Significance
Heterochromatin is a repressive mode of genetic storage that prevents cellular machineries from accessing DNA sequences. Here, we investigate how a protein machine, called SWI/SNF, can disrupt these heterochromatin structures and facilitate nuclear processes. We identify physical and functional interactions between SWI/SNF and a key heterochromatin protein, silent information regulator 3p (Sir3p).
Keywords: BAH, SWI/SNF, chromatin remodeling, Sir3, heterochromatin
Abstract
Heterochromatin is a specialized chromatin structure that is central to eukaryotic transcriptional regulation and genome stability. Despite its globally repressive role, heterochromatin must also be dynamic, allowing for its repair and replication. In budding yeast, heterochromatin formation requires silent information regulators (Sirs) Sir2p, Sir3p, and Sir4p, and these Sir proteins create specialized chromatin structures at telomeres and silent mating-type loci. Previously, we found that the SWI/SNF chromatin remodeling enzyme can catalyze the ATP-dependent eviction of Sir3p from recombinant nucleosomal arrays, and this activity enhances early steps of recombinational repair in vitro. Here, we show that the ATPase subunit of SWI/SNF, Swi2p/Snf2p, interacts with the heterochromatin structural protein Sir3p. Two interaction surfaces are defined, including an interaction between the ATPase domain of Swi2p and the nucleosome binding, Bromo-Adjacent-Homology domain of Sir3p. A SWI/SNF complex harboring a Swi2p subunit that lacks this Sir3p interaction surface is unable to evict Sir3p from nucleosomes, even though its ATPase and remodeling activities are intact. In addition, we find that the interaction between Swi2p and Sir3p is key for SWI/SNF to promote resistance to replication stress in vivo and for establishment of heterochromatin at telomeres.
All eukaryotic genomes are stored within the nucleoprotein structure of chromatin, the core subunit of which, the nucleosome, consists of 147 base pairs (bp) of DNA wrapped ∼1.7 times around an octamer of histone proteins (1). Over millions of years, eukaryotes have incorporated chromatin structure into the regulation of many aspects of DNA metabolism, from simple nuclear packaging to transcriptional control (2). This diversity of purpose is reflected in two general types of chromatin structures within the nucleus—euchromatin, which is decondensed and transcriptionally active, and heterochromatin, which is typically localized to the nuclear periphery and repressive for DNA recombination and transcription. Heterochromatin structures are commonly associated with centromeres and telomeres, and these domains package much of a genome’s repetitive DNA (3). Consequently, the maintenance of heterochromatin is key for genomic integrity, because it prevents illicit recombination among DNA repeats and promotes chromosome segregation during mitosis (4, 5).
On a molecular level, heterochromatic loci are marked by specific chromatin posttranslational modifications, which are recognized and bound by characteristic nonhistone proteins. In many vertebrates, heterochromatin is characterized by members of the heterochromatin protein 1 (HP1) family of proteins, whereas in budding yeast, the silent information regulator (Sir) proteins, Sir2p, Sir3p, and Sir4p, create heterochromatin structures at telomeres and the silent mating-type loci (6, 7). Sir3p is believed to be the key structural component of yeast heterochromatin—Sir3p contains numerous protein–protein interaction motifs (8–10), including an N-terminal Bromo-Adjacent Homology (BAH) domain that interacts with the nucleosomal surface (11–13). BAH domains are found in many other chromatin-associated factors, including the Rsc2p subunit of the remodels structure of chromatin (RSC) remodeling enzyme and the Orc1p subunit of the Origin Recognition Complex (ORC) (14). The stability of the Sir3p BAH–nucleosome complex requires deacetylated histone H4 lysine 16 (15); consequently, amino acid substitutions at H4-K16 disrupt Sir3p–nucleosome binding and eliminate heterochromatin assembly in vivo (15–17).
Despite the repressive structure of heterochromatin, these domains must be replicated and repaired, implying that mechanisms exist to regulate heterochromatin disassembly. Previously, we described an in vitro assay to monitor early steps of recombinational repair with recombinant nucleosomal array substrates (18). Whereas the repair machinery was not hindered by the simple presence of nucleosomes, we reported that the binding of the Sir proteins, or even Sir3p by itself, led to dramatic repression of recombinational repair events on nucleosomal arrays (18, 19). Surprisingly, we discovered that the ATP-dependent chromatin remodeling enzyme, SWI/SNF, was able to counteract these repressive effects of heterochromatin in vitro, stimulating early steps of homologous recombination. Intriguingly, these assays uncovered that SWI/SNF catalyzed the ATP-dependent eviction of Sir3p from nucleosomes, an activity not shared by several other remodeling enzymes (19). Thus, these studies suggested that the SWI/SNF enzyme may have a unique ability to disrupt heterochromatin structures.
In this work, we identify a physical interaction between SWI/SNF and the heterochromatin protein Sir3p. We identify a pair of interactions—between the Swi2p Helicase SANT Adjacent HSA domain and the Sir3p AAA+ domain and between the Swi2p ATPase domain and the Sir3p BAH domain. Surprisingly, the ATPase–BAH interaction is conserved between many Swi2p/Snf2p ATPase family members and between two classes of BAH domains, suggesting a common mode of binding between these domains. Mutations are generated that ablate the interaction between Swi2p and Sir3p, and we find that the Swi2p–Sir3p interaction surfaces are required for SWI/SNF to evict Sir3p from nucleosomal arrays in vitro. Furthermore, in vivo studies indicate that SWI/SNF–Sir3p interactions are important both for resistance to replication stress and for establishment of silenced heterochromatic domains.
Results
SWI/SNF Binds Sir3p.
