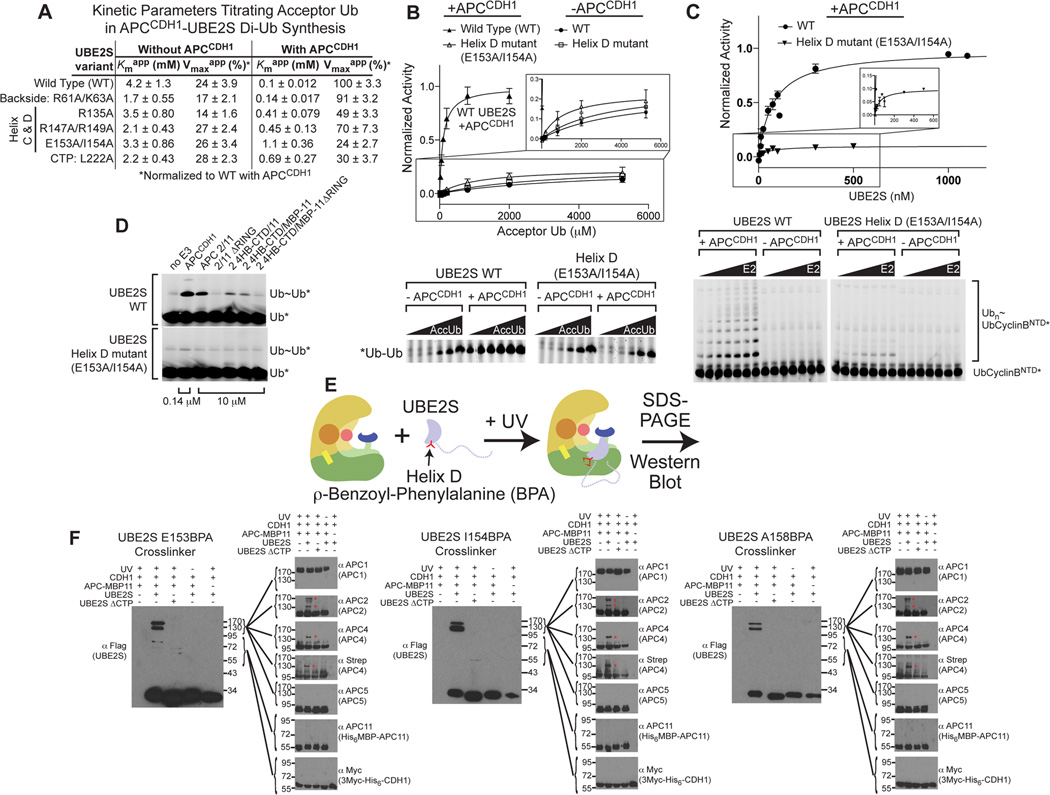
APC activates UBE2S-mediated Ub chain synthesis by lowering Kmapp for acceptor Ub
To investigate roles of APC and UBE2S toward an acceptor Ub, we examined di-Ub synthesis by monitoring transfer of a fluorescein-labeled donor Ub upon titrating unlabeled free Ub as the substrate, either in the absence or presence of APCCDH1 (Fig. 3A, S2E–F). Two interesting points emerged from comparing the kinetic parameters for the acceptor Ub substrate between the different reactions. First, there is a striking effect of adding APCCDH1: a ≈42-fold drop in Kmapp for the acceptor Ub and more than a 4-fold increase in Vmaxapp. Remarkably, APCCDH1 increased the overall catalytic efficiency of Ub chain synthesis by ≈175-fold in a manner predominantly mediated through delivery of the acceptor ubiquitin to UBE2S.
Figure 3. APC drives polyUb chain formation by activating a distinctive region of the UBE2S UBC domain and lowering Kmapp for acceptor Ub. (relates to Fig. S3).
A, Kinetic parameters from titrating acceptor Ub during di-Ub synthesis, to compare effects of APCCDH1 on catalytic efficiency of WT UBE2S or mutants. SEM, n ≥ 3.
B, Fits and representative SDS-PAGE for kinetic parameters in Fig. 3A, for activity as a function of acceptor Ub concentration, showing APCCDH1 activation of di-Ub synthesis by WT UBE2S and APCCDH1-specific defect for the helix D mutant (E153A/I154A).
C, Fits and representative SDS-PAGE for kinetic parameters in Fig. 1E, showing similar Kmapp for WT or Helix D mutant UBE2S, in Ub-CyclinBNTD* ubiquitination by APCCDH1 measured as a function of UBE2S concentration.
D, Comparison of APCCDH1 or indicated versions of cullin-RING-like APC2-APC11 complex (note 70-fold higher concentration) for activating UBE2S-mediated di-Ub synthesis. Reactions with UBE2S Helix D mutant are shown below. APC2/11 subcomplexes are described in Fig. S2.
E, Scheme for photocrosslinking FLAG-UBE2S with BPA (red symbol) in helix D with APC.
F, Western blots for FLAG-UBE2S or CTP mutant with indicated BPA substitutions, Platform subunits, and CDH1 after photocrosslinking as in Fig. 3E. Red asterisks indicate UBE2S-specific crosslinked species observed with APC2 and APC4.
Second, measuring kinetic parameters while titrating the acceptor Ub in assays with UBE2S mutants (Fig. 3A–B, S2E–F) revealed that APCCDH1 is almost invisible to both the CTP L222A and helix D E153A I154A mutants. APC’s inability to activate the CTP mutant is readily explained by greatly decreased interaction (Fig. 2C). However, the helix D E153A/I154A mutant showed substantial interaction with APC by coimmunoprecipitation (Fig. 2B) and in kinetic assays titrating UBE2S (Fig. 1E, data in 3C). Nonetheless, the UBC domain helix D mutant caused an 11-fold increase in Kmapp for the acceptor Ub, and a 4-fold decrease in Vmaxapp for synthesizing di-Ub linkages in the presence of APCCDH1 (Fig. 3A), which we attribute to the I154A substitution (Fig. S1A). Thus, the UBC domain helix D mutant can bind to but is not activated by APCCDH1 in vitro.
UBE2S helix D “senses” APC2/4 for activation
We took two approaches to localize regions of APC that activate UBE2S via its helix D. First, we identified a minimal APC subcomplex that stimulates UBE2S-mediated di-Ub synthesis in a helix D-dependent manner. Mutating UBE2S’s helix D ablated activation by all APC subcomplexes tested, including very low-level stimulation of di-Ub synthesis by a complex between APC11 and a truncated APC2 encompassing the 4HB and CTD (APC24HB-CTD) (Fig. 3D, S2). Surprisingly, deleting the APC11 RING domain eliminated activation of wild-type UBE2S, and this is explored in separate sections below.
