Abstract
AIM: To investigate the role of claudin 1 in the regulation of genes involved in cell migration and tumor necrosis factor alpha (TNF-α)-induced gene expression in human gastric adenocarcinoma cells.
METHODS: Knockdown experiments were conducted with claudin 1 small interfering RNA (siRNA), and the effects on the cell cycle, apoptosis, migration and invasion were analyzed in human gastric adenocarcinoma MKN28 cells. The gene expression profiles of cells were analyzed by microarray and bioinformatics.
RESULTS: The knockdown of claudin 1 significantly inhibited cell proliferation, migration and invasion, and increased apoptosis. Microarray analysis identified 245 genes whose expression levels were altered by the knockdown of claudin 1. Pathway analysis showed that the top-ranked molecular and cellular function was the cellular movement related pathway, which involved MMP7, TNF-SF10, TGFBR1, and CCL2. Furthermore, TNF- and nuclear frctor-κB were the top-ranked upstream regulators related to claudin 1. TNF-α treatment increased claudin 1 expression and cell migration in MKN28 cells. Microarray analysis indicated that the depletion of claudin 1 inhibited 80% of the TNF-α-induced mRNA expression changes. Further, TNF-α did not enhance cell migration in the claudin 1 siRNA transfected cells.
CONCLUSION: These results suggest that claudin 1 is an important messenger that regulates TNF-α-induced gene expression and migration in gastric cancer cells. A deeper understanding of these cellular processes may be helpful in establishing new therapeutic strategies for gastric cancer.
Keywords: Tumor necrosis factor alpha, Claudin 1, Cell migration, Microarray, Gene expression change
Core tip: The objectives of the present research were to investigate the role of claudin 1 in the regulation of genes involved in cell migration and tumor necrosis factor alpha (TNF-α)-induced gene expression in human gastric adenocarcinoma cells. Claudin 1 small interfering RNA transfection significantly inhibited cell migration and invasion in gastric cancer cells. Microarray analyses showed that down-regulation of claudin 1changed the expression levels of many genes related to cellular movement and TNF-α signal. We showed that TNF-α treatment induces the expression of claudin 1 in gastric carcinoma cells, and the latter controls gene expression and cell migration as the signal mediator.
INTRODUCTION
Claudin proteins play an essential role in the function of tight junction (TJ) and the maintenance of the polarity of epithelial cells, and 24 subtypes of the claudin have been identified[1-3]. On the other hand, it has been shown that claudin 1 may influence the biological behavior of tumor progression[4,5]. The expression of claudin 1 increases during tumor development of various types of neoplasm including gastrointestinal cancer[6-10]. Further, expression of claudin 1 has prognostic impact in colon cancer and is related to apoptosis in breast cancer cells[11-13]. In gastric cancer, it has been reported that regulation of claudin 1 is related to transformation in invasive front and metastatic lesion[6,13].
Tumor necrosis factor alpha (TNF-α) is involved in epithelial-mesenchymal transition (EMT)[14] and related to the malignant behavior of epithelial tumors by regulating cell migration and invasion. The progression of EMT usually decreases the expression of TJ and adherens junction proteins, such as claudins, occludins and E-Cadherin[15-20]. However, we recently found that TNF-α treatment increased the expression of claudin 1 in human lung carcinoma A549 cells[21]. Further studies showed that claudin 1 plays an important role in TNF-α-induced gene expression and cellular movement in A549 cells[21].
The objective of the present research was to investigate the role of claudin 1 in the regulation of genes involved in migration and TNF-α-induced gene expression in human gastric adenocarcinoma cells. Our results showed claudin 1 small interfering RNA (siRNA) transfection significantly inhibited cell migration and invasion in gastric cancer cells. Furthermore, microarray analyses showed that down-regulation of claudin 1changed the expression levels of many genes related to cellular movement and TNF-α signal. Our results indicate that claudin 1 plays an important role in TNF-α-induced gene expression and cellular movement in gastric carcinoma cells.
MATERIALS AND METHODS
Cell lines, antibodies, and other reagents
The human gastric adenocarcinoma cell lines MKN28, NUGC4, MKN45 and Kato-III were grown in plastic culture flasks (Corning Incorporated, NY, United States) and maintained in RPMI-1640 medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 μg/mL of streptomycin, as described previously[22]. The flasks were kept in a humidified incubator at 37 °C under 5.0% CO2[22].
The following antibodies were used in the study; claudin 1 antibody (Zymed Laboratories, San Francisco, CA), a horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Cell Signaling Technology, Beverly, MA), and a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Human TNF-α was from R&D Systemic Inc. (Minneapolis, MN).
Protein studies
Cells were harvested in M-PER lysis buffer supplemented with protease inhibitors (Pierce, Rockford, IL). The protein concentration was measured with a modified Bradford assay (Bio-Rad, Hercules, CA)[23]. Cell lysates containing equal amounts of total protein were separated by SDS-PAGE and then were transferred onto PVDF membranes (GE Healthcare, Piscataway, NJ)[23]. The membranes were probed with the indicated antibodies, and proteins were detected by the ECL Plus Western Blotting Detection System (GE Healthcare)[23,24].
siRNA transfection
Cells were transfected with 10 nmol/L claudin 1 siRNA (Santa Cruz) using the Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA). The medium containing the siRNA was replaced with fresh medium after 24 h[23,24]. The provided control siRNA (Santa Cruz) was used as a negative control. The siRNA transfected cells were harvested 48 h after transfection for protein studies[23,24]. Further, we used a second independent claudin 1 siRNA, Stealth RNAi siRNA targeting claudin 1 mRNA (Invitrogen) to exclude off target effects.
Real-time quantitative RT-PCR
Total RNA was extracted using an RNeasy Kit (Qiagen, Valencia, CA)[24]. Messenger RNA (mRNA) expression was measured by a quantitative real-time PCR (7300 Real-Time PCR System; Applied Biosystems, Foster City, CA) using TaqMan Gene Expression Assays (Applied Biosystems). The expression level of claudin-1 gene (Hs00221623_m1; Applied Biosystems) was measured, and normalized against the housekeeping gene beta-actin (ACTB, Hs01060665_g1; Applied Biosystems). Each assay was performed in triplicate.
Cell proliferation
Cells were seeded onto 6 well plastic plates at a density of 1.0 × 105 cells per well and incubated at 37 °C with 5% CO2. At 24 h after the cell seeding, siRNA transfection was performed. At 48 h after siRNA transfection, the cells were detached from the plates using a trypsin-EDTA and were counted using Countess® Automated Cell Counter (Invitrogen). Each sample was independently counted for three times, and each assay was performed in triplicate.
