Significance
Na+/Ca2+ exchangers (NCXs) have a key role in the homeostasis of cellular Ca2+ and consequently are implicated in diverse human-health disorders, including neurodegenerative and cardiovascular diseases. A detailed understanding of the molecular mechanisms of these membrane proteins is therefore of interest from fundamental and biomedical standpoints. Here, we establish the structural mechanism of Na+ recognition in a prokaryotic NCX homolog, using atomistic molecular-dynamics simulations based on recently reported crystallographic data, as well as experimental transport assays of wild-type and mutagenized exchangers. The results have general implications pertaining to the ion exchange stoichiometry and electrogenicity of the Na+/Ca2+ transport cycle across the NCX family, and provide the basis for future investigations of the conformational mechanism of these important transporters.
Keywords: secondary transporters, membrane antiporters, ion specificity, CaCA superfamily, molecular-dynamics simulations
Abstract
Na+/Ca2+ exchangers (NCXs) are ubiquitous membrane transporters with a key role in Ca2+ homeostasis and signaling. NCXs mediate the bidirectional translocation of either Na+ or Ca2+, and thus can catalyze uphill Ca2+ transport driven by a Na+ gradient, or vice versa. In a major breakthrough, a prokaryotic NCX homolog (NCX_Mj) was recently isolated and its crystal structure determined at atomic resolution. The structure revealed an intriguing architecture consisting of two inverted-topology repeats, each comprising five transmembrane helices. These repeats adopt asymmetric conformations, yielding an outward-facing occluded state. The crystal structure also revealed four putative ion-binding sites, but the occupancy and specificity thereof could not be conclusively established. Here, we use molecular-dynamics simulations and free-energy calculations to identify the ion configuration that best corresponds to the crystallographic data and that is also thermodynamically optimal. In this most probable configuration, three Na+ ions occupy the so-called Sext, SCa, and Sint sites, whereas the Smid site is occupied by one water molecule and one H+, which protonates an adjacent aspartate side chain (D240). Experimental measurements of Na+/Ca2+ and Ca2+/Ca2+ exchange by wild-type and mutagenized NCX_Mj confirm that transport of both Na+ and Ca2+ requires protonation of D240, and that this side chain does not coordinate either ion at Smid. These results imply that the ion exchange stoichiometry of NCX_Mj is 3:1 and that translocation of Na+ across the membrane is electrogenic, whereas transport of Ca2+ is not. Altogether, these findings provide the basis for further experimental and computational studies of the conformational mechanism of this exchanger.
Ca2+ signals control a variety of cellular processes essential for the basic function of multiple organs. In cardiac cells, for example, Ca2+ release from the sarcoplasmic reticulum is a necessary step for heart contraction, whereas Ca2+ extrusion from the cell is required for cardiac relaxation. These fluctuations in the cytosolic Ca2+ concentration underlie the initiation of the heartbeat (1, 2). Na+/Ca2+ exchangers (NCXs) play a central role in the homeostasis of cellular Ca2+ (3–5). These integral membrane proteins are ubiquitous in many types of tissues including the heart, brain, and kidney (4), and consequently their dysfunction is associated with numerous human pathologies such as cardiac arrhythmia, hypertension, skeletal muscle dystrophy, and postischemic brain damage (5). NCXs facilitate the translocation of either Ca2+ or Na+ across the membrane; thus, they can harness a transmembrane sodium motive force to energize Ca2+ transport against a concentration gradient. For example, the cardiac exchanger NCX1 mediates the extrusion of intracellular Ca2+ driven by a Na+ transmembrane gradient maintained by the Na+/K+ ATPase (3, 6).
Numerous electrophysiological studies over the past three decades have analyzed the functional and regulatory properties of these important exchangers. It is well established that NCXs are reversible and electrogenic, but it has been debated whether the Na+/Ca2+ exchange stoichiometry is 3:1 or 4:1 (3, 5, 7–12). In any case, NCXs can also facilitate Na+/Na+ and Ca2+/Ca2+ exchange, implying that the translocation of Na+ and Ca2+ are two distinct reactions (3, 5, 6, 13). NCXs are regulated by several factors, such as cytosolic Na+ and Ca2+ concentrations, pH, ATP, and PIP2 (5, 6). Ca2+ regulation in particular involves accessory cytoplasmic domains not directly implicated in the ion-exchange function of the transmembrane domain (5, 14, 15). It is in highly conserved regions within the latter domain, known as α1 and α2 (in transmembrane helices TM2/TM3 and TM7/TM8, respectively), where specific polar and carboxylic amino acids have been identified to be crucial for ion binding and transport (6, 16–18). Similar sequence motifs are found in related antiporters that exchange Na+ for K+ and Ca2+ (NCKX), and Ca2+ for H+ (CAX) (19–21).