As an orthogonal approach to map interactions with UBE2S helix D, we performed crosslinking (Fig. 3E). Amber stop codon suppression technology was used to incorporate the photoactivatable crosslinking amino acid benzoylphenylalanine (BPA) (Chin et al., 2002) into various positions in helix D of Flag-UBE2S, and into negative control variants that do not bind APC due to deletion of key CTP residues. Crosslinked complexes between UBE2S and APC2 and APC4 were identified in western blots as slower-migrating bands dependent on APCCDH1-UBE2S interaction and UV exposure (Fig. 3F). The same results were obtained with APCCDH1 in complex with an Ub-Hsl1 substrate, or with the isolated Platform subcomplex, so for simplification only results for APCCDH1 are shown. Together, the data suggested that the APC2/APC4 region of the Platform activates UBE2S by contacting the C-terminal helix of UBC domain.
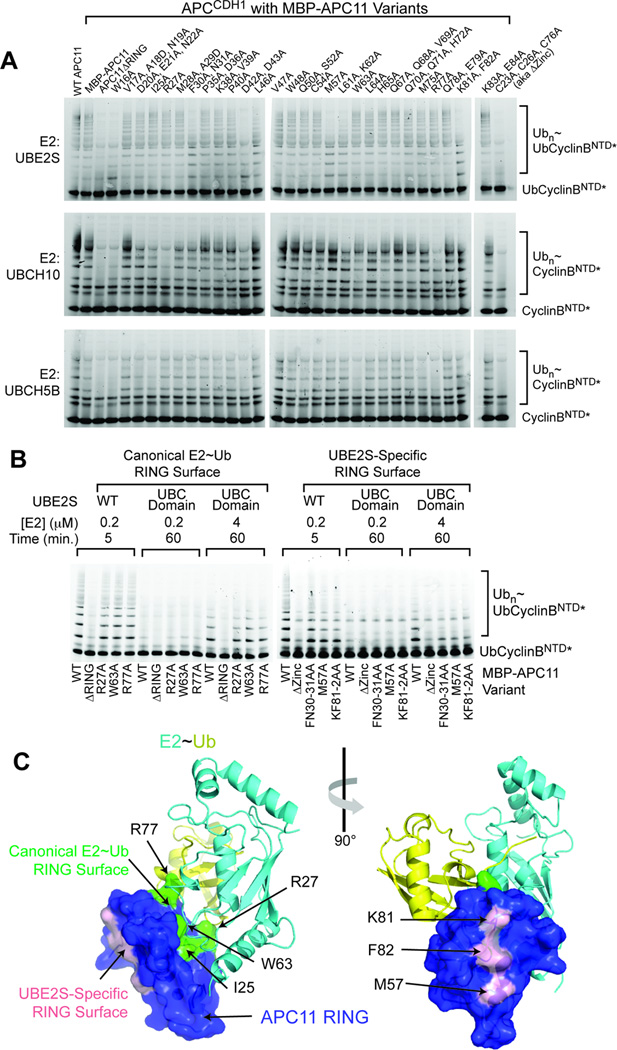
Evidence for unprecedented RING domain activation of Ub chain elongation
We were surprised that deleting the APC11 RING eliminated UBE2S activation by APC2–APC11 (Fig. 3D), as the RING does not influence UBE2S coimmunoprecipitation of APC (Fig. 2C–D), and our saturation mutagenesis did not implicate the surface of UBE2S expected to bind a RING as being important for ubiquitination (Fig. 1, S1A). Thus, we mutationally tested a role for APC11’s RING domain in the context of the whole APC. To facilitate detecting stoichiometric incorporation of mutants into recombinant APC, APC11 was expressed with an N-terminal His6-MBP tag, which did not influence ubiquitination activity (Fig. 4). As predicted, deleting the RING domain eliminated Ub-CyclinBNTD* ubiquitination with the initating E2s UBCH5 and UBCH10. Unexpectedly, however, deleting the APC11 RING domain also eliminated activity with UBE2S (Fig. 4).
Figure 4. Evidence for unprecedented RING E3 mechanism activating polyubiquitination by APC-UBE2S. (relates to Fig. S4).
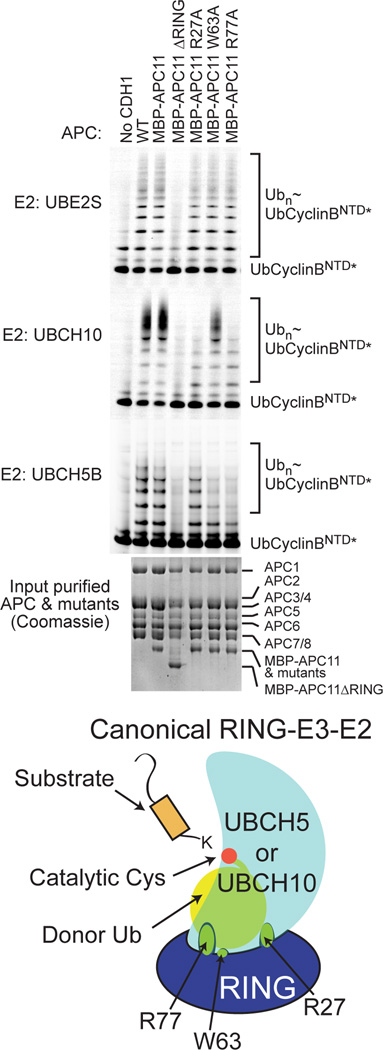
Comparison of Ub-CyclinBNTD* ubiquitination activity for UBE2S, or the initiating E2s UBCH10 or UBCH5, with WT APCCDH1 or versions incorporating His6-MBP-APC11 with indicated RING mutations (Δ - deletion; R27A, W63A, R77A – canonical RING-E2~Ub interaction surface as shown in cartoon). Coomassie-stained gel of APC variants shows stoichiometric incorporation of RING variants.
To test if APC acts as a canonical RING E3 toward UBE2S, we examined effects of Ala substitutions in place of a set of APC11 residues (Arg27, Trp63, and Arg77) that together correspond to anchors in various canonical RING E3-E2~Ub complex structures (Fig. 4) (Dou et al., 2012, 2013; Plechanovova et al., 2012; Pruneda et al., 2012; Scott et al., 2014). None of the canonical RING mutants affected polyubiquitination by UBE2S, despite decreasing ubiquitination with the initiating E2s UBCH5 or UBCH10. The results confirm that APC-mediated Ub-chain initiation occurs via a canonical RING mechanism activating UBCH5 or UBCH10, but indicate that the APC11 RING domain contributes to UBE2S-mediated Ub chain elongation in an atypical manner.