Analysis of apoptotic cells
As control, non-transfected cells were treated with staurosporine for 24 h. At 48 h after transfection, the siRNA transfected cells were harvested and stained with fluorescein isothiocyanate (FITC) conjugated annexin V and phosphatidylinositol using the annexin V Kit (Beckman Coulter, Brea, CA)[23]. A Becton Dickinson Accuri™ C6 Flow Cytometer was used to analyze the proportion of apoptotic cells.
Analysis of cell migration and invasion
The migration assay was conducted using a cell culture insert with 8 μm pore size (BD Biosciences, Bedford, MA). Biocoat Matrigel (BD Biosciences) was used to evaluate cell invasion potential. Briefly, at 24 h after siRNA transfection, cells (2.0 × 104 cells per well) were seeded in the upper chamber in serum free medium[25]. The lower chamber contained medium with 10% FBS. The chambers were incubated for 48 h for migration assay and 60 h for invasion assay at 37 °C in 5% CO2, and then, non-migrated or non-invaded cells were removed from the upper side of the membrane by scrubbing with cotton swabs[25]. Migrated or invaded cells were fixed on the membrane and stained with Diff-Quick staining reagents (Sysmex, Kobe, Japan)[25]. The migrated or invaded cells on the lower side of the membrane were counted in four independent fields of view at magnification × 100 of each insert[25]. Each assay was performed in triplicate.
Microarray sample preparation and hybridization
MKN28 cells were transfected with control siRNA and claudin 1 siRNA. At 48 h after siRNA transfection, total RNA was extracted using an RNeasy kit (Qiagen). Quality of RNA was monitored using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Cyanine-3 (Cy3)-labeled cRNA was prepared from 0.1 μg total RNA using the Low Input Quick Amp Labeling Kit (Agilent). Samples were purified using RNAeasy columns (Qiagen). A total of 0.60 μg of Cy3-labelled cRNA was fragmented and hybridized to an Agilent SurePrint G3 Human Gene Expression 8 × 60K Microarray for 17 h. After washing, slides were scanned on the Agilent DNA Microarray Scanner (G2565CA) using the one color scan setting for 8 K × 60 K array slides.
Processing of microarray data
The scanned images were analyzed with Feature Extraction Software 10.10 (Agilent) using default parameters to obtain background subtracted and spatially detrended Processed Signal intensities[26]. The fold change of each molecule was calculated by using raw signal data of two samples, and a fold change cutoff of 5 was set to identify molecules whose expression was significantly differentially regulated. The networks and functional analyses were generated using Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Inc., Redwood City, CA).
Immunofluorescent Staining and Confocal Microscopy
MKN28 cells were cultured on glass coverslips and stained as previously described[21]. After different stimulation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 1% BSA, stained with designated antibodies and rhodamine phalloidin. Slides were mounted with VECTASHIELD mounting medium and 4’,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, United States), and distribution of Claudin 1 was examined by confocal fluorescence microscopy (LSM510; Carl Zeiss Co. Ltd, Germany).
Statistical analysis
Student’s t-test was used to evaluate continuous variables. Differences were considered significant when the relevant P value was < 0.05. These analyses were performed using JMP version 10 (SAS Institute Inc., Cary, NC).
RESULTS
Claudin 1 controlled cell proliferation, apoptosis, migration and invasion in MKN28 cells
We conducted knockdown experiments with claudin 1 siRNA in MKN28 cells, and analyzed the effects of claudin 1 knockdown on cell proliferation, apoptosis, migration and invasion. Claudin 1 siRNA effectively reduced claudin 1 mRNA levels (Figure 1A) and claudin 1 protein levels (Figure 1B). The cell counts of claudin 1 siRNA transfected cells were significantly lower than those of control siRNA transfected cells 48 h after siRNA transfection (Figure 1C). Similar results were obtained by using another independent claudin 1 siRNA (Figure 1D, E). Down-regulation of claudin 1 increased both early (annexin V; positive and PI; negative) and late apoptosis (annexin V; positive and PI; positive) 48 h after siRNA transfection (Figure 2). Furthermore, down-regulation of claudin 1 significantly inhibited cell migration and invasion (Figure 3). These results suggest that claudin 1 plays a crucial role in regulating cell proliferation, apoptosis, migration and invasion in MKN28 cells.
Figure 1.
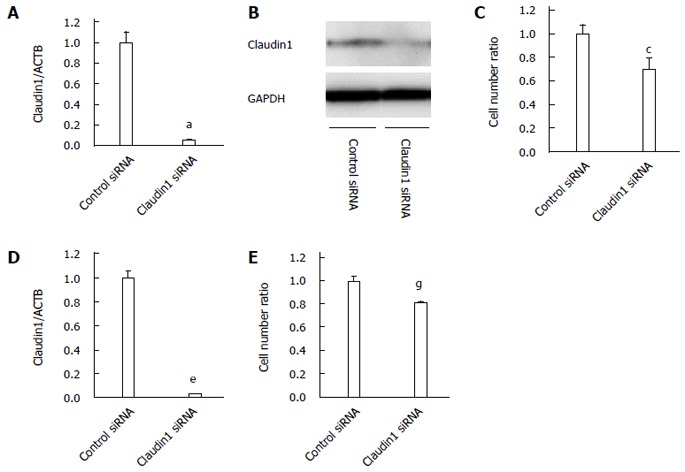
Claudin 1 controls cell proliferation in MKN28 cells. A: Claudin 1 siRNA effectively reduced the mRNA levels of claudin 1 in MKN28 cells. Mean ± SE. n = 3. aP < 0.05 vs control siRNA; B: Western blotting revealed that claudin 1 siRNA reduced the protein levels of claudin 1 in MKN28 cells; C: Downregulation of claudin 1 inhibited the proliferation of MKN28 cells. The number of cells was counted 48 h after siRNA transfection. Mean ± SE. n = 3. cP < 0.05 vs control siRNA; D: A second independent claudin 1 siRNA also reduced the mRNA levels of claudin 1 in the MKN28 cells. Mean ± SE. n = 3. eP < 0.05 vs control siRNA; E: A second independent claudin 1 siRNA also inhibited the proliferation of MKN28 cells. The number of cells was counted 48 h after siRNA transfection. Mean ± SE. n = 3. gP < 0.05 vs control siRNA.
Figure 2.