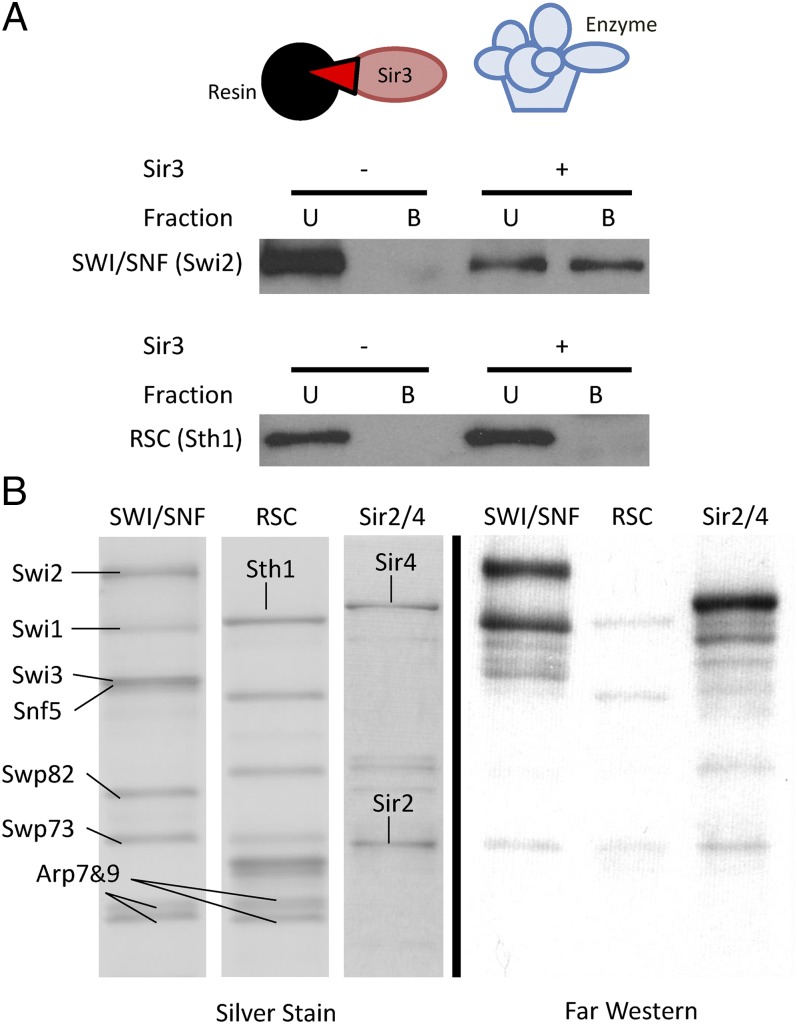
To investigate the unique ability of SWI/SNF to displace Sir3p from nucleosomes, we began by asking whether SWI/SNF and Sir3p physically interact. First, Sir3p–FLAG was affinity-purified from yeast and immobilized on anti-FLAG antibody resin. Purified SWI/SNF and RSC remodeling enzymes were incubated with Sir3p-bound beads, and bound and free fractions were analyzed by Western blotting (Fig. 1A). Strikingly, SWI/SNF, but not the highly related RSC complex, was able to interact with bead-bound Sir3p (Fig. 1A). This interaction was also apparent if SWI/SNF was immobilized on beads and incubated with purified Sir3p (Fig. S1A). To confirm the interaction and to gain insight into which SWI/SNF subunit might be involved, we used far Western analysis. Purified SWI/SNF, RSC, and Sir2p/Sir4p complexes were separated on an SDS/PAGE gel and transferred to a membrane. The membrane was incubated in buffer to stimulate protein renaturation and then incubated with purified Sir3p (Fig. 1B). Proteins bound to Sir3p were then detected by Western blotting, using antisera to Sir3p. As expected, Sir3p interacted strongly with Sir4p in this assay, but little interaction was detected with subunits of RSC (Fig. 1B, Right). In contrast, Sir3p interacted well with two polypeptides from SWI/SNF. The largest species comigrated with the Swi2p ATPase subunit (∼250 kDa), and the smaller species is either a proteolytic fragment of Swi2p or the Swi1p subunit (∼150 kDa).
Fig. 1.
SWI/SNF interacts with Sir3p. (A) SWI/SNF, but not RSC, interacts with resin-bound Sir3p. Purified remodeling enzyme was incubated with anti-FLAG resin that was prebound with (+) or without (−) Sir3p. B, bound fraction; U, unbound supernatant. (B) Subunits of SWI/SNF, but not RSC, interact with Sir3p by far Western. Equimolar amounts of SWI/SNF, RSC, and Sir2p/4p complex were separated on SDS/PAGE, electroblotted, renatured, and incubated with Sir3–FLAG. Sir3p-bound protein bands were visualized by anti-FLAG immunoblotting.