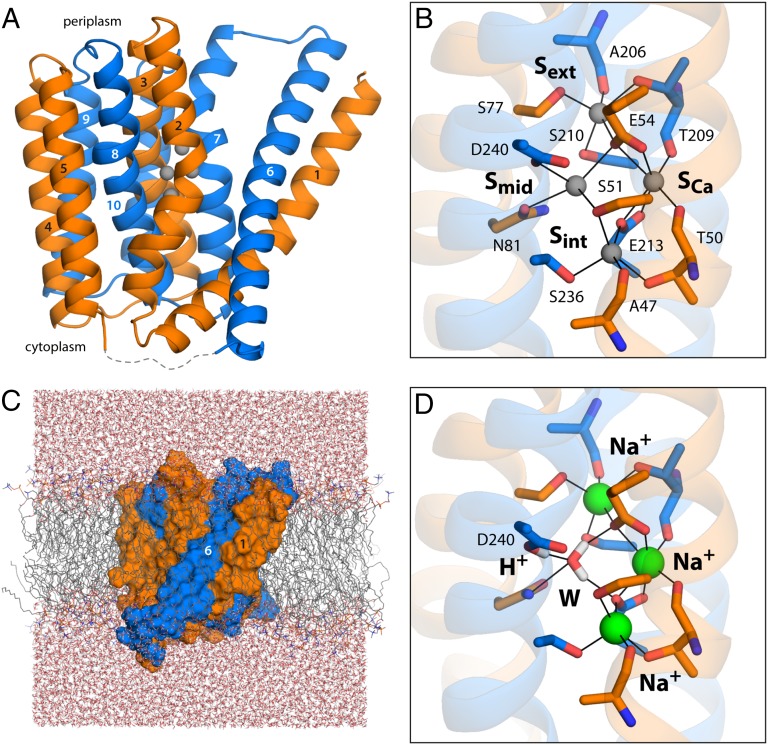
In a recent breakthrough, the NCX exchanger from the archaeobacterium Methanocaldococcus jannaschii was isolated and functionally reconstituted, and its crystal structure determined at 1.9-Å resolution (22). The structure of NCX_Mj revealed an intriguing architecture consisting of two inverted topological repeats of five transmembrane helices each (Fig. 1A), which adopt similar but not identical conformations (Fig. S1). This structural asymmetry is likely to underlie the ability of the exchanger to adopt outward- and inward-open conformations alternately, as is mandatory in secondary-transport processes (23, 24). Importantly, the electron density map for NCX_Mj also revealed clear peaks for four putative ion-binding sites in the α1 and α2 regions, involving several of the residues previously identified as essential (Fig. 1B). One of these sites, referred to as SCa, also produced an anomalous scattering signal, somewhat weak but indicative of the presence of a bound Ca2+. The sites referred to as Sext and Sint appeared to be Na+ sites, based on chemical and geometric considerations. The fourth site, Smid, was tentatively interpreted also as a Na+ site, although in this case the ion-coordination number and coordination geometry would be atypical (22).
Fig. 1.
Structure of the Na+/Ca2+ exchanger from M. jannaschii in an outward-facing conformation. (A) The architecture of NCX_Mj (PDB entry 3V5U) consists of two repeats of five transmembrane helices each (orange, marine cartoons), which adopt opposite orientations in the membrane (the cytoplasmic side is below). Four putative ion-binding sites (indicated by gray spheres) are found halfway across the transmembrane region, flanked by helices TM2–TM3 and TM7–TM8. The structures of the five-helix repeats are highly similar but not identical (Fig. S1). The major differences are the orientation of TM1/TM6 relative to the rest of the repeat, and a bend in the periplasmic half of TM7, not observed in the equivalent region of TM2. These structural asymmetries likely underlie the ability of the exchanger to alternatively adopt outward- and inward-facing conformations (24). (B) Close-up of the four putative ion-binding sites. The side-chain and backbone groups lining these sites are highlighted; the hypothetical ion coordination contacts are indicated in each case. (C) All-atom simulation model of NCX_Mj embedded in a phospholipid membrane. The membrane consists of 208 POPC molecules, and protein and membrane are hydrated by ∼14,900 water molecules. Chloride ions (omitted for clarity) were also included to counter the net charge of the ion–protein complex. The overall system size is ∼77,300 atoms. (D) Ion configuration that is most likely to correspond to the crystal structure of NCX_Mj, according to the structural and thermodynamic analyses described in Figs. 2 and 3. A water molecule (W) occupies the Smid site, while Na+ ions occupy Sext, SCa, and Sint. Note that D240 is protonated, and that the side-chain carboxamide group of N81 is inverted relative to the original assignment. The figure shows the conformation that is most frequently observed in a 200-ns MD simulation, according to an RMSD-based clustering analysis comprising the residues highlighted in B.