Distinctive RING-dependent interaction with acceptor Ub for APC/UBE2S-mediated Ub chain synthesis
To ascertain the function of the RING, APC complexes were purified containing either wild-type His6-MBP-APC11, or mutants with one to three Ala substitutions in the RING domain (Fig. S4A, 5). The APC RING variants were tested side-by-side for substrate ubiquitination with UBCH5, UBCH10, and UBE2S (Fig. 5A). Mutants showing defects toward all three E2s include a control eliminating zinc-binding cysteines (C23A/C26A/C76A), the W16A “pivot” that orients the RING in a cullin-RING ligase, and D42A/D43A that approach a zinc binding site and presumably influence RING domain stability. There are two other major classes of mutational effects. One class (I25A, R27A, W63A, R77A) maps to the canonical RING surface that binds and activates an E2~Ub intermediate. These are defective toward UBCH5 or UBCH10, but show no effect on UBE2S-mediated ubiquitination. We considered that the high-affinity binding mediated by the UBE2S tail might mask a role for the canonical RING binding site, but this was not the case: mutations in APC11’s canonical E2-binding surface also did not effect ubiquitination with a fragment of UBE2S corresponding to the isolated UBC domain that lacks the CTP but is still weakly stimulated by APC (Fig. 5B).
Figure 5. A distinctive region of APC11 RING domain functions with UBE2S catalytic UBC domain to stimulate polyubiquitination. (relates to Fig. S5).
A, Ala-scanning mutagenesis of APC11 RING domain of APCCDH1, comparing mutational effects on ubiquitination by UBE2S, or the initiating E2s UBCH10 or UBCH5, with Ub-CyclinBNTD* or CyclinBNTD* substrates. Confirmation of stoichiometric incorporation of RING variants was enabled by increasing the size of APC11 with a His6-MBP-APC11 for observation by Coomassie gels (not shown).
B, Similar mutational profiles for UBE2S and its isolated catalytic UBC domain with Ala mutants in APC11 RING domain of APCCDH1. The indicated high concentrations of UBE2S UBC domain and extended reaction times are required to observe ubiquitination of Ub-CyclinBNTD* by APCCDH1 and UBE2S’s isolated catalytic UBC domain due to lack of CTP-mediated binding.
C, UBE2S uses a noncanonical APC11 RING surface. APC11 RING structure (Fig. S5) is shown in blue surface, modeled as if in a canonical complex with E2 (cyan)~Ub (yellow) based on a prior RING-UBCH5~Ub structure (Plechanovova et al., 2012). Sites of mutations defective with UBE2S are shown in pink, and with UBCH10 or UBCH5 in green.
Intriguingly, the other class of RING mutations (F30A/N31A, M57A, K81A/F82A) preferentially impairs APC-UBE2S-mediated ubiquitination, with little or no effect on UBCH5 or UBCH10 (Fig. 5A). Notably, these also impair APCCDC20-mediated ubiquitination of Ub-CyclinBNTD* (Fig. S4B), and APCCDH1’s weak activity toward the UBE2S UBC domain (Fig. 5B), suggesting a role of the APC11 RING in influencing UBE2S’s fundamental catalytic function of generating Ub~Ub linkages. Strikingly, these mutations map to a distinctive surface on the structure of the APC11 RING domain, which we determined by X-ray crystallography and NMR (Fig. 5C, S5). Met57 and Phe82 together form an exposed hydrophobic patch on the opposite side of the RING domain from the canonical E2~Ub binding site (Fig. 5C). Lys81 is adjacent to the Met57/Phe82 surface. Although Asn31 is exposed, Phe30 is partially buried and supports the structural core positioning Met57. Thus, the F30A mutation may have a localized effect on the structure of Met57 that could explain the preferential loss of activity toward UBE2S, and could also impact RING stability, which may explain its minor effect on UBCH10.
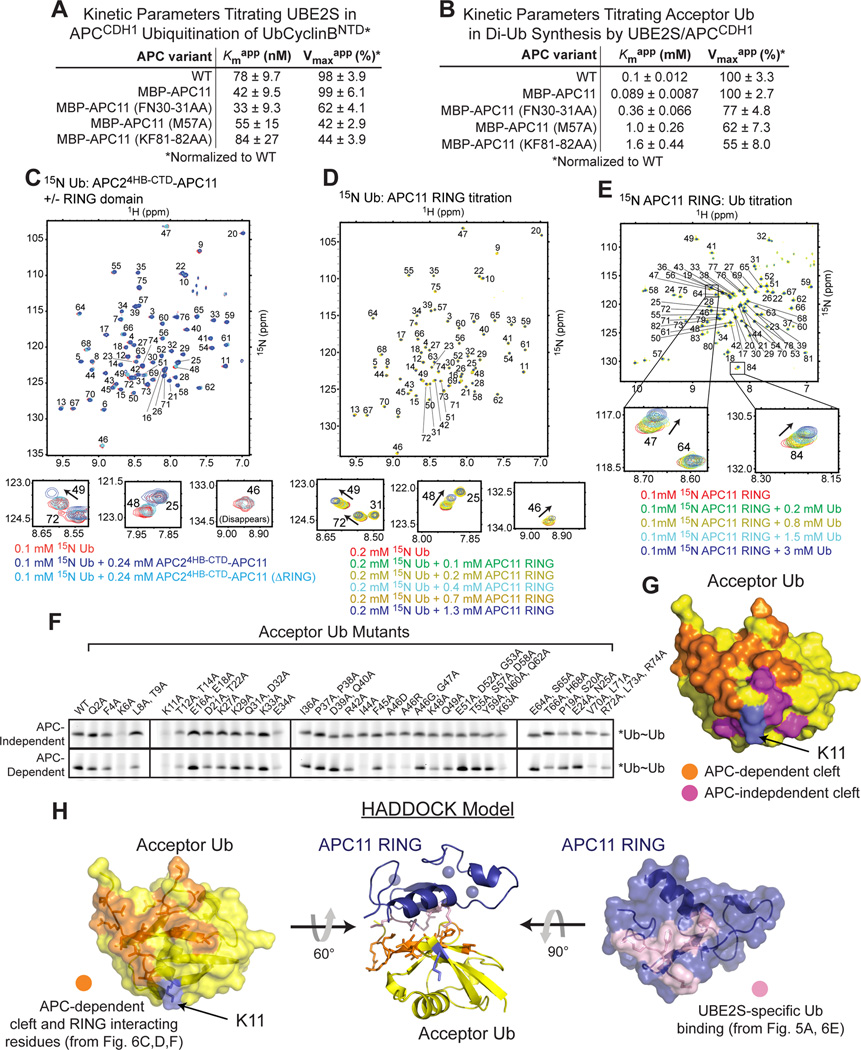
We used the mutants to address how the APC11 RING domain activates UBE2S-mediated Ub chain elongation. In reactions titrating UBE2S and monitoring ubiquitination of Ub-CyclinBNTD*, the distinctive APC11 hydrophobic patch mutants showed little effect on the Kmapp for UBE2S, but caused a roughly 2-fold decrease in Vmaxapp (Fig. 6A, S1G). Moreover, titrating the acceptor Ub in the di-Ub synthesis assay revealed that the distinctive APC11 RING hydrophobic patch mutants substantially increase the Kmapp for a free acceptor Ub, with 10- and 16-fold increases for M57A and K81A/F82A, respectively (Fig. 6B, S1H). At first glance this profound effect of RING mutants on the Kmapp for an acceptor Ub might seem reminiscent of the UBE2S UBC domain E153A/I54A mutant. However, there are differences. The UBE2S mutations appear to affect E2 activation and acceptor Ub interaction, whereas the APC11 RING mutants seem specifically impaired with respect to interacting with an acceptor Ub.
