Abstract
Background
Quantitation of cytomegalovirus (CMV) DNA using real-time PCR has been utilized for monitoring CMV infection. However, the CMV antigenemia assay is still the 'gold standard' assay. There are only a few studies in Korea that compared the efficacy of use of real-time PCR for quantitation of CMV DNA in whole blood with the antigenemia assay, and most of these studies have been limited to transplant recipients.
Method
479 whole blood samples from 79 patients, falling under different disease groups, were tested by real-time CMV DNA PCR using the Q-CMV real-time complete kit (Nanogen Advanced Diagnostic S.r.L., Italy) and CMV antigenemia assay (CINA Kit, ArgeneBiosoft, France), and the results were compared. Repeatedly tested patients were selected and their charts were reviewed for ganciclovir therapy.
Results
The concordance rate of the two assays was 86.4% (Cohen's kappa coefficient value=0.659). Quantitative correlation between the two assays was a moderate (r=0.5504, P<0.0001). Among 20 patients tested repeatedly with the two assays, 13 patients were transplant recipients and treated with ganciclovir. Before treatment, CMV was detected earlier by real-time CMV DNA PCR than the antigenemia assay, with a median difference of 8 days. After treatment, the antigenemia assay achieved negative results earlier than real-time CMV DNA PCR with a median difference of 10.5 days.
Conclusions
Q-CMV real-time complete kit is a useful tool for early detection of CMV infection in whole blood samples in transplant recipients.
Keywords: Cytomegalovirus, Real-time PCR, Antigenemia assay
INTRODUCTION
Cytomegalovirus (CMV) infection is a major cause of morbidity in recipients of solid organ and bone marrow transplants, in spite of significant advances resulting from preemptive therapy and early diagnosis, thus limiting the effectiveness of organ transplantation as a procedure for the treatment of end-stage diseases [1]. Many techniques are currently available for identifying and monitoring CMV infection, including shell viral culture, antigenemia assay, hybrid capture assay, and qualitative and quantitative PCR assays [2].
The CMV pp65 antigenemia test is an immunofluorescence-based assay that utilizes an indirect immunofluorescence technique for identifying the pp65 protein of CMV in peripheral blood leukocytes. The CMV pp65 assay is widely used as the gold standard for monitoring CMV infections and the response of CMV positive patients to antiviral treatment. Even though the results of the pp65 assay can be obtained in a few hours, it is labor-intensive and suffers from a significant inter-laboratory variation with respect to sensitivity (from 50% to 83%) and specificity (less than 80%) [3, 4, 5].
Quantitation of CMV DNA by real-time PCR is a useful diagnostic technique with its high detection sensitivity and simplicity of use [6]. However, consensus regarding the cut-off level for the diagnosis of CMV infection has not yet been established [7]. Q-CMV real-time complete kit (Cepheid, Nanogen Advanced Diagnostic S.r.L., Torino, Italy) is a real-time PCR kit used for the quantitation of CMV DNA in whole blood. So far, one study comparing this kit with the CMV antigenemia assay has been reported in transplant recipients [8]. The aim of this study was to compare the Q-CMV kit with the CMV antigenemia assay in different disease groups of patients and to investigate the clinical advantage of the use of real-time PCR for quantitation of CMV DNA in whole blood.
METHODS
1. Patients and samples
A retrospective study was conducted on a total of 79 patients who visited Korea University Anam Hospital from June 2011 to March 2013. The patients comprised of 41 stem cell transplant (SCT) recipients, 14 solid organ transplant recipients, 11 patients with hematologic or solid organ malignancies, 11 patients with inflammatory-related illnesses, one patient with diabetes mellitus (DM), and one patient with paroxysmal nocturnal hemoglobinuria (PNH) (Table 1). For the patients who were tested repeatedly, their medical records were reviewed to find if ganciclovir was treated. All patients signed an informed consent under the protocol for human use. The study was approved by the Human Use Ethical Committee of Korea University Anam Hospital. EDTA blood samples were collected simultaneously for the antigenemia assay and the real-time CMV DNA PCR.
Table 1.
