Abstract
Certain mitochondrial haplotypes (mthaps) are associated with disease, possibly through differences in oxidative phosphorylation and/or immunosurveillance. We explored whether mthaps are associated with allogeneic HCT outcomes. Recipient (n=437) and donor (n=327) DNA was genotyped for common European mthaps (H, J, U, T, Z, K, V, X, I, W, K2). HCT outcomes for mthap matched siblings (n=198), all recipients, and all donors were modeled using relative risks (RR) and 95% confidence intervals and compared to mthap H, the most common. Siblings with I and V were significantly more likely to die within 5 years (RR=3.0; 1.2–7.9 and 4.6; 1.8–12.3, respectively). W siblings experienced higher aGVHD II–IV events (RR=2.1; 1.1–2.4) with no events for K or K2. Similar results were observed for all recipients combined, although J recipients experienced lower GVHD and higher relapse. Patients with I donors had a 2.7 fold (1.2–6.2) increased risk of death in five years, while few patients with K2 or W donors died. No patients with K2 donors and few patients with U donors relapsed. Mthap may be an important consideration in HCT outcomes, although validation and functional studies are needed. If confirmed, it may be feasible to select donors based on mthap to increase positive or decrease negative outcomes.
Keywords: Mitochondria, HCT, GVHD, polymorphism
INTRODUCTION
Mitochondria (mt) are essential organelles of bacterial origin captured by eukaryotic cells through endosymbiosis billions of years ago [reviewed in [1]]. Mt provide cells energy through oxidative phosphorylation (OXPHOS) and electron transport, regulate cell survival and death, and are increasingly thought to play a key role in innate and adaptive immune system responses [2–4]. While most mitochondrial DNA (mtDNA) was incorporated into human nuclear DNA throughout evolution, there remains a 16.6kb closed double-stranded circular mitochondrial genome that contains 37 genes encoding 22 tRNAs, 2 rRNAs and 13 proteins that play an integral role in OXPHOS [5, 6]. Unlike nuclear DNA, which is inherited from both maternal and paternal sources, paternal mtDNA is degraded by ubiquitination during fertilization; thus mtDNA inheritance is almost exclusively maternal.
Like nuclear DNA, mtDNA can experience deletions and mutations that lead to a variety of rare diseases including dystonia, myopathies, myoclonic epilepsy and ragged red fibers, lactic acidosis, Leber’s hereditary optic neuropathy, aminoglycoside-induced sensorineuronal hearing loss and Pearson’s syndrome, among others [1, 7–9]. In contrast to nuclear DNA, the mt genome can accumulate mutations much more readily since it contains no introns or histones, thus mtDNA heteroplasmy is often seen with aging and may also contribute to disease risk or modify disease severity [9–11].
The mt genome is inherited solely through the maternal line, it is a useful marker of human migration around the globe [12]. Approximately 30 mt haplotypes (mthaps) have been identified by restriction fragment length polymorphism worldwide [13, 14], with additional subdivisions among haplotype groups. Select mthaps are found in indigenous populations within specific regions of the world, suggesting either climatic selection or genetic drift [15]. Haplotype L is the oldest, with origins in Africa, and gave rise to macrohaplogroups (M, N, R) in Europe and Asia, and subsequent descendant sub lineage haplotypes (www.mitomap.org). Haplotype H is the most common in Western Europe. Nevertheless, within any one self-identified race/ethnicity group or geographic region, population admixture can be variable, making it sometimes difficult to strongly associate race or ethnicity with specific mthaps [16].
Using cytoplasmic hybrids (cybrids), which are cells consisting of identical nuclear DNA but containing different mt haplotypes, there is increasing evidence of OXPHOS, reactive oxygen species (ROS) and other functional differences among mthaps [17, 18], supporting a role for these polymorphisms in cell function and/or disease susceptibility. Related, several association studies have linked mthaps with longevity [19], specific cancers [20], Leber’s hereditary optic neuropathy [21], survival after sepsis [22], progression to heart disease [23] and progression from HIV to AIDS [24], while others have found no associations [25, 26]. While differences between donor and recipient mtDNA have been used to quantitate donor engraftment after allogeneic transplantation [27], no study to our knowledge has investigated mthaps in relation to HCT outcomes.
Allogeneic HCT is a time of high metabolic demand: there is a need for hematopoietic restoration and response to febrile and septic events, as well as response to challenges of graft versus host disease (GVHD) and graft versus leukemia (GVL) reactions [28]. Given increasing evidence of functional differences in mthap cellular energetic [2–4, 23], mthap variations could be important in HCT outcomes. We explored associations between mthaps of recipients and donors on patient outcomes following HCT. Our data suggest that certain mthaps may be important independent predictors of mortality, GVHD and relapse.
METHODS
Patient Demographics
Pre-transplant DNA was available from 437 adult and pediatric patients who received an allogeneic HCT at the University of Minnesota for a hematological malignancy between 1995 and 2005, along with DNA from 327 donors (DNA was not available from 110 umbilical cord blood (UCB) donors). Of the transplants, 213 were related donor (198 siblings, 15 other related), 73 adult unrelated, and 151 UCB (Table 1). Clinical and laboratory data were systematically and prospectively collected on all patients and entered into the University of Minnesota Blood and Marrow Transplant Database. All patients and/or their parents or guardians provided signed consent to participate in institutional review board approved transplant protocols; outcomes were reviewed retrospectively.
Table 1.