Figure 6. APC-mediated Ub chain elongation involves RING domain presentation of the acceptor Ub to UBE2S.
A, UBE2S-specific APC11 RING surface does not recruit UBE2S. Kinetic parameters upon titrating UBE2S in ubiquitination of Ub-Cyclin BNTD*, with WT or indicated APC11 RING mutant versions of APCCDH1. SEM, n≥3.
B, UBE2S-specific APC11 RING surface influences interaction with acceptor Ub. Kinetic parameters comparing effects of APC11 RING mutations upon titrating acceptor Ub during APCCDH1-UBE2S-mediated di-Ub synthesis. SEM, n ≥ 3.
C, Chemical shift perturbations indicate RING-dependent interactions between Ub and the APC2 4HB-CTD-APC11 subcomplex. TROSY spectra for 15N-labeled Ub alone (red), with APC24HB-CTD-APC11 that can weakly activate Ub-chain synthesis by UBE2S (blue), or a mutant deleted for the RING domain (ΔRING, cyan).
D, Specific chemical shift perturbations upon titrating the isolated APC11 RING into 15N Ub indicate interaction surface. TROSY spectra for 15N Ub alone (red), and titration with 0.1, 0.2, 0.4, 0.7, and 1.3 mM APC11 RING domain in green, yellow, cyan, dark yellow and blue, respectively.
E, Specific chemical shift perturbations upon titrating Ub into 15N-labeled APC11 RING domain indicate interaction surface. TROSY spectra for 15N APC11 RING alone in red, and titration with 0.2, 0.8, 1.5, and 3 mM Ub and colored green, yellow, cyan, dark and blue, respectively.
F, Scanning mutagenesis to identify acceptor Ub surface required for APC-dependent UBE2S-mediated di-Ub synthesis. Fluorescent scans of *Ub-Ub chain formation by UBE2S alone (top) or by UBE2S activated with APCCDH1 (bottom), with indicated acceptor Ub mutants.
G, Results from Fig. 6F shown on surface of Ub structure. Acceptor Ub residues required for APC-dependent UBE2S-mediated Ub~Ub chain formation are orange. Acceptor Ub residues required for UBE2S-mediated chain synthesis both with or without APC are magenta, with acceptor Lys11 in purple.
H, Center - HADDOCK-derived NMR-based model of complex between acceptor Ub (yellow) and APC11 RING (blue). Acceptor Ub residues activated by APC or interacting with APC11 RING identified in Fig. 6 C, D, and F are orange, and their localizing to APC11 RING-binding surface is shown on the left. APC11 RING residues activating the acceptor Ub or interacting with Ub identified in Figs. 5A and 6E are pink, and their localizing to Ub-binding surface is shown on the right.
To explore the role of the APC11 RING, we performed NMR and mutational experiments that together are consistent with a mechanism in which the RING domain would recruit an acceptor Ub for UBE2S-mediated chain elongation. First, adding APC24HB-CTD-APC11, which is minimally sufficient to stimulate Ub chain synthesis by UBE2S (Fig. 3D), to15N-labeled Ub caused selective chemical shift perturbations in15N-1H TROSY spectra (Fig. 6C). The most strongly shifted resonances map to Ub’s Ala46, Lys48, Gln49, His68, and C-terminal region. Notably, this is not the RING-binding surface for the donor Ub in a canonical RING-E2~Ub intermediate (Dou et al., 2012, 2013; Plechanovova et al., 2012; Pruneda et al., 2012). Second, in an experiment performed side-by-side, there was no effect of adding a mutant APC24HB-CTD-APC11 complex lacking the RING domain (ΔRING) (Fig. 6C). Thus, the RING is required for the chemical shift perturbations to Ub. Third, because of the weak interaction, we wished to do a titration experiment. It was necessary to use the isolated APC11 RING domain because we could not obtain high concentrations of the APC24HB-CTD-APC11 complex, produced from insect cells. Although high concentrations were required, titrating the isolated APC11 RING domain progressively shifted the same Ub resonances as APC24HB-CTD-APC11, with predominant effects on Ub’s Thr9, Ile13, Arg42, Ala46, Lys48, Gln49, His68, and C-terminal region (Fig. 6D). Fourth, in converse experiments, adding unlabeled Ub to15N-labeled APC11 RING domain revealed selective chemical shift perturbations for Val47 and Glu84 (Fig. 6E), which are adjacent to the side-chains of Met57 and Phe82 identified in the APC11 Ala scan as critical for reducing the Kmapp for the acceptor Ub (Fig. S5B). Fifth, given the correlation between NMR and mutagenesis experiments for the APC11 RING, we also performed mutational analysis for the acceptor Ub in UBE2S-catalyzed Ub-chain synthesis (Fig. 6F–G). Acceptor Ub mutants impaired for APC-independent catalysis surround the target Lys11, and correspond to those identified previously (Wickliffe et al., 2011). Importantly, acceptor Ub mutants with specific defects in APC-dependent ubiquitination map to the RING-binding surface identified by NMR.