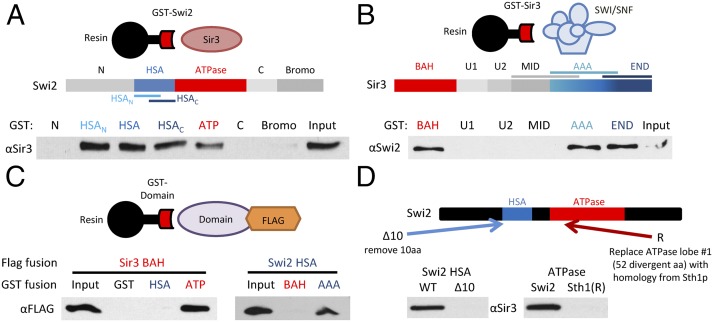
To directly monitor interactions between Swi2p and Sir3p, each protein was divided into several domains, expressed as GST fusion proteins in bacteria, and used in interaction studies (Fig. 2). First, GST–Swi2p fusions were tested for binding to full-length, purified Sir3p (Fig. 2A). Two regions of Swi2p were found to interact with Sir3p, the HSA domain and the central ATPase domain (20). Likewise, two regions of Sir3p bound to SWI/SNF complex, the N-terminal BAH domain and a region at the C terminus of the AAA+ domain (Fig. 2B). Each domain was then expressed as a FLAG fusion protein and used in GST interaction assays. Interestingly, these domains were found to interact in a pairwise manner—the Swi2p ATPase domain bound the Sir3p BAH domain, and the Swi2p HSA domain bound the Sir3p AAA+ domain (Fig. 2C). Progressive N- and C-terminal truncations of the GST–HSA fusion protein (Fig. S1B) defined a region of 10 amino acids in the Swi2p HSA domain that is required for interaction with Sir3p (Fig. 2D). Likewise, dissection of the Swi2p ATPase domain identified a 49-amino-acid fragment within the first RecA-like fold that retained Sir3p binding activity (Fig. S1C). Interestingly, the analogous residues from the ATPase domain of the RSC catalytic subunit, Sth1p, were unable to bind to Sir3p (Fig. 2D).
Fig. 2.
Swi2p and Sir3p have multiple interaction domains. (A) Schematic shows Swi2p domains. GST–Swi2 fusion proteins were used in pull-down assays with full-length Sir3p. GST-bound fractions were analyzed by Western blot. Shown is 10% of Input. (B) Schematic shows Sir3p domains. GST–Sir3 fusions were used in pull-down studies with the SWI/SNF complex. Bound fractions were assayed by Western blot as in A. (C) GST–Swi2 or GST–Sir3 fusion proteins were incubated with FLAG-tagged Swi2p or Sir3p domains, and interactions were identified by GST pull-down and Western analyses. (D) Swi2p alterations that disrupt Sir3p interactions. Schematic depicts alterations within either the Swi2p HSA or ATPase domain. The Δ10 derivative removes Swi2p residues 613–623; the Sth1(R) derivative replaces Swi2p residues 836–885 with the homologous region from Sth1 (residues 539–588). GST–Swi2 fusions harboring the indicated alterations were used in GST pull-downs with full-length Sir3p. Note that these binding assays used the individual HSA and ATPase regions of Swi2p.
SWI/SNF and RSC Interact with Core BAH Domains.
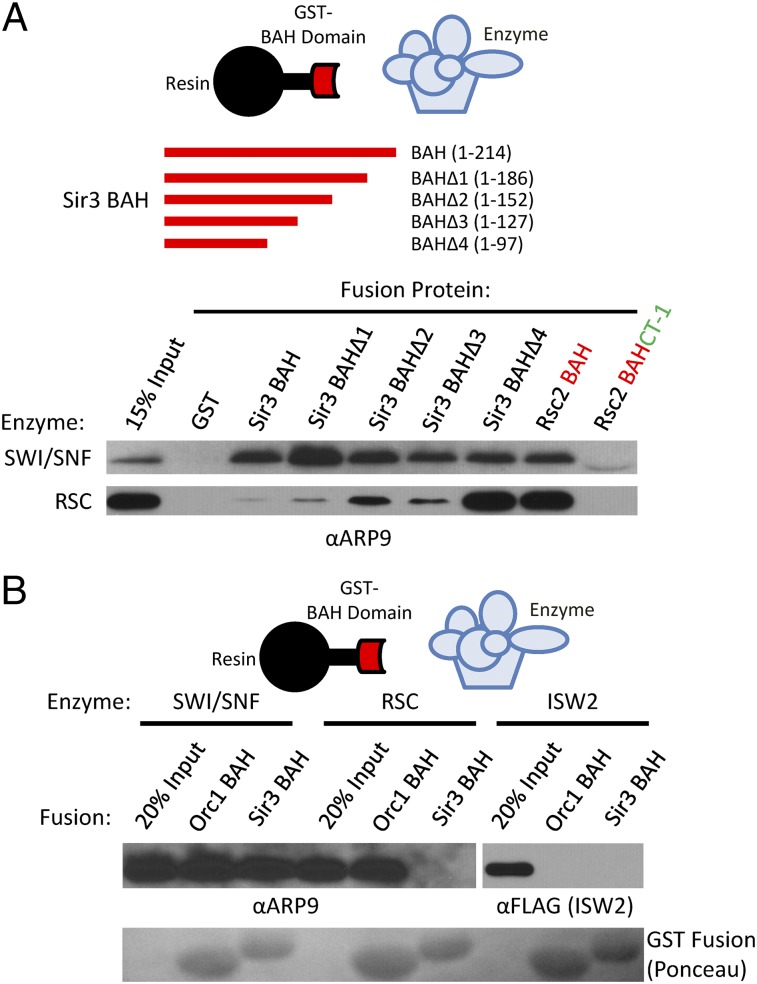
Progressive C-terminal truncations were used to delimit the SWI/SNF-interacting sequences within the Sir3p BAH domain (Fig. 3A). Each deletion construct retained SWI/SNF binding, and a GST fusion that contained only the 97-amino acid core BAH domain was sufficient to interact with SWI/SNF. Surprisingly, this core BAH domain also interacted strongly with the RSC remodeling enzyme, whereas larger BAH-containing fragments were either unable to interact or interacted only weakly with RSC (Fig. 3A). To test whether a BAH core domain might generally be sufficient for interaction with SWI/SNF-like enzymes, the core BAH domain of Rsc2p was assayed for interactions. Indeed, both SWI/SNF and RSC interacted well with the Rsc2p BAH core domain; however, inclusion of the conserved C-terminal (CT-1) domain eliminated interactions with both SWI/SNF and RSC (21). Furthermore, SWI/SNF also bound to the BAH domain from Orc1p, a subunit of the ORC (Fig. 3B). The RSC remodeling enzyme was also able to bind to the Orc1p BAH, despite being unable to interact with Sir3p BAH. Both SWI/SNF and RSC were also competent to bind to the human ORC1 BAH (Fig. S1D). In contrast, the Isw2 remodeling enzyme did not interact at detectable levels with either the Sir3 or yORC1 BAH domain, suggesting that BAH interactions may be a general feature of only the SWI/SNF subfamily of chromatin-remodeling enzymes (Fig. 3B). These data also suggest that sequences C-terminal to BAH core domains may govern the specificity of remodeling enzyme interactions.