Clinical diagnosis of patients
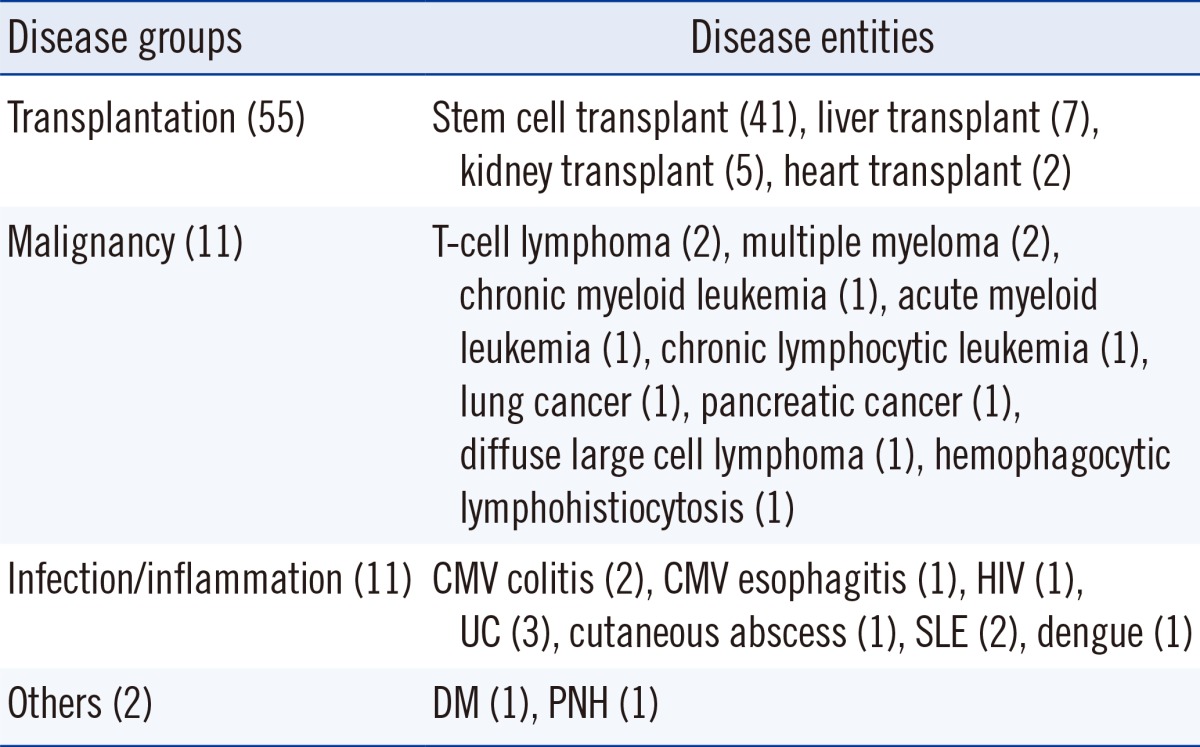
Abbreviations: UC, ulcerative colitis; SLE, systemic lupus erythematosus; DM, diabetes mellitus; PNH, paroxysmal nocturnal hemoglobinuria.
2. The CMV pp65 antigenemia assay
The CMV pp65 antigenemia assay was carried out within 4 hours of specimen collection using the CINA Kit system (ArgeneBiosoft, Varilhes, France). Briefly, the cytospin slides, with 200,000 cells per glass slide, were prepared, fixed, and permeabilized. The presence of CMV pp65 antigen was then detected using a monoclonal antibody against the CMV pp65 antigen and visualized with a fluorescent secondary antibody. The results were expressed as the number of positive cells per slide, with each slide containing 200,000 leukocytes. The test was considered positive when ≥1 fluorescent cell was observed for every 200,000 leukocytes.