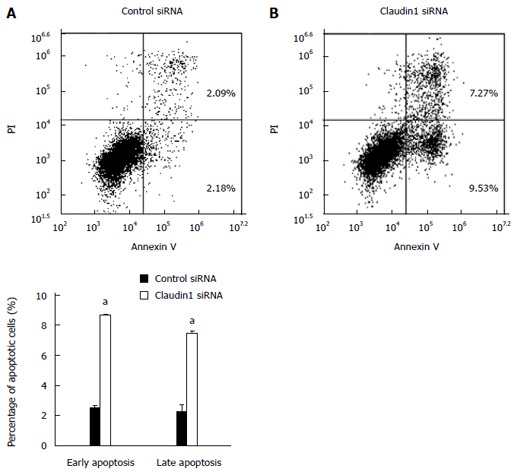
Claudin 1 controls apoptosis in MKN28 cells. Down-regulation of claudin 1 induced both early (annexin V positive/PI negative) and late apoptosis (annexin V/PI double positive) in MKN28 cells 48 h after siRNA transfection. Mean ± SE. n = 3. aP < 0.05 vs control siRNA.
Figure 3.
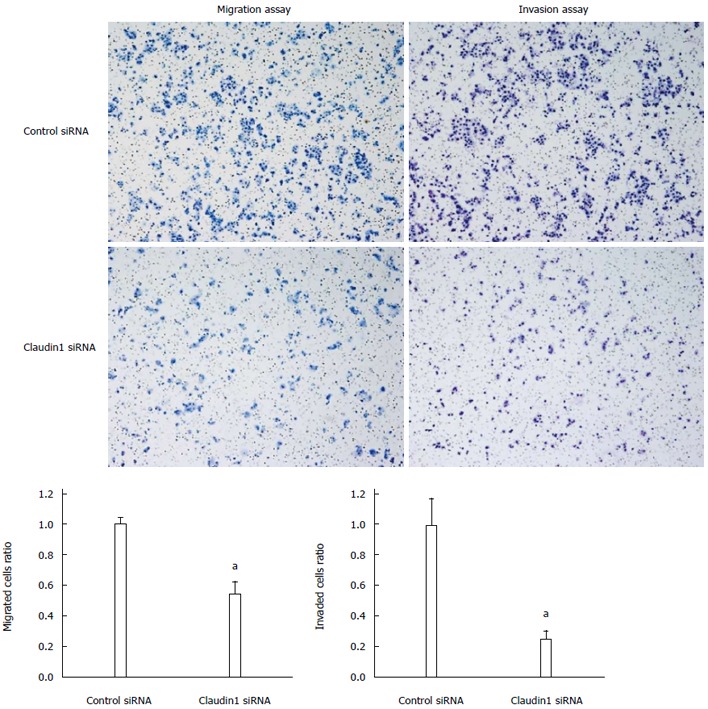
Claudin 1 controlls cell migration and invasion in MKN28 cells. Down-regulation of claudin 1 significantly inhibited cell migration and invasion in MKN28 cells. Cell migration and invasion were determined by Boyden chamber assay. Mean ± SE. n = 3. aP < 0.05 vs control siRNA.
Gene expression profile of claudin 1 siRNA transfected cells
We analyzed the gene expression profiles of claudin 1 siRNA transfected MKN28 cells with microarray and bioinformatics. Microarray analysis showed that the expression levels of 245 genes displayed fold changes of > 5.0 in MKN28 cells subjected to claudin 1 knockdown. Of these genes, 76 were upregulated, and 169 were downregulated in claudin 1 siRNA transfected cells. The 20 genes whose expression levels were the most strongly up- or down-regulated in claudin 1 siRNA transfected cells are shown in Table 1. Claudin 1 expression was down-regulated in claudin 1 siRNA transfected cells (fold change: -26.87; Table 1). Ingenuity Pathway Analysis showed that “Cellular movement” was the top-ranked molecular and cellular functions (Table 2(. Furthermore, TNF- and nuclear factor (NF)-κB were the top-ranked upstream regulators related to claudin 1 (Table 2). We then examined the signal transduction networks induced by the knockdown of claudin 1 expression (Table 2). One of the top-ranked signal networks was related to the cellular movement (Table 2, Figure 4A). These results indicate that the expression level of claudin 1 influences genes related to cellular movement, and that TNF-α signal may be its important upstream regulator.
Table 1.
Genes that displayed the greatest changes in their expressions in the claudin 1 small interfering RNA transfected MKN28 cells
| Gene symbol | Gene ID | Gene name | Fold change |
| Up-regulated genes | |||
| FGF6 | NM_020996 | Fibroblast growth factor 6 | 151.34 |
| TM4SF20 | NM_024795 | Transmembrane 4 L six family member 20 | 72.03 |
| B4GALNT2 | NM_153446 | Beta-1,4-N-acetyl-galactosaminyl transferase 2 | 47.30 |
| SLC2A13 | BC047507 | Solute carrier family 2 (facilitated glucose transporter), member 13 | 43.02 |
| HS3ST2 | NM_006043 | Heparan sulfate (glucosamine) 3-O-sulfotransferase 2 | 28.39 |
| ZNF596 | NM_001042416 | Zinc finger protein 596 | 26.36 |
| IL2RA | NM_000417 | Interleukin 2 receptor, alpha | 22.00 |
| CCL2 | NM_002982 | Chemokine (C-C motif) ligand 2 | 18.05 |
| SGOL2 | BC048349 | Shugoshin-like 2 | 17.64 |
| SYT4 | NM_020783 | Synaptotagmin IV | 17.29 |
| FGF19 | NM_005117 | Fibroblast growth factor 19 | 16.44 |
| ARID5B | NM_032199 | AT rich interactive domain 5B | 14.47 |
| DEFB105B | NM_001040703 | Defensin, beta 105B | 14.34 |
| DSG3 | NM_001944 | Desmoglein 3 | 12.99 |
| RSPH1 | NM_080860 | Radial spoke head 1 homolog (Chlamydomonas) | 12.82 |
| COL5A2 | NM_000393 | Collagen, type V, alpha 2 | 12.72 |
| NRG2 | NM_013982 | Neuregulin 2 | 12.63 |
| TMEM26 | NM_178505 | Transmembrane protein 26 | 12.22 |
| CCL3L3 | NM_001001437 | Chemokine (C-C motif) ligand 3-like 3 | 11.54 |
| OR51E1 | NM_152430 | Olfactory receptor, family 51, subfamily E, member 1 | 9.43 |
| Down-regulated genes | |||
| LY6G6F | NM_001003693 | Lymphocyte antigen 6 complex, locus G6F | -332.04 |
| WT1 | NM_024426 | Wilms tumor 1 | -68.28 |
| MADCAM1 | NM_130760 | Mucosal vascular addressin cell adhesion molecule 1 | -60.84 |
| SLC2A9 | NM_001001290 | Solute carrier family 2 (facilitated glucose transporter), member 9 | -48.05 |
| SULT4A1 | NM_014351 | Sulfotransferase family 4A, member 1 | -39.30 |
| SYNPR | NM_144642 | Synaptoporin | -36.73 |
| C10orf47 | NM_153256 | Chromosome 10 open reading frame 47 | -29.96 |
| MAS1L | NM_052967 | MAS1 oncogene-like | -29.67 |
| CLDN1 | NM_021101 | Claudin 1 | -26.87 |
| CBX1 | NM_006807 | Chromobox homolog 1 | -26.37 |
| MPPED2 | NM_001145399 | Metallophosphoesterase domain containing 2 | -25.00 |
| SCUBE2 | NM_020974 | Signal peptide, CUB domain, EGF-like 2 | -22.13 |
| CHI3L2 | NM_001025199 | Chitinase 3-like 2 | -21.67 |
| SLIT2 | NM_004787 | Slit homolog 2 | -21.60 |
| HFM1 | NM_001017975 | HFM1, ATP-dependent DNA helicase homolog | -20.56 |
| CR1 | NM_000651 | Complement component (3b/4b) receptor 1 | -18.04 |
| PLA2G10 | NM_003561 | Phospholipase A2, group X | -16.70 |
| GRIP2 | NM_001080423 | Glutamate receptor interacting protein 2 | -16.45 |
| XIST | NR_001564 | X (inactive)-specific transcript | -15.79 |
| TNF-SF10 | NM_003810 | Tumor necrosis factor (ligand) superfamily, member 10 | -15.26 |
Table 2.
