Abstract
Rapid identification of traumatic intracranial hematomas following closed head injury represents a significant health care need because of the potentially life-threatening risk they present. This study demonstrates the clinical utility of an index of brain electrical activity used to identify intracranial hematomas in traumatic brain injury (TBI) presenting to the emergency department (ED). Brain electrical activity was recorded from a limited montage located on the forehead of 394 closed head injured patients who were referred for CT scans as part of their standard ED assessment. A total of 116 of these patients were found to be CT positive (CT+), of which 46 patients with traumatic intracranial hematomas (CT+) were identified for study. A total of 278 patients were found to be CT negative (CT−) and were used as controls. CT scans were subjected to quanitative measurements of volume of blood and distance of bleed from recording electrodes by blinded independent experts, implementing a validated method for hematoma measurement. Using an algorithm based on brain electrical activity developed on a large independent cohort of TBI patients and controls (TBI-Index), patients were classified as either positive or negative for structural brain injury. Sensitivity to hematomas was found to be 95.7% (95% CI=85.2, 99.5), specificity was 43.9% (95% CI=38.0, 49.9). There was no significant relationship between the TBI-Index and distance of the bleed from recording sites (F=0.044, p=0.833), or volume of blood measured F=0.179, p=0.674). Results of this study are a validation and extension of previously published retrospective findings in an independent population, and provide evidence that a TBI-Index for structural brain injury is a highly sensitive measure for the detection of potentially life-threatening traumatic intracranial hematomas, and could contribute to the rapid, quantitative evaluation and treatment of such patients.
Key words: : classifier function, mild TBI (mTBI), neuroimaging, quantitative EEG (QEEG), traumatic hematomas
Introduction
Traumatic intracranial hematomas presenting to the Emergency Department (ED) following closed head injury must be rapidly diagnosed because of the potentially life-threatening risk they present. Such hematomas can be seen in cases that appear mild by presenting symptoms and apparent neurological status (e.g., high Glasgow Coma Score [GCS]). The current “gold standard” for initial assessment and triage is noncontrast CT scan, performed as soon as possible following injury. The use of head CT scan has several significant drawbacks including time, radiation exposure, and patient safety. That is, many patients are unnecessarily exposed to CT radiation, considering that specificity is poor for existing symptom-based guidelines for CT scanning (3.0–12.7% in mild traumatic brain injury [mTBI] populations1,2). Further, the increased use of CT scanning in emergency settings often imposes a delay on making clinically important triage decisions.
The use of the BrainScope device in development for the identification of traumatic intracranial hematomas does not have these drawbacks. Prichep and colleagues3,4 described the development of TBI-Index using a binary classification algorithm based on selected quantitative features of brain electrical activity recorded from five electrodes placed on the forehead. In a prospective validation study (funded in part by the Department of Defense, contract #W911QY-12-C-0004, Assessment of Head Injury in the Emergency Department) using such an algorithm (Genetic algorithm, GA), high sensitivity (85.3%, 95% CI 78.8, 90.4) and negative predictive value (NPV) (96.5–92.2% at 10–20% prevalence) were obtained for identification of CT positive (CT+) TBI in a large population (n=552) of mTBI patients. Clinical utility has been demonstrated in multiple publications that have reported high sensitivity in discriminating between CT+ and CT negative (CT−) ED mTBI patients (92.4–94.7%)5,6 and have shown to have specificity more than double that seen using the New Orleans Criteria (NOC) in such populations (TBI-Index was 49.4% and NOC was 23.5%).6
In these studies, CT scans were evaluated by subjective visual inspection by an experienced neuroradiologist. Although studies of inter-rater reliability are few, they report poor diagnostic concordance between raters, especially in the identification of different etiologies of TBI (up to 50% failure),7,8 which raises concerns because the CT scan is generally regarded as the “gold standard.” Additional quantitative measurements of brain injury have been used to more objectively define the magnitude of injury,9 as well as to aid in more objective selection of patients for research protocols.10 Quantitative measurements of the volume of blood within the hematoma, performed with operator-defined region of interest segmentation,11 is one such method.
Using the TBI-Index, Hanley and colleagues12 reported the accuracy of identification of traumatic hematomas in a retrospective sample of brain-injured patients (n=38). These results showed high sensitivity (100%) and specificity (66%). Results were not influenced by the distance of the bleed from the recording electrodes, type of hematoma, or volume of the bleed. Other methods described in the literature (such as those based on near-infrared spectroscopy [NIS]) have significant clinical limitations of use based on depth, volume, and type of hematoma.13–16
The present study prospectively evaluates the clinical utility of the TBI-Index of brain function for the identification of the presence of intracranial hematomas in an independent ED brain-injured population.