Patient (n=437) Demographics Across Entire Study Population
| Variable | Total Study Group | |
|---|---|---|
| Patient Age (Years) | 0–17 | 83 (19.0%) |
| ≥ 18 | 354 (81.0%) | |
| Year of Transplant | 1995–2000 | 166 (38.0%) |
| 2000–2005 | 271 (62.0%) | |
| Patient Sex | Male | 250 (57.2%) |
| Female | 187 (42.8%) | |
| Patient Race | Caucasian | 376 (86.0%) |
| Hispanic | 16 (3.7%) | |
| Asian | 14 (3.2%) | |
| Unknown | 12 (2.7%) | |
| Mixed | 9 (2.1%) | |
| African American | 7 (1.6%) | |
| American Indian | 3 (0.7%) | |
| Donor Type | Sibling | 198 (45.3%) |
| Unrelated Donor (URD) | 73 (16.7%) | |
| Single UCB | 44 (10.1%) | |
| Double UCB | 107 (24.5%) | |
| Related Other | 15 (3.4%) | |
| Conditioning | Myeloablative | 302 (69.1%) |
| RIC | 135 (30.9%) | |
| Recipient CMV Serostatus | R+ | 219 (50.1%) |
| R−/D− | 177 (40.5%) | |
| R−/D+ | 41 (9.4%) | |
| Conditioning | Cy/TBI +/− ATG | 418 (95.7%) |
| Bu/Cy/Melphalan | 2 (0.5%) | |
| Bu/Flu/TBI | 10 (2.3%) | |
| Cy/TBI/VP16 | 1 (0.2%) | |
| GvHD Prophylaxis | CNI + MTX (calcineurin inhibition) | 225 (51.5%) |
| CsA/MMF | 182 (41.6%) | |
| CsA +/− MPD +/− ATG | 27 (6.2%) | |
| CsA/MTX/Pred | 2 (0.5%) | |
| Diagnosis | AML | 122 (27.9%) |
| NHL | 81 (18.5%) | |
| ALL | 78 (17.8%) | |
| CML | 64 (14.6%) | |
| MDS/MPS | 40 (9.2%) | |
| Hodgkins | 23 (5.3%) | |
| CLL | 13 (3.0%) | |
| JMML | 8 (1.8%) | |
| Other Malignancy | 8 (1.8%) | |
| Disease Risk | Std Risk | 227 (51.9%) |
| High Risk | 210 (48.1%) |
Mthap DNA Testing
Participants were assessed for the eleven most common European mthaps (in descending order of frequency: H, J, U, T, Z, K, V, X, I, W, K2), which required genotyping each DNA sample for eight different mt single nucleotide polymorphisms (SNPs) including mt1719, mt4580, mt7028, mt8251, mt9055, mt10398, mt12308, and mt13368 using Taqman [29]. For each Taqman SNP assay performed, a master mix was made using 1× concentration of Taqman 2× Genotyping Master Mix, 5uM of each forward and reverse primer and 1uM of each VIC and FAM probe (Life Technologies, Grand Island, NY) aliquoted to a final volume of 11.5ul per well in a 96 well plate. Using a multi-channel pipette, 1ul of DNA (average concentration, 2.5ng/ul) was pipetted from stock DNA into the assay plate for a final volume reaction of 12.5ul. Each plate included 4 negative controls, 4 positive controls for the VIC labeled allele, 4 positive control wells for the FAM labeled allele, and 5% duplicates to verify calls. After plating was complete, an adhesive cover was securely applied and the plate was placed in a thermal cycler for the following: 2 minutes at 50°C followed by 10 minutes at 95°C, then 40 cycles of 15 seconds at 95°C followed by 1 minute at 60°C. After cycling, the plate was read on a 7900 HT Prism Sequence Detection System (Life Technologies, Grand Island, NY) using the allelic discrimination feature and analyzed using Sequence Detection Software v2.1.1. This procedure was performed for all 8 SNPs separately. Following an allele call for all 8 SNPs for a single sample, one of the 11 mthaps was determined using a checkerboard approach [29]. A total of 29 samples (19 patient, 10 donor) had an mthap that did not correspond to one of the 11 European groups defined by the above primer sets and were labeled as ‘other’.
Transplant Regimens, GVHD Prophylaxis and Data Collection
Data on characteristics of transplantation, post transplantation complications and outcomes were prospectively collected by the Biostatistical Support Group at the University of Minnesota using standardized collection procedures. Details regarding graft selection, conditioning regimens, and GVHD prophylaxis have been previously described [30–34]. All patients were followed longitudinally until death or last follow-up. Endpoints considered included disease-free survival (DFS) through 5 years, non-relapse mortality (NRM), relapse, and GVHD.
Statistical Analysis
The analysis focused on the influence of mthap on HCT outcomes in matched siblings, all recipients (including matched sibling patients, other related donor patients, and patients with URD and UBC HCTs) and all donors (including matched sibling donors, other related donors, URDs and UBCs) with available DNA. The all recipient and all donor groups were strongly influenced by the dominant sibling group, since, unfortunately, we had insufficient sample size to evaluate unrelated donors or UCB donors separately from the siblings.
Comparisons between donor sources were completed by the general Wilcoxon test for continuous factors and the chi-square test for categorical factors. Factors considered for adjustment included stem cell source (marrow vs. peripheral blood), age (<18 vs. ≥ 18 years), disease risk (standard vs. high), conditioning (reduced intensity conditioning (RIC) vs. myeloablative), recipient cytomegalovirus (CMV) serostatus (positive vs. negative) and patient sex. Disease risk at the time of HCT was classified into standard risk or high risk based on the ASBMT RFI 2006 risk scoring schema (http://www.asbmt.org). Acute leukemia in first or second complete remission; CML in first chronic phase; Hodgkin's or non-Hodgkin's lymphoma in complete or partial chemotherapy sensitive remission and CLL in first remission; myelodysplastic syndrome or myeloproliferative disorder without excess blasts were considered standard risk and all others high risk at the time of transplantation. Association between HLA and haplotypes was assessed by Fisher’s exact test. DFS was estimated by Kaplan-Meier curves [35]. NRM was analyzed using cumulative incidence treating relapse as a competing risk. Relapse and GVHD were analyzed using cumulative incidence treating non-event death as a competing risk [36]. Comparisons were completed with the simple log rank test. Cox regression was used to assess the independent effect of the indices on five-year overall DFS [37] and Fine and Gray proportional hazards regression was used to assess the independent effect of the indices on NRM, relapse and GVHD [38]. The most common mthap, H, was used as the referent group. Forest plots were used to facilitate visualization.
RESULTS
Mthap data were available from pre-transplant specimens collected from 437 patients and 327 donors. Table 1 shows patient demographic data among the entire study population. Over 80% of patients were 18 years or older at transplant (range 6 months to 69.6 years, median: 39.9 years). The majority (45%) of transplants were matched related siblings, followed by double UBC (24.5%) and URD transplants (16.7%). Overall, the distribution of mthaps in both patients and donors was comparable (Table 2) with H being the most frequent. Below we present transplant outcomes based on mthap of siblings, all recipients, and all donors, with the latter two groups combining matched related donors and unrelated donors.