3. Quantitation of CMV DNA by real-time PCR in whole blood
The real-time CMV DNA PCR was carried out alongside the CMV antigenemia assay using the Q-CMV real-time complete kit according to the manufacturer's instructions. This test was based on the simultaneous amplification of the exon 4 region of the CMV MIEA (Major Immediate Early Antigen HCMVUL123) gene and the human β-globin gene DNA that was used as the internal control. Briefly, CMV DNA was isolated from 200 µL of EDTA-treated whole blood samples using the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany). Five microliters of the extracted DNA sample and 20 µL of the reaction mix were added to each microplate well. Sterile water containing the reaction mix was used as a negative control. The PCR conditions were as follows: decontamination at 50℃ for 2 min, initial denaturation at 95℃ for 10 min, followed by 45 cycles at 95℃ for 15 sec each, and at 60℃ for 1 min. PCR reactions were performed on an Applied C1000 thermal cycler with a CFX96 real-time system (Bio-Rad, Hercules, CA, USA). The laboratory-determined limit of detection (LOD) of the assay was 65 copies/mL. The assay detected CMV DNA in a linear range from 790 copies/mL to 5×106 copies/mL. The value of LOD and the linear detection range were determined according to the manufacturer's package insert instructions.
4. Definition of CMV infection and CMV disease
CMV infection was defined as the detection of CMV DNA in blood leukocytes in the absence of clinical manifestations or organ function abnormalities [9]. CMV disease was defined as the association of documented CMV infection with clinical symptoms, such as unexplained fever and leukopenia (<4×109/L in two consecutive samples) and/or thrombocytopenia (<150×109/L) not developing in any patient during the follow-up [9].
5. Statistical analysis
A proportion of the positive and negative results were compared by using the chi-square test. Agreement between the real-time CMV DNA PCR and the antigenemia assay results was assessed by Cohen's kappa coefficient value with 95% confidence intervals (CI). Cohen's kappa coefficient values were interpreted as follows: 0-0.2 as slight agreement, 0.21-0.40 as fair, 0.41-0.60 as moderate, 0.61-0.80 as substantial, and 0.81-1 as excellent. The correlation between the two assays was analyzed using the Pearson's correlation coefficient. P values less than 0.05 were considered significant. MedCalC (MedCalc Software, Mariakerke, Belgium) and SPSS version 20 (SPSS Inc., Chicago, IL, USA) were used to analyze the data.
RESULTS
1. Qualitative results of the real-time CMV DNA PCR and the antigenemia assay
479 samples were obtained from 79 patients; the test was done only once in twenty-nine patients and repeatedly done in fifty patients. Out of a total of 479 samples, CMV was detected in 156 (32.6%) samples by the real-time CMV DNA PCR and in 99 (20.7%) samples by the antigenemia assay. Substantial concordance of the two assays was observed in 414 (86.4%) samples (Cohen's kappa coefficient value=0.659, 95% CI=0.585-0.732, Table 2). Of the 65 discrepant samples, 61 samples were antigenemia-negative but real-time CMV DNA PCR-positive. Among these, 45 were below 790 copies/mL and 16 were above 790 copies/mL. Four samples were antigenemia-positive but real-time CMV DNA PCR-negative. There were no clinical particularities for the four patients who were PCR-negative and antigenemia-positive. The number of pp65-positive cells was between 1 and 6 cells/200,000 leukocytes in all four samples.
Table 2.
Comparison of the qualitative results of the Q-CMV real-time complete kit and the CINA Kit
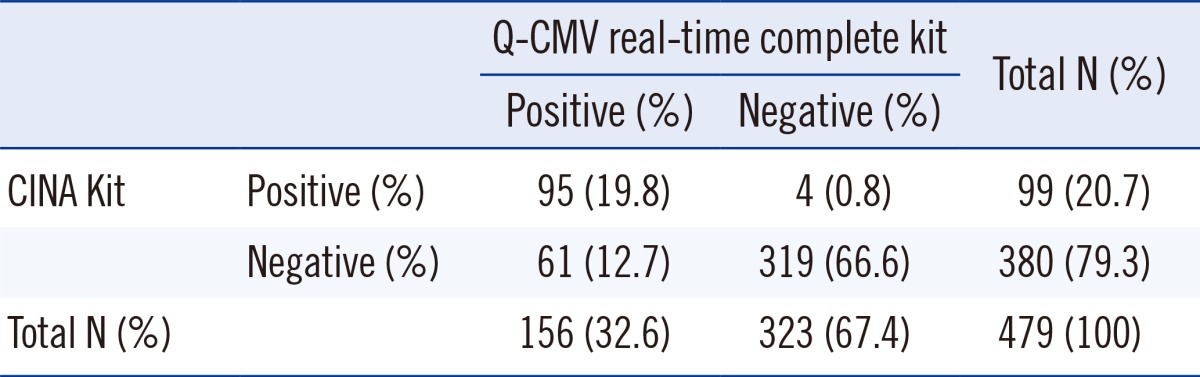
Concordance rate 86.4% (414/479). Cohen's kappa coefficient value=0.659 (95% confidence intervals=0.585-0.732).