Methods
Patient population
Eleven ED sites participated in patient recruitment for the study from a consecutive sample of patients presenting following a closed head injury and meeting the inclusion/exclusion criteria described subsequently. Clinical sites included: Barnes Jewish Medical Center, Detroit Receiving Hospital, University of Virginia Medical Center, University of Maryland Medical Center (R. Adams Cowley Shock Trauma Center), Sinai-Grace Hospital, Brooke Army Medical Center, University of Rochester Strong Memorial Hospital, Duke University Hospital, Hartford Hospital, Sinai Grace Hospital, and Inova Fairfax Hospital. From this sample, 46 patients who were confirmed to have intracranial bleeding (≥1 cc), based on blinded, adjudicated measurements of ED CT scans, were selected for study. Those with such hematomas represented 40% of the the CT+ population (n=116) identified in the 394 cases referred for CT. A total of 278 CT− mTBI patients were selected from the same sample of ED brain-injured patients, and used as controls. mTBI was defined as a closed head injury, with or without loss of consciousness or amnesia, and with a GCS >8, (with all but one patient with GCS between 14 and 15).
All sites received approval from their respective Human Subjects Research Committees, and written informed consent was obtained prior to testing of all subjects. Assessment of the capacity of the subject to give informed consent was performed using the Conley criteria.17
Inclusion/exclusion criteria
Male and female patients between the ages of 18 and 80 years who presented to the ED after a closed head injury and who had a CT scan of the head ordered as part of their clinical evaluation were eligible for inclusion. Patients were excluded if clinical conditions would not allow placement of the electrodes, or if they were obtunded as a result of intoxication to the point where they could not provide informed consent. In addition, patients with known psychiatric disorders, including chronic drug or alcohol dependence disorders, chronic seizure history, or mental retardation, or who were currently taking medication affecting the central nervous system (CNS) on a daily basis for a chronic psychiatric condition, were not eligible for the study. If the head injury occurred following a seizure, the patient was not a candidate for this study.
CT scans
All head CT scans were noncontrast, and were reviewed by a three member panel of independent experienced neuroradiologists (blinded to clinical and electroencephalographic [EEG] data), and positive CT findings were determined by majority rule. The CT+ reports in these patients included all intracranial hematomas, contusions, subarachnoid hemorrhages, cortical edema, and combinations of these etiologies. All but one CT scan were performed within 23 h of the EEG data acquisition, with a mean of 5.03 h, (85% were <12 h, and were in the time frame of the standard of care practiced at the ED site of acquisition).
The neuroradiological adjudicators also scored all CT scans using the Marshall Scale (MS) scoring system.18 The MS, used largely by neurosurgeons, rates the severity of CT abnormality using six categories (I–VI), based largely on volume of blood and midline shift.19 Although the MS is relatively insensitive to gradations of neurological severity in cases in which neurosurgical intervention is not indicated, and is not used routinely in ED assessment, it is applied here as a measure of the mild nature of the CT abnormalities in the population of study.
Blood volume measurements
Because of concerns about the subjective nature of CT scan readings when evaluated conventionally by visual inspection and the poor inter-rater and within-rater reliability reported in the literature, in this study we used a method for quantification of the severity of the hematoma.