Table 2.
Mitochondrial Haplotype (mthap) Frequencies for Recipients and Donors
| mthap (Recipient) | H | 156 (35.7%) |
| I | 7 (1.6%) | |
| J | 59 (13.5%) | |
| K | 22 (5.0%) | |
| K2 | 7 (1.6%) | |
| T | 39 (8.9%) | |
| U | 55 (12.6%) | |
| V | 16 (3.7%) | |
| W | 7 (1.6%) | |
| X | 15 (3.5%) | |
| Z | 35 (8.0%) | |
| “Other” | 19 (4.3%) | |
| mthap (Donor) | H | 127 (37.6%) |
| I | 7 (2.1%) | |
| J | 36 (11.0%) | |
| K | 18 (5.5%) | |
| K2 | 7 (2.1%) | |
| T | 32 (9.8%) | |
| U | 34 (9.2%) | |
| V | 9 (2.8%) | |
| W | 4 (1.2%) | |
| X | 8 (2.4%) | |
| Z | 35 (10.7%) | |
| “Other” | 10 (3.1%) |
Sibling HCT Outcomes
All 198 sibling donors matched their recipients on mthap, as expected given they have the same biological mother. The majority of sibling recipients received myeloablative conditioning for a variety of malignant hematological diseases (data not shown). Most (73%) received peripheral blood stem cells; GVHD prophylaxis was mainly with cyclosporine/methotrexate (CSA/MTX) (75%). MtHap did not significantly differ by patient age, year of transplant, sex, donor type, CMV serostatus, conditioning regimen, GVHD prophylaxis, disease risk, HLA mismatch, or source of cells (data not shown). Eighty-nine percent of patients across the European mthaps self-identified as non-Hispanic white (NHW), with the exceptions of Z (n=15; 53% NHW, 20% Hispanic, 20% Asian, 7% mixed), V (n=5; 60% NHW, 40% unknown), and K (n=8, 75% NHW, 13% mixed, 13% mixed).
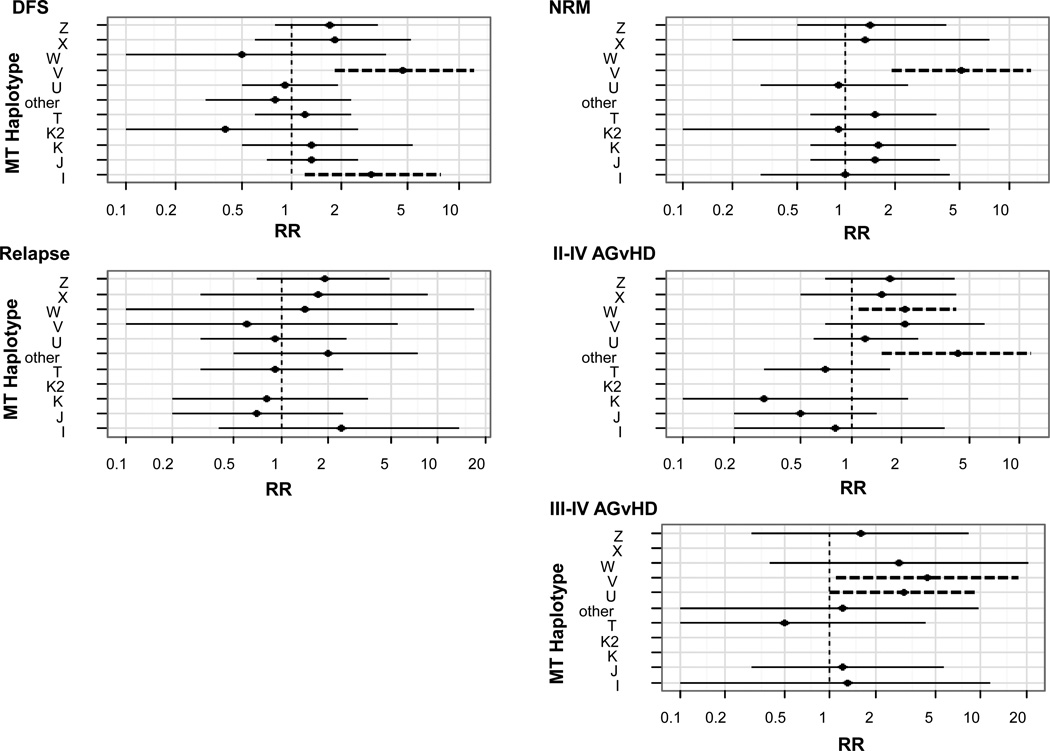
A total of 109/198 (55%) sibling recipients died within five years of transplant, but there were significant differences depending on mthap. Compared to haplotype H (43 events/83 patients, 52%), recipients with I and V were significantly more likely to die within 5 years from HCT (n=5/6 (83%), RR=3.0, 95% CI=1.2–7.9; n=5/5 (100%), 4.6, CI=1.8–12.3), respectively, after controlling for stem cell source, age, disease risk, conditioning, CMV status and sex (Figure 1). A total of 55/198 (28%) sibling recipients died of NRM events in two years. Compared to H (21/83, 26%), 80% of patients with V experienced NRM within two years following transplant (RR=5.1, 1.9–13.7), while no patients with W (n=4) or ‘other’ (n=8) died in that time period. Overall, 48/198 (25%) sibling recipients relapsed within 5 years of transplant (Figure 1). There were no statistically significant differences among mthap compared to H (24% had events).
Figure 1. Multivariate analysis of transplant outcomes for mthap matched sibling donors and recipients based on European mthaps.
All outcomes were normalized to mthap H, which was the most common. Outcomes are shown for disease free survival (DFS) (relative risk (RR) reflects risk of death), non-relapse mortality (NRM), relapse and graft versus host disease (GvHD). Factors which we attempted to adjust for included stem cell source (marrow vs. pbsc), age (<18 vs. ≥ 18), disease risk (standard vs. high), conditioning (RIC vs. myeloablative), recipient CMV serostatus (positive vs. negative) and patient sex (male vs. female). Dashed lines represent significance at the p<0.05 level.