If one assumes that the exchange of Na+ and Ca2+ mediated by NCX_Mj proceeds according to a “ping-pong” mechanism, in analogy with other NCXs (13), binding of Na+ and Ca2+ ought to be mutually exclusive (22), even if the exchange stoichiometry is 3:1 rather than 4:1. Thus, the detection of concurrent electron density signals for three Na+ and one Ca2+ must reflect the coexistence of protein molecules with either of these two ion configurations in the crystal lattice. On the other hand, configurations in which Na+ and Ca2+ are simultaneously bound cannot be entirely ruled out; indeed, a minor transport mode has been reportedly observed for cardiac NCX1 whereby one Na+ and one Ca2+ are cotranslocated (9). Either way, it is puzzling that the local structure of the binding sites revealed by the electron density is uniquely defined, as it seems unlikely that this structure is identical irrespective of which ion type is bound. This lack of disorder could be explained if only one of the ion configurations that hypothetically coexist in the protein crystal was significantly populated, but it is not immediately apparent which of these configurations is represented by the data. Thus, notwithstanding the groundbreaking insights provided by the NCX_Mj structure, the interpretation of the electron density in regard to the ion configuration is, in our view, not straightforward: could the proposed Na+ occupancy of the suboptimal Smid site result from the concurrent binding of Ca2+, explaining the minor transport mode observed for NCX1? If so, would Smid remain occupied by Na+ in the absence of Ca2+? Or is the SCa site alternatively occupied by Na+ and Ca2+? If so, does the electron density signal detected in Smid reflect a fourth Na+ ion? Or could it be a water molecule, or another cation, such as K+? It is worth noting that the amino acid composition of the binding sites in NCX_Mj is more akin to that of NCKX exchangers, which cotransport K+ along with Ca2+, than to NCX homologs (Fig. S2), including those of Escherichia coli and Methanosarcina acetivorans (25). The functional reconstitution of NCX_Mj showed that Na+/Ca2+ exchange in NCX_Mj is not influenced by a K+ gradient (22), but it is conceivable that K+ is constitutively bound throughout the transport cycle.
Here, we use all-atom molecular-dynamics (MD) simulations and free-energy calculations of NCX_Mj to systematically assess a series of possible ion configurations, both in terms of their consistency with the protein conformation observed in the experimental crystal structure, and from a thermodynamic standpoint. This computational analysis yields a clear-cut testable hypothesis, which we examine experimentally via Na+/Ca2+ and Ca2+/Ca2+ exchange assays for wild-type and mutagenized NCX_Mj.
Results and Discussion
Valence Screening Indicates Either Na+ or Ca2+ Bind to SCa, but Not to Smid.
As previously mentioned, the NCX_MJ X-ray structure [Protein Data Bank (PDB) entry 3V5U] shows clear spherical densities at four putative ion-binding sites referred to as Sext, Smid, Sint, and SCa, all located near the center of the membrane (Fig. 1B). If ions were indeed bound to these sites, they would be coordinated by a series of polar side chains and backbone carbonyl groups (Fig. 1B), as well as three acidic residues, namely E54, E213, and D240. Even at high resolution, however, a clear interpretation of crystallographic data for ion-binding sites is often not straightforward. The electron density may reflect partial occupancies, and different chemical species may yield comparable signals. In such cases, analysis of the coordination geometry of a putative binding site in reference to an empirical model, as in the valence screening method, is sometimes helpful (26). The outcome of such an analysis for the four putative sites in NCX_Mj is summarized in Table 1. The results indicate that Sext and Sint reflect prototypical Na+ sites, as was deduced initially, rather than sites for, e.g., K+ or Li+ or Ca2+. However, SCa seems to be similarly compatible with Na+ or Ca2+. In contrast, the Smid site seems to be inadequate not only for Na+, but also for Li+, Ca2+, or Cd2+.
Table 1.
Valences of different ionic species for each of the putative ion-binding sites in the crystal structure of the NCX_Mj Na+/Ca2+ exchanger (22)
| Site | Na+ | Ca2+ | K+ | Li+ | Cd2+ |
| Sext | 0.96 | 1.48 | 3.19 | 0.54 | 1.28 |
| Sint | 0.95 | 1.48 | 3.17 | 0.54 | 1.27 |
| Smid | 0.55* | 0.68* | 0.76* | 0.33* | 0.40* |
| SCa | 1.15 | 1.81 | 4.07 | 0.64 | 1.60 |
The valence is defined as follows: ν = ∑i (Ri / R0)−N, where R0 and N are empirical ion-dependent parameters, i denotes each of the coordinating oxygen atoms within 4 Å of a hypothetical ion, and Ri are the distances between the ion and those atoms. The values used for R0 and N were as follows: Na+, 1.622 Å and 4.29; Ca2+, 1.909 Å and 5.4; K+, 2.276 Å and 9.1; Li+, 1.378 Å and 4.065; Cd2+, 1.99 Å and 7.4 (26).
If the coordinates of the N and O atoms in the side-chain carboxamide group of N81 are exchanged (these are not distinguishable at 1.9-Å resolution), the Smid valences become ν(Na+) = 0.50, ν(Ca2+) = 0.61, ν(K+) = 0.67, ν(Li+) = 0.29, and ν(Cd2+) = 0.36.
