Significance
There are few laboratory models that recapitulate human cardiac disease. Here, we created human cell models for Jervell and Lange-Nielsen syndrome (JLNS) in vitro, based on human induced pluripotent stem cells (hiPSCs). JLNS is one of the most severe disorders of heart rhythm and can cause sudden death in young patients. JLNS is inherited recessively and is caused by homozygous mutations in the slow component of the delayed rectifier potassium current, IKs. Cardiomyocytes (CMs) from two independent sets of patient-derived and engineered hiPSCs showed electrophysiological defects that reflect the severity of the condition in patients. Our work allowed better understanding of the mechanisms of recessive inheritance. Furthermore, JLNS-CMs showed increased sensitivity to proarrhythmic drugs, which could be rescued pharmacologically, demonstrating the potential of hiPSC-CMs in drug testing.
Keywords: Jervell and Lange-Nielsen syndrome, long QT syndrome, human induced pluripotent stem cells, disease modeling, KCNQ1
Abstract
Jervell and Lange-Nielsen syndrome (JLNS) is one of the most severe life-threatening cardiac arrhythmias. Patients display delayed cardiac repolarization, associated high risk of sudden death due to ventricular tachycardia, and congenital bilateral deafness. In contrast to the autosomal dominant forms of long QT syndrome, JLNS is a recessive trait, resulting from homozygous (or compound heterozygous) mutations in KCNQ1 or KCNE1. These genes encode the α and β subunits, respectively, of the ion channel conducting the slow component of the delayed rectifier K+ current, IKs. We used complementary approaches, reprogramming patient cells and genetic engineering, to generate human induced pluripotent stem cell (hiPSC) models of JLNS, covering splice site (c.478-2A>T) and missense (c.1781G>A) mutations, the two major classes of JLNS-causing defects in KCNQ1. Electrophysiological comparison of hiPSC-derived cardiomyocytes (CMs) from homozygous JLNS, heterozygous, and wild-type lines recapitulated the typical and severe features of JLNS, including pronounced action and field potential prolongation and severe reduction or absence of IKs. We show that this phenotype had distinct underlying molecular mechanisms in the two sets of cell lines: the previously unidentified c.478-2A>T mutation was amorphic and gave rise to a strictly recessive phenotype in JLNS-CMs, whereas the missense c.1781G>A lesion caused a gene dosage-dependent channel reduction at the cell membrane. Moreover, adrenergic stimulation caused action potential prolongation specifically in JLNS-CMs. Furthermore, sensitivity to proarrhythmic drugs was strongly enhanced in JLNS-CMs but could be pharmacologically corrected. Our data provide mechanistic insight into distinct classes of JLNS-causing mutations and demonstrate the potential of hiPSC-CMs in drug evaluation.
Jervell and Lange-Nielsen syndrome (JLNS) is a rare, autosomal recessive disease characterized by congenital bilateral deafness, severe QT interval prolongation on the electrocardiogram (ECG), polymorphic ventricular arrhythmias, syncope, and high risk of sudden death (1, 2). JLNS results from homozygous (or compound heterozygous) mutations in the KCNQ1 or KCNE1 genes. These encode the α- and β-subunits, respectively, of the ion channel conducting the slow component of the delayed rectifier K+ current (IKs) (3, 4). Another long QT condition termed Romano–Ward syndrome (RWS) is, by contrast, an autosomal-dominant form of QT interval prolongation without deafness, caused by heterozygous mutations in 16 different genes, including KCNQ1 (LQT1) and KCNE1 (LQT5) (5–7). However, the recessive JLNS is among the most severe forms of the disease, together with Timothy syndrome and a long QT syndrome variant caused by calmodulin mutations (8, 9). JLNS patients usually have severe clinical symptoms, early disease onset (∼12 mo old), and require aggressive interventions because of the limited efficacy of β-receptor blockers (2).
JLNS patients with KCNQ1 mutations usually display longer QT intervals and higher risk for arrhythmic events than those with KCNE1 mutations (2). Attempts to associate the type of mutation (e.g., missense, nonsense, frameshift) in KCNQ1 with the RWS or JLNS phenotype have proven challenging. In general, however, missense mutations with a dominant-negative effect on the tetrameric KCNQ1 channel tend to cause RWS, whereas JLNS is frequently caused by nonsense and frameshift mutations (10–13). However, exceptions exist in that missense mutations can also result in JLNS (14). Furthermore, there are rare but well-documented cases of symptoms in heterozygous carriers of JLNS mutations (11, 15–17).
Human induced pluripotent stem cells (hiPSCs) are already proving to provide powerful cellular models to study both genetic and sporadic diseases in humans (18). Several cardiac ion channel diseases have been investigated by using hiPSC-derived cardiomyocytes (hiPSC-CMs), including distinct subtypes of RWS (LQT1, LQT2, LQT3, and LQT8) (19–21). Here, we report and analyze independent hiPSC models for the severe and recessively inherited JLNS. Two JLNS-causing mutations were investigated: the novel c.478-2A>T and the previously described c.1781G>A single nucleotide exchanges (22). Compared with heterozygous and wild-type (wt) controls, cardiomyocytes (CMs) of both JLNS models showed severe functional abnormalities caused by complete or near-complete loss of IKs. Although disease phenotypes in the homozygous c.478-2A>T and c.1781G>A cells were similar, distinct loss-of-function molecular mechanisms (strictly recessive and gene dosage-dependent, respectively) were mediated by the two mutations. JLNS-CMs were also highly sensitive to adrenergic and proarrhythmic stress, which could be exploited in future drug safety pharmacology for identifying high-risk individuals. Conversely, arrhythmia phenotypes could be prevented by pharmacological treatment, highlighting the value of hiPSC-CMs in drug testing.
Results
Generation of hiPSC Lines from Patients with KCNQ1 Mutations.
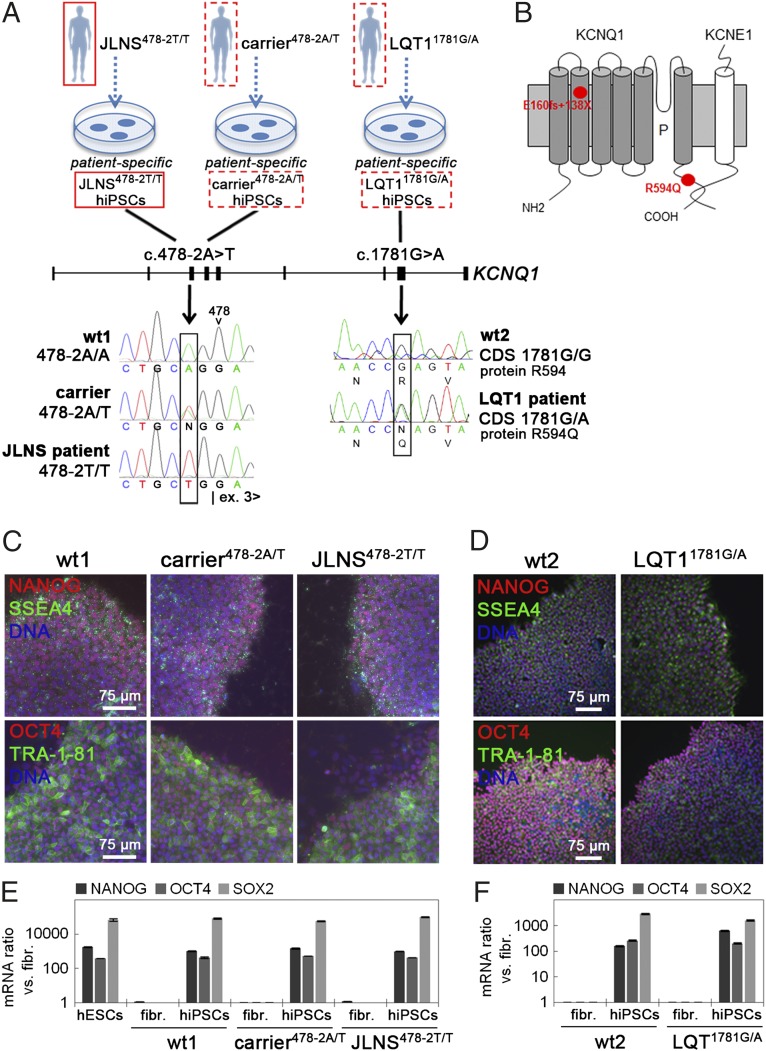
Fibroblasts were obtained from patients with different KCNQ1 mutations, as follows: (i) a JLNS patient with the homozygous single nucleotide c.478-2A>T mutation at the 3′ end of intron 2; (ii) an asymptomatic heterozyous carrier from the same family; (iii) a LQT1 patient with the heterozygous missense c.1781G>A mutation in exon 15 (Fig. 1A and Fig. S1 A–D). Fibroblasts were also derived from two gender-matched healthy individuals (wt1 and wt2; Fig. S1 C and D and SI Materials and Methods). The newly described c.478-2A>T mutation would possibly lead to a splice defect, resulting in the skipping of exon 3 with concomitant frame shift and appearance of a premature stop codon (E160fs+138X). The c.1781G>A KCNQ1 mutation causes substitution of an arginine with a glutamine residue at position 594 of the coding sequence (R594Q) (Fig. 1B). Sendai viruses carrying the four transcription factors OCT4, SOX2, KLF4, and MYC (23) were used to generate hiPSCs. The resulting lines showed typical human embryonic stem cell (hESC) morphology and growth characteristics, with erasure of the Sendai vectors upon passage (Fig. S1 E–H). One line per genotype was selected for further experiments. hiPSCs showed activation of endogenous pluripotency markers (Fig. 1 C–F), were pluripotent as assessed by spontaneous differentiation into derivatives of the three germ layers (Fig. S2 A and B), displayed hESC-like transcriptomes (24) (Fig. S2 C and D), and had normal karyotypes (Fig. S2 E and F). These results indicated successful integration-free reprogramming of all lines.