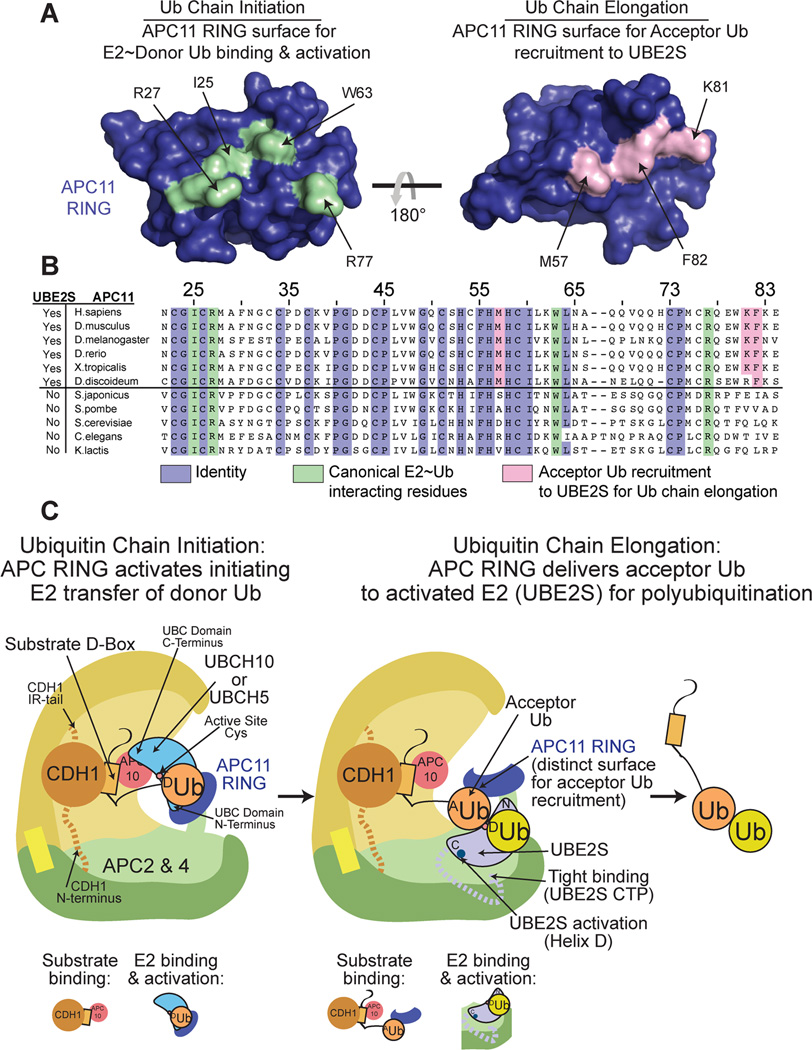
To further examine the potential for direct interactions between the APC11 RING and Ub, we generated models based on the NMR data using the program HADDOCK (de Vries et al., 2007). Clustering the 200 resultant models revealed a dominant cluster representing 87% of all docked models. Although the actual interactions may differ in the context of the whole APC-UBE2S complex mediating polyubiquitination, a representative NMR-based HADDOCK model suggests how an acceptor Ub might bind the distinct APC11 hydrophobic patch, and agrees with our mutational data (Fig. 6H). Notably, sequence alignments of APC11 across several organisms also show striking correlation between conservation of the RING region (Met57, Lys81, Phe82) that lowers the Kmapp for the acceptor Ub and APC’s use of UBE2S for polyubiquitination (Fig. 7).
Figure 7. Repurposing the APC RING to coordinate the acceptor Ub for polyubiquitination by human APC-UBE2S. (relates to Fig. S6).
A, Separate RING domain surfaces activate Ub chain initiation and elongation by the APC E3. APC11 RING structure is shown as blue surface. Canonical APC11 RING surface activating initiating E2~Ub intermediate is green. The APC11 RING surface involved in interacting with and presenting the acceptor Ub to UBE2S for chain elongation maps to the opposite side of the RING domain and is in pink.
B, Sequence alignment of APC11 RING domain from different organisms, and whether or not UBE2S is present in the organism, shows that APC11 surface mediating acceptor Ub interaction during Ub chain elongation is conserved across organisms where UBE2S is present. Conserved, canonical E2~Ub binding, and acceptor Ub interacting residues are colored slate, green and pink, respectively.
C, Model of 2-step/2-E2 polyubiquitination by human APC, highlighting dual RING E3 mechanisms. Both Ub chain initiation with a canonical E2 activation mechanism (left) and Ub chain elongation with UBE2S (right) require the APC11 RING domain. In comparison to canonical RING-E2~Ub mechanisms, human APC activates Ub chain elongation via unique E3-E2 interactions (APC Platform binding UBE2S’s C-terminal peptide), distinctive E3-activation of E2 (the APC2/APC4-region activating helix D of UBE2S’s UBC domain), and by the APC11 RING coordinating the acceptor Ub with UBE2S.
Discussion
Distinctive E2 and RING E3 mechanisms for APC-mediated Ub chain initiation and elongation
Despite fundamental importance, mechanisms of RING E3-mediated polyubiquitination have remained elusive. Here we show how the critical E3 human APC mediates 2-step/2-E2 polyubiquitination through two completely different RING-dependent mechanisms (Fig. 7C). APC initiates Ub ligation to substrates by a mechanism involving canonical RING E3 activity, which involves 1) binding a specific substrate motif, 2) the RING domain recruiting an E2~Ub intermediate and stabilizing the reactive conformation, and 3) crosstalk between the substrate, E2, and E3 to stimulate ligation (Metzger et al., 2014; Scott et al., 2014). APC latches onto substrates via motifs such as a “D-box” co-recruited by CDH1 and APC10, and our mutagenesis showed that the canonical RING E3 surface of APC11 is required with the chain initiating E2s UBCH5 and UBCH10 (Fig. 4, 5). APC substrates drive their own initial modification, apparently by allosterically modulating APC structure to enhance binding of chain initiating E2s (Chang et al., 2014; Van Voorhis and Morgan, 2014).