Seventy-five (38%) patients experienced grade II–IV acute GVHD (aGVHD) within 100 days of transplant (data not shown). Compared to H (39% total events), however, there were significantly higher events among W (3/4 (75%), RR=2.1, 1.1–4.2) and the ‘other’ group (6/8 (75%), RR=4.3, 1.5–11.7).. Twenty-four (12%) patients experienced Grade III–IV aGVHD by day 100. Compared to H (9/83, 11%), patients with U (5/19 (26%)) and V (2/5 (40%)) experienced significantly higher events (RR=3.1, 1.0–9.3; 4.4, 1.1–17.7, respectively); there were no events for K, K2, or X.
Lastly, a total of 82 (41%) patients experienced chronic GVHD (cGVHD). However, after controlling for the competing risk of non-GVHD death within two years, there were no mthaps that were significantly different than H.
All Recipients Combined and HCT Outcomes
As the above comparison within siblings could not distinguish whether recipient or donor mtDNA played differing roles in outcomes, we combined the sibling cohort with the remaining group of transplants, which added 239 recipients (15 non-sibling related donor, 73 adult URD and 151 UCB donors).
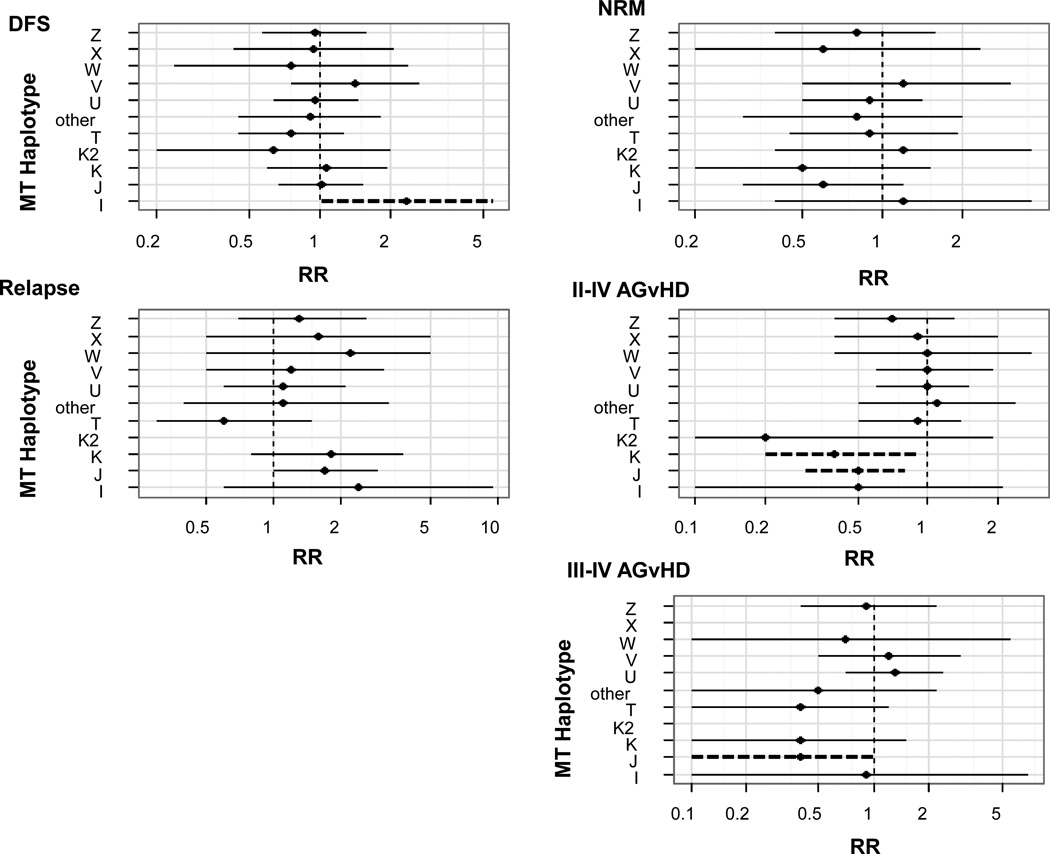
Of the 437 recipients, 241 (55%) died within five years of transplant (data not shown). Compared to haplotype H with 89 events/156 (57%), recipients with I were more likely to die in 5 years (n=6/7 (86%), RR=2.4, 95% CI=1.0–5.4), whereas in contrast to the 198 matched full siblings, individuals in the recipients overall group having mthap V did not experience a significantly increased risk of death (12/16 (75%) RR=1.4, 0.8–2.6) after controlling for donor type, age, disease risk, conditioning, CMV status and sex (Figure 2). Overall, 118/437 (27%) patients experienced NRM in two years. Compared to H (31%), recipients with W (n=7) experienced no events (p=0.10).
Figure 2. Multivariate analysis of transplant outcomes for all recipients based on European mthaps.
All outcomes were normalized to mthap H, which was the most common. Outcomes are shown for disease free survival (DFS) (relative risk (RR) reflects risk of death), non-relapse mortality (NRM), relapse and graft versus host disease (GvHD). Factors which we attempted to adjust for included donor type (hla sibling match vs. sibling mm+URD vs UCB), age (0–18 vs. 18+), disease risk (standard vs. high), conditioning (RIC vs. myeloablative), recipient CMV serostatus (positive vs. negative) and patient sex (male vs. female). Dashed ines represent significance at the p<0.05 level.
Relapse occurred in 115/437 patients within 5 years of transplant (data not shown). There were no significant differences among mthaps compared to H (37 events/156 (24%)), although no patients with K2 (n=7) experienced relapse (Figure 2).
A total of 195 (45%) patients experienced grade II–IV aGVHD within 100 days of transplant (data not shown). Compared to H (78/156 (50%) events), recipients with J and K experienced significantly fewer events (17/59 (29%), RR=0.50 (0.3–0.8) and 6/22 (27%), RR=0.4 (0.2–0.9), respectively (Figure 2). Seventy-one (16%) patients experienced Grade III–IV aGVHD by day 100. Compared to H (30/156, 19%), patients with J (5/59, 8%) experienced fewer events (RR=0.4, 0.1–1.0), while no patients with K2 (n=7) or X (n=15) experienced Grade III–IV aGVHD. In contrast to the matched siblings, recipients overall with U (30/50, 55%) and V (12/16, 75%) experienced more events compared to H (89/156, 57%), but they did not reach statistical significance (Figure 2).