Fig. 1.
Generation of hiPSCs from patients with KCNQ1 mutations. (A) Sequencing of the KCNQ1 gene identified the c.478-2A>T mutation at the splice acceptor site of intron 2 in the JLNS patient and the heterozygous carrier and the heterozygous c.1781G>A mutation in exon 15 of the LQT1 patient. The genotypes of the two healthy controls wt1 and wt2 are shown as reference. (B) The c.478-2A>T mutation is predicted to result in skipping of exon 3, with a concomitant reading frame shift from position 160 onwards, giving rise to a premature stop codon (E160fs+138X). The c.1781G>A mutation results in the substitution of an arginine with a glutamine at position 594 (R594Q) in the C-terminal domain of the KCNQ1 protein. (C and D) Immunofluorescence analysis of the pluripotency-associated markers NANOG, SSEA4, OCT4, and TRA1-81 in wt1, carrier478-2A/T, and JLNS478-2T/T (C), and in wt2 and LQT11781G/A hiPSCs (D). (E and F) qPCR expression analysis of the endogenous pluripotency genes NANOG, OCT4, and SOX2 in the indicated hiPSC lines.
Generation of Isogenic Pairs of JLNS Human Pluripotent Stem Cells.
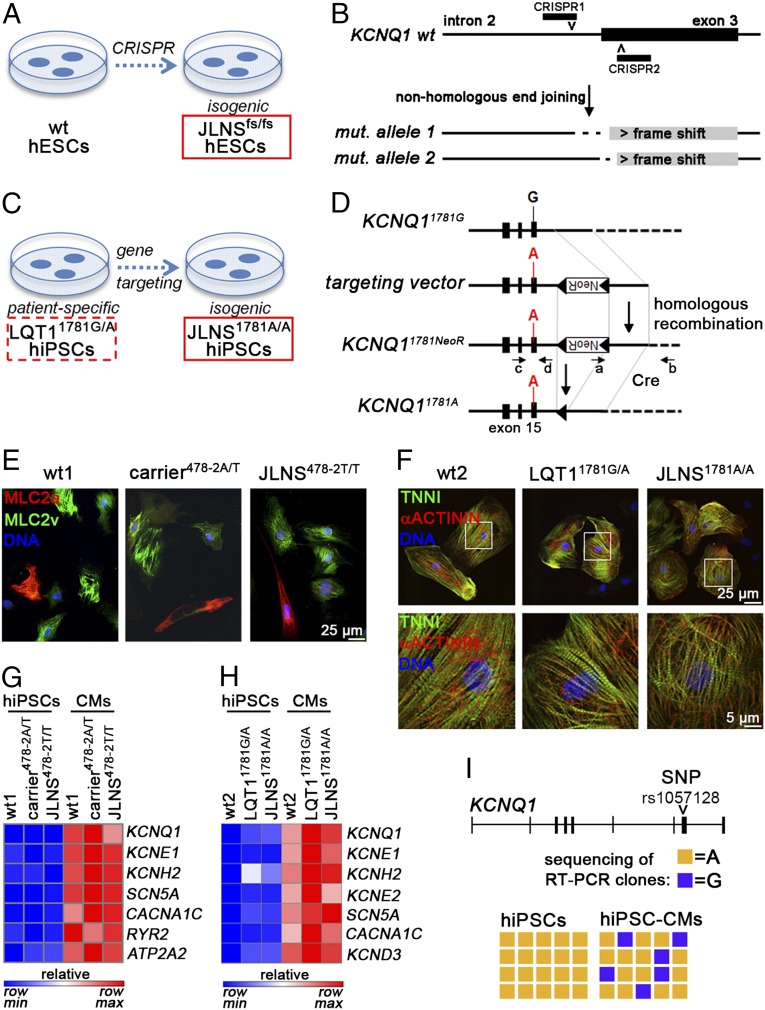
To be able to assess the impact of the c.478-2A>T mutation on an independent genetic background, we used the CRISPR/Cas9 system to generate isogenic pairs of wt and homozygous c.478-2A>T hESCs (JLNSfs/fs) by disrupting the intron 2-exon 3 boundary of KCNQ1 (25) (Fig. 2 A and B). Moreover, we sought to generate a JLNS model from the heterozygous c.1781G>A hiPSCs, by inserting this mutation into the wt allele of the LQT11781G/A line by using homologous recombination (Fig. 2 C and D). This approach resulted in the isogenic homozygous mutated JLNS1781A/A hiPSC line. Correct targeting in this homozygous clone was confirmed by PCR and DNA sequencing (Fig. S1I). The JLNS1781A/A hiPSCs were further characterized as above, with respect to self-renewal and pluripotency features (Fig. S2 B, D, and F). These cell lines complemented the patient-derived hiPSCs for further investigation.
Fig. 2.
Generation of isogenic JLNS hiPSC and hESC lines and general characterization of CMs. (A and B) CRISPR/Cas9-mediated homozygous disruption of the c.478-2 site in wt hESCs. Clones harboring frame shift-causing mutations on both alleles were isolated and expanded. (C and D) Schematic of the gene targeting strategy to introduce the c.1781G>A mutation in LQT11781G/A hiPSCs. Black boxes indicate exons (D). The wt allele of LQT11781G/A hiPSCs is shown with the black G in exon 15. The targeting vector has the mutation (A nucleotide, red) and a loxP-flanked G418-resistance cassette (NeoR). PCR primers a+b and c+d served to identify correctly targeted clones. (E and F) Immunofluorescence images of cardiac sarcomeric proteins MLC2a, MLC2v, TNNI, and α-actinin in the indicated lines following ∼4 wk of in vitro differentiation and maturation. F, Lower is a magnification of the framed area in Upper. (G and H) Heat map representation of qPCR gene expression of cardiac ion channels, before and after cardiomyocyte differentiation. Data are relative to wt hiPSCs. (I) Loss of KCNQ1 imprinting in wt1-CMs compared with undifferentiated hiPSCs. The SNP rs1057128 was used to discriminate between the two KCNQ1 alleles in wt1 cells. Data denote sequencing results of 20 RT-PCR clones.
Differentiation into the Cardiac Lineage.
The two independent, validated sets of cell lines (wt1/carrier478-2A/T/JLNS478-2T/T hiPSC and wt2/LQT11781G/A/JLNS1781A/A hiPSC) were induced to differentiate into CMs by using directed differentiation protocols (SI Materials and Methods). Beating areas were visible from day 6–10 onwards. CMs stained positively for the cardiac sarcomeric proteins MLC2a, MLC2v, troponin I, and α-actinin (Fig. 2 E and F) and expressed genes encoding cardiac ion channels (Fig. 2 G and H). KCNQ1 is an imprinted gene that is monoallelically expressed during early development, but later, in the heart, expression becomes biallelic (26). Hence, KCNQ1 imprinting could potentially interfere with proper manifestation of the disease phenotype in hiPSC-CMs. We therefore examined the imprinting status of KCNQ1 upon cardiac differentiation, by taking advantage of a single-nucleotide polymorphism allowing the two KCNQ1 alleles in wt1 hiPSCs to be distinguished. Undifferentiated hiPSCs displayed a strictly monoallelic expression pattern, confirming the imprinted status of the gene. However, after differentiation into CMs, expression became biallelic (Fig. 2I). These data suggested that the differentiation methods used here should allow faithful monitoring of differences between heterozygous and homozygous CMs.