2. Quantitative results of the real-time CMV DNA PCR and the antigenemia assay
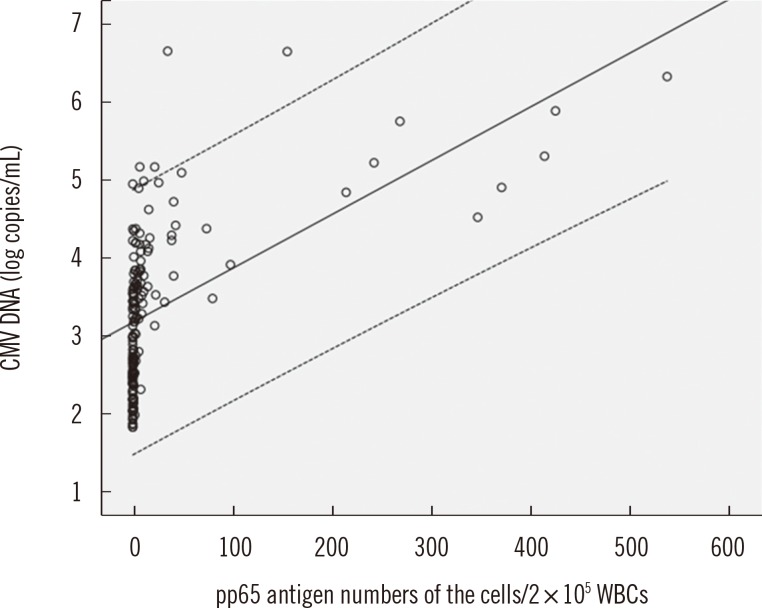
The Pearson's correlation analysis was performed to assess the correlation between the quantitation of CMV DNA by real-time PCR, and the number of positive cells as determined by the antigenemia assay. There was a statistically significant linear correlation between the real-time PCR and the antigenemia assay for 156 real-time CMV DNA PCR-positive samples (P<0.0001), but the Pearson's coefficient value was moderately correlated (r=0.5504, Fig. 1).
Fig. 1.
Correlation between the cytomegalovirus (CMV) DNA copy number and the number of pp65-antigen-positive cells in 156 real-time CMV DNA PCR-positives. Results from these tests are expressed as follows: pp65 assay as positive cells/200,000 examined white blood cells (WBCs); CMV DNA as log10 copies/mL (Pearson's correlation coefficient; n=156, r=0.5504, P<0.0001). The linear regression (solid line) with 95% confidence intervals (dotted line) is shown.
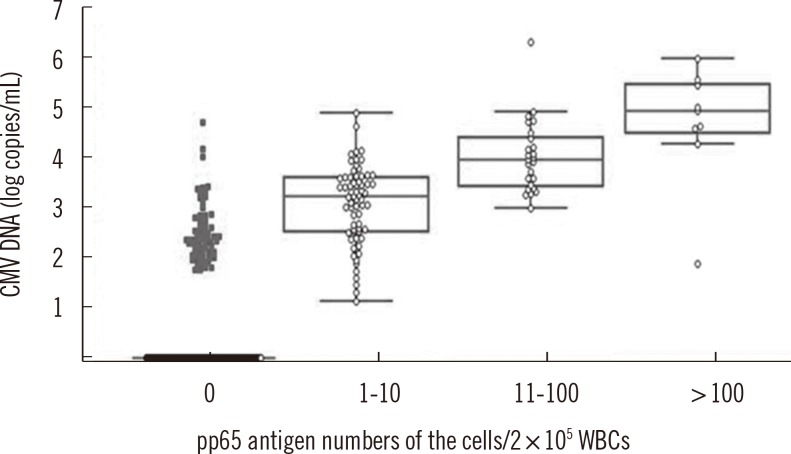
Antigenemia values were divided into four groups: group I having negative values, group II having low values (1-10 positive/200,000 leukocytes), group III having intermediate values (11-100 positive/200,000 leukocytes) and group IV having high values (>100 positive/200,000 leukocytes). Median CMV viral load of the real-time PCR was calculated for each antigenemia category. As shown in Fig. 2, when the antigenemia result was negative, the median CMV viral load was 0.1 log10 copies /mL. In groups II, III and IV, the median viral load increased to 3.1 log10 copies/mL, 4.1 log10 copies/mL, and 5.1 log10 copies/mL, respectively.