How do RING E3s target a substrate-linked Ub to build a polyUb chain? For APCCDH1, the D-box of a Ub-modified substrate is thought to remain bound to CDH1 and APC10. However, how APC and UBE2S extend the chain on a substrate-linked Ub has been unclear. Unexpectedly, we found that APC specifically stimulates Ub chain elongation by recruiting and activating UBE2S by a novel multimodal RING-dependent mechanism (Fig. 7C). This involves distinctive surfaces from APC and UBE2S, remote from previously defined RING E3-E2~Ub interaction sites. First, the terminus of UBE2S’s flexible CTP is anchored to the APC Platform in a RING-independent manner (Fig. 2). This tethers UBE2S to APC, and could facilitate E1 reloading of donor Ub molecules onto APC-bound UBE2S to enhance processive polyubiquitination. Second, helix D from UBE2S’s catalytic UBC domain is tweaked by the APC Platform to greatly stimulate polyubiquitination (Fig. 3). Although the activation mechanism remains unknown, one possibility may be inferred from regulation of another E2: helix D from the E2 Pex4 binds an allosteric activator to stimulate peroxisomal ubiquitination (Williams et al., 2012) (Fig. S6). Alternatively, UBE2S’s helix D could coordinate multisite interactions involving the APC2–APC4 region of the Platform, the acceptor Ub, and the APC11 RING. Finally, we showed that APC lowers the Km of an acceptor Ub reacting with the UBE2S~Ub intermediate. In addition to APC activation of UBE2S’s helix D, this involves the I44/A46/K48/Q49/C-terminal region of the acceptor Ub and surprisingly, a distinctive hydrophobic patch from the APC11 RING domain. Although it is possible that future studies will show an indirect role for the RING, at this point the simplest explanation for our data would be that during Ub chain elongation, the APC11 RING domain directly captures the substrate-linked acceptor Ub to enhance its interaction with the UBE2S active site, while the APC2–APC4 region recruits and activates UBE2S in proximity (Fig. 6, 7).
Importantly, there is potential for synergy between key features of this mechanism, UBE2S’s active site, and the Lys11-linked chains produced and extended on substrates. APC binding to the acceptor Ub could synergize with UBE2S’s inherent active site preference for Lys11, by simultaneously directing the acceptor Ub’s Lys11 toward the active site and masking Lys48 from serving as an alternative target (Bremm et al., 2010). Furthermore, whereas the I44/A46/K48/Q49/C-terminal region of Ub is buried in di-Ub chains with some other linkages, this surface is exposed in Lys11-linked chains (Bremm et al., 2010). Thus, as a Lys11-linked chain grows on a substrate, the I44/A46/K48/Q49/C-terminal surface of the distal Ub is available to bind APC for delivery of its Lys11 to UBE2S’s active site.
APC increasing UBE2S access to the acceptor Ub could massively increase polyubiquitination
The importance of enhancing UBE2S access to an acceptor Ub is underscored by results of an earlier study performed without knowledge of the APC mechanism, where tethering UBE2S’s catalytic UBC domain to a heterologous Ub-binding domain increased formation of polyUb chains (Bremm et al., 2010). APC achieves this function naturally, by multiple elements converging to increase interactions between the UBE2S~Ub intermediate and the acceptor Ub, manifested as a ≈40-fold decrease in apparent Km for a free acceptor Ub and an overall ≈175-fold increase in catalytic efficiency under the conditions of our assay (Fig. 3A).
In the context of a highly interconnected APC-coactivator-Substrate~AcceptorUb-UBE2S~DonorUb complex, avidity effects that culminate in bringing an acceptor Ub to the active site could massively stimulate polyubiquitination in many ways. The accompanying study by Rape and coworkers shows that APC binding to an acceptor Ub can serve as a means to track the acceptor at the tip of a single Ub chain growing on a substrate (Kelly et al., 2014). Indeed, we found that deleting the APC11 RING domain, which is required for lowering the Km for a free acceptor Ub, completely eliminated generation of a chain on the Ub-CyclinBNTD* substrate (Fig. 4). Interestingly, point mutations in the RING domain that partially impair acceptor Ub interaction caused a striking loss of long chains on Ub-CyclinBNTD* in qualitative assays (Fig. 5A–B). Although future studies will be required to determine the precise basis for this effect, one possibility is that APCCDH1 binding to both a D-box and to an acceptor Ub could increase the residence time of Ub-modified substrates on APC, thereby enhancing processivity of UBE2S-mediated polyubiquitination. It is also possible that the position of an acceptor Ub influences its capture by APC, with preferential effects on long chain formation in the context of the Ub-CyclinBNTD* substrate. More complicated scenarios may arise with natural substrates that are initially decorated with multiple individual Ub molecules (Dimova et al., 2012). Here, APC may preferentially guide a particular priming Ub to the UBE2S active site, and essentially recapture the tip of one selected chain as it grows. At some point, the length of a given chain may exceed topological constraints for the substrate to remain anchored to a coactivator and APC10, and for the acceptor Ub to simultaneously engage the APC11 RING and UBE2S’s active site. When the optimal length is exceeded for one chain, another substrate-linked Ub may be preferentially guided for extension by APC/UBE2S. As a result, multiple Ub chains may ultimately be produced on a substrate. In the context of the cellular milieu, the APC-Ub interaction could also protect a growing chain and prevent its premature disassembly or degradation, by blocking access of deubiquitinating enzymes, proteasome receptors, or other destabilizing Ub-binding machineries. Thus, by delivering the acceptor Ub to UBE2S, the distinctive APC RING mechanism would influence the linkage, length, nature, positions, and density of polyUb chains on a substrate, with the net effect of driving proteasomal turnover of cell cycle regulatory proteins to control cell division.
Implications of APC/UBE2S mechanism for cell cycle regulation
The distinct mechanism of APC/UBE2S-mediated Ub chain elongation may contribute to ordering of events during the cell cycle. A prior study indicated that APC substrates processively modified by chains in a single binding event are degraded earlier in mitosis compared to those substrates needing more cycles of APC binding to receive multiple ubiquitins (Rape et al., 2006). In a related vein, substrates modified by UBE2S are likely more extensively modified with polyUb chains. Thus, substrates that require long Ub chains for proteasomal turnover might be degraded only during phases of the cell cycle when UBE2S is most active (Dimova et al., 2012; Matsumoto et al., 2010).
UBE2S activity in vivo is positively regulated by assembly with APC in a CDC20-dependent manner (Kelly et al., 2014), and inhibited by EMI1 and EMI2 mimicking UBE2S’s CTP and impeding Ub chain elongation during interphase and meiosis, respectively (Frye et al., 2013; Sako et al., 2014; Wang and Kirschner, 2013). It is conceivable that EMI1/2 inhibition could also involve blocking the distinctive APC11 RING surface and acceptor Ub interaction. Timing is further influenced by autocatalytic ubiquitin-dependent proteolysis of UBE2S (Garnett et al., 2009; Rape and Kirschner, 2004; Williamson et al., 2009; Wu et al., 2010b). Notably, the accompanying work suggests UBE2S specifically regulates inactivation of the spindle assembly checkpoint (Kelly et al., 2014). Future studies will be required to identify key roles of APC/UBE2S-generated Lys11-linked polyUb chains in the checkpoint and other aspects of cell cycle regulation.