Electrophysiological Phenotype of JLNS478-2T/T and carrier478-2A/T hiPSC-CMs.
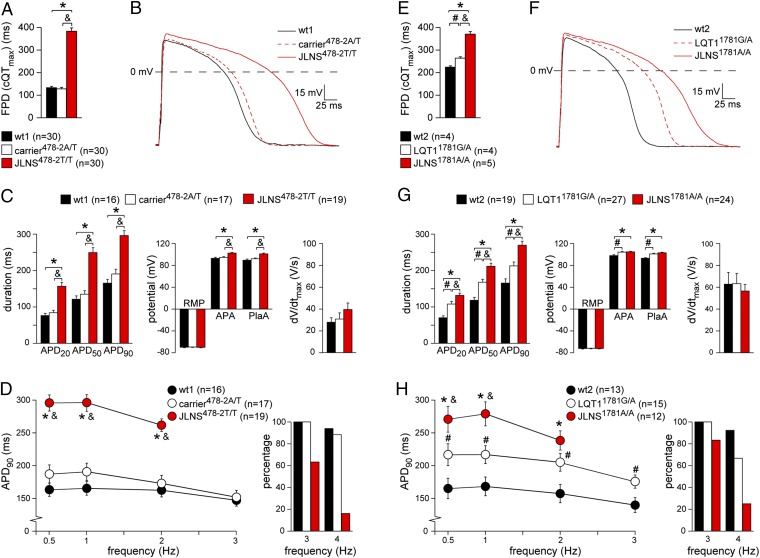
We first characterized the electrophysiological properties of homozygous and heterozygous c.478-2A>T KCNQ1 mutated hiPSC-CMs. Using multielectrode arrays (MEAs) that allow field potential duration (FPD) measurements in cell aggregates, JLNS478-2T/T-CMs showed pronounced FPD prolongation compared with wt1 and carrier478-2A/T CMs, although wt1- and carrier478-2A/T-CMs did not differ significantly from each other (Fig. 3A). The same “recessive” manifestation of the electrophysiological phenotype was also observed in an independent set of (retrovirally) reprogrammed cell lines, ruling out cell clone-specific effects (Fig. S3A). To investigate the phenotype at the single-cell level, action potentials (APs) were measured by using the amphotericin-perforated patch-clamp technique. Fig. 3 B and C show typical APs and average AP properties, respectively, measured at 1 Hz. AP duration at 20, 50, and 90% repolarization (APD20, APD50, and APD90, respectively) in JLNS478-2T/T-CMs were significantly increased compared with wt1 and heterozygous controls. In addition, AP amplitude (APA) and plateau amplitude (PlaA) were significantly increased in JLNS478-2T/T-CMs. Maximal upstroke velocity (dV/dtmax) and resting membrane potential (RMP) were unchanged between cell lines. Of note, AP properties did not differ significantly between wt1- and carrier478-2A/T-CMs. The changes in AP duration between JLNS478-2T/T-CMs and wt1- and carrier478-2A/T-CMs were also detected at slower (0.5 Hz) and faster (2 Hz) pacing rates (Fig. 3D, Left). Interestingly, most of the JLNS-CMs could not be paced at 3 and 4 Hz (Fig. 3D, Right). These data are in agreement with the severe and recessive disease phenotype observed in JLNS.
Fig. 3.
Electrophysiological characterization of hiPSC-CMs. A–D show results for the wt1-, carrier478-2A/T-, and JLNS478-2T/T-CMs, and E–H for the wt2-, LQT11781G/A-, and JLNS1781A/A-CMs. (A and E) FPD quantification on MEAs. (B and F) Representative APs at 1 Hz. (C and G) Average APD20, APD50, APD90, dV/dtmax, RMP, APA, and PlaA. (D and H) Frequency dependence of APD90 (Left). Most JLNS-CMs could not be paced at high frequencies (Right). Symbols *, &, and # indicate statistical significance based on pairwise comparisons (P < 0.05).
Electrophysiological Phenotype of JLNS1781A/A and LQT11781G/A hiPSC-CMs.
Next, we investigated the electrophysiological properties of the c.1781G>A KCNQ1 mutated CMs. Similarly to JLNS478-2T/T CMs, FPD in homozygous JLNS1781A/A-CMs was significantly increased compared with wt2-CMs. However, in contrast to the first set of hiPSC lines, the FPD in LQT11781G/A-CMs displayed an intermediate phenotype compared with wt2- and JLNS1781A/A-CMs (Fig. 3E). APs measured at 1 Hz revealed significant AP prolongation in the JLNS1781A/A-CMs compared with both LQT11781G/A- and wt2-CMs (Fig. 3 F and G). Of note, and in agreement with MEA measurements, heterozygous LQT11781G/A-CMs displayed prolonged AP duration with respect to wt2-CMs. APA and PlaA were slightly increased in the JLNS1781A/A- and LQT11781G/A-CMs compared with wt2-CMs, whereas dV/dtmax and RMP did not differ significantly between the lines. Significant differences in AP duration were also observed between the different hiPSC-CMs when they were stimulated at other frequencies (0.5 and 2 Hz) (Fig. 3H, Left). Much like JLNS478-2T/T CMs, fewer JLNS1781A/A-CMs could be paced at 3 and 4 Hz compared with control CMs (Fig. 3H, Right). These results suggest a gene-dosage effect for the KCNQ1 c.1781G>A mutation, in contrast to the strictly recessive c.478-2A>T nucleotide exchange. The more pronounced AP prolongation in the homozygous—compared with the heterozygous—state is in agreement with a more severe repolarization abnormality in JLNS than in LQT1.
The c.478-2A>T KCNQ1 Mutation Abolishes IKs in Homozygous CMs.
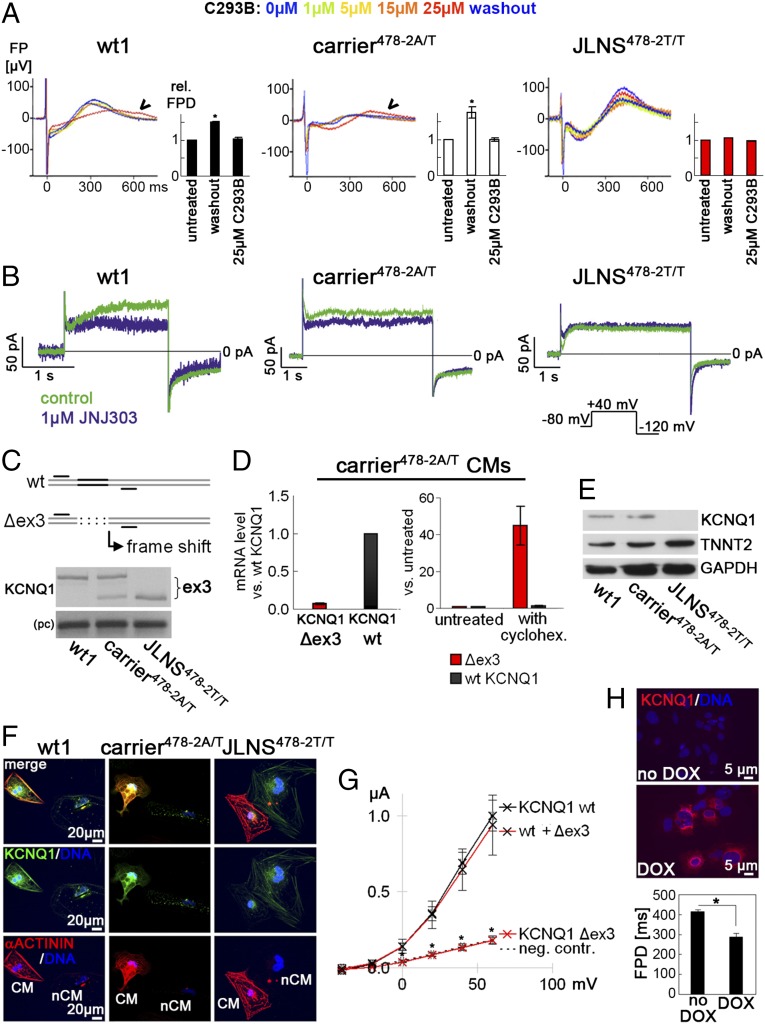
We next investigated the role of IKs in AP and FPD prolongation. Both KCNQ1 and KCNE1 were expressed at similar levels in the three cell lines (Fig. S3B). However, FPD in JLNS478-2T/T-CMs was completely insensitive to the IKs blocker chromanol 293B (C293B) (Fig. 4A), whereas wt1- and carrier478-2A/T-CMs showed FPD prolongation when IKs was blocked. Similarly, at the single-cell level, IKs was clearly detected in wt1- and carrier478-2A/T-CMs, but virtually absent in JLNS478-2T/T-CMs, as revealed by patch clamp electrophysiology using the IKs blocker JNJ303 (Fig. 4B). Furthermore, when E4031 was added to inhibit the rapid component of the delayed rectifier K+ current, IKr, which cooperates with IKs in the repolarization of the AP, FPD was prolonged in wt1- and carrier478-2A/T-CMs without altering spontaneous beating, as expected (27) (Fig. S4A). However, in JLNS478-2T/T-CMs, E4031 reproducibly induced arrhythmia already at moderate doses (100 nM), in agreement with reduced repolarization reserve in these cells due to defective IKs. Collectively, these data indicate that the homozygous c.478-2A>T KCNQ1 mutation is amorphic (nonfunctional).