Fig. 3.
SWI/SNF ATPases interact with BAH core domains. (A) Schematic shows C-terminal truncations within the Sir3p BAH domain. The indicated GST–BAH fusion proteins were incubated with either SWI/SNF or RSC, and bound fractions were assayed by Western. The Rsc2p BAH fusion contains only the core BAH domain; the BAH–CT-1 fusion also contains the C-terminal conserved CT-1 domain from Rsc2p. Western analyses used sera to the Arp9p subunit, common to both remodeling enzymes. (B) SWI/SNF, RSC, or Isw2 complexes were incubated with GST–BAH fusions from yeast Orc1p or Sir3p. Bound fractions were assayed by Western to the indicated subunits. Lower shows Ponceau-stained membrane, depicting levels of GST fusions.
Swi2/Snf2–Sir3p Interactions Are Required for Sir3p Eviction in Vitro.
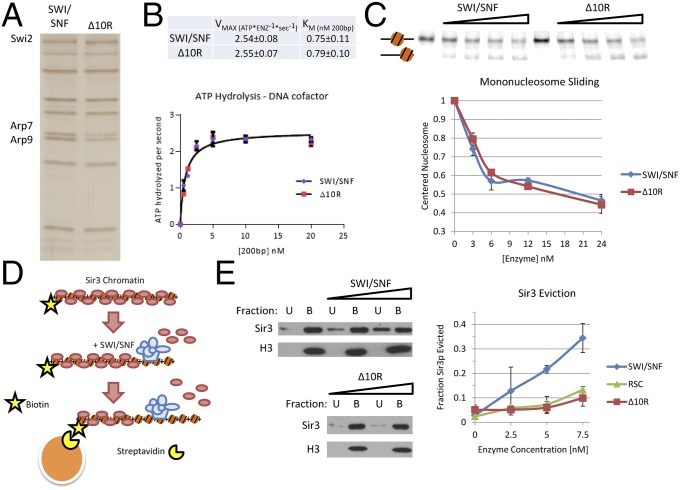
Having identified Sir3p-interaction domains within Swi2p, we asked whether they were required for the ATP-dependent eviction of Sir3p by SWI/SNF. To this end, a SWI2 gene was created that contains a 10-amino-acid deletion within the HSA domain (Δ10) as well as a 197-amino-acid swap between the Sth1p and Swi2p ATPase domains (Sth1[R]) (termed swi2–Δ10R; Fig. 2D). This region of Sth1p encompasses the first RecA-like lobe of the ATPase domain. This region is nearly homologous to that of Swi2p, with the exception of a central, 52-amino-acid divergent region. A C-terminal, TAP-tagged version of Swi2–Δ10R was then expressed in yeast from its normal promoter on a low-copy CEN/ARS plasmid, and SWI/SNF complex (SWI/SNF–Δ10R) that harbors Swi2p–Δ10R was isolated by tandem affinity purification. The concentration of active enzyme was determined by ATPase assays, and equal ATPase units of wild-type and SWI/SNF–Δ10R complexes were analyzed by SDS/PAGE and silver staining. The subunit composition of the SWI/SNF–Δ10R complex was nearly identical to that of wild-type SWI/SNF, with the exception of an approximately twofold depletion of the Arp7p and Arp9p subunits (Fig. 4A). Because Arp subunits have been implicated in the regulation of ATPase kinetic parameters (22), we characterized the ATPase activity of the SWI/SNF–Δ10R complex. Importantly, the SWI/SNF–Δ10R complex exhibited kinetic parameters for DNA-stimulated ATPase activity indistinguishable from the wild-type complex (Fig. 4B).
Fig. 4.
Swi2p–Sir3p contacts are required for eviction of Sir3p from nucleosomes. (A) SDS/PAGE analysis of SWI/SNF and SWI/SNF–Δ10R complexes, visualized by silver staining. Equal levels of ATPase activity were loaded for each enzyme. (B) DNA-stimulated ATPase kinetics of SWI/SNF and SWI/SNF–Δ10R are equivalent. ATPase reactions were performed with varying concentrations of DNA cofactor, and hydrolysis rates were fit to Michaelis–Menten kinetic parameters. (C) Mononucleosome mobilization by SWI/SNF and SWI/SNF–Δ10R enzymes is equivalent. Varying concentrations of enzymes were incubated with a mononucleosome positioned in the center of a radiolabeled, 282-bp DNA fragment harboring a 601 positioning sequence. Predicted positions of mononucleosomes are indicated to the left. (Upper) Gel. (Lower) Quantification (error bars reflect SD). (D) Schematic of the chromatin capture assay. Biotinylated nucleosomal arrays are bound to Sir3p, incubated with chromatin-remodeling enzyme and ATP and captured on streptavidin-coated magnetic beads. Chromatin-bound B and unbound U are assayed by Western blotting. (E) SWI/SNF–Δ10R is defective for Sir3p eviction from nucleosomes. Increasing amounts of chromatin-remodeling enzyme were incubated with Sir3p-bound nucleosomal array, and Sir3p eviction into the chromatin-unbound fraction U was measured by Western blotting. (Left) Representative blots. (Right) Quantification.