General implications for RING E3-E2-mediated polyubiquitination
Acceptor Ub recruitment and/or positioning is emerging as a fundamental component of RING-E3-E2-mediated polyubiquitination. Prior studies showed potential for the acceptor Ub to be recruited by domains embedded within E2s or their partners, or by specialized polyubiquitinatining E4 enzymes (Choi et al., 2010; Eddins et al., 2006; Koegl et al., 1999; Liu et al., 2014; Spratt and Shaw, 2011). Even the chain elongating E2 functioning with yeast APC, Ubc1, which is thought to be activated by a canonical RING mechanism, can recruit Ub via its own UBA domain (Merkley and Shaw, 2004). During evolution and transfer of chain elongating activity to UBE2S, the job of acceptor Ub recruitment apparently was shifted to APC. RING domain repurposing may be an ideal mechanism for recruiting an acceptor Ub. Indeed, with its location adjacent to the catalytic center, and the notorious ability of RING domains to rotate relative to the rest of E3 enzymes (Duda et al., 2008), the APC11 RING domain may be ideally poised to capture a terminal Ub immediately after its ligation (Fig. 7A,B).
It is appealing to speculate that the APC11 RING serves as a hub to integrate Ub chain initiation and elongation (Williamson et al., 2009). Although we do not have any evidence for cooperativity between UBCH10 and UBE2S, this is in principle possible based on mutagenic and NMR studies showing separation between the canonical RING site for E2~Ub activation and the surface delivering the acceptor Ub to UBE2S (Fig. 5–6). Future studies will be required to determine if, how, when, and why the two steps of Ub chain formation by APC occur simultaneously or synergistically.
At this point, we do not know if other E3s utilize their RING domains to deliver an acceptor Ub to the E2 for polyubiquitination. Nonetheless, our findings with APC and UBE2S expand our knowledge of RING E3 and E2 mechanisms. We speculate that future studies will show that some of the massive number of RING proteins associated with Ub pathways have alternative functions like human APC, providing specialized points regulating ubiquitination.
Experimental Procedures
Other than yeast Hsl1, proteins are human, and were expressed and purified for enzyme and interaction assays, NMR, and EM largely as described (Frye et al., 2013), with some alterations. In particular, recombinant APC was purified to greater homogeneity by a protocol similar to that used by (Chang et al., 2014), with affinity purification based on a C-terminal twin-Strep tag on APC4, followed by anion exchange chromatography and gel filtration. Although our previous protocol for purifying recombinant APC led to a Km for UBE2S in Ub-CyclinBNTD* ubiquitination assays (Frye et al., 2013) that matched well to values obtained in similar reactions with endogenous APC (Wang and Kirschner, 2013), we attribute improved catalytic efficiency herein to improved homogeneity from our revised APC purification scheme.
NMR and crystallography were performed much as described previously (Duda et al., 2008; Frye et al., 2013) (Tables 1 and 2). Samples for cryo-EM were prepared using the Grafix protocol (Kastner et al., 2008), blotted and vitrified (Vitrobot, FEI Company). Images were recorded at a magnification of 74,000× (2 Å/pixel) under cryogenic conditions in a Cs corrected Titan Krios (FEI Company) electron microscope on a Falcon II direct electron detector (FEI Company).
Table 1.
Crystallographic Data and Refinement Statistics.
| Data Collection | |
|---|---|
| Wavelength (λ) | 1.2827 |
| Space Group | P21 |
| Unit cell parameters | |
| a, b, c (Å) | 52.45, 39.88, 65.04 |
| α, β, ϒ(°) | 90.00, 108.49, 90.00 |
| Resolution (Å) | 61.58 – 1.76 |
| No. of measured reflections | 123,853 |
| No. of unique reflections | 24,444 |
| Overall Rsym (%) | 5.5 (16.7) |
| Completeness (%) | 95.1 (80.2) |
| Overall I/σI | 24.0 (7.6) |
| Mean Redundancy | 5.1 |
| Refinement | |
| Rwork/Rfree | 0.192/0.229 |
| rmsd bond lengths (Å) | 0.008 |
| rmsd bond angles (°) | 1.2 |
| Subunits in asymmetric unit | 4 |
| No. of atoms | |
| Protein | 2030 |
| Zinc | 12 |
| Water | 166 |
| Ramachandran statistics | |
| Residues in most favored regions (%) | 98.3 |
| Residues in disallowed regions (%) | 0.0 |
Highest resolution shell is shown in parenthesis. Rfree is the cross-validation of R-factor, with >8% of the total reflections omitted in model refinement.
Table 2.
Statistics for the NMR structure calculation of APC11 RING.
| Constraints | |
|---|---|
| No. of upper distance limits | 1033 |
| Intraresidue | 408 |
| Short range (|i-j| ≤ 1) | 260 |
| Medium range (2 ≤ |i-j| ≥ 5) | 118 |
| Long range (5 < |i-j|) | 247 |
| Zinc Restraints | 12 |
| No. of dihedral angle constraints | 32 |
| No. of Hydrogen Bonds | 18 |
| Residual target function (Å2) | 0.33 ± 0.04 |
| Distance violations > 0.2 Å | |
| Minimum violation (Å) | 0.02 ± 0.004 |
| Maximum violation (Å) | 0.03 ± 0.004 |
| Angle violation (deg) | |
| Minimum | 0.04 ± 0.03 |
| Maximum | 0.14 ± 0.03 |
| Atomic pairwise rmsd (Å)a | |
| Backbone atoms | 0.42 ± 0.08 |
| Heavy atoms | 1.01 ± 0.01 |
| Structural analysis | |
| Residues in allowed region (%) | 98.3 |
| Residues in generously allowed region (%) | 1.7 |
| Residues in disallowed region (%) | 0.0 |
Backbone and heavy atom rms deviations are obtained by superimposing residues 8–15, 30–70 of the APC11 RING. Residues 1 to 6 and 16 to 30 did not show any long range NOEs and hence were unstructured. Backbone heavy atom rmsd for 3–70 is 1.07 ± 0.2.