Fig. 4.
Molecular mechanism underlying the c.478-2A/T mutation. (A) Effect of the IKs-blocker C293B on FPD. Single representative FPDs and average quantification are shown (n = 3). JLNS478-2T/T-CMs are insensitive to the drug. (B) Representative current traces before (control) and after JNJ303 treatment in the indicated hiPSC-CMs. IKs is virtually absent in JLNS478-2T/T-CMs. (Inset) Voltage protocol. (C) KCNQ1 RT-PCR analysis in the indicated hiPSC-CMs. (Top) Schematic showing primers used to reveal the skipping of exon 3 in mutant hiPSC-CMs. (Bottom) Upper band, wt transcript; lower band, Δex3 mRNA; pc, positive control. (D) qPCR analysis of relative abundance of Δex3 and wt KCNQ1 transcripts in carrier478-2A/T-CMs under baseline conditions (Left) and upon cycloheximide treatment (Right); cycloheximide (30 µg/mL, 3 h) selectively induces the Δex3 transcript. (E) Western blot analysis in hiPSC-CMs by using an antibody detecting the C terminus of KCNQ1. TNNT2 and GAPDH are shown as cardiac and loading controls, respectively. (F) Immunofluorescence analysis of KCNQ1 and α-actinin in hiPSC-CMs. Noncardiomyocyte cells are shown next to CMs to indicate specificity of staining. (G) Average I-V relationship for IKs measured upon injection of wt, Δex3, and wt+Δex3 mRNA into Xenopus oocytes (n = 10 each). (H) Genetic rescue of JLNS478-2T/T-CMs by using DOX-inducible KCNQ1 overexpression (Top); FPD shortening after 48 h of DOX treatment (Bottom, n = 4).
The c.478-2A>T mutation Abolishes KCNQ1 Protein Expression Through a Splicing Defect.
We next sought to understand the molecular mechanisms underlying the recessive c.478-2A>T mutation. RT-PCR analysis showed that KCNQ1 mRNA was smaller in JLNS478-2T/T-CMs compared with wt1-CMs (Fig. 4C). Sequencing of the PCR product revealed skipping of exon 3 at the RNA level (Δex3), in agreement with the disruption of the intron 2 splice acceptor site at the DNA level. As expected, heterozygous carrier478-2A/T-CMs expressed both alleles, which also confirmed the loss of KCNQ1 imprinting (Fig. 4C and Fig. S4B). Interestingly, isoform-specific quantitative real-time PCR (qPCR) revealed that in carrier478-2A/T-CMs, Δex3 KCNQ1 was strongly diminished with respect to the wt transcript in the same cells, whereas the wt KCNQ1 was expressed at levels comparable to wt1-CMs (Fig. 4D, Left and Fig. S4C). The exon 3 deletion causes a frame shift and premature stop codon in the KCNQ1 ORF. The low Δex3 expression level may therefore be a result of nonsense-mediated mRNA decay (NMD) (28). To test this hypothesis, carrier478-2A/T-CMs were transiently treated with cycloheximide, which inhibits the highly protein synthesis-dependent NMD pathway (29). Cycloheximide specifically up-regulated Δex3- but not wt-KCNQ1 mRNA in carrier478-2A/T-CMs, suggesting that indeed, the mutant transcript is targeted by NMD (Fig. 4D, Right).
These data predict that the splice defect caused by the c.478-2A>T mutation abolishes KCNQ1 protein in homozygous JLNS478-2T/T-CMs, whereas in carrier478-2A/T cells KCNQ1 may be unaffected. Western blot and immunofluorescence analysis, using an antibody recognizing the C terminus of KCNQ1, confirmed this hypothesis (Fig. 4 E and F). That the absence of KCNQ1 correlates with significant FPD prolongation on MEAs was also evident in engineered isogenic JLNSfs/fs-CMs, again indicating that the disease phenotype correlates with the loss of KCNQ1 (Fig. S4 D and E). Moreover, to test whether residual truncated KCNQ1 could still exert a dominant negative effect on the wt channel, wt- and Δex3-KCNQ1 mRNA isoforms were injected, alone and in combination, into Xenopus oocytes. Quantification of the corresponding currents confirmed complete loss of function of Δex3-KCNQ1, without any dominant negative effect on wt-KCNQ1 (Fig. 4G). Finally, to demonstrate functionally that AP and FPD prolongation in JLNS478-2T/T-CMs was due to the absence of KCNQ1, a doxycycline (DOX)-inducible transgene was used to rescue the phenotype (Fig. S4F, Left). wt-KCNQ1 transgene activation significantly reduced FPD prolongation and restored sensitivity to C293B (Fig. 4H and Fig. S4F, Right).
The c.1781G>A KCNQ1 Mutation Reduces IKs in Heterozygous and Homozygous CMs.
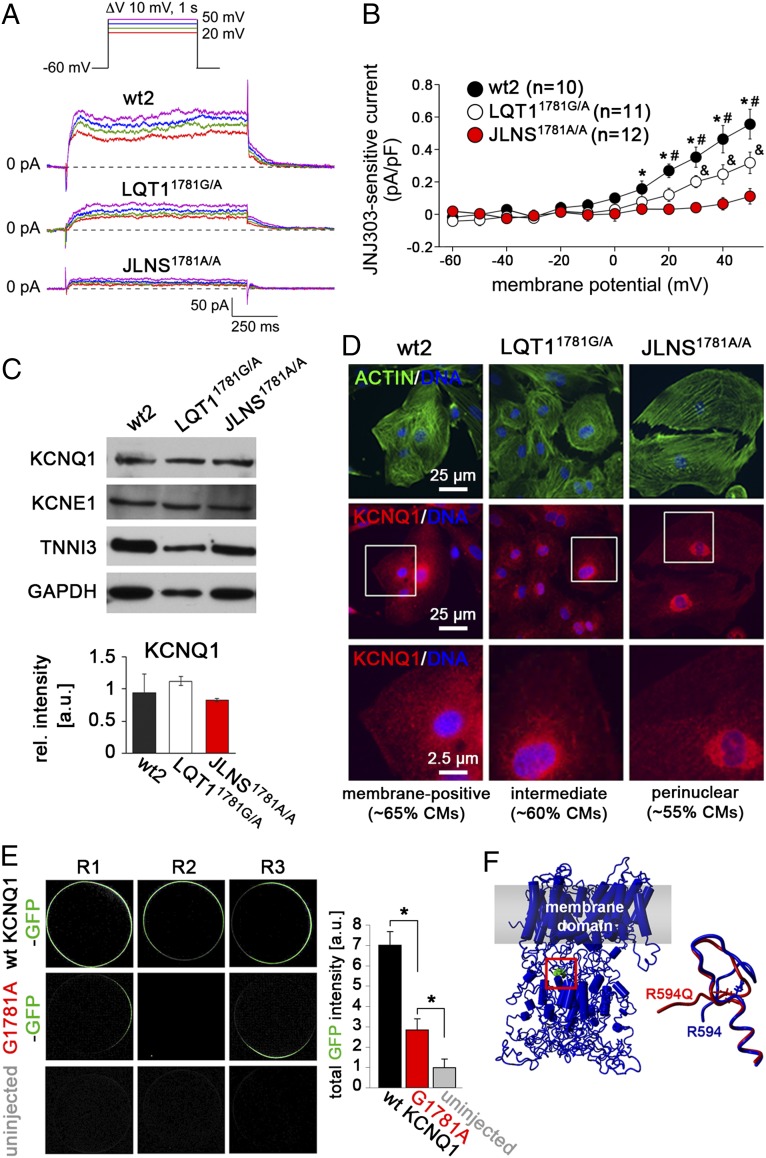
We then investigated the effect of the c.1781G>A KCNQ1 mutation on IKs, measured as JNJ303-sensitive current (Fig. S5A). Fig. 5 A and B show typical IKs and average JNJ303-sensitive tail current densities in wt2-, LQT11781G/A-, and JLNS1781A/A-CMs. IKs tail current density was significantly smaller in JLNS1781A/A-CMs compared with LQT11781G/A-CMs, whereas both mutated lines showed reduced IKs compared with wt2-CMs. By contrast, IKr tail density, measured as E4031-sensitive current, was comparable between the three lines (Fig. S5 A and B), indicating a specific effect of the c.1781G>A mutation on IKs only. Taken together, these data suggest that the c.1781G>A mutation causes a larger reduction of IKs in the homozygous compared with the heterozygous genotype, in agreement with it being responsible for the more severe phenotype observed in JLNS1781A/A-CMs (Fig. 3).