The activity of the SWI/SNF–Δ10R complex was also monitored in several chromatin-remodeling assays. First, equal ATPase units of wild-type and SWI/SNF–Δ10R complexes were incubated with mononucleosomes positioned in the center of a radiolabeled 282-bp DNA fragment by a 601-nucleosome positioning sequence. The ATP-dependent movement of the nucleosome toward the DNA ends leads to faster mobility on native PAGE, and in this assay, the SWI/SNF–Δ10R enzyme was equivalent to wild type (Fig. 4C). Chromatin remodeling was also assessed by a nucleosomal array accessibility assay (23). This quantitative assay uses a positioned array of 11 nucleosomes, where the central nucleosome of the array occludes a unique SalI restriction enzyme recognition site. As the array is remodeled by SWI/SNF, this central nucleosome is repositioned or removed, increasing the rate of SalI cleavage. Similar to the ATPase and mononucleosome remodeling assays, the SWI/SNF–Δ10R enzyme showed equivalent activity compared with the wild-type complex (Fig. S2).
Finally, we assayed the ability of the SWI/SNF–Δ10R enzyme to catalyze the ATP-dependent eviction of Sir3p protein from nucleosomal arrays (Fig. 4D). In this assay, 12-mer nucleosomal arrays were assembled with recombinant histone octamers, and ∼15% of the octamers contained histone H2A biotinylated at an engineered cysteine within the exposed C-terminal domain (18). Purified Sir3p protein was bound to these arrays at a ratio of two Sir3p monomers per nucleosome (24) and then incubated with chromatin-remodeling enzyme in the presence of ATP. Reactions were captured on streptavidin-coated magnetic beads, and chromatin-bound (B) and unbound (U) fractions were subjected to Western blotting, probing for both histone H3 and Sir3p. In these reactions, wild-type SWI/SNF was able to evict ∼35% of the Sir3p into the unbound fraction, whereas the SWI/SNF–Δ10R complex was defective at Sir3p eviction (Fig. 4E). Indeed, the SWI/SNF–Δ10R complex resembled the activity of RSC, in that it only evicted small amounts of Sir3p at high concentrations (Fig. 4E). We conclude that the Sir3p interaction surfaces within Swi2p are dispensable for chromatin remodeling, but they are required for Sir3p eviction.
SWI/SNF–Sir3p Interactions Are Important in Vivo.
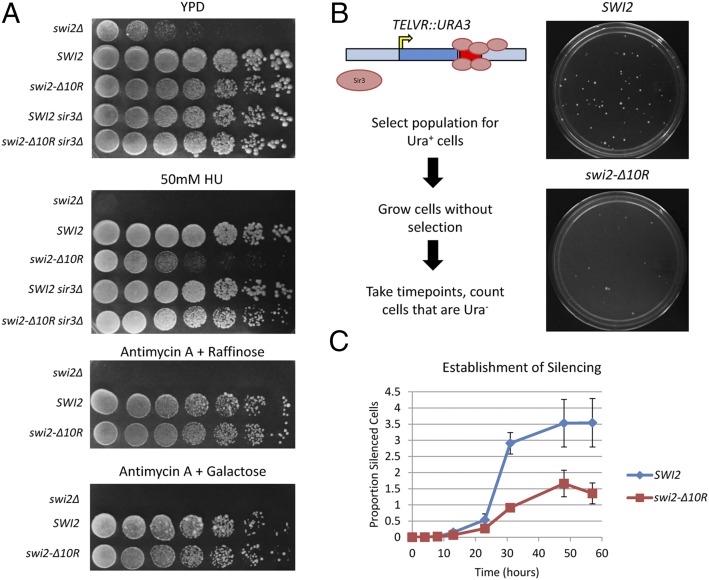
To identify potential phenotypes for the swi2–Δ10R allele that might be linked to Sir3p function, a plasmid-borne copy of swi2–Δ10R was introduced into swi2Δ and swi2Δ sir3Δ strains, and growth was assayed by spot dilution on several media. In the absence of SWI2, cells grow poorly on rich medium or on medium containing galactose or raffinose as carbon sources (25). In these cases, the swi2–Δ10R allele fully complemented these phenotypes, behaving like a wild-type strain (Fig. 5A). In contrast, the swi2–Δ10R allele showed a marked sensitivity to the replication stress agent hydroxyurea (HU; Fig. 5A). Previous studies have suggested that the HUs phenotype of swi/snf mutants may be due to a defect in transcriptional induction of ribonucleotide reductase (RNR) genes (26); however, the swi2–Δ10R strain exhibited wild-type levels of RNR3 transcriptional induction (Fig. S3A). Indeed, no significant changes in RNA expression were observed between wild-type and swi2–Δ10R strains when assayed by RNA sequencing (RNA-seq) (Fig. S3 B and C and Dataset S1). Consistent with previous work (27), swi2–Δ10R did not affect SIR2 or SIR3 expression (Fig. S3D and Dataset S1). Interestingly, the HUS phenotype of the swi2–Δ10R was suppressed by deletion of SIR3, consistent with a functional interaction between SWI/SNF and Sir3p during replication stress.
Fig. 5.