Fig. 2.
Evaluation of cytomegalovirus (CMV) DNA in whole blood samples for different groups of pp65-antigenemia (Pearson's correlation coefficient; n=156, r=0.3877, P=0.0013). The CMV DNA load was plotted for four pp65-antigenemia groups. For each pp65 group, the median CMV DNA load (log10 copies/mL), the interquartile 50% range, and the range of values are represented. Abbreviation: WBC, white blood cell.
3. Longitudinal analysis in patients with CMV disease
Among 23 patients showing discrepant results between the two assays, 20 patients were tested repetitively. Thirteen CMV-positive patients out of 20 were treated with ganciclovir and CMV disease was observed in 5 out of 13 patients. The median number of days for obtaining the first positive result was 15.5 days (range, 0-56 days) and 23.5 days (range, 8-252 days) for CMV viral load of the real-time CMV DNA PCR and positive cells of the antigenemia assays, respectively. The positive CMV viral load was detected prior to the antigenemia assay positively in 9 of the 13 ganciclovir-treated patients and the results of the antigenemia assay reached the threshold earlier in one patient and simultaneously in three patients. At the start of ganciclovir therapy, the median CMV viral load was 2,716 copies/mL (range, negative to 93,918 copies/mL) and the median number of the antigenemia-positive cells was 3/200,000 WBCs (range, 0-351/200,000 WBCs), respectively. Clinical monitoring during treatment revealed that the median number of days to obtain negative results after ganciclovir therapy was 36.0 days (range, 11-57 days) and 25.5 days (range, 3-53 days) by the real-time CMV DNA PCR and antigenemia assays, respectively. The results of the antigenemia assay were observed to be negative before the real-time CMV DNA PCR in 8 of the 13 ganciclovir-treated patients. The real-time CMV DNA PCR achieved negative results earlier in 3 patients, and simultaneous results were obtained in 2 patients.
DISCUSSION
Antigenemia assay and real-time CMV DNA PCR are widely used for monitoring viral infection, for tracking its recurrence, and for initiating preemptive CMV therapy [1, 10]. However, guidelines for preemptive therapy based on these methods have yet to be established owing to the lack of standardization [11]. Clinically relevant cut-off values based on CMV diagnosis test differ among patient populations and institutional settings. Griffiths et al. [12] suggested guidelines for preemptive therapy based on the antigenemia test: thresholds of >10 positive cells/2×105 WBCs and of ≥1 or 2 positive cells/2×105 WBCs in solid organ and SCT recipients, respectively. Lilleri et al. [13] suggested a cut-off value for initiation of preemptive therapy on a real-time CMV DNA PCR: 300,000 copies/mL and 10,000 copies/mL in solid organ and SCT recipients, respectively. In our center, different assays are adopted for solid organ transplantation and SCT. In solid organ transplant, antigenemia assay results guide preemptive therapy, and treatment is started when more than 1 positive cell/2×105 WBCs is detected. In SCT, real-time CMV DNA PCR guides preemptive therapy, and treatment is started when more than 65 copies/mL is detected. Therefore, consensus regarding the cut-off level for diagnosis of CMV infection has to be defined for the real-time CMV DNA PCR and antigenemia assay.