Top molecular and cellular functions, upstream regulators, and networks of claudin1 according to ingenuity pathway analysis
| Name | P value | Molecules | Score |
| Top molecular and cellular functions | |||
| Cellular movement | 2.77 × 10-9-1.01 × 10-2 | 51 | |
| Cell-to-cell signaling and interaction | 7.24 × 10-7-1.02 × 10-2 | 55 | |
| Molecular transport | 1.23 × 10-6-9.98 × 10-3 | 51 | |
| Small molecule biochemistry | 1.23 × 10-6-9.98 × 10-3 | 55 | |
| Cellular development | 3.80 × 10-6-9.50 × 10-3 | 59 | |
| Top upstream regulators | |||
| TNF-RSF1A | 1.80 × 10-7 | ||
| TNF- | 1.70 × 10-6 | ||
| NF-κB (complex) | 2.01 × 10-6 | ||
| Escherichia coli B5 lipopolysaccharide | 2.09 × 10-6 | ||
| NF-κB1 | 3.23 × 10-6 | ||
| Top networks | |||
| Associated network functions | |||
| Cellular assembly and organization, hair and skin development and function, cellular function and maintenance | 33 | ||
| Cell-to-cell signaling and interaction, cellular growth and proliferation, tissue morphology | 32 | ||
| Cellular movement, hematological system development and function, immune cell trafficking | 31 | ||
| Cardiovascular system development and function, organismal development, cellular movement | 22 | ||
| Hereditary disorder, respiratory disease, tissue development | 22 |
TNF-: Tumor necrosis factor; NF: Nuclear factor.
Figure 4.
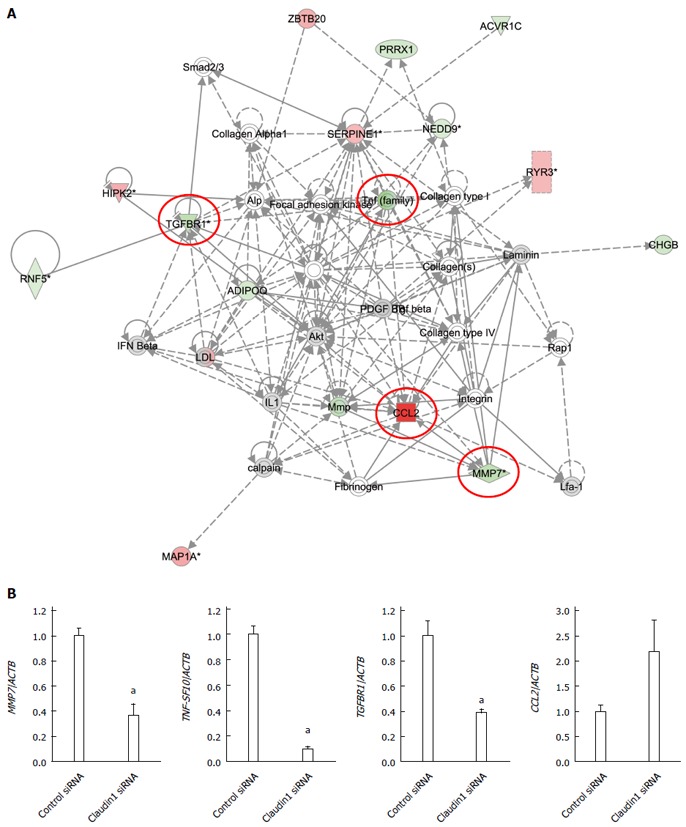
Analysis of gene expression change in claudin 1 siRNA transfected MKN28 cells. A: One of the top-ranked signaling networks related to claudin 1 down-regulation according to ingenuity pathway analysis. Red and green indicate genes whose expression levels were higher or lower, respectively, than reference RNA levels. Genes analyzed for verification were highlighted by red circles; B: Verification of gene expression by real-time quantitative RT-PCR. The expression levels of for selected genes MMP7, TNF-SF10, TGFBR1, and CCL2 in claudin 1 siRNA transfected MKN28 cells were compared with those in control siRNA transfected cells. Gene expression levels were normalized to the level of ACTB. Mean ± SE. n = 3. aP < 0.05 vs control siRNA.
Verification of gene expression by real-time quantitative RT-PCR
Four genes (MMP7, TNF-SF10, TGFBR1, and CCL2) were examined further using quantitative RT-PCR. All these genes were chosen from Figure 4A. The expression levels of MMP7, TNF-SF10, and TGFBR1 mRNA were significantly lower in claudin 1 siRNA transfected cells than in control siRNA transfected ones (Figure 4B). The expression levels of CCL2 tended to be increased in claudin 1 siRNA transfected cells although the difference was not statistically significant (Figure 4B). These changes were in agreement with the microarray results.