Fig. 5.
Molecular mechanism underlying the c.1781G>A mutation. (A) Representative IKs measured as JNJ303-sensitive current in the indicated hiPSC-CMs. (Inset) Voltage protocol. (B) Average JNJ303-sensitive tail currents in hiPSC-CMs. Symbols *, &, and # indicate statistical significance based on pairwise comparisons (P < 0.05). (C) Western blot analysis of KCNQ1 and KCNE1 in hiPSC-CMs (Top). TNNI3 and GAPDH are used as cardiac and loading controls, respectively. (Bottom) Densitometric quantification of the KCNQ1 band, normalized to TNNI3 (n = 2). (D) Immunofluorescence analysis of KCNQ1 and actin in hiPSC-CMs. Bottom are a magnification of the area framed in Middle. Percentages of CMs showing the indicated staining patterns are indicated. (E) Representative confocal images showing membrane localization of KCNQ1 following injection of GFP-tagged wt and G1781-mutated KCNQ1 mRNA into Xenopus oocytes (Left). Image-based quantification of total GFP intensity at the outer cell membrane (Right, n = 10 each). (F) Structural model of KCNQ1 highlighting the position of the R594 residue in the C-terminal assembly domain (Left) and change in predicted local structure induced by the R594Q mutation (Right).
The c.1781G>A Mutation Results in a Partial KCNQ1 Trafficking Defect.
To investigate the molecular mechanisms underlying the electrophysiological phenotype of the c.1781G>A mutation, KCNQ1 RNA and protein levels were determined in mutated and wt2-CMs. Channel expression at the protein and RNA levels was not reduced in either LQT11781G/A-CMs or in JLNS1781A/A-CMs, ruling out a disease mechanism based on reduced KCNQ1 abundance (Fig. 5C and Fig. S5C, Top). The β-subunit–encoding KCNE1 was also expressed at similar levels in CMs of all lines, both at the protein and RNA levels (Fig. 5C, Top and Fig. S5 C–E). To investigate the effect of the missense c.1781G>A mutation further, we examined ion channel expression by immunofluorescence. In all lines, KCNQ1 was detected both on the cell surface and in intracellular compartments through which the channel normally transits during its biogenesis (Fig. 5D). However, both JLNS1781A/A- and LQT11781G/A-CMs showed a reduction of KCNQ1 at the outer cell membrane, with a more pronounced perinuclear signal in the majority of homozygous JLNS1781A/A-CMs. These observations suggest that the c.1781G>A mutation possibly interferes with the normal trafficking of the mutated channel to the cell membrane.
To confirm this result independently, we expressed GFP-tagged wt- or G1781A-mutated KCNQ1 mRNA in Xenopus oocytes. Although the wt channel was abundant at the oocyte membrane, the mutated protein was greatly reduced in this compartment, although it was not completely absent (Fig. 5E). These data support a partial trafficking defect associated with the c.1781G>A KCNQ1 mutation, as shown in other systems (30). To better understand the consequences of the R594Q amino acid exchange in the KCNQ1 protein, we used a described in silico model (31). Molecular dynamic simulations suggested that the R594Q mutation may induce local structural rearrangements in the C-terminal domain of KCNQ1, with potential impact on tetrameric assembly (Fig. 5F and Fig. S5F). Taken together, these data suggest that the c.1781G>A mutation causes a partial KCNQ1 trafficking defect, giving rise to LQT1 and JLNS phenotypes in a gene dosage-dependent manner.
JLNS-CMs Are Sensitive to β-Adrenergic and Proarrhythmic Stress but Benefit from β-Blockade and IKr Activation.
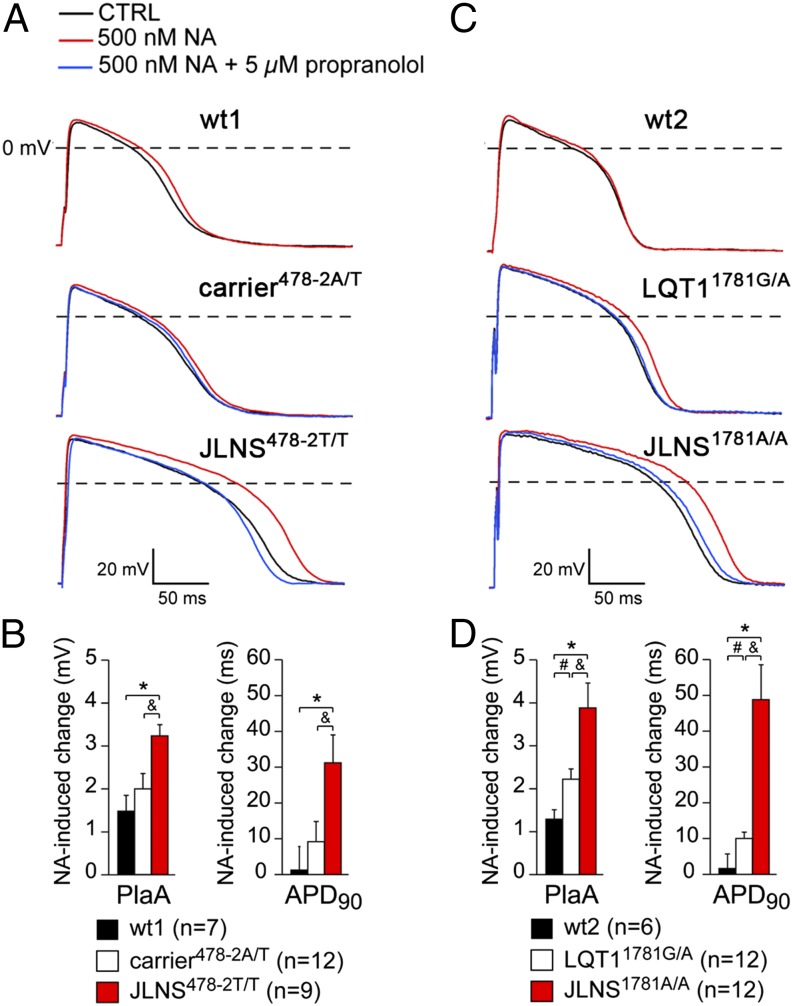
Because JLNS patients are at high risk for arrhythmic events, we further examined the cellular models for effects of β-adrenergic stimulation and proarrhythmic drugs. APs were measured at 1 Hz in the absence or presence of 500 nM noradrenaline in CMs derived from all hiPSC lines. Typical APs and average effects on PlaA and APD90 are shown in Fig. 6. β-adrenergic stimulation significantly increased the PlaA in all cases but only significantly prolonged the APD90 in the JLNS478-2T/T-, JLNS1781A/A-, and LQT11781G/A-CMs. The increase in PlaA and in APD90 was most pronounced in JLNS-CMs. These effects were partially reversed by treatment with the β-blocker propranolol (Fig. 6 A and C). Neither early and delayed afterdepolarizations (EADs and DADs, respectively) nor spontaneous activity was observed at a pacing frequency of 1 Hz in the presence of noradrenaline.
Fig. 6.