A total of 139 (32%) patients experienced cGVHD. After controlling for the competing risk of non-GVHD death within two years, there were no recipient haplotypes that were significantly different than mthap H (59/156, 38%).
All Donors Combined and HCT Outcomes
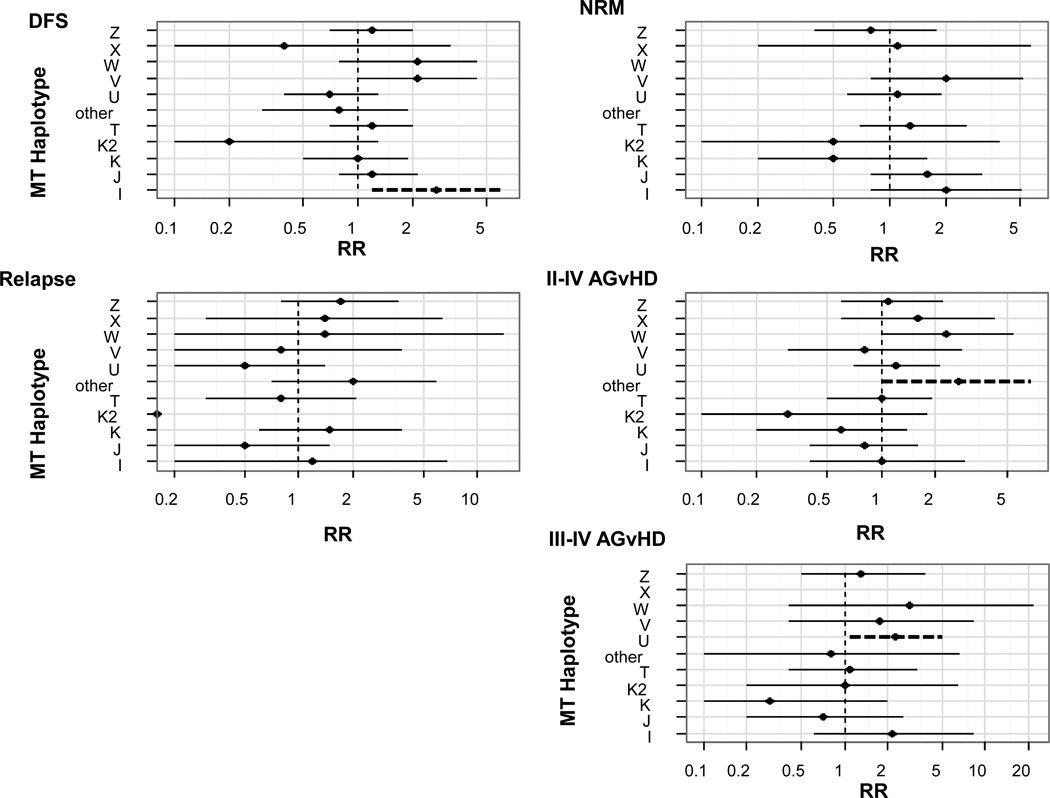
Finally, we combined the 198 sibling donors with the remaining group of 129 donors with DNA available. Of the 327 recipients with known donor mthap, 176 (54%) died within five years of transplant. Compared to H donors (68/127, 54%), patients who received transplants from I donors (6/7, 86%) were 2.7 times (1.2–6.2) more likely to die after controlling for donor type, age disease risk, CMV status and sex (Figure 3); V donors were also associated with higher risk of death in recipients (8/9, 89%, RR=2.1, 1.0–4.5). Overall, 97 (30%) patients experienced NRM in two years (data not shown); there were no statistically significant differences for any of the mthaps, although there was no NRM among recipients from W (n=4) or ‘other’ (n=10) donors (Figure 3).
Figure 3. Multivariate analysis of transplant outcomes based on donor European mthaps.
All outcomes were normalized to mthap H, which was the most common. Outcomes are shown for disease free survival (DFS) (relative risk (RR) reflects risk of death), non-relapse mortality (NRM), relapse and graft versus host disease (GvHD). Factors which we attempted to adjust for included donor type (hla sibling match vs. sibling mm+URD vs UCB), age (0–18 vs. 18+), disease risk (standard vs. high), conditioning (RIC vs. myeloablative), recipient CMV serostatus (positive vs. negative) and patient sex (male vs. female). Dashed lines represent significance at the p<0.05 level.
Overall, 70/327 (21%) patients relapsed within 5 years of transplant. While there were no significant differences among haplotype groups compared to H donors (27/127, 23%), no patients receiving transplants from K2 donors (n=7) experienced relapse. Further, patients who received transplants from J donors experienced a 49% reduction in relapse (CI=0.2–1.5), with only 4/36 (11%) events (Figure 3).
A total of 127 (39%) patients experienced grade II–IV aGVHD within 100 days of transplant. Compared to H donors (48/127, 38%), there were significantly higher events among the ‘other’ donor group (7/10 (70%), RR=2.7, 1.0–6.8), and borderline significantly higher events among recipients of W donors (3/4 (75%), RR=2.3, 95% CI=1.0–5.4) (Figure 3). For grade III–IV aGVHD, compared to H donors (16/127, 13%), patients who received transplants from U donors experienced significantly higher events (10/34 (29%), RR=2.3, 1.1–4.9), while patients receiving transplants from donors with haplotype X and K experienced few events (0/8 and 1/18, respectively).
Lastly, a total of 107 (33%) patients experienced cGVHD. After controlling for the competing risk of non-GVHD death within two years (majority of patients who received transplants from donors with haplotype V and I died), there were no haplotype donors that resulted in significantly different outcomes from H donors (48/127, 38%).