Claudin 1 plays important roles in TNF-α-induced cell migration in gastric cancer cells
We previously showed that claudin 1 mediates TNF-α-induced gene expression and cellular movement in human lung carcinoma A549 cells[21]. In the present study, our microarray data showed TNF- and NF-κB were the top-ranked upstream regulators related to claudin 1 in MKN28 cells. To determine the effect of TNF-α on claudin 1 expression in MKN28 cells, we analyzed protein levels of claudin 1, using western blotting. The basal expression level of claudin 1 was increased in a time-dependent manner at 24 and 48 h (Figure 5A). And, it was further increased by TNF-α treatment in MKN28 cells (Figure 5A). Similar trends that the expression level of claudin 1 was increased by TNF-α stimulation were found in several gastric cancer cell lines, including NUGC4, MKN45 and Kato-III (Figure 5B). Furthermore, results of immunofluorescent staining showed that TNF-α-induced claudin 1 expression was mainly detected in the cytoplasm of MKN28 cells (Figure 5C). TNF-α treatment increased cell migration in MKN28 cells (Figure 6A). Similar trends were found in several gastric cancer cell lines, including NUGC4, MKN45 and Kato-III (Figure 6B).We then transfected MKN28 cells with claudin 1 siRNA and found that both the basal and TNF-α-induced expression of claudin 1 mRNA and protein were effectively reduced (Figure 7A, B). To examine TNF-α-induced gene expression, we performed microarray to determine gene expression profiles in cells treated with or without TNF-α and in the presence of claudin 1 siRNA or control siRNA. In control siRNA transfected MKN28 cells, TNF-α stimulation changed expression of 169 genes, of which changes of 135 genes were not seen in claudin 1 siRNA transfected cells. For example, of the top 10 genes up-regulated by TNF-α, the fold of change was reduced in 7 of them, and all the top 10 genes down-regulated by TNF-α, the fold of changes was dramatically reduced in claudin 1 siRNA transfected MKN28 cells (Table 3). Moreover, TNF-α did not enhance cell migration in the claudin 1 siRNA transfected MKN28 cells (Figure 7C). These results show that claudin 1 has crucial role in mediating TNF-α-induced gene expression and migration in gastric cancer cells.
Figure 5.
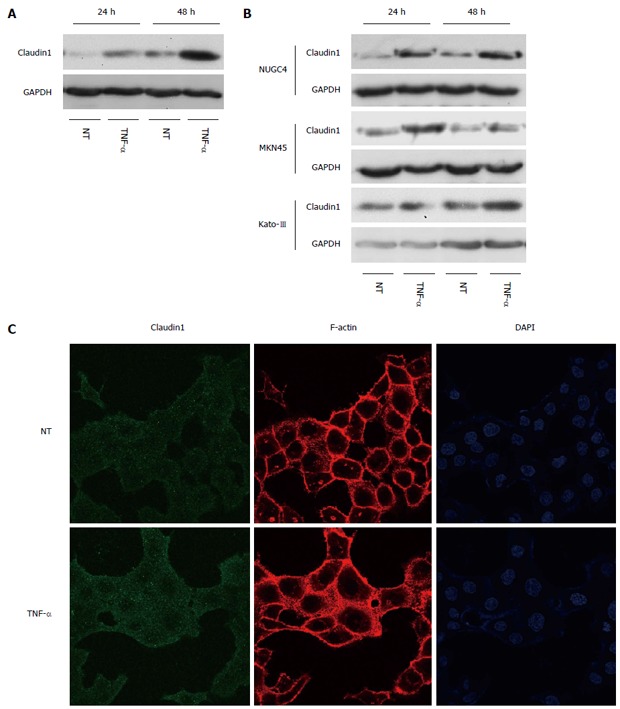
Tumor necrosis factor α induces claudin 1 expression in gastric cancer cells. A: Western blotting revealed that the basal expression level of claudin 1 was increased in a time-dependent manner at 24 and 48 h, which was further increased by tumor necrosis factor α (TNF-α) (20 ng/mL) stimulation in MKN28 cells; B: Western blotting revealed that the expression level of claudin 1 was increased by TNF-α (20 ng/mL) stimulation in several gastric cancer cell lines, including NUGC4, MKN45 and Kato-III; C: At 24 h after treating the cells with TNF-α, expression of claudin 1 protein was increased mainly in cytoplasm. MKN28 cells were immunostained with an anti-claudin 1 antibody and counterstained F-actin and nuclei with rhodamine phalloidin and DAPI, respectively.
Figure 6.
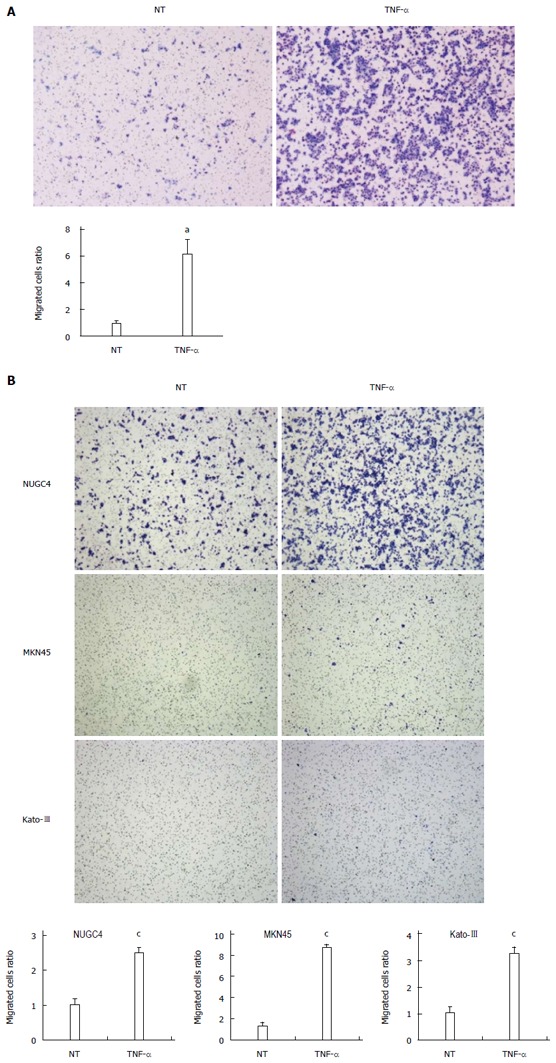
Tumor necrosis factor α induces cell migration in gastric cancer cells. A: Tumor necrosis factor α (TNF-α) stimulation (20 ng/mL, 24 h) significantly increased cell migration in MKN28 cells. Cell migration was determined by Boyden chamber assay. Mean ± SE. n = 3. aP < 0.05 vs control siRNA, NT; B: TNF-α stimulation (20 ng/mL, 48 h) significantly increased cell migration in several gastric cancer cell lines, including NUGC4, MKN45 and Kato-III. Cell migration was determined by Boyden chamber assay. Mean ± SE. n = 3. cP < 0.05 vs control siRNA. NT: No treatment.
Figure 7.