SWI/SNF–Sir3p interactions regulate resistance to replication stress and the establishment of telomeric silencing. (A) Growth assays. CEN/ARS plasmids containing SWI2 (CP1410), swi2–Δ10R (CP1413), or no insert (CP1250; pRS410) were introduced into swi2Δ or swi2Δ sir3Δ strains. WT and swi2–Δ10R complement swi2Δ growth and transcriptional defects, but swi2–Δ10R does not complement HU sensitivity. Fivefold serial dilutions of yeast cultures were spotted onto the indicated plates and allowed to grow for 3 d [yeast extract/peptone/dextrose (YPD)] or 6 d (all others) at 30 °C. (B, Left) Schematic of the subtelomeric silencing establishment assay. CY1755 (L1088; swi2Δ TELVR::URA3) was transformed with plasmids containing either SWI2 (CP1410) or swi2–Δ10R (CP1413), and transformant colonies were grown on SD–URA+G418 plates to select for Ura+ cells. Colonies were then cultured in medium lacking uracil for the indicated times and plated on 5-FOA plates to score establishment of silencing (Ura−). (Right) Representative 5-FOA plates after 24 h of growth on 5-FOA. (C) Quantitation of the assay from B. Five independent transformants were analyzed; error bars reflect SD.
To test whether SWI/SNF regulates the dynamics of heterochromatin assembly, wild-type and swi2–Δ10R strains were assayed in a transcriptional silencing establishment assay (28). This assay was performed in strains with a URA3 gene integrated adjacent to the telomere on right arm of chromosome V (TELVR::URA3). In this location, URA3 expression is repressed by the spreading of adjacent subtelomeric heterochromatin, creating a biphasic population of Ura− and Ura+ cells. To monitor the establishment of the silenced state, cells were first grown in medium lacking uracil, to enrich for cells in which URA3 is in the ON state (Ura+). Cells were then grown in the presence of uracil for increasing time and then plated onto plates that contain 5-fluoroorotic acid (5-FOA), scoring for cells that have silenced URA3 (Ura−). Compared with the wild type, the swi2–Δ10R mutant had a delayed onset of silencing and achieved a lower final level of silencing (Fig. 5D). Furthermore, the swi2–Δ10R strain formed much smaller colonies, suggesting that silencing was inherited less stably (Fig. 5D). Thus, these results suggest that interactions between SWI/SNF and Sir3p impact heterochromatin dynamics in vivo.
SWI/SNF Is Not Required for Heterochromatic Recombinational Repair.
Yeast mating-type switching requires that a double-strand break (DSB) induced at the MAT locus is repaired by homologous recombination with sequences from a heterochromatic HM locus (29). Previously, in vivo studies suggested that SWI/SNF is essential for mating-type switching and that SWI/SNF promotes repair only when the donor sequences are heterochromatic (19, 30). As an initial test for whether the swi2–Δ10R allele impacts heterochromatic mating-type switching, a plasmid expressing a galactose-inducible homothallic (HO) endonuclease was introduced into isogenic wild-type, swi2Δ, and swi2–Δ10R strains. The strand-invasion step of mating-type switching was then assayed by a PCR-based assay following a switch to galactose medium (31). Surprisingly, neither the swi2–Δ10R nor swi2Δ strains showed a significant defect in strand invasion (Fig. S4A).
To confirm this observation, a swi2Δ strain was created by tetrad dissection in a strain harboring a chromosomal, galactose-inducible HO gene. Notably, this is the same background as used in previous studies (30). Multiple swi2Δ segregants from independently created diploids showed severe growth defects (Fig. S4B) and delayed galactose induction kinetics that precluded kinetic analyses of strand invasion. However, after growth for 4 h in galactose medium, swi2Δ strains were competent to switch mating types with efficiencies similar to the wild-type strain (Fig. S4C). To circumvent the galactose induction defects of a swi2Δ and to study the kinetics of strain invasion, an auxin-inducible degron system was used to conditionally deplete Swi2p (32). After a 2-h treatment with synthetic auxin [1-naphthaleneacetic acid (NAA)] to deplete Swi2p, galactose was added to cultures, and PCR was used to monitor DSB formation and strand invasion. Consistent with the results from the swi2Δ strain, depletion of Swi2p did not alter DSB repair kinetics (Fig. S4D). Because the Swi2p ATPase is essential for SWI/SNF function, these results indicate that SWI/SNF is dispensable for mating-type switching, even with a heterochromatic donor.
Discussion
Here, we have defined two distinct protein–protein interfaces between the Sir3p heterochromatin protein and the Swi2p subunit of the SWI/SNF chromatin remodeling enzyme. The HSA domain from Swi2p interacts with a region of Sir3p that contains its AAA+ domain, and an N-terminal portion of the Swi2p ATPase domain interacts with the nucleosome-binding, BAH domain of Sir3p. Intriguingly, Sth1p, the related ATPase from the RSC remodeling enzyme, can also bind to the Sir3p BAH domain, but only after elimination of flanking sequence elements. Furthermore, both Swi2p and Sth1p are able to bind to the central core of the Rsc2p and Orc1p BAH domains, suggesting that SWI/SNF-like ATPase domains may harbor a general affinity for BAH domains. Importantly, elimination of Sir3p interaction surfaces within Swi2p (Swi2p–Δ10R) disrupts the ability of SWI/SNF to catalyze the ATP-dependent eviction of Sir3p from nucleosomal arrays in vitro, without impairing its ATPase or more canonical chromatin-remodeling activities. Furthermore, these alterations led to specific phenotypes in vivo, consistent with functional interactions between SWI/SNF and Sir3p-dependent heterochromatin structures.