Adrenergic stress and β-block in hiPSC-CMs. A and B show results for the wt1-, carrier478-2A/T-, and JLNS478-2T/T-CMs; and C and D for the wt2-, LQT11781G/A-, and JLNS1781A/A-CMs. (A and C) Representative APs at 1 Hz in presence of noradrenaline (NA, red) or NA+propranolol (blue). (B and D) Average PlaA and APD90 changes in presence of NA. Symbols *, &, and # indicate statistical significance based on pairwise comparisons (P < 0.05).
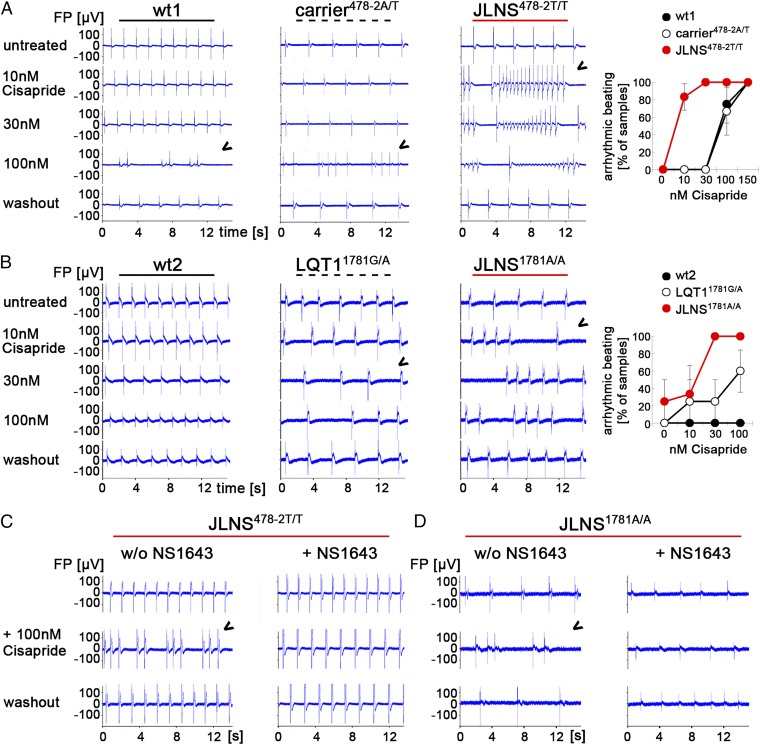
To further assess whether mutated CMs were more sensitive to stress caused by proarrhythmic compounds not targeting KCNQ1 itself, CMs from all lines were exposed to Cisapride, a gastrointestinal drug that has been withdrawn as a pharmaceutical because of arrhythmogenic side effects in humans (32, 33). Interestingly, although wt controls were rather insensitive to this drug, JLNS-CMs of both genotypes studied reproducibly developed arrhythmias at Cisapride doses of only 10–30 nM (Fig. 7 A and B). Similar observations were made with engineered JLNSfs/fs-CMs (Fig. S6A). Heterozygous CMs behaved like wt controls or showed an intermediate response, depending on the underlying mutation (Fig. 7 A and B). Hence, these results revealed a strong correlation between genotype-dependent IKs reduction and risk of arrhythmic responses. Because IKr cooperates with IKs in the repolarization of cardiac AP, we investigated whether drug-based IKr activation could rescue disease phenotypes in mutated hiPSC-CMs. Indeed, NS1643, a small molecule that activates IKr by reducing hERG channel inactivation (34, 35), reduced the FPD in hiPSC-CMs (Fig. S6B) and effectively protected JLNS478-2T/T, JLNS1781A/A, and LQT11781G/A-CMs from Cisapride-induced arrhythmias (Fig. 7 C and D and Fig. S6C).
Fig. 7.
Drug-induced arrhythmia phenotypes in JLNS-CMs and rescue by a hERG activator. (A and B) Cisapride causes arrhythmias in spontaneously beating wt1-, carrier478-2A/T-, and JLNS478-2T/T-CMs (A) as well as in LQT11781G/A- and JLNS1781A/A- CMs. (B, Left) Representative MEA traces. Arrowheads indicate first appearance of arrhythmia. (B, Right) Statistics of arrhythmic beatings at different Cisapride doses from independent experiments (n = 2–6). (C and D) Pretreatment with the hERG channel activator NS1643 (30 µM) protects JLNS478-2T/T- (C) and JLNS1781A/A-CMs (D) from high-dose Cisapride-induced arrhythmia. Representative MEA traces from independent experiments are shown.
Discussion
Reprogramming somatic cells from patients with genetic diseases has already advanced understanding of human maladies, particularly for diseases difficult to model in mice in vivo, or where primary tissues are difficult to access (36). Channelopathies have been particularly successfully modeled in hiPSC-CMs, in part because differing ion channel use in human and mouse heart often precludes direct extrapolation of phenotypes (37). Most striking has been their value in demonstrating the causality of specific LQTS mutations (20, 36). In the present study, we report a side-by-side analysis of two distinct KCNQ1 mutations causing JLNS, the recessive and one of the most severe forms of LQTS (2). We showed that both patient-derived and genetically engineered hiPSC-CMs recapitulated the aggressive cardiac phenotype of the disease, although the effect was mediated by different mechanisms.
Although previous JLNS studies in heterologous systems, where single ion channels are expressed in noncardiomyocytes, and mice have contributed significantly to understanding this disease, it has not always been possible to recapitulate the severity of cardiac phenotype caused by homozygous and heterozygous loss of function mutations in the same gene. Although the hearing loss observed in patients was recapitulated in KCNQ1 or KCNE1 knockout mice, clear cardiac phenotypes were not always evident (38, 39), illustrating the value of the human models here.
The c.478-2A>T mutation resulted in a splice defect at the mRNA level, causing exon 3 skipping and a reading frame shift (E160fs+138X). Consequently, KCNQ1 protein was not detectable in JLNS478-2T/T-CMs and, accordingly, these cells did not produce any IKs, providing an explanation for the severity of the observed cardiac phenotype. Indeed, the FPD was significantly shortened by transgenic KCNQ1 overexpression in JLNS478-2T/T cells. Moreover, in the heterozygous carrier478-2A/T-CMs, Δex3 KCNQ1 mRNA underwent NMD, whereas nonmutated KCNQ1 mRNA was elevated above the expected 50% of the wt level. This latter observation may indicate a gene-dosage compensatory effect warranting further investigation. Furthermore, Xenopus oocyte assays revealed that Δex3 KCNQ1 mRNA was nonfunctional and did not interfere with the wt form. In sum, these effects result in normal KCNQ1 expression and function in heterozygous carrier478-2A/T-CMs, explaining why the disease is recessive at the molecular and cellular level. This scenario likely has universal validity for nonsense and frame shift lesions in KCNQ1 (10) and contrasts with dominant-negative effects of point mutations causing LQT1 (40, 41).
The c.1781G>A KCNQ1 mutation, however, caused AP prolongation even when expressed heterozygously (LQT11781G/A-CMs), whereas the effect on AP prolongation was additive in the homozygous JLNS1781A/A-CMs. This phenotype is in contrast with the strictly recessive loss-of-function phenotype caused by the c.478-2A>T KCNQ1 lesion. These observations correspond well with clinical data from the patients from which the lines were derived: the heterozygous LQT11781G/A patient had a prolonged QT interval (QTc: 506 ms), whereas the carrier478-2A/T had a normal ECG (QTc: 400 ms). Accordingly, molecular characterization of the c.1781G>A mutation revealed a distinct, gene dosage-dependent mechanism. Even as a heterozygote, the c.1781G>A point mutation reduced ion channel expression at the plasma membrane. This result could be a direct consequence of the localization of the c.1781G>A mutation in the C-terminal assembly domain of the protein (13) that might affect assembly of the mutated α-subunits and, consequently, the amount of protein reaching the membrane. Our results are in agreement with a recent study in which the same mutation was overexpressed in a heterologous system and a trafficking defect was evident (30, 42). In line with this mechanism, the homozygous mutation in JLNS1781A/A-CMs caused a large reduction in IKs, albeit not a complete loss. The fact that in the homozygous mutant genotype some residual IKs current was present (∼20%) would also support the possibility that homozygous patients harboring the c.1781G>A mutation could present with autosomic recessive LQTS (i.e., without deafness) (14). However, a larger number of such patients, together with their clinical data, would help provide a clear classification of this mutation. In heterozygous LQT11781G/A-CMs, IKs was reduced by ∼40%, indicating haploinsufficiency as the mechanism of action of this mutation, in agreement with a prolonged QT interval in our LQT1 patient. Of note, the lack of a dominant-negative effect is in agreement with the general mechanism proposed for the JLNS-causing mutations (11, 13, 43, 44).