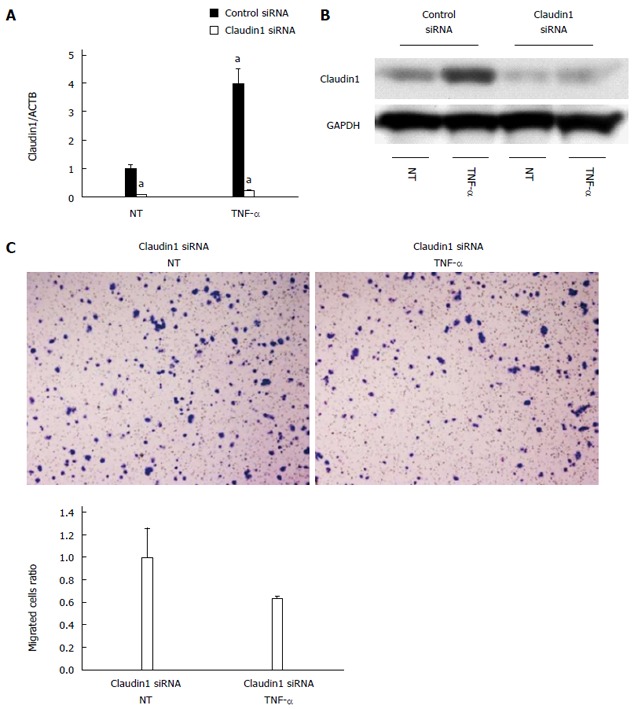
Claudin 1 is important for tumor necrosis factor α-induced cell migration in MKN28 cells. A: Claudin 1 siRNA effectively reduced both the basal and tumor necrosis factor α (TNF-α)-induced expression of claudin 1 mRNA. Mean ± SE. n = 3. aP < 0.05 vs control siRNA, NT; B: Western blotting revealed that transfection of claudin 1 siRNA effectively reduced both the basal and TNF-α-induced expression of claudin 1 protein; C: TNF-α stimulation (20 ng/mL, 48 h) did not enhance cell migration in the claudin 1 siRNA transfected cells. Cell migration was determined by Boyden chamber assay. Mean ± SE. n = 3. NT: No treatment.
Table 3.
Top 10 up or down-regulated genes induced by tumor necrosis factor α in control small interfering RNA or claudin 1 small interfering RNA transfected MKN28 cells
| Gene symbol | Gene ID | Gene name | Control siRNA | Claudin1 siRNA |
| Fold change | Fold change | |||
| Up-regulated genes | ||||
| IL6 | NM_000600 | Interleukin 6 | 11.16 | 7.16 |
| MMP9 | NM_004994 | Matrix metallopeptidase 9 | 10.49 | 18.13 |
| BMP2 | NM_001200 | Bone morphogenetic protein 2 | 10.12 | 5.62 |
| VSTM1 | NM_198481 | V-set and transmembrane domain containing 1 | 9.83 | 2.53 |
| CLEC1A | NM_016511 | C-type lectin domain family 1, member A | 9.71 | -1.22 |
| CXCL10 | NM_001565 | Chemokine (C-X-C motif) ligand 10 | 9.33 | 1.82 |
| IL2RG | NM_000206 | Interleukin 2 receptor, gamma | 9.14 | 15.56 |
| TFPI | NM_001032281 | Tissue factor pathway inhibitor | 8.98 | 11.79 |
| KLHDC7B | NM_138433 | Kelch domain containing 7B | 8.90 | 1.79 |
| CCL5 | NM_002985 | Chemokine (C-C motif) ligand 5 | 8.85 | 7.84 |
| Down-regulated genes | ||||
| LY6G6F | NM_001003693 | Lymphocyte antigen 6 complex, locus G6F | -311.81 | 1.05 |
| RFTN2 | NM_144629 | Raftlin family member 2 | -291.41 | 1.39 |
| WT1 | NM_024426 | Wilms tumor 1 | -75.96 | 1.05 |
| MADCAM1 | NM_130760 | Mucosal vascular addressin cell adhesion molecule 1 | -67.39 | 1.06 |
| SLC2A9 | NM_001001290 | Solute carrier family 2, member 9 | -49.12 | -1.21 |
| C10orf47 | NM_153256 | Chromosome 10 open reading frame 47 | -44.11 | 1.85 |
| SYNPR | NM_144642 | Synaptoporin | -40.64 | 1.08 |
| GGT7 | NM_178026 | Gamma-glutamyltransferase 7 | -40.33 | 1.24 |
| LRRC36 | NM_018296 | Leucine rich repeat containing 36 | -38.25 | 1.06 |
| MAS1L | NM_052967 | MAS1 oncogene-like | -32.62 | 1.07 |
DISCUSSION
Recent reports have indicated that claudin proteins were up-regulated and mis-located in cancer cells[2]. Although there are several reports on the expression of claudin 1 in gastric cancer, no consensus has been reached about the relationship between claudin 1 expression and clinicopathological parameters[5,6,13,27]. Some reports have indicated that claudin 1 was highly expressed in intestinal subtype of adenocarcinomas[5,27], while others found that it was highly expressed in diffuse subtype of adenocarcinomas[6,13]. Several studies reported claudin 1 expression at invasive front of gastric carcinomas, which suggests that over expression of claudin 1 is related to carcinogenesis in invasive and metastatic gastric cancer[6,13,27]. The up-regulation of claudin 1 in colon cancer cells increased tumor growth and metastasis in vivo, whereas depletion of claudin 1 in metastatic colon cancer cells with siRNA inhibited cellular invasion[7]. Furthermore, over expression of claudin 1 increased cellular movement in melanoma[8], oral squamous cell carcinoma[9], hepatocellular carcinoma[10] and lung carcinoma cells[21]. In the present study, our results also showed that knocking down of claudin 1 decreased cell proliferation, cell migration and invasion, and increased apoptosis in gastric cancer cells, which suggests the importance of claudin 1 in the progression of gastric carcinomas.
Our data showed that “Cellular Movement” was the top-ranked molecular and cellular functions related to claudin 1 down-regulation according to Ingenuity Pathway Analysis in gastric cancer cells. MMP7 was found in the signal network related to cellular movement, strongly suggesting that the expression of claudin 1 may involve in migration, invasion and metastasis of gastric cancer. Furthermore, TNF- and NF-κB were the top-ranked upstream regulators related to claudin 1. Tumor necrosis factor superfamily member 10 (TNF-SF10), was significantly down-regulated by claudin 1 siRNA in MKN28 cells. These data show the important role of claudin 1 in TNF-α-induced cell migration.