In the reported X-ray structure (22), four side chains define the Smid site, namely D240, E54, N81, and S210, and each contributing an oxygen atom to the coordination sphere (Fig. 1B). This coordination number would be suboptimal for Na+, as would the hypothetical ion–oxygen distances, which would range from 2.6 to 2.9 Å. By contrast, Sext and Sint each feature a coordination number of 5, with distances of 2.3–2.5 Å (Fig. 1B). Thus, it seems possible that the species bound to Smid is not an ion, but a water molecule (which has the same number of electrons as Na+), and that, accordingly, D240 is not ionized, but protonated. Interestingly, although E54, S210, and N81 are conserved in the NCX family, the conserved residue at position 240 is an asparagine (Fig. S2). However, an aspartate at this position is typical of K+-dependent Na+/Ca2+ exchangers (NCKX), which exchange four Na+ for one K+ and one Ca2+ (Fig. S2). Thus, it is also conceivable that D240 in NCX_Mj is ionized, and that the species bound to the Smid site is K+. The valence analysis shows Smid is suboptimal for K+, but less so than for the other ions (Table 1). Either way, this empirical model indicates that neither Na+ nor Ca2+ is likely to bind stably to Smid.
It should be noted, however, that the valence screening method is necessarily imprecise, as it neglects the natural flexibility of protein structures and their ability to adapt to different ion occupancy states; the underlying empirical model may also not be representative of situations where multiple ions bind in close proximity and interact with one another. Therefore, we set out to further inspect a range of possible ion configurations, including those suggested by the valence screening, using fully atomistic MD simulations of NCX_Mj in a lipid membrane (Fig. 1C).
Simulations Identify Three Ion Configurations Similarly Consistent with the X-Ray Structure.
Although conventional MD simulations are at the present time unable to probe the timescales spanned by the conformational cycles of membrane transporters, there is ample evidence that this approach may be used to characterize the structural dynamics and energetics involved in the recognition of the transported substrates, for well-defined protein conformations (27, 28). We therefore reasoned that sufficiently long simulations ought to reveal which of several possible ion configurations is most likely to be reflected in the experimental electron density map for NCX_Mj, if these alternative configurations are ranked in terms of the perturbation they induce on the geometry of the binding sites. To quantify this geometric perturbation for each of the configurations considered, we monitored the arrangement of the side chains and backbone groups that line the four putative binding sites, using a set of 92 interatomic distances (Fig. 1B). We then computed the percentage of these distances that are inconsistent with the X-ray structure, which we refer to as violations. Specifically, a distance violation occurs when the averaged value of this distance over the length of the MD simulation differs from that in the X-ray structure by more than two times the SD of the simulation average. According to this metric, the ion configurations most likely to be represented by the experimental structure must have a small percentage of violations.
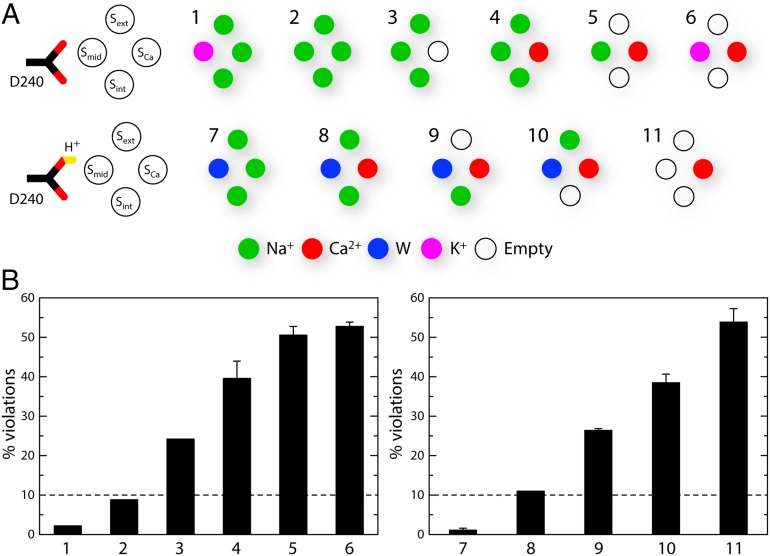
The specific ion configurations considered are shown schematically in Fig. 2A. This set includes the configurations tentatively proposed originally, as well as those suggested by the valence screening analysis. We postulate that D240 is ionized in configurations in which an ion is bound to Smid, whereas D240 is considered to be protonated when Smid is empty, or occupied by a water molecule. E213 and E54 are, however, always ionized, because in all considered cases they coordinate a cation directly. In addition, we explore the configurations that might be expected for a K+-dependent exchanger, whether or not K+ is transported, as well as mixed Na+/Ca2+ configurations that might correspond to atypical stoichiometries, as noted for cardiac NCX1 (9).