JLNS and other LQTS subtypes are complex diseases in which phenotypes may be influenced by epigenetics and gene modifiers (44, 45). In addition, the type of mutation and its localization, and variable penetrance, have an impact on phenotype severity and complexity (43). These characteristics might explain the variability sometimes observed between patients even harboring the same mutation, and why individuals with heterozygous KCNQ1 mutations can be either carriers or symptomatic patients (44). In line with these findings, engineered JLNSfs/fs CMs showed strong but somewhat less pronounced FPD prolongation and arrhythmia phenotypes compared with their isogenic controls than patient-derived hiPSC-CMs compared with nonisogenic wt or heterozyous carrier CMs. In comparison, our data on the c.1781G>A KCNQ1 mutation, where isogenic heterozygous and homozygous mutated CMs were compared, confirmed that precise gene targeting in hiPSC-CMs is a powerful tool to study the role of such mutations and evaluate dosage effects on disease severity (46).
One important characteristic of LQT1 and JLNS patients is their increased sensitivity to β-adrenergic triggers that are often the cause of sudden death (2, 47). Consistently, our data revealed significant AP prolongation in LQT1- and JLNS-CMs upon β-adrenergic stimulation, whereas AP prolongation was not observed in wt-CMs. These results are likely due to β-adrenergic–induced increases in L-type Ca2+ current (44) being counteracted by a prominent increase in IKs. The noradrenaline-induced AP prolongation in LQT1- and JLNS-CMs shows similarities with findings in freshly isolated ventricular CMs of human failing hearts (44), which have a reduced IKs (48).
Our data also revealed a particularly high sensitivity of JLNS-CMs to proarrhythmic stress. The increased susceptibility to drug-induced arrhythmias of JLNS1781A/A- and JLNS478-2T/T-CMs is likely a direct consequence of impaired IKs and concomitant reduction of the repolarization reserve (49). One promising application of hiPSC-CMs is screening for proarrhythmic side effects of certain drugs (50, 51). JLNS hiPSC-CMs may be particularly suited to this purpose because of their extraordinary sensitivity. It will be interesting to evaluate whether this effect holds true only for inhibitors of IKr—such as E4031 and Cisapride used here (32)—or if it also applies to other classes of cardiotoxic compounds.
Collectively, our data exemplify how distinct mechanisms may lead to similar JLNS phenotypes. These findings may also have implications for potential therapeutic strategies. For example, in the case of point mutations, the KCNQ1 channel itself could in principle be considered as a drug target (52, 53), whereas in the case of frameshift mutations, it cannot. Moreover, LQT1 patients are often responsive to β-blocker therapy, but for the majority of JLNS patients, this treatment is only partially effective (2). Alternatively, compromised KCNQ1 function might be compensated by drug-induced IKr activation (54). Using the hERG activator NS1643, FPDs were shortened in hiPSC-CMs, although this effect required high drug concentrations (30 µM). Interestingly, the high susceptibility of JLNS-CMs to drug-induced arrhythmia could also be balanced by IKr activator treatment. This finding may also suggest alternative therapeutic perspectives for patients, although more potent compounds will be needed to make this strategy a realistic option.
Materials and Methods
Detailed procedures are outlined in SI Materials and Methods.
Patients.
The patients described in this study include the following: (i) a child with congenital deafness and QT interval prolongation (JLNS478-2T/T, QTc: 570 ms; Fig. S1A), harboring the homozygous c.478-2A>T KCNQ1 mutation—he is being treated with β-blockers and implantable cardioverter defibrillator implantation; (ii) his heterozygous and asymptomatic father (carrier478-2A/T, QTc: 400 ms); and (iii) an independent woman presenting with LQT1 (LQT11781G/A, QTc: 506 ms; Fig. S1B) carrying the heterozygous c.1781G>A mutation in KCNQ1—she is asymptomatic but treated with β-blockers.
Cell Culture and Molecular Biology.
hiPSC were generated, cultured, and differentiated into cardiomyocytes by using procedures specified in SI Materials and Methods. Genetic engineering was achieved by using either a CRISPR/Cas-9 or a conventional homologous recombination strategy. Functional complementation was performed by using PiggyBac transposition. Sequencing, gene expression, and imprinting analysis were performed by using standard procedures. All primers used in this study are listed in Table S1. Immunofluorescence and Western blot analysis were performed according to standard procedures, using antibodies given in SI Materials and Methods.
Cellular Electrophysiology.
Experimental details about FPD, APs, IKs, and IKr measurements and analysis in hiPSC-CMs and Xenopus laevis oocytes are described in SI Materials and Methods.
Statistics.
Data are presented as mean ± SEM. Groups were compared by using one-way ANOVA followed by Tukey’s test or two-way repeated measures ANOVA followed by pairwise comparison by using the Student–Newman–Keuls test. MEAs data were analyzed on the basis of two-sided unpaired Student t tests. P < 0.05 was considered statistically significant and is indicated by *, &, and #.
Supplementary Material
Acknowledgments
We thank M. Ohtaka, K. Nishimura, and M. Nakanishi (National Institute of Advanced Industrial Science and Technology, Japan) for providing the Sendai virus and reprogramming protocol; the hiPSC core facility (Leiden University Medical Center) for hiPSCs spontaneous differentiation; F. Stewart (Technical University of Dresden) for providing the R6K plasmid; M. J. M. van der Burg, K. Szuhai, H. Tanke (Leiden University Medical Center) for karyotyping; M. J. Goumans (Leiden University Medical Center) for providing adult heart samples; K. Adachi (Max Planck Institute Münster) for providing the KA0717 vector; and A. Wilde and N. Hofman (both Academic Medical Center Amsterdam) for constructive discussion. We are especially thankful to all the patients for tissue donation. This work was supported by European Research Council Grant 323182 (to C.L.M.); ZonMW Animal Alternatives Grant 114000101 (to C.L.M.); Netherlands Proteomics Consortium Grant 050-040-250 (to C.L.M.); the Netherlands Institute of Regenerative Medicine (C.L.M. and C.D.); European Union Marie Curie FP7-people-2011-IEF Programme, HPSCLQT 29999 (to M.B.) and a short-term EMBO Fellowship, ASTF 387.00-2011 (to C.D.); Foundation Leducq (E.S.-B.), Interdisciplinary Center for Clinical Research (E.S.-B.); Eva Luise and Horst Köhler Foundation (B.G.); Bundesinstitut für Risikobewertung, FK-3-1329-471 (to B.G.); and Chemical Genomics Centre of the Max Planck Society (B.G.).
Footnotes
Conflict of interest statement: C.L.M. is cofounder and advisor of Pluriomics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419553111/-/DCSupplemental.