Our study revealed an 86.4% concordance rate between the real-time CMV DNA PCR and the antigenemia assays from whole blood samples of diverse patient groups, which is similar to the results obtained from other studies (Table 3). The design of this study differed from those of other studies in a number of ways, such as the type of samples, the different primer sets, and the diverse patient groups. In this study, whole blood specimen was selected over plasma or serum since several prior studies suggested that whole blood-based real-time PCR has a higher sensitivity compared to plasma or serum-based assays [14, 15]. In this study, the CMV MIEA gene was chosen for the Q-CMV Kit assay, but Heo et al. [16] targeted the CMV glycoprotein B (gB). No standardization of target genes is yet agreed on by the larger research community, so further work is needed to establish standardization to encourage comparison of assays across various laboratories using different target genes. In the current study, transplant recipients as well as diverse patients groups were investigated, including those with hematologic or solid organ malignancies, inflammatory-related illnesses, DM and PNH. Contrary to this study, other studies were comprised only of transplant recipients.
Table 3.
Summary of results from the real-time quantitative PCR assay and the antigenemia assay in Korea
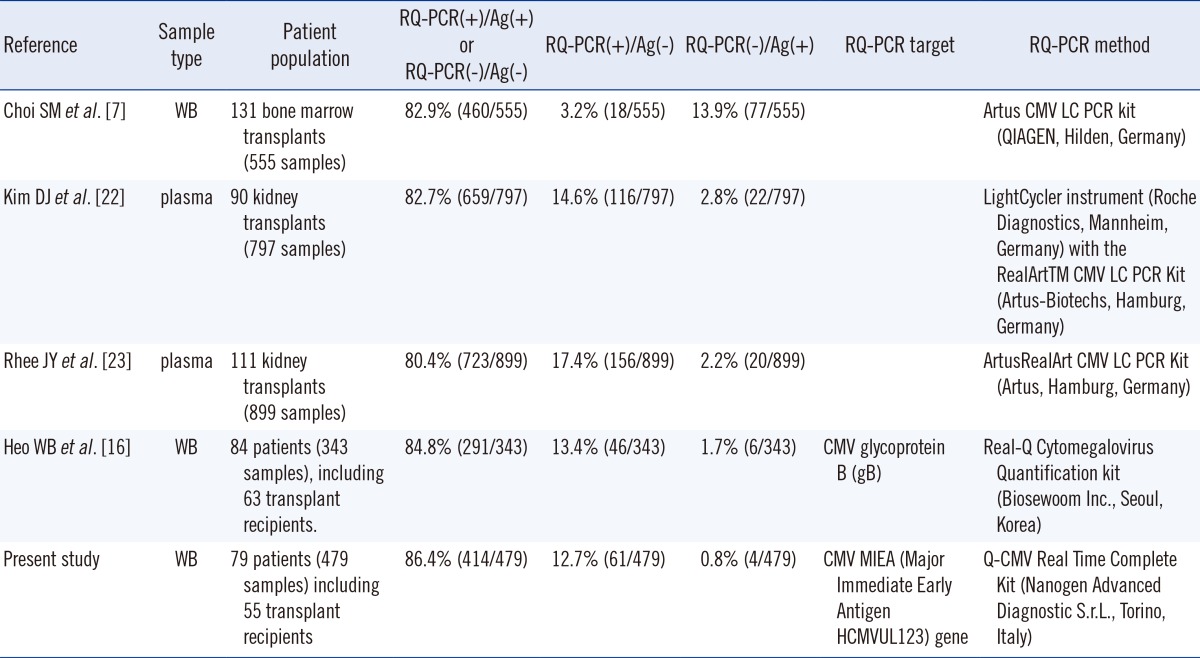
Abbreviations: WB, whole blood; RQ-PCR, real-time quantitative PCR assay; Ag, antigenemia assay; CMV, cytomegalovirus.
Majority of the discrepancy in qualitative results (93.8%, 61/65) was real-time CMV DNA PCR detection in antigenemia-negative and 45 out of 61 samples were below lower linear range (790 copies/mL). This discrepancy could be explained by the increased sensitivity of real-time CMV DNA PCR compared to the antigenemia assay.
In this study, the correlation between the real-time CMV DNA PCR and antigenemia assay was moderate (r=0.5504, P<0.0001), as shown in a previous study [17]. In samples showing antigenemia levels of 0, 1-10, 10-100, and >100 positive cells, the corresponding median viral load measured by real-time CMV DNA PCR was 0.1, 3.1, 4.1, and 5.1 log10 copies/mL, respectively. Significant increases in viral load were observed in samples with more than 1 antigenemia-positive cell, which was above the cut-off level for starting preemptive therapy as chosen by the solid organ transplant centers. When real-time CMV DNA PCR results were compared with groups classified according to the results of the antigenemia assay, the results were moderately correlated (r=0.3877, P=0.0013), which is in agreement with previous studies [18].