DISCUSSION
To our knowledge, this is the first study to explore associations between patient and donor mthaps and HCT outcomes. We first focused on biological siblings, given they were the most homogenous group with respect to mtDNA since they were all matched. We found that compared with haplotype H (see Supplemental Table 1), there were significantly increased risks of 1) death at five years from HCT for mthaps I and V, 2) NRM for V, 3) aGVHD II–IV for W and the ‘other’ group, and 4) aGVHD III–IV for U and V. In contrast, we observed lower risks of 1) death five years from HCT for K2 and W, 2) NRM for W, 3) relapse for K2, 4) aGVHD II–IV for J, K and K2, and 5) aGVHD III–IV for K, K2, and X. However, because siblings are matched on mthap, it was impossible to distinguish the contribution of recipient or donor mthaps to these HCT outcomes, and, unfortunately, we had insufficient sample size to evaluate other recipient and donor groups separately from siblings. Thus, to evaluate which mthap (donor or recipient) might be driving sibling associations, we combined the other related, adult URD and UCB recipients and donors with the siblings to determine whether the associations in the matched siblings became stronger, weaker, or new associations emerged.
For five year overall survival, similar to siblings, mthap I was associated with a significantly increased risk of death over mthap H for all recipients and all donors (Supplemental Table 1). However, donors with mthap V were associated with an increased risk of patient death, but recipients with V had no increased risk, suggesting V donors may be contributing to the increased risk of death in siblings. Similar to siblings, recipients or donors with K2 or W were associated with fewer deaths, albeit with very small numbers of patients in each group. For two year NRM, again V donors rather than V recipients appeared to be driving the significant association in siblings. Identical to siblings, recipients or donors with mthap W were associated with no patient NRM events. For 5 year relapse, J recipients experienced borderline significantly higher events compared to H, which was not observed in either the sibling or donor groups. Identical to siblings, K2 donors and patients with K2 recipients experienced no relapse. For aGVHD II–IV, W donors appeared to be contributing to the significantly increased risk in siblings, whereas J recipients experienced a significantly lower number of events that was not observed with the J donor group. Similar to siblings, patients with K or K2 mthaps, or, receiving HCT from K or K2 donors, experienced fewer aGVHD II–IV events than H. Finally, for aGVHD III–IV, U donors appeared to be driving the significantly increased risk in matched siblings, whereas V was only significantly associated with an increased risk in the siblings. J recipients experienced significantly fewer aGVHD III–IV events, which were not observed in siblings or donors. K, K2 and X experienced few aGVHD III–IV events regardless of donor or recipient status.
MtDNA lacks introns, thus genetic variation can lead to amino acid substitutions and functional changes in the proteins they encode resulting in variation in OXPHOS, ROS production or other essential mitochondrial functions [39, 40]. While the function of many of the 11 European mthaps remains to be determined, cybrid studies show notable differences in oxygen consumption (higher in H compared to J [41]), susceptibility to ROS (higher in H compared to T [18] or J [42]), mt copy number (higher in J compared to H [43]), and ATP production (lower in J compared to H [42]). As functional studies of mthaps in donors and recipients have not yet been done in the context of HCT, we can only speculate on underlying mechanisms for our observations.
For recipients, mthaps could influence the cellular energetics needed for tissue repair following chemotherapy and radiation, and thus contribute to increased mortality, aGVHD and/or relapse. In our study, we observed significantly lower risks of GVHD but higher risk of relapse in J recipients compared to H recipients. Mthap J is associated with lower OXPHOS and ATP production [41, 42]; it is possible that such a response in recipients could contribute to a lower GVHD response, which has been associated with higher relapse [44].
For donors, mthaps could also influence cellular energetics [45] and/or the relative magnitude and duration of an alloreactive response. Recent murine studies show that during GVHD, T cells shift from aerobic glycolysis to a combination of glycolysis and OXPHOS [46]. As well, T cells have a hyperpolarized mt membrane potential and elevated superoxide production. Given these observations, and that mthaps have been associated with time to progression for multiple sclerosis and HIV [47, 48], various donor mthaps might contribute to aGVHD (through high OXPHOS) and perhaps, relapse (through low OXPHOS). Here, even with small numbers, we found patients of donors with W and U had an increased risk of GVHD. In further support of immune function effects, mt cybrids have differential sensitivity to NK cell mediated attack, apparent in a murine orthotropic tumor model where tumor cybrids showed differential growth rates based on NK cell recognition [4]. Moreover, it is also possible that the genetic differences between donor and recipient mtDNA may serve as a minor histocompatibility antigen, leading to T cell recognition and increased aGVHD. We were unable to examine this question since among our URD and UCB recipient-donor pairs (n=223), only 15 (6.7%) were mthap matched. In a previous study, two mt proteins (MTAP8 and MTND3) that have predicted HLA-A2 binding motifs did not increase GVHD in mismatched donors and recipients [49]. However, others have shown that peptides derived from mtDNA can be presented in the context of MHC class I [50]. There is also evidence from somatic cell nuclear transfer studies in cattle that mthap incompatibility between oocytes and donor cells leads to less developmental competence of constructed embryos [51]. Therefore, variations in donor and recipient mtDNA serving as minor histocompatibility antigens remain a distinct possibility in the context of HCT and require further study.
There are several limitations to these preliminary analyses. We had very small cell sizes for many of the donor mthaps that show the most promise for favorable HCT outcomes (K, K2, X). There were many comparisons and it is possible that our observations (especially among the smaller mthap groups) were due to chance. The patient group was heterogeneous with respect to disease, which could influence observed associations between disease recurrence and mthap. However, while we adjusted for a number of potential confounders, we found (ad hoc) that restricting our analysis to only highly significant confounders did not change results. We also were not able to evaluate the non-sibling recipient and donor groups separately, nor could we evaluate mismatch of mthap between donor and recipient. Nevertheless, despite our small patient numbers, we demonstrated potential differences in HCT outcomes based on mthap, especially with regard to NRM, GVHD and DFS. There is also growing evidence of a biological basis for such observations. We will be extending this work to the URD setting in the National Marrow Donor Program and conducting a large study in over 4200 recipients and 4200 donors to validate our findings. We will also investigate the relative contribution of mthap of donor compared to recipient and HCT outcomes following mismatch. Further, we will be conducting functional studies of the extreme mthaps in an HCT setting.