For UBE2S-APC co-immunoprecipitation binding assays, APC complexes and UBE2S variants were mixed at concentrations of 0.1 µM and 6 µM, respectively, with Anti-FLAG M2 affinity gel (Sigma) in 20 mM HEPES pH 8.0, 200 mM NaCl, 0.25 mg/mL BSA, and 0.1% Tween20. After three hours of gentle mixing, the beads were washed repeatedly with cycles of resuspending in buffer, spinning down beads, and re-washing prior to adding SDS gel-loading buffer to the beads. The samples were then boiled and analyzed by SDS-PAGE and Coomassie staining.
Enzyme assays were performed largely as described previously (Frye et al., 2013). In the figures, the position of the * indicates the position of the fluorescent label on a substrate, with * before the name indicating an N-terminal label, and * after the name indicating a C-terminal label. The assays measuring enzyme kinetics of APC-dependent ubiquitination have two substrates, the UBE2S~DonorUb intermediate and the substrate being modified by the donor Ub, which here was either Ub-CyclinBNTD* or various versions of free Ub optimal for observing di-Ub synthesis in the different conditions. Thus, titrating UBE2S concentrations yielded kinetic constants for UBE2S and titrating free acceptor Ub yielded kinetic constants for the acceptor Ub.
Detailed methods are provided in Supplementary Information.
Supplementary Material
Highlights.
-
-
APC uses canonical RING mechanism for initial substrate ubiquitin (Ub) modification
-
-
APC uses unprecedented mechanisms to activate the E2 UBE2S for polyubiquitination
-
-
APC drives UBE2S-mediated Ub chain elongation by lowering Kmapp for the acceptor Ub
-
-
Noncanonical APC11 RING surface delivers acceptor Ub for chain elongation by UBE2S
Acknowledgments
We thank M Rape and colleagues for communicating results prior to publication; KP Wu, A Dick and D Gerlich for advice, discussions, and collaboration; and S Bozeman, DW Miller, and J Bollinger for support. For funding, we thank Jane Coffin Childs Foundation (NGB); Deutsche Forschungsgemeinschaft Sonder forschungsbereich 860 (HS); Boehringer Ingelheim, the Laura Bassi Centre for optimized structural studies, EU-FP7 grant MitoSys and the Austrian Research Fund (JMP); ALSAC, NIH R37GM065930 and P30CA021765, and HHMI (BAS); and NIH P41GM103403 and DOE DE-AC02-06CH11357 (NECAT and APS). BAS is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
NGB, BAS, JMP and HS planned and supervised the project. NGB, ERW, MAJ, RQ and IFD performed biochemical and biophysical analyses. FW, RV, GP and JJF developed recombinant APC system used here. NGB and RV prepared EM samples. HS performed EM with help from PD. NGB and CRRG performed NMR analyses. NGB, SEC and OA prepared samples for biochemical and biophysical studies. AN performed AUC. JB and JZ performed HADDOCK modeling. NGB, OA, and IK performed x-ray crystallography. NGB, JJF, ERW, HS and BAS prepared the manuscript with input from all authors.
Accession codes
4R2Y.pdb, 2MT5.pdb, BMRB-25149, EMD-2775, EMD-6084
References
- Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. The Journal of biological chemistry. 1996;271:2823–2831. doi: 10.1074/jbc.271.5.2823. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Wiener R, Yu IW, Ringel AE, Wolberger C. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat Chem Biol. 2013;9:154–156. doi: 10.1038/nchembio.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nature structural & molecular biology. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Molecular cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nature structural & molecular biology. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014 doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Wu K, Jeong K, Lee D, Jeon YH, Choi BS, Pan ZQ, Ryu KS, Cheong C. The human Cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. The Journal of biological chemistry. 2010;285:17754–17762. doi: 10.1074/jbc.M109.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nature cell biology. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nature structural & molecular biology. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nature structural & molecular biology. 2013;20:982–986. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P, Herzog F, Gieffers C, Sander B, Riedel D, Muller SA, Engel A, Peters JM, Stark H. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Molecular cell. 2005;20:867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature structural & molecular biology. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Frye JJ, Brown NG, Petzold G, Watson ER, Grace CR, Nourse A, Jarvis MA, Kriwacki RW, Peters JM, Stark H, et al. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nature structural & molecular biology. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nature cell biology. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science (New York, NY. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Molecular cell in revision. 2014 doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Molecular cell. 2008;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Liu W, Shang Y, Zeng Y, Liu C, Li Y, Zhai L, Wang P, Lou J, Xu P, Ye Y, et al. Dimeric Ube2g2 simultaneously engages donor and acceptor ubiquitins to form Lys48-linked ubiquitin chains. The EMBO journal. 2014;33:46–61. doi: 10.1002/embj.201385315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Molecular cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nature structural & molecular biology. 2014;21:308–316. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- Merkley N, Shaw GS. Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. The Journal of biological chemistry. 2004;279:47139–47147. doi: 10.1074/jbc.M409576200. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochimica et biophysica acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. The Journal of cell biology. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2 approximately Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-box Ligases. Molecular cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Molecular cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako K, Suzuki K, Isoda M, Yoshikai S, Senoo C, Nakajo N, Ohe M, Sagata N. Emi2 mediates meiotic MII arrest by competitively inhibiting the binding of Ube2S to the APC/C. Nature communications. 2014;5:3667. doi: 10.1038/ncomms4667. [DOI] [PubMed] [Google Scholar]
- Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA. Structure of a RING E3 Trapped in Action Reveals Ligation Mechanism for the Ubiquitin-like Protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Hong JH, Doherty R, Srikumar T, Shloush J, Avvakumov GV, Walker JR, Xue S, Neculai D, Wan JW, et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol Cell Proteomics. 2012;11:329–341. doi: 10.1074/mcp.O111.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt DE, Shaw GS. Association of the disordered C-terminus of CDC34 with a catalytically bound ubiquitin. Journal of molecular biology. 2011;407:425–438. doi: 10.1016/j.jmb.2011.01.047. [DOI] [PubMed] [Google Scholar]
- Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nature structural & molecular biology. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhis VA, Morgan DO. Activation of the APC/C Ubiquitin Ligase by Enhanced E2 Efficiency. Curr Biol. 2014 doi: 10.1016/j.cub.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nature cell biology. 2013;15:797–806. doi: 10.1038/ncb2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Panjikar S, Stanley WA, Distel B, Wilmanns M. Insights into ubiquitin-conjugating enzyme/ co-activator interactions from the structure of the Pex4p:Pex22p complex. The EMBO journal. 2012;31:391–402. doi: 10.1038/emboj.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Molecular cell. 2010a;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2010b;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.