References
- 1.Jervell A, Lange-Nielsen F. Congenital deaf-mutism, functional heart disease with prolongation of the Q-T interval and sudden death. Am Heart J. 1957;54(1):59–68. doi: 10.1016/0002-8703(57)90079-0. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz PJ, et al. The Jervell and Lange-Nielsen syndrome: Natural history, molecular basis, and clinical outcome. Circulation. 2006;113(6):783–790. doi: 10.1161/CIRCULATIONAHA.105.592899. [DOI] [PubMed] [Google Scholar]
- 3.Jespersen T, Grunnet M, Olesen SP. The KCNQ1 potassium channel: From gene to physiological function. Physiology (Bethesda) 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 4.Schulze-Bahr E, et al. KCNE1 mutations cause jervell and Lange-Nielsen syndrome. Nat Genet. 1997;17(3):267–268. doi: 10.1038/ng1197-267. [DOI] [PubMed] [Google Scholar]
- 5.Romano C, Gemme G, Pongiglione R. [Rare cardiac arrythmias of the pediatric age. Ii. syncopal attacks due to paroxysmal ventricular fibrillation. (Presentation of 1st case in Italian pediatric literature)] Clin Pediatr (Bologna) 1963;45:656–683. [PubMed] [Google Scholar]
- 6.Ward OC. A New Familial Cardiac Syndrome in Children. J Ir Med Assoc. 1964;54:103–106. [PubMed] [Google Scholar]
- 7.Schwartz PJ, Ackerman MJ, George AL, Jr, Wilde AA. Impact of genetics on the clinical management of channelopathies. J Am Coll Cardiol. 2013;62(3):169–180. doi: 10.1016/j.jacc.2013.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Splawski I, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Crotti L, et al. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation. 2013;127(9):1009–1017. doi: 10.1161/CIRCULATIONAHA.112.001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Splawski I, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102(10):1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 11.Tyson J, et al. Mutational spectrum in the cardioauditory syndrome of Jervell and Lange-Nielsen. Hum Genet. 2000;107(5):499–503. doi: 10.1007/s004390000402. [DOI] [PubMed] [Google Scholar]
- 12.Jackson H, et al. LQTS in Northern BC: Homozygosity for KCNQ1 V205M presents with a more severe cardiac phenotype but with minimal impact on auditory function. Clin Genet. 2014;86(1):85–90. doi: 10.1111/cge.12235. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt N, et al. A recessive C-terminal Jervell and Lange-Nielsen mutation of the KCNQ1 channel impairs subunit assembly. EMBO J. 2000;19(3):332–340. doi: 10.1093/emboj/19.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhuiyan ZA, Wilde AA. IKs in heart and hearing, the ear can do with less than the heart. Circ Cardiovasc Genet. 2013;6(2):141–143. doi: 10.1161/CIRCGENETICS.113.000143. [DOI] [PubMed] [Google Scholar]
- 15.Neyroud N, et al. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet. 1997;15(2):186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- 16.Splawski I, Timothy KW, Vincent GM, Atkinson DL, Keating MT. Molecular basis of the long-QT syndrome associated with deafness. N Engl J Med. 1997;336(22):1562–1567. doi: 10.1056/NEJM199705293362204. [DOI] [PubMed] [Google Scholar]
- 17.Tranebjaerg L, Bathen J, Tyson J, Bitner-Glindzicz M. Jervell and Lange-Nielsen syndrome: A Norwegian perspective. Am J Med Genet. 1999;89(3):137–146. [PubMed] [Google Scholar]
- 18.Inoue H, Nagata N, Kurokawa H, Yamanaka S. iPS cells: A game changer for future medicine. EMBO J. 2014;33(5):409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moretti A, Laugwitz KL, Dorn T, Sinnecker D, Mummery C. Pluripotent stem cell models of human heart disease. Cold Spring Harb Perspect Med. 2013;3(11) doi: 10.1101/cshperspect.a014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller M, Seufferlein T, Illing A, Homann J. Modelling human channelopathies using induced pluripotent stem cells: A comprehensive review. Stem Cells Int. 2013;2013:496501. doi: 10.1155/2013/496501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, et al. Modeling type 3 long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. Int J Cardiol. 2013;168(6):5277–5286. doi: 10.1016/j.ijcard.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Huang L, Bitner-Glindzicz M, Tranebjaerg L, Tinker A. A spectrum of functional effects for disease causing mutations in the Jervell and Lange-Nielsen syndrome. Cardiovasc Res. 2001;51(4):670–680. doi: 10.1016/s0008-6363(01)00350-9. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura K, et al. Development of defective and persistent Sendai virus vector: A unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem. 2011;286(6):4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller FJ, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8(4):315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MP, Hu RJ, Johnson LA, Feinberg AP. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet. 1997;15(2):181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 27.Braam SR, et al. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res (Amst) 2010;4(2):107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Maquat LE. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5(2):89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 29.Carter MS, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270(48):28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 30.Harmer SC, et al. Cellular mechanisms underlying the increased disease severity seen for patients with long QT syndrome caused by compound mutations in KCNQ1. Biochem J. 2014;462(1):133–142. doi: 10.1042/BJ20140425. [DOI] [PubMed] [Google Scholar]
- 31.Strutz-Seebohm N, et al. Structural basis of slow activation gating in the cardiac I Ks channel complex. Cell Physiol Biochem. 2011;27(5):443–452. doi: 10.1159/000329965. [DOI] [PubMed] [Google Scholar]
- 32.Rampe D, Roy ML, Dennis A, Brown AM. A mechanism for the proarrhythmic effects of cisapride (Propulsid): High affinity blockade of the human cardiac potassium channel HERG. FEBS Lett. 1997;417(1):28–32. doi: 10.1016/s0014-5793(97)01249-0. [DOI] [PubMed] [Google Scholar]
- 33.Bran S, Murray WA, Hirsch IB, Palmer JP. Long QT syndrome during high-dose cisapride. Arch Intern Med. 1995;155(7):765–768. [PubMed] [Google Scholar]
- 34.Hansen RS, et al. Activation of human ether-a-go-go-related gene potassium channels by the diphenylurea 1,3-bis-(2-hydroxy-5-trifluoromethyl-phenyl)-urea (NS1643) Mol Pharmacol. 2006;69(1):266–277. doi: 10.1124/mol.105.015859. [DOI] [PubMed] [Google Scholar]
- 35.Casis O, Olesen SP, Sanguinetti MC. Mechanism of action of a novel human ether-a-go-go-related gene channel activator. Mol Pharmacol. 2006;69(2):658–665. doi: 10.1124/mol.105.019943. [DOI] [PubMed] [Google Scholar]
- 36.Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: The new patient? Nat Rev Mol Cell Biol. 2012;13(11):713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- 37.Davis RP, et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125(25):3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- 38.Rivas A, Francis HW. Inner ear abnormalities in a Kcnq1 (Kvlqt1) knockout mouse: A model of Jervell and Lange-Nielsen syndrome. Otol Neurotol. 2005;26(3):415–424. doi: 10.1097/01.mao.0000169764.00798.84. [DOI] [PubMed] [Google Scholar]
- 39.Casimiro MC, et al. Targeted disruption of the Kcnq1 gene produces a mouse model of Jervell and Lange-Nielsen Syndrome. Proc Natl Acad Sci USA. 2001;98(5):2526–2531. doi: 10.1073/pnas.041398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egashira T, et al. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res. 2012;95(4):419–429. doi: 10.1093/cvr/cvs206. [DOI] [PubMed] [Google Scholar]
- 41.Moretti A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363(15):1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 42.Wilson AJ, Quinn KV, Graves FM, Bitner-Glindzicz M, Tinker A. Abnormal KCNQ1 trafficking influences disease pathogenesis in hereditary long QT syndromes (LQT1) Cardiovasc Res. 2005;67(3):476–486. doi: 10.1016/j.cardiores.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 43.Moss AJ, et al. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115(19):2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saenen JB, Vrints CJ. Molecular aspects of the congenital and acquired Long QT Syndrome: Clinical implications. J Mol Cell Cardiol. 2008;44(4):633–646. doi: 10.1016/j.yjmcc.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Seebohm G, et al. Long QT syndrome-associated mutations in KCNQ1 and KCNE1 subunits disrupt normal endosomal recycling of IKs channels. Circ Res. 2008;103(12):1451–1457. doi: 10.1161/CIRCRESAHA.108.177360. [DOI] [PubMed] [Google Scholar]
- 46.Bellin M, et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32(24):3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz PJ, et al. Genotype-phenotype correlation in the long-QT syndrome: Gene-specific triggers for life-threatening arrhythmias. Circulation. 2001;103(1):89–95. doi: 10.1161/01.cir.103.1.89. [DOI] [PubMed] [Google Scholar]
- 48.Janse MJ. Electrophysiological changes in heart failure and their relationship to arrhythmogenesis. Cardiovasc Res. 2004;61(2):208–217. doi: 10.1016/j.cardiores.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 49.Braam SR, et al. Repolarization reserve determines drug responses in human pluripotent stem cell derived cardiomyocytes. Stem Cell Res (Amst) 2013;10(1):48–56. doi: 10.1016/j.scr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Rajamohan D, et al. Current status of drug screening and disease modelling in human pluripotent stem cells. BioEssays. 2013;35(3):281–298. doi: 10.1002/bies.201200053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarrete EG, et al. Screening drug-induced arrhythmia [corrected] using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation. 2013;128(11, Suppl 1):S3–S13. doi: 10.1161/CIRCULATIONAHA.112.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu X, et al. Increasing I(Ks) corrects abnormal repolarization in rabbit models of acquired LQT2 and ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2002;283(2):H664–H670. doi: 10.1152/ajpheart.00076.2002. [DOI] [PubMed] [Google Scholar]
- 53.Nattel S, Carlsson L. Innovative approaches to anti-arrhythmic drug therapy. Nat Rev Drug Discov. 2006;5(12):1034–1049. doi: 10.1038/nrd2112. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, et al. Modulation of hERG potassium channel gating normalizes action potential duration prolonged by dysfunctional KCNQ1 potassium channel. Proc Natl Acad Sci USA. 2012;109(29):11866–11871. doi: 10.1073/pnas.1205266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.