It has been shown that TNF-α stimulation induced EMT in colonic organoids[14], renal carcinoma[28-30] and skin cells[31]. TJ and adherens junction proteins are usually down-regulated during the progression of EMT[15-20]. In this manner, claudin 1 expression is generally decreased by TNF-α stimulation, and the decreased protein expression leads to the increase in the paracellular permeability of epithelial cells[32,33]. On the other hand, TNF-α was reported to increase claudin 1 protein expression in pancreatic cancer cells[34] and airway smooth muscle cells[35]. Our recent report showed that TNF-α stimulation remarkably increased claudin 1 protein expression in the cytoplasm in A549 cells, and that depletion of claudin 1 inhibited the TNF-α-induced gene expression and cell migration[21]. Similarly, in the present study, TNF-α induced over expression of claudin 1 in the cytoplasm in gastric carcinoma MKN28 cells, and knocking down of claudin 1 blocked the TNF-α-induced gene expression and cellular movement in human gastric cancer cells. Generally, claudin 1 participates in cell-to-cell adhesion as TJ proteins, and its down-regulation may promote cell migration. However, our findings showed claudin 1 induced by TNF-α is mainly in cytoplasm, and regulates TNF-α-induced gene expression[21]. In addition, many of these claudin 1 dependent genes are related to cellular movement, suggesting that claudin 1 mediates TNF-α-initiated cell migration with various mechanisms[21].
In conclusion, we found that claudin 1 played a role in the proliferation, apoptosis, cell migration and invasion in gastric carcinoma cells, suggesting the importance of claudin 1 expression in the progression of gastric carcinomas. Our microarray results also suggest that claudin 1 has marked effects on the expression of genes related to cellular movement. Furthermore, we showed that TNF-α induces the gene expression of claudin 1 in gastric carcinoma cells, and the latter acts as the signal mediator to regulate gene expression and cellular movement. A deeper understanding of this pathway may serve as a mean to establish a new therapeutic target for gastric carcinoma.
COMMENTS
Background
Recent reports have indicated that claudin proteins were up-regulated and mis-located in cancer cells, and influenced the biological behavior of tumor progression. Although there are several reports on the expression of claudin 1 in gastric cancer, no consensus has been reached about the relationship between claudin 1 expression and clinicopathological features.
Research frontiers
The authors recently found that the expression of claudin 1 was increased in response to tumor necrosis factor alpha (TNF-α) stimulation, and that claudin 1 played an important role in TNF-α-induced gene expression and cellular movement in human lung carcinoma A549 cells. The objectives of the present research were to investigate the role of claudin 1 in the control of genes involved in cell migration and TNF-α-induced gene expression in human gastric adenocarcinoma cells.
Innovations and breakthroughs
The authors showed that the knockdown of claudin 1 significantly inhibited cell migration and invasion in gastric cancer cells. Microarray analyses showed that down-regulation of claudin 1changed the expression levels of many genes related to cellular movement and TNF-α signal. They found that TNF-α stimulation induced the gene expression of claudin 1 in gastric carcinoma cells, and the latter acted as the signal mediator to regulate gene expression and migration.
Applications
The results of the present study suggest that claudin 1 acts as a crucial signal mediator in TNF-α induced gene expression and cell migration in gastric carcinoma cells. A deeper understanding of these cellular processes may be helpful in establishing new therapeutic strategies for gastric cancer.
Terminology
Claudin proteins play an essential role in the function of TJ, and 24 subtypes of the claudin have been identified. They interact with each other through homo- and heterophilic interactions and are crucial for the maintenance of cellular polarity of epithelial cells.
Peer review
This is a good descriptive study in which the role of claudin 1 in the regulation of genes involved cell migration and TNF-α-induced gene expression in human gastric cancer cells. They reported that claudin 1 knock down significantly inhibited cell migration and invasion in gastric carcinoma cells. And the depletion of claudin 1 changed the expression level of TNF-α signal. The results are interesting and meaningful for further understand the role of claudin 1 on cancer development.
Footnotes
Supported by Grants-in-Aid for Young Scientists (B); No. 22791295, No. 23791557 and No. 24791440; and Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science, No. 22591464 and No. 24591957
P- Reviewer: Gazouli M, Kuramitsu Y, Li QQ, Miyazaki T, Ricci-Vitiani L S- Editor: Ma YJ L- Editor: A E- Editor: Liu XM
References
- 1.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kominsky SL. Claudins: emerging targets for cancer therapy. Expert Rev Mol Med. 2006;8:1–11. doi: 10.1017/S1462399406000056. [DOI] [PubMed] [Google Scholar]
- 3.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57:919–928. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Turksen K, Troy TC. Barriers built on claudins. J Cell Sci. 2004;117:2435–2447. doi: 10.1242/jcs.01235. [DOI] [PubMed] [Google Scholar]
- 5.Soini Y, Tommola S, Helin H, Martikainen P. Claudins 1, 3, 4 and 5 in gastric carcinoma, loss of claudin expression associates with the diffuse subtype. Virchows Arch. 2006;448:52–58. doi: 10.1007/s00428-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 6.Wu YL, Zhang S, Wang GR, Chen YP. Expression transformation of claudin-1 in the process of gastric adenocarcinoma invasion. World J Gastroenterol. 2008;14:4943–4948. doi: 10.3748/wjg.14.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, Neff J, Washington MK, Beauchamp RD. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT, Dissanayake SK, Earley R, Indig FE, Nickoloff BJ, et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene. 2007;26:3846–3856. doi: 10.1038/sj.onc.1210155. [DOI] [PubMed] [Google Scholar]
- 9.Dos Reis PP, Bharadwaj RR, Machado J, Macmillan C, Pintilie M, Sukhai MA, Perez-Ordonez B, Gullane P, Irish J, Kamel-Reid S. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer. 2008;113:3169–3180. doi: 10.1002/cncr.23934. [DOI] [PubMed] [Google Scholar]