Mhiri et al. [19] reported that the rate of viral load increase is significantly associated with the development of the disease; antigenemia assay does not give an accurate indication of viral load rate increase, since it only counts the number of infected cells. In the present study, the time for detection by the real-time PCR was earlier (median of 15.5 days) compared with that by the antigenemia assay (median of 23.5 days), consistent with the data of Ghaffari et al. [20]. Furthermore, the time to become undetectable by the real-time PCR was slower (median of 36 days) than the antigenemia assay (median of 25.5 days), which is similar to the results by Mhiri et al. [19]. The clinical application of real-time CMV DNA PCR for monitoring response to therapy remains unclear, because PCR used for detection of viral DNA cannot distinguish between CMV-infected cell destruction and the genome of the defective virus [21].
Although the real-time CMV DNA PCR has high sensitivity, currently there is no clear agreement on the ideal cut-off for the diagnosis of CMV infection. Thus, treatment decisions should be done with caution, considering trends in viral load and the sensitivity of the methods used in individual laboratories and not just relying on the absolute value recorded in a single test. Even though moderate correlation was observed in this study, this study included a diverse group of patients with a small number of patients in each group. Thus, larger group studies are needed to confirm the optimal cut-off value of real-time PCR for CMV infection.
In conclusion, quantitative results of the Q-CMV real-time complete kit had good correlation with results of the CMV antigenemia assay, suggesting that the real-time CMV DNA PCR can be effective in early detection of CMV infection and guiding therapy.
Acknowledgments
The authors would like to thank the assistance of Ace Cho and Yong Hyun Yu, medical technologists of Korea University Anam Hospital.
Footnotes
No potential conflicts of interest relevant to this article were reported.
References
- 1.Paya CV. Prevention of cytomegalovirus disease in recipients of solid-organ transplants. Clin Infect Dis. 2001;32:596–603. doi: 10.1086/318724. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998;11:533–554. doi: 10.1128/cmr.11.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allice T, Cerutti F, Pittaluga F, Varetto S, Franchello A, Salizzoni M, et al. Evaluation of a novel real-time PCR system for cytomegalovirus DNA quantitation on whole blood and correlation with pp65-antigen test in guiding pre-emptive antiviral treatment. J Virol Methods. 2008;148:9–16. doi: 10.1016/j.jviromet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Guiver M, Fox AJ, Mutton K, Mogulkoc N, Egan J. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation. 2001;71:1609–1615. doi: 10.1097/00007890-200106150-00021. [DOI] [PubMed] [Google Scholar]
- 5.Camargo LF, Uip DE, Simpson AA, Caballero O, Stolf NA, Vilas-Boas LS, et al. Comparison between antigenemia and a quantitative-competitive polymerase chain reaction for the diagnosis of cytomegalovirus infection after heart transplantation. Transplantation. 2001;71:412–417. doi: 10.1097/00007890-200102150-00013. [DOI] [PubMed] [Google Scholar]
- 6.Heid CA, Stevens J, Livak KJ, Williams PM. Realtime quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 7.Choi SM, Lee DG, Lim J, Park SH, Choi JH, Yoo JH, et al. Comparison of quantitative cytomegalovirus real-time PCR in whole blood and pp65 antigenemia assay: clinical utility of CMV real-time PCR in hematopoietic stem cell transplant recipients. J Korean Med Sci. 2009;24:571–578. doi: 10.3346/jkms.2009.24.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchetti S, Santangelo R, Manzara S, D'onghia S, Fadda G, Cattani P. Comparison of real-time PCR and pp65 antigen assays for monitoring the development of Cytomegalovirus disease in recipients of solid organ and bone marrow transplants. New Microbiol. 2011;34:157–164. [PubMed] [Google Scholar]