Fig. 2.
Comparative analysis of alternative ion occupancy configurations of the binding sites in NCX_Mj, based on the consistency between simulation and crystallographic data. (A) Specific ion configurations considered in this study. Note that when an ion occupies Smid, D240 is ionized; if Smid is empty or occupied by water, D240 is protonated. (B) Geometric perturbation of the experimental structure induced by each of the ion configurations simulated. For each simulation, we analyzed a set of 92 interatomic distances between side-chain and backbone atoms lining the binding sites (Fig. 1B), and identified those that are inconsistent with the X-ray structure, which we refer to as violations. A distance violation occurs when the time-averaged value of this distance over the length of the simulation differs from that in the X-ray structure by more than two times the SD of the simulation average. A lower percentage of violations therefore implies better agreement with the experimental structure. The values plotted were obtained with simulations using the standard CHARMM27 force field. The three configurations with a percentage of distance violations smaller than 10 were also simulated with an optimized version of the force field (SI Methods and Figs. S3 and S4). The resulting distance violations were 1.1%, 2.2%, and 4.4%.
The results of this systematic analysis are summarized in Fig. 2B. A first conclusion from these data is that the conformation of the binding sites observed in the X-ray structure of NCX_Mj is not consistent either with a configuration in which three Na+ occupy the Sext, Smid, and Sint sites, with SCa unoccupied (and D240 ionized) (#3), or with a configuration in which Ca2+ occupies SCa, while Sext, Smid, and Sint are vacant (and D240 is protonated) (#11). Thus, the true Na+ and Ca2+ states of NCX_Mj must adopt a different ion occupancy configuration, or a different conformation of several of the side chains that define the binding sites. The experimental structure is also inconsistent with all of the configurations with Ca2+ bound to Sca concurrently with one, two, or three Na+ or water molecules in Sext, Smid, and Sint (and D240 protonated or not accordingly) (#4, #5, #8, #9, and #10), ruling out that these mixed states are significantly populated in the protein crystal. A configuration with K+ at Smid and Ca2+ in SCa (#6), which as mentioned might be expected for an NCKX-like exchanger, also induces a significant perturbation of the experimental structure. However, a configuration with four Na+ bound (#2) preserves the structure relatively well (using a 10% violation cutoff), and so does a configuration with K+ at Smid and three Na+ at Sext, SCa, and Sint (#1). Thus, this analysis alone does not conclusively rule out a NCKX-like transport cycle in which one Ca2+ and one K+ are exchanged for four Na+, or a variant thereof in which a K+ ion is constitutively bound; either way, however, the hypothetical state with Ca2+ and K+ bound (#6) would feature a conformation of the binding sites that differs from that observed in the NCX_Mj structure.
These results notwithstanding, the ion configuration that is most consistent with the experimental crystal structure, with only ∼1% of violations, is that with three Na+ ions bound to Sext, SCa, and Sint, while one water molecule is bound to Smid and D240 is protonated (#7, Fig. 1D). That is, this systematic analysis indicates that the specific arrangement of the ion-binding sites in the available outward-facing structure of NCX_Mj (22) most likely represents the Na+-bound form of the exchanger. The Smid site is, however, not one of the three primary Na+ sites. Instead, the simulation results indicate that the electron density detected at Smid is most probably a water molecule, coordinated by D240 in the neutral form, or alternatively, with less likelihood, a K+ or an additional Na+, coordinated by D240 in the ionized form.
Admittedly, this analysis does not provide a conclusive explanation for the detection of an anomalous scattering signal attributable to Ca2+ at the SCa site (22). As mentioned, all of the ion configurations considered here that include Ca2+ at this site appear to be inconsistent with the crystal structure, because the arrangement of the side chains lining Sext, Smid, and Sint sites changes significantly in the course of the simulations. We conclude, therefore, that if one of these configurations were indeed present in the protein crystal, alongside the three-Na+ state described above, it would be minimally populated and thus would not be reflected in the electron density detected for the protein (although it may be sufficiently populated to yield a weak anomalous scattering signal). However, it is also possible that Ca2+ binds in a configuration not considered here, for example, involving multiple water molecules mediating ion–protein contacts, as observed in a recent X-ray structure of a Ca2+/H+ exchanger in the inward-facing conformation (29). The experimental electron density for NCX_Mj, however, reveals no indication of such a water network, so this configuration too would be marginally populated in the crystal. Either way, it is worth noting that, although Ca2+ was available during the purification of NCX_Mj in large amounts (10 mM), no Ca2+ was present in the crystallization buffer, which contained instead 100–200 mM Na+ (22). It is therefore plausible that Na+ replaces Ca2+ in most, although apparently not all, of the protein molecules in the crystal.
Free-Energy Calculations Reveal the Most Likely Ion Configuration in NCX_Mj.