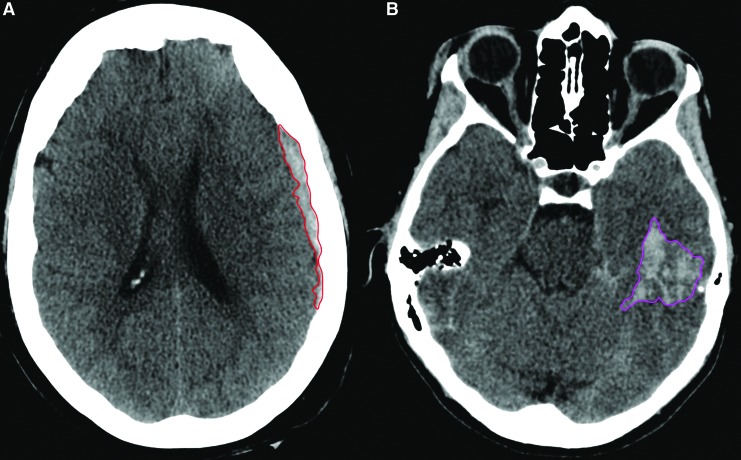
The volume of blood and distance from recording electrodes were measured by blinded independent experts using a published method for volume measurement,11 in the same manner as has been done in the previously published work, and which was described in detail by Hanley and colleagues, cited earlier.12 Measurements of intracranial hematomas were calculated by two independent neuroradiologically trained technician readers (A.M.), blinded to the EEG results and adjudicated by a third reader (D.H.) when required by discrepancy. Each hematoma was classified in terms of the lobe(s) compromised (frontal, parietal, temporal, or occipital), hemisphere affected (left or right), and hematoma type (epidural [EDH]; subdural [SDH]; or intracerebral [ICH]). Quantitative measurements of the volume of blood within the hematoma were performed with operator-defined region of interest segmentation as described by OsiriX11 (see Fig. 1). In instances of multiple hematomas, a single volume was attributed to each unique bleeding location. When a single bleed spanned multiple lobes, the volume was indicated in the lobe in which the epicenter was localized. It is important to note that subarachnoid hemorrhages (SAH) were not assigned a volume. The distance of the bleeding event from the location of the EEG electrodes (midpoint between FP1 and FP2) was also evaluated, to assess whether using only frontal electrodes influenced accuracy of detection. Two linear measurements (mm) were performed for each unique bleeding event, one to the shortest distance from the electrodes to the bleed, (using FP1 or FP2, depending upon the lobe of the bleed); and the second corresponded to the distance from the same sensor to the epicenter of the bleed. Only patients with hematomas with blood volumes ≥1 cc were considered candidates for this study.
FIG. 1.
Representation of the perimeter of a subdural (left panel, red) and an intracranial hematoma (right panel, pink). The volume was calculated by multiplying the area of each region of interest by the slice thickness.
Electrophysiological data acquisition and analysis
Five to ten minutes of eyes closed resting EEG was recorded on a handheld device. EEG recordings were made from five frontal electrode sites of the International 10/20 system referenced to linked ears, using a disposable headset that optimized ease and standardization of placement. Recording sites included: FP1, FP2, AFz (located just anterior to Fz on the forehead, below the hairline), F7, and F8. All electrode impedances were below 10 kΩ. Amplifiers had a bandpass from 0.3 to 250 Hz (3 dB points). The EEG data were subjected to automatic artifact rejection algorithms for identification and removal of any biologic and nonbiologic contamination, such as that from eye movement or muscle movement. Details of these algorithms are given elsewhere.3 When an artifact was identified in any channel, data from all channels at that time point were removed. Artifact-free data (60–120 sec) were concatenated after removal of artifact, with the minimum for any artifact-free segment being 2.5 sec. Sufficient artifact-free data were obtained from all study subjects. EEG recording was made as early as was practically possible without interfering with the clinical workup of the patient, with the vast majority (>85%) tested within 12 h of injury.
The artifact-free EEG data were submitted to offline feature extraction, and the features were input to an independently developed linear discriminant classifier function described previously and in detail elsewhere (see Prichep et al.4), which had high sensitivity and specificity for distinguishing patients with structural brain damage (CT+). This binary discriminant classification function was derived using an evolutionary algorithm.20 An evolutionary algorithm performs a stochastic search (which involves randomness from one iteration to the next) and evaluates a series of candidate solutions, in which each new candidate is informed by high-performing predecessors, similar to genetic evolution.21–29 The final classifier function consists of a weighted combination of selected linear and nonlinear features that reflect brain electrical activity, which mathematically describes traumatic structural brain injury as distinguished from normal or concussed brain activity. The patients in this study were completely independent of the database used to develop the classification algorithm. Each patient's brain electrical activity data were submitted to the algorithm off line, without any information about the patient's clinical status. In addition to the binary classification output (TBI-Index), there was a continuous discriminant score, which was used in analyses that explored the correlation between such scores and blood volume and distance measurements described previously.