What is the functional role for Sir3p eviction by SWI/SNF? A previous study from Laurent and colleagues (30) was consistent with this activity playing an essential role in recombinational repair events that involve heterochromatin. Specifically, they used strains harboring a galactose-inducible HO endonuclease to create a single DNA DSB at the euchromatic MAT locus. The recombinational repair of this DSB requires a successful homology search and strand invasion of a homologous, but heterochromatic, HM locus. In these assays, they reported that inactivation of the Snf5p subunit of SWI/SNF had no effect on early steps of HR, but that snf5Δ eliminated capture of the heterochromatic donor sequences, and repair was blocked (30). Subsequently, we showed that SWI/SNF is not required for recombinational repair of these same sequences when they are euchromatic, suggesting that this role for SWI/SNF might be specific for the heterochromatic context (19). To our surprise, however, our studies presented here do not support this key role for SWI/SNF in heterochromatic recombinational repair. We created swi2Δ strains that harbor a GAL–HO gene by tetrad dissection, and we found that these strains are competent to repair an HO-induced DSB, leading to mating-type switching with efficiencies similar to wild type. Furthermore, we used an inducible degron strategy to remove Swi2p from these GAL–HO strains, but in this case as well, the loss of Swi2p, and thus SWI/SNF, had no impact on repair of a DSB at the MAT locus. Why our results differ from those of Laurent and colleagues in not clear. Unfortunately, the original snf5Δ strain is no longer available. The most likely explanation is that the previously observed phenotype was specific to this particular snf5Δ isolate that was created by direct cell transformation, rather than tetrad dissection. Alternatively, it could represent a phenotype that is unique to a snf5Δ mutant and does not reflect a role for SWI/SNF per se.
Yeast strains that lack SWI/SNF show a variety of phenotypes, including growth defects on rich medium or medium containing alternative carbon sources (e.g., galactose or raffinose), inositol auxotrophy (25, 33) and sensitivity to DNA-damaging and replication stress agents (26, 30). Consistent with the intact chromatin-remodeling activities of the SWI/SNF–Δ10R enzyme, strains harboring the swi2–Δ10R allele showed normal growth on nearly every condition tested. The lone exception, however, was sensitivity to the replication stress agent HU. Furthermore, this phenotype was suppressed by deletion of the SIR3 gene, consistent with a role for ATP-dependent Sir3p eviction during replicative stress. This phenotype was not due to a defect in transcriptional induction of the RNR genes, and the swi2–Δ10R allele did not lead to significant transcriptional changes that could be detected by RNA-seq. Thus, this HU phenotype is likely to reflect a transcription-independent role of SWI/SNF action in antagonizing Sir3p during DNA replication. One simple model posits that SWI/SNF is required for efficient replication through SIR heterochromatin and that HU-induced fork stress heightens the need for SWI/SNF to remove Sir3p. Alternatively, Taddei and colleagues have shown that Sir proteins can be recruited to stalled replication forks (34). Perhaps SWI/SNF plays a role in removing Sir proteins from stalled forks, alleviating the negative consequences of this Sir recruitment. This model may also provide an explanation for the defect in heterochromatin establishment observed in the swi2–Δ10R strain, because an accumulation of Sir3p at stalled forks may titrate Sir proteins from heterochromatic domains, interfering with heterochromatin assembly.
The ATP-dependent eviction of Sir3p from chromatin is reminiscent of the ability of the yeast Mot1p ATPase to catalyze the eviction of the general transcription factor TATA-binding protein (TBP) from DNA. Mot1p is a member of the Swi2p/Snf2p family of DNA-stimulated ATPases and DNA translocases, and the ability of Mot1p to disrupt TBP–DNA interactions appears to be key for redistributing TBP from TATA-containing binding sites to less-preferred, TATA-less promoter elements (35, 36). Similar to the SWI/SNF-dependent eviction of Sir3p from nucleosomes, Mot1p evicts TBP from a preformed TBP–DNA complex in an ATP-dependent reaction. Mot1p binds to TBP using two distinct interaction domains—a region containing multiple HEAT domains binds to the convex surface of the TBP–DNA complex, whereas a distinct “latch” domain interacts with the surface of TBP that is bound to DNA (37). These structural studies have led to a model in which Mot1p binds to DNA adjacent to the TBP–DNA complex, allowing its HEAT domain to make extensive contacts with the exposed, convex surface of TBP. As Mot1p hydrolyzes ATP, DNA translocation leads to the removal of TBP from DNA, and the latch domain of Mot1p interacts with the DNA-binding surface of TBP, preventing reassociation with promoter DNA (37). By analogy, we propose that the HSA domain of Swi2p may interact with the Sir3p–nucleosome complex, facilitating Sir3p removal during the DNA translocation reaction. Likewise, sequences within the N-terminal lobe of the ATPase domain may function as latches that bind the Sir3 BAH domain, preventing reassociation with the nucleosome (Fig. 6).
Fig. 6.
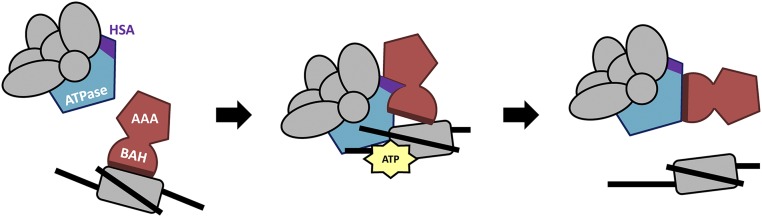
Model for eviction of Sir3 from nucleosomes by SWI/SNF. See text for description. Sir3 is in red; Swi2 is in blue.