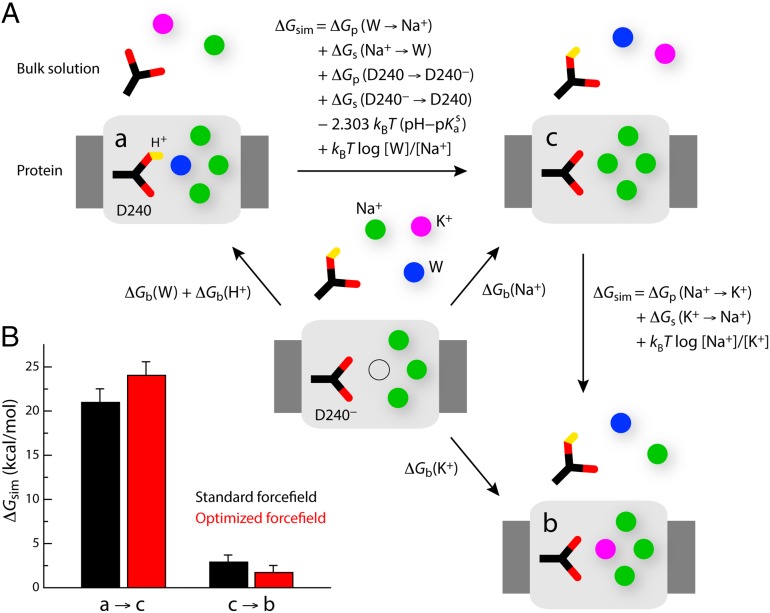
To attest to the plausibility of a hypothetical ion configuration, consistency with the experimental protein geometry is a necessary, but not sufficient, condition; it is also necessary to assess its thermodynamic stability relative to other geometrically similar alternatives. Thus, to identify the most probable ion configuration reflected in the crystal structure of NCX_Mj, we carried out an additional series of simulations designed to evaluate the relative free energy of the three configurations for which the distance violations relative to the experimental geometry is 10% or less. As mentioned, these are the following: (a) three Na+ bound to Sext, SCa, and Sint, a water bound to Smid and D240 protonated (Fig. 2A, #7); (b) three Na+ bound to Sext, SCa, and Sint, a K+ bound to Smid, and D240 deprotonated (Fig. 2A, #1); and (c) four Na+ bound, with D240 deprotonated (Fig. 2A, #2).
The free-energy differences between these configurations can be deduced from the thermodynamic cycles depicted in Fig. 3A. In principle, these relative free energies could be obtained by comparing the calculated binding affinities (ΔGb) for the species that are distinct in each of the configurations. However, it is more computationally convenient, and formally equivalent, to calculate the free energy associated with the “alchemical” transformation of one configuration into another (ΔGsim), provided this free energy is adjusted by the value obtained for the opposite transformation in bulk water, as well as by the assumed bulk concentration of all species involved, and the solution pKa of the aspartate side chain. Note that this approach permits us to evaluate the energetics of protonation of D240 under a classical-mechanics framework.
Fig. 3.
Thermodynamic analysis of the three ion occupancy configurations with best structural consistency with the NCX_Mj crystal structure. (A) Thermodynamic cycles used to deduce the alchemical transformations (ΔGsim) that are required to evaluate the relative free energies of the three ion configurations considered. (B) Calculated free-energy differences between configurations a and c, and between configurations c and b. The sum of these is the free-energy difference between a and b. It was assumed that pH = 7, [Na+] = [K+] = 100 mM, [W] = 55 M, and pKas = 3.86. The last two terms in the expression of the free-energy difference between a and c are thus -4.28 and 3.73 kcal/mol, respectively; and the last term in the expression of the free-energy difference between c and b is zero. Note that the volumes occupied by Na+, K+, or water at the Smid site are approximately equivalent, and thus the related entropic contributions are omitted. Free-energy values are reported for the standard CHARMM27 force field, as well as for a modified version in which the Lennard–Jones Rmin parameter for the Na+–carboxylate and K+–carboxylate interactions are optimized so as to reproduce osmotic pressure and binding affinity measurements (SI Methods and Figs. S3 and S4). The values shown are means of two independent free-energy calculations carried out in opposite directions, e.g., a to c and c to a. The differences between the calculated values in each direction are shown as error bars.
This thermodynamic analysis demonstrates that configuration a, which is the most consistent with the experimental X-ray structure from a geometric standpoint, is also the most energetically favored, by a large margin (Fig. 3B). Specifically, configuration a is over 20 kcal/mol more favorable than, for example, configuration c. Configuration b is also energetically favored over configuration c, but only by ∼2 kcal/mol. Note that comparable results are obtained with the standard CHARMM27 force field and with a modified version in which the parameters describing the van der Waals interactions between Na+/K+ and aspartate/glutamate are optimized to reproduce osmotic pressure measurements for concentrated solutions of Na+/K+−acetate/propionate, as well as experimental dissociation constants for these ion pairs (SI Methods and Figs. S3 and S4). Therefore, from both geometric and thermodynamic standpoints, we conclude that the recently determined outward-facing structure of NCX_Mj (22) reflects a state in which three Na+ ions are bound to the protein, occupying the Sext, SCa, and Sint sites, while a water molecule occupies Smid; D240, which flanks this site, is protonated (Fig. 1D). Taken together with the geometric analysis reported above, these free-energy calculations also indicate that neither the Na+ nor the Ca2+ half-cycles in NCX_Mj are coupled to K+ translocation.