Results
The 46 hematoma subjects (28 males and 18 females) were compared with 278 control CT− mTBI patients. The mean age for the hematoma group was 48.5 years (19.1–79.1) and the mean age for the CT− controls was 41.4 years (18.5–80.5). Eighty-three percent (38/46) of the CT+ hematoma group had GCS scores of 15 (normal), with a mean of 14.7 (SD=0.96), with only one patient's GCS <14, and 98.9% (274/277) of the CT− control group had GCS scores of 15, with a mean of 14.88 (SD=1.38), and there were no cases with a GCS <14. All cases were scored using the Marshall Score for grading the severity of the CT+ finding. Forty cases (87%) were Marshall Score II, three had a score of III, two had a score of IV, and one had a score of V (requiring neurosurgery). CT− controls all had Marshall Scores of I.
Neuroradiological findings
The 46 hematomas were located throughout lobes and hemispheres, with 33% (15/46) involving only one lobe, 30% (14/46) involving two lobes, 26% (12/46) involving three lobes, and 11% (5/46) involving all four lobes. It was of note that the frontal lobes were involved most often, in 67% (31/46) of the cases, and occipital lobes were involved least often, only in 20% (9/61) of the cases. Seventy-six percent (35/46) involved bleeds in only one hemisphere and 24% (11/46) were bilateral bleeds. The 46 hematomas included: 1 EDH, which was accompanied by a SAH; 23 SDH, 14 of which also had a SAH; 14 additional subdural hematomas that also had an ICH, 11 of which also had a SAH; and 8 ICH, 5 of which also had a SAH. Figure 2 shows the distribution (as a percentage of all hematomas) of the classifications of the hematomas in the study population.
FIG. 2.
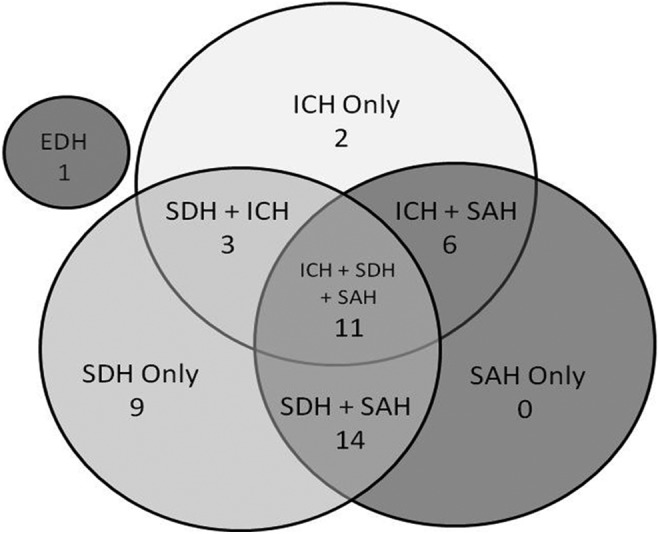
Venn diagram of relative distribution of CT findings (location/etiology) in the hematoma population. Numbers shown are the percentage of the total hematoma population. The small gray circle represents the one epidural hematoma (EDH) in this population. ICH, intracerebral hematoma; SAH, subarachnoid hemorrhage; SDH, subdural hematoma.
Total blood volumes <1cc were not included in this sample. The average volume of blood summed across regions was 16.4 cc (SD=36.43, range 1.14–235.38). The largest single bleed with a volume equal to 235.38 cc, was an outlier to the distribution of volumes measured. Without this individual the mean for summed volumes across regions was 11.63 cc (SD=15.60, range 1.26–74.96). The mean distance from the recording electrodes to the closest blood measured was 42.07 mm (SD=28.03) and to the epicenter of the bleed was 73.25 mm (SD=36.41), with a range of 17.29–160.10 mm. The mean difference between edge and epicenter of the bleed was 30.72 mm (SD=31.23, range 0.65–125.35mm).
TBI-Index
Table 1 shows the contingency table for he accuracy of the TBI-Index for discriminating the hematoma patients from the mTBI CT− patients.
Table 1.
Contingency Table for Classification by TBI-Index
| Structural abnormality | ||
|---|---|---|
| Classification | Present (CT+) | Absent (CT−) |
| Positive | 44 | 156 |
| Negative | 2 | 122 |
TBI, traumatic brain injury.
Of the 46 subjects with hematomas, 44 were classified as positive by the TBI-Index. Of the 278 CT− mTBI controls, 122 were classified as negative by the TBI-Index. Therefore, using the TBI-Index to determine classification, sensitivity to hematomas was 95.7% (95% CI=85.2, 99.5), and specificity was 43.9% (95% CI=38.0, 49.9). Because CT+ hematoma patients were an enriched population, the NPV and positive predictive value (PPV) were not calculated.