If validated, it would be feasible to select donor mthaps associated with less GVHD and/or relapse (e.g. J, K2, U) or avoid use of donor mthaps associated with adverse outcomes (e.g., I, V). Notably, while we performed Taqman to genotype individual SNPs for mthap calls, we have been provided an estimate of ~$11.50/DNA sample to completely sequence the mt genome and analyze for mthaps with a turnaround time of only weeks (personal communication, Dr. Kenneth Beckman, Director of the University of Minnesota Genomics Center). Thus, it would be economically feasible and expedient to genotype DNA samples on a large scale housed at donor centers for mthaps. Moreover, with over 11 million potential donors in the NMDP pool (www.bethematch.org) and more than 22.5 million potential donors available worldwide, it is likely that additional matching by mthap is feasible.
Nevertheless, donor selection is becoming increasingly complex, with multiple factors needing to be considered. We would speculate that there are a relatively small number of patients with very common HLA types who have many donors. For these patients, or for recipients of UCB, assessment of donor mtDNA may lead to improved outcomes by selecting donors associated with lower rates of GVHD, etc. However, for the majority of recipients of adult donor transplantation, we would speculate that there are relatively few donors (or no donors). For these patients, it is possible that mtDNA assessment will not guide donor selection per se, but may lead to changes in supportive care, such as higher doses (or levels) of immune suppression and/or more extensive antibiotics prophylaxis. So, while donor selection may be further complicated, we envision these data could ultimately improve outcomes.
Supplementary Material
Highlights.
There is increasing evidence that mitochondrial DNA polymorphisms (haplotypes, “mthap”) have functional effects on cellular energetics and immune function
No study to our knowledge has explored the potential role of mthaps in HCT outcomes
We show preliminary evidence that specific mthaps (both recipient and donor) may play an important role in patient outcomes following HCT
If our data our confirmed, it may be feasible to additionally select donors on mthap to increase positive HCT outcomes
Acknowledgements
Supported by Children’s Cancer Research Fund Hodder Chair (JAR); NIH T32 CA099936 (JAR); NIH K05 CA157439 (JAR), R21 CA135180 (KR), and the Tulloch Chair in Stem Cell Biology, Genetics and Genomics (JT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure Statement: The authors have nothing to disclose.
REFERENCES
- 1.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 2.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol. 2012;13:780–788. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa K, Toyama-Sorimachi N, Nakada K, et al. The innate immune system in host mice targets cells with allogenic mitochondrial DNA. J Exp Med. 2010;207:2297–2305. doi: 10.1084/jem.20092296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 6.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 7.Chinnery PF, Hudson G. Mitochondrial genetics. Br Med Bull. 2013;106:135–159. doi: 10.1093/bmb/ldt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace DC, Murdock DG. Mitochondria and dystonia: the movement disorder connection? Proc Natl Acad Sci U S A. 1999;96:1817–1819. doi: 10.1073/pnas.96.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu DP, Economou EP, Antonarakis SE, Maumenee IH. Mitochondrial DNA mutation and heteroplasmy in type I Leber hereditary optic neuropathy. Am J Med Genet. 1992;42:173–179. doi: 10.1002/ajmg.1320420208. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 11.Brown DT, Herbert M, Lamb VK, et al. Transmission of mitochondrial DNA disorders: possibilities for the future. Lancet. 2006;368:87–89. doi: 10.1016/S0140-6736(06)68972-1. [DOI] [PubMed] [Google Scholar]
- 12.Singh KK, Kulawiec M. Mitochondrial DNA polymorphism and risk of cancer. Methods Mol Biol. 2009;471:291–303. doi: 10.1007/978-1-59745-416-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 14.Brotherton P, Haak W, Templeton J, et al. Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nat Commun. 2013;4:1764. doi: 10.1038/ncomms2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace DC. Mitochondria, bioenergetics, and the epigenome in eukaryotic and human evolution. Cold Spring Harbor Symp Quant Biol. 2009;74:383–393. doi: 10.1101/sqb.2009.74.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell SL, Goodloe R, Brown-Gentry K, Pendergrass SA, Murdock DG, Crawford DC. Characterization of mitochondrial haplogroups in a large population-based sample from the United States. Human Genet. 2014;133:861–868. doi: 10.1007/s00439-014-1421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney MC, Chwa M, Atilano SR, et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: Implications for population susceptibility to diseases. Biochim Biophys Acta. 2014;1842:208–219. doi: 10.1016/j.bbadis.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller EE, Brunner SM, Mayr JA, Stanger O, Sperl W, Kofler B. Functional differences between mitochondrial haplogroup T and haplogroup H in HEK293 cybrid cells. PloS One. 2012;7:e52367. doi: 10.1371/journal.pone.0052367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Benedictis G, Rose G, Carrieri G, et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 20.Verma M, Naviaux RK, Tanaka M, Kumar D, Franceschi C, Singh KK. Meeting report: mitochondrial DNA and cancer epidemiology. Cancer Res. 2007;67:437–439. doi: 10.1158/0008-5472.CAN-06-4119. [DOI] [PubMed] [Google Scholar]
- 21.Torroni A, Petrozzi M, D'Urbano L, et al. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 22.Baudouin SV, Saunders D, Tiangyou W, et al. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366:2118–2121. doi: 10.1016/S0140-6736(05)67890-7. [DOI] [PubMed] [Google Scholar]