- 10.Yoon CH, Kim MJ, Park MJ, Park IC, Hwang SG, An S, Choi YH, Yoon G, Lee SJ. Claudin-1 acts through c-Abl-protein kinase Cdelta (PKCdelta) signaling and has a causal role in the acquisition of invasive capacity in human liver cells. J Biol Chem. 2010;285:226–233. doi: 10.1074/jbc.M109.054189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol. 2005;18:511–518. doi: 10.1038/modpathol.3800301. [DOI] [PubMed] [Google Scholar]
- 12.Hoevel T, Macek R, Swisshelm K, Kubbies M. Reexpression of the TJ protein CLDN1 induces apoptosis in breast tumor spheroids. Int J Cancer. 2004;108:374–383. doi: 10.1002/ijc.11571. [DOI] [PubMed] [Google Scholar]
- 13.Jung H, Jun KH, Jung JH, Chin HM, Park WB. The expression of claudin-1, claudin-2, claudin-3, and claudin-4 in gastric cancer tissue. J Surg Res. 2011;167:e185–e191. doi: 10.1016/j.jss.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Bates RC, Mercurio AM. Tumor necrosis factor-alpha stimulates the epithelial-to-mesenchymal transition of human colonic organoids. Mol Biol Cell. 2003;14:1790–1800. doi: 10.1091/mbc.E02-09-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi E, Nagano O, Ishimoto T, Yae T, Suzuki Y, Shinoda T, Nakamura S, Niwa S, Ikeda S, Koga H, et al. Tumor necrosis factor-alpha regulates transforming growth factor-beta-dependent epithelial-mesenchymal transition by promoting hyaluronan-CD44-moesin interaction. J Biol Chem. 2010;285:4060–4073. doi: 10.1074/jbc.M109.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamauchi Y, Kohyama T, Takizawa H, Kamitani S, Desaki M, Takami K, Kawasaki S, Kato J, Nagase T. Tumor necrosis factor-alpha enhances both epithelial-mesenchymal transition and cell contraction induced in A549 human alveolar epithelial cells by transforming growth factor-beta1. Exp Lung Res. 2010;36:12–24. doi: 10.3109/01902140903042589. [DOI] [PubMed] [Google Scholar]
- 17.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–724. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 18.Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell. 2006;17:1871–1879. doi: 10.1091/mbc.E05-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 20.Ohkubo T, Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci. 2004;117:1675–1685. doi: 10.1242/jcs.01004. [DOI] [PubMed] [Google Scholar]
- 21.Shiozaki A, Bai XH, Shen-Tu G, Moodley S, Takeshita H, Fung SY, Wang Y, Keshavjee S, Liu M. Claudin 1 mediates TNF-α-induced gene expression and cell migration in human lung carcinoma cells. PLoS One. 2012;7:e38049. doi: 10.1371/journal.pone.0038049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iitaka D, Shiozaki A, Ichikawa D, Kosuga T, Komatsu S, Okamoto K, Fujiwara H, Ishii H, Nakahari T, Marunaka Y, et al. Blockade of chloride ion transport enhances the cytocidal effect of hypotonic solution in gastric cancer cells. J Surg Res. 2012;176:524–534. doi: 10.1016/j.jss.2011.10.039. [DOI] [PubMed] [Google Scholar]
- 23.Shiozaki A, Shen-Tu G, Bai X, Iitaka D, De Falco V, Santoro M, Keshavjee S, Liu M. XB130 mediates cancer cell proliferation and survival through multiple signaling events downstream of Akt. PLoS One. 2012;7:e43646. doi: 10.1371/journal.pone.0043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeshita H, Shiozaki A, Bai XH, Iitaka D, Kim H, Yang BB, Keshavjee S, Liu M. XB130, a new adaptor protein, regulates expression of tumor suppressive microRNAs in cancer cells. PLoS One. 2013;8:e59057. doi: 10.1371/journal.pone.0059057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welte G, Alt E, Devarajan E, Krishnappa S, Jotzu C, Song YH. Interleukin-8 derived from local tissue-resident stromal cells promotes tumor cell invasion. Mol Carcinog. 2012;51:861–868. doi: 10.1002/mc.20854. [DOI] [PubMed] [Google Scholar]
- 26.Waki H, Gouraud SS, Bhuiyan ME, Takagishi M, Yamazaki T, Kohsaka A, Maeda M. Transcriptome of the NTS in exercise-trained spontaneously hypertensive rats: implications for NTS function and plasticity in regulating blood pressure. Physiol Genomics. 2013;45:58–67. doi: 10.1152/physiolgenomics.00074.2012. [DOI] [PubMed] [Google Scholar]
- 27.Resnick MB, Gavilanez M, Newton E, Konkin T, Bhattacharya B, Britt DE, Sabo E, Moss SF. Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol. 2005;36:886–892. doi: 10.1016/j.humpath.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 28.Kim MS, Takahashi T, Lee JY, Hwang GW, Naganuma A. Methylmercury induces CCL2 expression through activation of NF-κB in human 1321N1 astrocytes. J Toxicol Sci. 2012;37:1275–1278. doi: 10.2131/jts.37.1275. [DOI] [PubMed] [Google Scholar]
- 29.Chuang MJ, Sun KH, Tang SJ, Deng MW, Wu YH, Sung JS, Cha TL, Sun GH. Tumor-derived tumor necrosis factor-alpha promotes progression and epithelial-mesenchymal transition in renal cell carcinoma cells. Cancer Sci. 2008;99:905–913. doi: 10.1111/j.1349-7006.2008.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu ST, Sun GH, Hsu CY, Huang CS, Wu YH, Wang HH, Sun KH. Tumor necrosis factor-α induces epithelial-mesenchymal transition of renal cell carcinoma cells via a nuclear factor kappa B-independent mechanism. Exp Biol Med (Maywood) 2011;236:1022–1029. doi: 10.1258/ebm.2011.011058. [DOI] [PubMed] [Google Scholar]
- 31.Yan C, Grimm WA, Garner WL, Qin L, Travis T, Tan N, Han YP. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-alpha through bone morphogenic protein-2. Am J Pathol. 2010;176:2247–2258. doi: 10.2353/ajpath.2010.090048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker OJ, Camden JM, Redman RS, Jones JE, Seye CI, Erb L, Weisman GA. Proinflammatory cytokines tumor necrosis factor-alpha and interferon-gamma alter tight junction structure and function in the rat parotid gland Par-C10 cell line. Am J Physiol Cell Physiol. 2008;295:C1191–C1201. doi: 10.1152/ajpcell.00144.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poritz LS, Garver KI, Tilberg AF, Koltun WA. Tumor necrosis factor alpha disrupts tight junction assembly. J Surg Res. 2004;116:14–18. doi: 10.1016/s0022-4804(03)00311-1. [DOI] [PubMed] [Google Scholar]
- 34.Kondo J, Sato F, Kusumi T, Liu Y, Motonari O, Sato T, Kijima H. Claudin-1 expression is induced by tumor necrosis factor-alpha in human pancreatic cancer cells. Int J Mol Med. 2008;22:645–649. [PubMed] [Google Scholar]
- 35.Fujita H, Chalubinski M, Rhyner C, Indermitte P, Meyer N, Ferstl R, Treis A, Gomez E, Akkaya A, O’Mahony L, et al. Claudin-1 expression in airway smooth muscle exacerbates airway remodeling in asthmatic subjects. J Allergy Clin Immunol. 2011;127:1612–1621.e8. doi: 10.1016/j.jaci.2011.03.039. [DOI] [PubMed] [Google Scholar]