Although the algorithm is a binary classifier function that compares discriminant scores to a previously independently determined threshold, we used the discriminant scores for additional analyses. The mean discriminant score for the hematoma group was 68.10 (SD=25.98), and the mean for the CT− controls was 33.52 (SD=28.37). Discriminant scores for the hematoma CT+ patients were significantly higher than those for the CT− controls (t=−7.49, p<0.0001).
A regression analysis among the continuous discriminant score and the volume of the bleed and the distance from the recording electrodes and the presence or absence of accompanying SAH was also studied. No significant correlations were found between the continuous discriminant score and the volume of blood present for the largest single region (F=0.179, p=0.674) or for the sum or total volume across regions (F=0.868, p=0.354). There were also no significant differences found between the discriminant score and the shortest distance to the bleed (F=0.045, p=0.833), or to the epicenter of the bleed (F=0.862, p=0.358). The lack of correlation suggests that the TBI-Index was not influenced by the distance of the hematoma from the recording electrodes, or by the volume of the bleed. There were no significant differences between discriminant scores in hematomas with or without accompanying SAH (t=0.48, p=0.64).
Discussion
The present study is a prospective validation of retrospective results previously reported by Hanley and colleagues,12 in an entirely independent population of ED closed head injured patients with traumatic hematomas. These results lend strong support to the important conclusions of the prior study, showing that a TBI-Index of brain function/dysfunction can be used to correctly identify ED mTBI patients with traumatic intracranial hematomas seen on CT scan. The current study goes beyond the prior report in several important ways, foremost being the importance of using a prospective independent population; and further, this population was also independent of the training of the algorithm used to determine the classification of the patients. Further, the target population in this study was a subset of brain-injured mTBI patients who were found to have a traumatic hematoma with blood volume ≥1cc, but who were referred for CT scan with relatively low suspicion of brain injury, utilizing current standards based on subjective clinical criteria. The use of a minimum criterion (above the level that could be considered artifact) for the presence of blood volume in these cases helps to target the assessment of performance on the correct identification of TBI patients who are at risk for potentially life-threatening bleeds and who would likely require further clinical action. The stratification of risk in such cases requires high sensitivity, low tolerance for false negatives at the expense of false positives and lower specificity. It is noted that specificity in the high 40s represents the potential to reduce CT scans in those in whom scans would be found to be negative, and is much higher than the reported specificity of symptom-based guidelines for selection of those in need of CT scans, which are reportedly in the teens.30
Quantitative features of brain electrical activity (QEEG) used in the BrainScope technology have been reported in the literature to be sensitive to changes in brain activity associated with TBI.31–33 Further, changes in connectivity reported in TBI using diffusion tensor imaging (DTI) are consistent with the phase synchrony abnormalities reported using QEEG.34 The features contributing most to classification algorithms used in this study included those representative of measures that reflect changes in power and frequency distributions, as well as those features that measure disturbances in connectivity between regions (including coherence, phase synchrony and asymmetry), and ratios of these quantities such that the numerator and denominator may come from different bands and channels, in order to capture temporal and spatial relationships in brain activity among different regions and frequency bands. It is of note that because this study used an existing classification algorithm which was a weighted combination of specific features, other features were not explored. The fact that the vast majority of the subjects (∼95%) in this study were classified as abnormal on this discriminant algorithm suggests that these features are useful in describing the changes in brain electrical activity that occur with such traumatic structural injuries, and well reflect the underlying pathophysiology hypothesized in the scientific literature for structural and functional brain injuries sustained in TBI. This sensitivity to both structural and functional brain injury may also relate to the high number of false positives identified by the algorithm.
Other attempts to identify traumatic hematomas use near-infrared probes held to regions of the scalp. The sensitivity of such devices is dependent on the depth (e.g., <2.5 cm from the brain surface) and volume (e.g., >3.5 mL) of the bleed, and are therefore limited in clinical utility.36 When bleeds do not meet the criteria, sensitivity is drastically decreased. In contrast, strong evidence was presented herein that the TBI-Index was not influenced by the distance of the bleed from the EEG recording electrodes. It was also sensitive to a wide and clinically important range of bleed volumes. Therefore, this radiographic evaluation of bleed type and location is consistent with the hypothesis that the TBI algorithm can identify all types of intracranial bleeding.