- 23.Strauss KA, DuBiner L, Simon M, et al. Severity of cardiomyopathy associated with adenine nucleotide translocator-1 deficiency correlates with mtDNA haplogroup. Proc Natl Acad Sci U S A. 2013;110:3453–3458. doi: 10.1073/pnas.1300690110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, et al. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadjixenofontos A, Schmidt MA, Whitehead PL, et al. Evaluating mitochondrial DNA variation in autism spectrum disorders. Ann Hum Genet. 2013;77:9–21. doi: 10.1111/j.1469-1809.2012.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson G, Panoutsopoulou K, Wilson I, et al. No evidence of an association between mitochondrial DNA variants and osteoarthritis in 7393 cases and 5122 controls. Ann Rheum Dis. 2013;72:136–139. doi: 10.1136/annrheumdis-2012-201932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao YG, Childs RW, Kajigaya S, McCoy JP, Jr, Young NS. Mitochondrial DNA sequence heterogeneity of single CD34+ cells after nonmyeloablative allogeneic stem cell transplantation. Stem Cells. 2007;25:2670–2676. doi: 10.1634/stemcells.2007-0269. [DOI] [PubMed] [Google Scholar]
- 28.Welniak LA, Blazar BR, Murphy WJ. Immunobiology of allogeneic hematopoietic stem cell transplantation. Annu Rev Immunol. 2007;25:139–170. doi: 10.1146/annurev.immunol.25.022106.141606. [DOI] [PubMed] [Google Scholar]
- 29.Raby BA, Klanderman B, Murphy A, et al. A common mitochondrial haplogroup is associated with elevated total serum IgE levels. J Allergy Clin Immunol. 2007;120:351–358. doi: 10.1016/j.jaci.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 30.Wagner JE, Barker JN, DeFor TE, et al. Transplantation of unrelated donor umbilical cord blood in 102 patients with malignant and nonmalignant diseases: influence of CD34 cell dose and HLA disparity on treatment-related mortality and survival. Blood. 2002;100:1611–1618. doi: 10.1182/blood-2002-01-0294. [DOI] [PubMed] [Google Scholar]
- 31.Oran B, Dolan M, Cao Q, Brunstein C, Warlick E, Weisdorf D. Monosomal karyotype provides better prognostic prediction after allogeneic stem cell transplantation in patients with acute myelogenous leukemia. Biol Blood Marrow Transplant. 2011;17:356–364. doi: 10.1016/j.bbmt.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Bachanova V, Brunstein CG, Burns LJ. Fewer infections and lower infection-related mortality following non-myeloablative versus myeloablative conditioning for allotransplantation of patients with lymphoma. Bone Marrow Transplant. 2009;43:237–244. doi: 10.1038/bmt.2008.313. [DOI] [PubMed] [Google Scholar]
- 33.Warlick ED, Tomblyn M, Cao Q, et al. Reduced-intensity conditioning followed by related allografts in hematologic malignancies: long-term outcomes most successful in indolent and aggressive non-Hodgkin lymphomas. Biol Blood Marrow Transplant. 2011;17:1025–1032. doi: 10.1016/j.bbmt.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisdorf D, Zhang MJ, Arora M, Horowitz MM, Rizzo JD, Eapen M. Graft-versus-host disease induced graft-versus-leukemia effect: greater impact on relapse and disease-free survival after reduced intensity conditioning. Biology Blood Marrow Transplant. 2012;18:1727–1733. doi: 10.1016/j.bbmt.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 36.Lin DY. Non-parametric inference for cumulative incidence functions in competing risks studies. Stat Med. 1997;16:901–910. doi: 10.1002/(sici)1097-0258(19970430)16:8<901::aid-sim543>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 37.Cox D. Regression models and life tables. J Royal Stat Soc B. 1972;34:187–220. [Google Scholar]
- 38.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 39.Lenaz G, Genova ML. Structural and functional organization of the mitochondrial respiratory chain: a dynamic super-assembly. Int J Biochem Cell Biol. 2009;41:1750–1772. doi: 10.1016/j.biocel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Rachek LI, Grishko VI, Alexeyev MF, Pastukh VV, LeDoux SP, Wilson GL. Endonuclease III and endonuclease VIII conditionally targeted into mitochondria enhance mitochondrial DNA repair and cell survival following oxidative stress. Nucleic Acids Res. 2004;32:3240–3247. doi: 10.1093/nar/gkh648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez-Duran A, Pacheu-Grau D, Martinez-Romero I, et al. Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber's hereditary optic neuropathy. Biochim Biophys Acta. 2012;1822:1216–1222. doi: 10.1016/j.bbadis.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Kenney MC, Chwa M, Atilano SR, et al. Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1 genes: implications for age-related macular degeneration. PloS One. 2013;8:e54339. doi: 10.1371/journal.pone.0054339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suissa S, Wang Z, Poole J, et al. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan KM, Weiden PL, Storb R, et al. Influence of acute and chronic graft-versus-host disease on relapse and survival after bone marrow transplantation from HLA-identical siblings as treatment of acute and chronic leukemia. Blood. 1989;73:1720–1728. [PubMed] [Google Scholar]
- 45.Battersby BJ, Redpath ME, Shoubridge EA. Mitochondrial DNA segregation in hematopoietic lineages does not depend on MHC presentation of mitochondrially encoded peptides. Hum Mol Genet. 2005;14:2587–2594. doi: 10.1093/hmg/ddi293. [DOI] [PubMed] [Google Scholar]
- 46.Gatza E, Wahl DR, Opipari AW, et al. Manipulating the bioenergetics of alloreactive T cells causes their selective apoptosis and arrests graft-versus-host disease. Sci Ttransl Med. 2011;3:67ra68. doi: 10.1126/scitranslmed.3001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzman-Fulgencio M, Jimenez JL, Garcia-Alvarez M, et al. Mitochondrial haplogroups are associated with clinical pattern of AIDS progression in HIV-infected patients. J Acquir Immune Defic Syndr. 2013;63:178–183. doi: 10.1097/QAI.0b013e3182893f74. [DOI] [PubMed] [Google Scholar]
- 48.Campbell GR, Ziabreva I, Reeve AK. Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann Neurol. 2011;69:481–492. doi: 10.1002/ana.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa Y, Kashiwase K, Okai M, et al. Polymorphisms in the coding region of mtDNA and effects on clinical outcome of unrelated bone marrow transplantation. Bone Marrow Transplant. 2001;28:603–607. doi: 10.1038/sj.bmt.1703199. [DOI] [PubMed] [Google Scholar]
- 50.Duvvuri B, Duvvuri VR, Wang C, et al. The human immune system recognizes neopeptides derived from mitochondrial DNA deletions. J Immunol. 2014;192:4581–4591. doi: 10.4049/jimmunol.1300774. [DOI] [PubMed] [Google Scholar]
- 51.Yan ZH, Zhou YY, Fu J, et al. Donor-host mitochondrial compatibility improves efficiency of bovine somatic cell nuclear transfer. BMC Dev Biol. 2010;10:31. doi: 10.1186/1471-213X-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.