Experimental Measurements Validate Computational Interpretation.
To test experimentally our computational interpretation of the NCX_Mj structure, the exchanger was heterologously overexpressed in E. coli cells, and its functionality was compared with that of three mutants in which the aspartate at position 240 was substituted by either asparagine, alanine, or glutamine. Because the asparagine side chain mimics the protonated form of D240, the transport activity of NCX_Mj should not be inhibited in D240N, if the ion configuration identified computationally as the most probable is indeed physiologically relevant. However, the alanine and glutamine substitutions ought to have a more pronounced structural effect, and thus lead to changes in activity.
More specifically, a protocol was developed to produce membrane vesicles in which NCX_Mj is expressed at high levels (10–12% of total membrane protein content) and predominantly (95%) in the right-side-out orientation, i.e., with the periplasmic side exposed to the extravesicular space (30). The transport activity of the exchanger was then analyzed by measuring the kinetics of 45Ca2+ uptake into the vesicles in conditions conducive to either Ca2+/Ca2+ or Na+/Ca2+ exchange (30–32). We reasoned that, if the mutations have a deleterious effect on binding and/or transport of Na+, this will be reflected on the rate of 45Ca2+ uptake in conditions of Na+/Ca2+ exchange, whereas the influence of the mutations on Ca2+ binding and/or transport will be revealed in conditions of Ca2+/Ca2+ exchange.
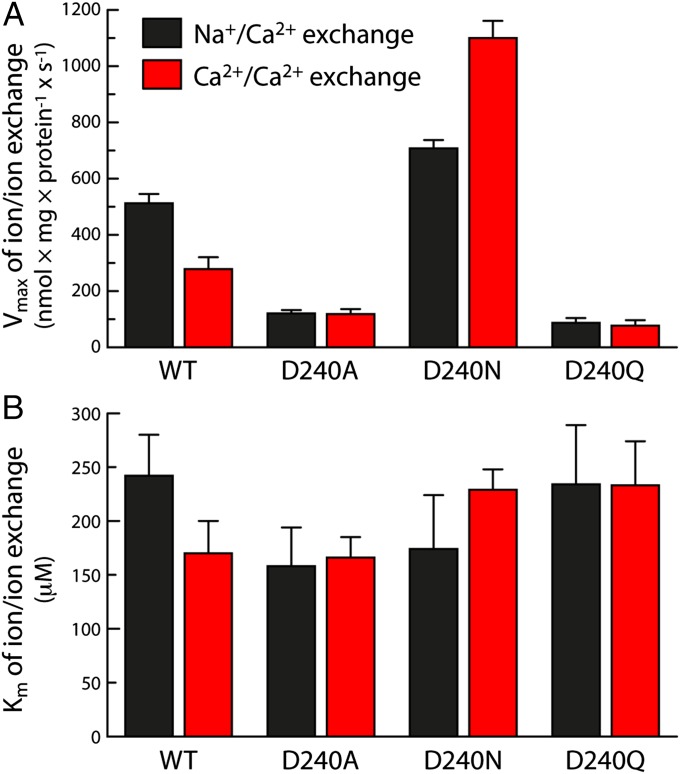
Fig. 4A shows the measured Vmax values for 45Ca2+ uptake by wild-type and mutant NCX_Mj in these two experiments. The similarity between wild-type and D240N in conditions of Na+/Ca2+ exchange is consistent with the notion that D240 is protonated in the Na+-translocation step of the transport cycle. Indeed, the 45Ca2+ uptake rate in D240N is slightly higher than in the wild-type exchanger, suggesting that deprotonation of D240 is inhibitory. This effect is more apparent in the Ca2+/Ca2+ exchange experiment, in which 45Ca2+ uptake is significantly enhanced by the D240N mutation. It is clear, then, that D240 is protonated both in the Ca2+ and Na+-translocations steps, and therefore, that neither Ca2+ nor Na+ occupies the Smid site in the functional transporter. The three transported Na+ ions must therefore occupy the Sext, SCa, and Sint sites, as suggested by the computational analysis.
Fig. 4.
Experimental transport kinetics for wild-type NCX_Mj and D240 mutants. To characterize the exchange of Na+/Ca2+ and Ca2+/Ca2+ mediated by NCX_Mj, E. coli-derived vesicles in which the protein had been overexpressed were loaded with either Na+ (160 mM) or Ca2+ (250 µM), and diluted in an assay medium containing 20–2,000 µM 45CaCl2. Initial 45Ca2+-uptake rates (t = 5 s) were measured as described in Methods. (A) Vmax and (B) Km values for Na+/Ca2+ and Ca2+/Ca2+ exchange were obtained by fitting the experimental data to the Michaelis equation. The bars represent mean ± SE values obtained from three independent experiments.