Limitations
The fact that there was only one epidural EDH in the study population is a limitation on the ability to study relationships to EDH location. In addition, as has been noted in previous work, the lack of a “gold standard” for the CT− group is another weakness of such studies. Questions related to generalizability exist as a consequence of exclusion rules and the requirement that all subjects be capable of providing informed consent. Although obtunded subjects were excluded, the presence of drugs and/or alcohol per se were not criteria for exclusion, and such subjects were not found to perform differently on the algorithm. Although not the study group targeted, the performance of the algorithm in more obtunded patients, including those with lower GCS scores, would be important for further study. Future studies might explore the potential utility in serial recordings over time to access sensitivity to progressive bleeding events.
Conclusions
This study was an independent replication of a prior retrospective report demonstrating the accuracy of using a classification algorithm based on brain electrical activity to identify traumatic hematomas in the mTBI population in the ED. This independent prospective validation of such technology is critical in the assessment its potential clinical utility. Employing a blinded quanititative measurement of blood volume, a group of traumatic hematomas with blood volume ≥1 cc were selected for study and compared with a group of CT− controls, thus focusing the study on those mildly presenting cases (98% with GCS of 14–15) in whom clinical interventions or more intensive follow-ups are indicated. High classification sensitivity (95.7%) and specificity (43.9%) were obtained using this algorithm. Further, performance was independent of the distance of the hematoma from the recording electrodes and the volume of the bleed (≥1 cc). Specificity, although in the 40s, was substantially higher than that obtained using existing symptom-based guildelines and could result in a large reduction in referrals for scanning compared with the current standard of care. The potential clinical utility of such a technology is high in situations with limited access to CT, situations requiring rapid triage and situations with concurrent severe multisystem injury, which are all areas requiring rapid identification of traumatic intracranial bleeds.
Acknowledgments
The authors acknowledge the contributions of those who made this research possible, including the primary investigators and research technicians at all the clinical sites and the patients who volunteered. This research was supported in part by DoD contract #W911QY-12-C-0004, and by BrainScope Co., Inc., Bethesda Maryland, who granted expenses related to data acquisition.
Author Disclosure Statement
Data acquisition for this study was supported in part by Department of Defense contract #W911QY-12-C-0004, Assessment of Head Injury in the Emergency Department, and by research grants from BrainScope Co., Inc. to the clinical sites. Drs. Naunheim and Bazarian were Prinicipal Investigators at clinical data acquisition sites. Dr. Prichep serves as a consultant to BrainScope Company, Inc., and holds potential financial interest through patented technology licensed by BrainScope from NYU School of Medicine. Dr. Hanley sits on the Medical Advisory Board of BrainScope Company, Inc. W. A. Mould was a paid consultant who performed the hematoma measurements, blinded to all other data. BrainScope had no role in the conduct of the studies, analysis of the data, or preparation of manuscripts. Strict adherence to ethical concerns was followed, and all subjects signed written informed consents for participation in the study.
References
- 1.Stiell I.G., Clement C.M., Rowe B.H., Schull M.J., Brison R., Cass D., Eisenhauer M. A., McKnight R.D., Bandiera G., Holroyd B., Lee J.S., Dreyer J., Worthington J.R., Reardon M., Greenberg G., Lesiuk H., MacPhail I., and Wells G. A. (2005). Comparison of the Canadian CT Head Rule and the New Orleans Criteria in Patients with Minor Head Injury. JAMA 294, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 2.Smits M., Dippel D.W.J., de Haan G.G., Dekker H.M., Vos P.E., Kool D.R., Nederkoorn P.J., Hofman P.A.M., Twijnstra A., Tanghe H.L.J., and Hunink M.G.M. (2005). External validation of the Canadian CT Head Rule and the New Orleans Criteria for CT Scanning in Patients With Minor Head Injury. JAMA 294, 1519–1525 [DOI] [PubMed] [Google Scholar]