- 9.Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34:1094–1097. doi: 10.1086/339329. [DOI] [PubMed] [Google Scholar]
- 10.Paya CV, Wilson JA, Espy MJ, Sia IG, DeBernardi MJ, Smith TF, et al. Preemptive use of oral ganciclovir to prevent cytomegalovirus infection in liver transplant patients: a randomized, placebo-controlled trial. J Infect Dis. 2002;185:854–860. doi: 10.1086/339449. [DOI] [PubMed] [Google Scholar]
- 11.Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Danziger-Isakov L, et al. Updated international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2013;96:333–360. doi: 10.1097/TP.0b013e31829df29d. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths PD, Whitley RJ, editors. Recommendations from the International Herpes Management Forum (IHMF) Managements Strategies Workshop and 8th Annual Meeting of the IHMF. The challenge of CMV infection and disease in transplantation. UK: Cambridge Medical Publications; 2000. [Google Scholar]
- 13.Lilleri D, Baldanti F, Gatti M, Rovida F, Dossena L, De Grazia S, et al. Clinically-based determination of safe DNAemia cutoff levels for preemptive therapy or human cytomegalovirus infections in solid organ and hematopoietic stem cell transplant recipients. J Med Virol. 2004;73:412–418. doi: 10.1002/jmv.20107. [DOI] [PubMed] [Google Scholar]
- 14.Razonable RR, Brown RA, Wilson J, Groettum C, Kremers W, Espy M, et al. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation. 2002;73:968–973. doi: 10.1097/00007890-200203270-00025. [DOI] [PubMed] [Google Scholar]
- 15.Von Müller L, Hampl W, Hinz J, Meisel H, Reip A, Engelmann E, et al. High variability between results of different in-house tests for cytomegalovirus (CMV) monitoring and a standardized quantitative plasma CMV PCR assay. J Clin Microbiol. 2002;40:2285–2287. doi: 10.1128/JCM.40.6.2285-2287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heo WB, Won DI, Kim YL, Kim MH, Oh HB, Suh JS. Evaluation of Biosewoom Real-Q Cytomegalovirus Quantification kit for Cytomegalovirus viral load measure. Korean J Lab Med. 2007;27:298–304. doi: 10.3343/kjlm.2007.27.4.298. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Dummer JS, Estes WR, Meng S, Wright PF, Tang YW. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J Clin Microbiol. 2003;41:187–191. doi: 10.1128/JCM.41.1.187-191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalpoe JS, Kroes AC, de Jong MD, Schinkel J, de Brouwer CS, Beersma MF, et al. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J Clin Microbiol. 2004;42:1498–1504. doi: 10.1128/JCM.42.4.1498-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mhiri L, Kaabi B, Houimel M, Arrouji Z, Slim A. Comparison of pp65 antigenemia, quantitative PCR and DNA hybrid capture for detection of cytomegalovirus in transplant recipients and AIDS patients. J Virol Methods. 2007;143:23–28. doi: 10.1016/j.jviromet.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 20.Ghaffari SH, Obeidi N, Dehghan M, Alimoghaddam K, Gharehbaghian A, Ghavamzadeh A. Monitoring of cytomegalovirus reactivation in bone marrow transplant recipients by real-time PCR. Pathol Oncol Res. 2008;14:399–409. doi: 10.1007/s12253-008-9030-3. [DOI] [PubMed] [Google Scholar]
- 21.Pillet S, Roblin X, Cornillon J, Mariat C, Pozzetto B. Quantification of cytomegalovirus viral load. Expert Rev Anti Infect Ther. 2014;12:193–210. doi: 10.1586/14787210.2014.870887. [DOI] [PubMed] [Google Scholar]
- 22.Kim DJ, Kim SJ, Park J, Choi GS, Lee S, Kwon CD, et al. Real-time PCR assay compared with antigenemia assay for detecting cytomegalovirus infection in kidney transplant recipients. Transplant Proc. 2007;39:1458–1460. doi: 10.1016/j.transproceed.2007.01.088. [DOI] [PubMed] [Google Scholar]
- 23.Rhee JY, Peck KR, Lee NY, Song JH. Clinical usefulness of plasma quantitative polymerase chain reaction assay: diagnosis of cytomegalovirus infection in kidney transplant recipients. Transplant Proc. 2011;43:2624–2629. doi: 10.1016/j.transproceed.2011.05.054. [DOI] [PubMed] [Google Scholar]