Although the Swi2 ATPase domain is uniquely able to interact with the Sir3p BAH, the ATPase domains from both Swi2p and Sth1p can interact with the yeast Orc1p BAH domain. Likewise, both the SWI/SNF and RSC complexes can bind to the BAH domain of human Orc1. These latter interactions are surprising given that the primary sequence of the yeast and human Orc1 BAH domains have diverged considerably, although the overall structures are homologous (Fig. S5A). Orc1p is a highly conserved member of the ORC that is essential for cell viability and important for DNA replication (38–40). Orc1p and Sir3p are paralogs, and as such they display domain and primary sequence conservation, particularly in their N-terminal BAH domains (47% identical sequence). In Kluyveromyces lactis, Orc1p has been shown to function analogously to the role of Sir3 in heterochromatin formation, in addition to its traditional role in replication (41). We postulate that the binding interaction between SWI/SNF-family enzymes and Orc1-like BAH domains is ancestral and that specificity for Sir3p and Swi2p arose following the silencing subfunctionalization of Sir3p. Indeed, the sequences within the Swi2p ATPase domain that diverge from Sth1p and that appear to provide specificity for Sir3p are not well conserved in mammalian Swi2p/Snf2p homologs (Fig. S5B). The specificity for different BAH domains seems to be imparted by regions within BAH domains that surround and regulate access to the core BAH fold. In line with this hypothesis, we found that the truncated, core BAH domains of Rsc2p and Sir3p were able to interact with both the SWI/SNF and RSC enzymes, but inclusion of C-terminal regions that wrap about the folds inhibited RSC and SWI/SNF binding. Given the plethora of BAH domains associated with chromatin (14), this theme of BAH accessibility and gating might help regulate ATP-dependent chromatin-remodeling enzyme activities in a context-dependent manner.
Materials and Methods
Detailed information on reagent preparation, biochemical assays, and yeast culture work is located in SI Materials and Methods. Oligonucleotides, plasmids, and yeast strains used in this study are listed in Tables S1, S2, and S3, respectively.
Supplementary Material
Acknowledgments
We thank Fred Winston (Harvard Medical School) for providing the telomeric silencing reporter strain, Or Gozani (Stanford University) for providing the human Orc1 expression plasmid, Shinya Watanabe [University of Massachusetts Medical School (UMMS)] for providing Isw2 complex, Nicholas Adkins (UMMS) for providing additional RSC complex, Mayuri Rege (UMMS) for key insights, and other members of the C.L.P. laboratory for discussion. This research was supported by National Institute of General Medical Sciences Grant GM49650 (to C.L.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420096111/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics. 2012;190(2):351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 4.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9(1):25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5(3):e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canzio D, Larson A, Narlikar GJ. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014;24(6):377–386. doi: 10.1016/j.tcb.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grunstein M, Gasser SM. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol. 2013;5(7):a017491–a017491. doi: 10.1101/cshperspect.a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norris A, Boeke JD. Silent information regulator 3: The Goldilocks of the silencing complex. Genes Dev. 2010;24(2):115–122. doi: 10.1101/gad.1865510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrentraut S, et al. Structural basis for the role of the Sir3 AAA+ domain in silencing: Interaction with Sir4 and unmethylated histone H3K79. Genes Dev. 2011;25(17):1835–1846. doi: 10.1101/gad.17175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oppikofer M, et al. Dimerization of Sir3 via its C-terminal winged helix domain is essential for yeast heterochromatin formation. EMBO J. 2013;32(3):437–449. doi: 10.1038/emboj.2012.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armache K-J, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334(6058):977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, et al. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proc Natl Acad Sci USA. 2013;110(21):8495–8500. doi: 10.1073/pnas.1300126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnaudo N, et al. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat Struct Mol Biol. 2013;20(9):1119–1121. doi: 10.1038/nsmb.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callebaut I, Courvalin J-C, Mornon J-P. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446(1):189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 15.Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87(16):6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onishi M, Liou G-G, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28(6):1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Johnson A, et al. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell. 2009;35(6):769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha M, Peterson CL. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol Cell. 2008;30(6):803–810. doi: 10.1016/j.molcel.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138(6):1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szerlong H, et al. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15(5):469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers AL, Pearl LH, Oliver AW, Downs JA. The BAH domain of Rsc2 is a histone H3 binding domain. Nucleic Acids Res. 2013;41(19):9168–9182. doi: 10.1093/nar/gkt662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12(1):147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 23.Logie C, Peterson CL. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16(22):6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swygert SG, et al. Solution-state conformation and stoichiometry of yeast Sir3 heterochromatin fibres. Nat Commun. 2014;5:4751. doi: 10.1038/ncomms5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108(4):845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma VM, Li B, Reese JC. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 2003;17(4):502–515. doi: 10.1101/gad.1039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenstra TL, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42(4):536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dror V, Winston F. The Swi/Snf chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24(18):8227–8235. doi: 10.1128/MCB.24.18.8227-8235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63(2):349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19(14):1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugawara N, Haber JE. 2012. DNA Repair Protocols, Methods in Molecular Biology, ed Bjergbæk L (Humana, New York), Vol 920, pp 349–370.
- 32.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6(12):917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 33.Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68(3):573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 34.Dubarry M, Loïodice I, Chen CL, Thermes C, Taddei A. Tight protein-DNA interactions favor gene silencing. Genes Dev. 2011;25(13):1365–1370. doi: 10.1101/gad.611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auble DT. The dynamic personality of TATA-binding protein. Trends Biochem Sci. 2009;34(2):49–52. doi: 10.1016/j.tibs.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zentner GE, Henikoff S. Mot1 redistributes TBP from TATA-containing to TATA-less promoters. Mol Cell Biol. 2013;33(24):4996–5004. doi: 10.1128/MCB.01218-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wollmann P, et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011;475(7356):403–407. doi: 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fox CA, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995;9(8):911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 39.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88(4):493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 40.Bell SP. The origin recognition complex: From simple origins to complex functions. Genes Dev. 2002;16(6):659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 41.Hickman MA, Rusche LN. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc Natl Acad Sci USA. 2010;107(45):19384–19389. doi: 10.1073/pnas.1006436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.