- 3.Prichep L.S., Jacquin A., Filipenko J., Ghosh Dastidar S., Zabele S., Vodencarevic A., and Rothman N.S. (2012). Classification of traumatic brain injury severity using informed data reduction in a series of binary classification algorithms. IEEE Trans. Neural Syst. Rehabil. Eng. 20, 806–822 [DOI] [PubMed] [Google Scholar]
- 4.Prichep L.S., Huff S., O'Neil B., Naunheim R.S., Jacquin A., Radman T., Miller J., and Ghosh Dastidar S. (2014). Classification algorithms for the identification of structural injury in TBI using brain electrical activity. Comput. Biol. Med. 53C:125–133 [DOI] [PubMed] [Google Scholar]
- 5.Naunheim R.S., Treaster M., English J., Casner T., and Chabot R. (2010). Use of Brain Electrical Activity to Quantify Traumatic Brain Injury in the Emergency Department. Brain Inj. 24, 1324–1329 [DOI] [PubMed] [Google Scholar]
- 6.O'Neil B., Naunheim R.S., Prichep L.S., and Chabot R.J. (2012). Can quantitative brain electrical activity aid in the initial screening of mild traumatic brain injured patients. West. J. Emerg. Med. 13, 394–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina D.K., Nichols J.J., and DiMaio V. J. (2007). The sensitivity of computed tomography (CT) scans in detecting trauma: are CT scans reliable enough for courtroom testimony? J. Trauma 63, 625–629 [DOI] [PubMed] [Google Scholar]
- 8.Laalo J.P., Kurki T.J., Sonninen P.H., and Tenovuo O.S. (2009). Reliability of diagnosis of traumatic brain injury by computed tomography in the acute phase. J. Neurotrauma 26, 2169–2178 [DOI] [PubMed] [Google Scholar]
- 9.Saatman K.E., Duhaime A.C., Bullock R., Maas A.I.R., Valadka A., and Manley G.T. (2008). Classification of traumatic brain injury for targeted therapies. J. Neurotrauma 25, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs B., Beems T., van der Vliet T.M., Diaz–Arrastia R., Borm G.F., and Vos P.E. (2011). Computed tomography and outcome in moderate and severe traumatic brain injury: hematoma volume and midline shift revisited. J. Neurotrauma 28, 203–215 [DOI] [PubMed] [Google Scholar]
- 11.Rosset A., Spadola L., and Ratib O. (2004). OsiriX: An open-source software for navigating in multidimensional DICOM images. J. Digit. Imaging 17, 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanley D.F., Chabot R.J., Mould W.A., Morgan T., Naunheim R.S., Sheth K., Chiang W., and Prichep L.S. (2013). Use of brain electrical activity for the identification of hematomas in mild traumatic brain injury. J. Neurotrauma 30, 2051–2056 [DOI] [PubMed] [Google Scholar]
- 13.Kahraman S., Kayali H., Atabey C., Acar F., and Gocmen S. (2006). The accuracy of near-infrared spectroscopy in detection of subdural and epidural hematomas. J. Trauma Acute Care Surg. 61, 1480–1483 [DOI] [PubMed] [Google Scholar]
- 14.Kessel B., Jeroukhimov I., Ashkenazi I., Khashan T., Oren M., Haspel J., Medvedev M., Nesterenko V., Halevy A., and Alfici R. (2007). Early detection of life-threatening intracranial haemorrhage using a portable near-infrared spectroscopy device. Injury 38, 1065–1068 [DOI] [PubMed] [Google Scholar]
- 15.Leon–Carrion J., Dominguez–Roldan J. M., Leon–Dominguez U., and Murillo–Cabezas F. (2010). The Infrascanner, a handheld device for screening in situ for the presence of brain haematomas. Brain Inj. 24, 1193–1201 [DOI] [PubMed] [Google Scholar]
- 16.Robertson C.S., Zager E.L., Narayan R.K., Handley L., Sharma A., Hanley D.F., Garza H., Maloney–Wilensky E., Plaum J.M., Koenig C.H., Johnson A., and Morgan T. (2010). Clinical evaluation of a portable near-infrared device for detection of traumatic intracranial hematomas. J. Neurotrauma 27, 1597–1604 [DOI] [PubMed] [Google Scholar]
- 17.DeRenzo E. G., Conley R. R., and Love R. (1998). Assessment of capacity to give consent to research participation: state-of-the-art and beyond. Journal of Health Care and Law Policy 1, 66–87 [PubMed] [Google Scholar]
- 18.Marshall L.F., Marshall S.B., Klauber M., VanBerkum C., Eisenberg H., Jane J., Luerssen T., Marmarou A., and Foulkes M. (1992). The diagnosis of head injury requires a classification based on computed axial tomography. J. Neurotrauma 9, S287–S292 [PubMed] [Google Scholar]
- 19.Marshall L.F., Bowers–Marshall S., Klauber M.R., van Berkum Clark M., Eisenberg H.M., Jane J.A., Luerssen T.G., Marmarou A., and Foulkes M.A. (1991). A new classification of head injury based on computerized tomography. J. Neurosurgery 75 (Suppl.), S14–S30 [Google Scholar]
- 20.de la Iglesia B. (2013). Evolutionary computation for feature selection in classification problems. WIREs Data Mining Knowl Discov 3, 381–407 [Google Scholar]
- 21.Tomassini M. (1999). Parallel and distributed evolutionary algorithms: a review, in: Evolutionary Algorithms in Engineering and Computer Science. K. Miettinen M.M.M., P. Neittaanmaki J.P. (eds.). J. Wiley and Sons: Chichester [Google Scholar]
- 22.Syswerda G., and Palmucci J. (1991). The application of genetic algorithms to resource scheduling. Presented at Proceedings of the Fourth International Conference on Genetic Algorithms, San Diego [Google Scholar]
- 23.Jakob W., Gorges–Schleuter M., and Blume C. (1992). Application of Genetic Algorithms to Task Planning and Learning. Proceedings of Parallel Problem Solving from Nature (PPSN II), North Holland, Amsterdam. Elsevier: Amsterdam, pp. 291–300 [Google Scholar]
- 24.Jones G., Brown R.G., Clark D.E., Willett P., and Glen R.C. (1993). Searching Databases of Two-Dimensional and Three-Dimensional Chemical Structures Using Genetic Algorithms. Morgan Kaufmann Publishers, Inc., San Francisco, CA, pp. 597–602 [Google Scholar]
- 25.Mitchell M. (1996). An Introduction to Genetic Algorithms (Complex Adaptive Systems). First MIT Press: Cambridge [Google Scholar]
- 26.Siedlecki W., and Sklansky J. (1989). A note on genetic algorithms for large-scale feature selection. Pattern Recognit. Lett. 10, 335–347 [Google Scholar]
- 27.Yang J.H. and Honavar V. (1998). Feature subset selection using a genetic algorithm. IEEE Intell. Syst. 13, 44–49 [Google Scholar]
- 28.Raymer M.L., Punch E.D., Goodman E.D., Kuhn L.A., and Jain A.K. (2000). Dimensionality reduction using genetic algorithms. IEEE Trans. Evolutionary Computation 4, 164–171 [Google Scholar]
- 29.Oh I.S., Lee J.S., and Moon B.R. (2004). Hybrid genetic algorithms for feature selection. IEEE Trans. Pattern Anal. Mach. Intell. 26, 1424–1437 [DOI] [PubMed] [Google Scholar]
- 30.Jagoda A.S., Bazarian J.J., Bruns J.J., Jr, Cantrill S.V., Gean A.D., Howard P.K., Ghajar J., Riggio S., Wright D.W., Wears R.L., Bakshy A., Burgess P., Wald M.M., and Whitson R.R. (2008). Clinical policy: neuroimaging and decisionmaking in adult mild traumatic brain injury in the acute setting. Ann. Emerg. Med. 52, 714–748 [DOI] [PubMed] [Google Scholar]
- 31.Tebano M.T., Cameroni M., Gallozzi G., Loizzo A., Palazzino G., Pezzini G., and Ricci G.F. (1988). EEG spectral analysis after minor head injury in man. Electroencephalogr. Clin. Neurophysiol. 70, 185–189 [DOI] [PubMed] [Google Scholar]
- 32.Thatcher R.W., Biver C., Mc Alaster R., Camacho M., and Salazar A. (1998). Biophysical Linkage betweeen MRI and EEG amplitude in closed head I. Neuroimage 7, 352–367 [DOI] [PubMed] [Google Scholar]
- 33.Thatcher R.W., North D.M., Curtin R.T., Walker R.A., Biver C.J., Gomez J.F., and Salazar A.M. (2001). An EEG severity index of traumatic brain injury. J. Neuropsychiatry Clin. Neurosci. 13, 77–81 [DOI] [PubMed] [Google Scholar]
- 34.Sponheim A.R., McGuire K.A., Kang S.S., Davenport N.D., Aviyente S., Bernat E.M., and Lim K.L. (2011). Evidence of disrupted functional connectivity in the brain after combat-related blast injury. Neuroimage 54, s21–s29 [DOI] [PubMed] [Google Scholar]
- 35.United States Food and Drug Administration, Center for Devices and Radiological Health. (2011). Infrascanner Model 1000, Evaluation of Automatic Class III Designation Petition (de novo) K080377 Classification Order