In contrast to D240N, the D240A and D240Q mutations have a marked deleterious effect on the 45Ca2+ uptake rates, in both the Na+/Ca2+ and the Ca2+/Ca2+ experiments. Nevertheless, the measured Km values (Fig. 4B) indicate that this effect is not due to a reduction in the apparent binding affinity for Ca2+ (which is comparable to wild type, and to D240N), consistent with the notion that Ca2+ does not occupy the Smid site during translocation. Instead, these mutations appear to somehow impair one or more of the subsequent states in the transition between outward- and inward-facing conformations of the exchanger.
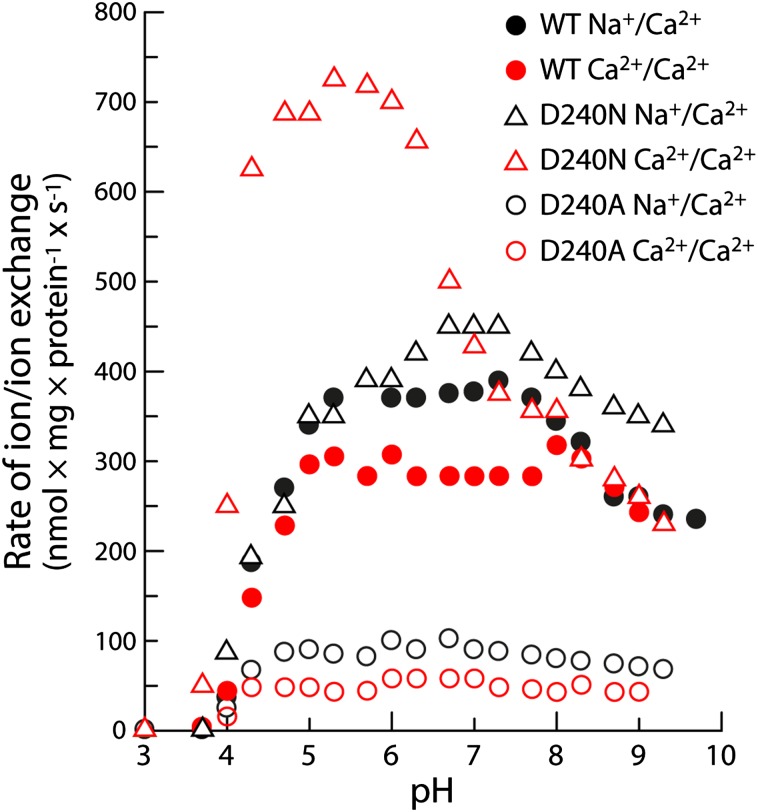
Fig. 5 shows measurements of the initial 45Ca2+ uptake rates in Na+/Ca2+ and Ca2+/Ca2+ exchange conditions, as a function of pH. These data underscore the conclusions drawn above in regard to D240 and provide additional mechanistic insights. Both wild-type and D240N NCX_Mj are inactive at pH 4 or lower, are maximally active between pH 5.5 and 8, and become slightly less active, reaching a plateau, at higher pH values. We envisage that the lack of activity at low pH values is caused by the protonation of E54 and E213 (in addition to D240), which would preclude binding of both Na+ and Ca2+. The steep rise in activity as the pH is raised would thus reflect the increasing probability of ionization of these side chains, and thus the increased likelihood of Na+ or Ca2+ binding. Interestingly, however, in this optimal pH range the D240N mutant is significantly more active than the wild-type protein, particularly in conditions of Ca2+/Ca2+ exchange. As mentioned, this result confirms that a proton must be bound to D240 for transport to proceed, but also indicates that this proton is not completely localized on D240 early on in the ion recognition process. This might be due to the existence of transient and/or partial ion-bound configurations not conducive to transport, mediated by the deprotonated form of D240. Indeed, a crystal structure of NCX_Mj bound to Cd2+ (22), which inhibits the exchanger, shows the ion occupying the Smid site, directly coordinated by D240, which is therefore deprotonated. These experimental results indicate that such “inhibitory” configurations, which may be thought as kinetic traps, might also occur under normal transport conditions, particularly for Ca2+. In the D240N mutant, however, the formation of transport-ready ion-bound configurations would be faster because the proton is effectively locked in the correct place throughout the cycle, and this kinetic advantage would be manifested in the overall transport rate.
Fig. 5.
pH dependence of the ion exchange rates for wild-type NCX_Mj and D240 mutants. The initial rates of Na+/Ca2+ and Ca2+/Ca2+ exchange were measured as in the experiments shown in Fig. 4, in an assay medium with 500 µM 45CaCl2 (for additional details, see Methods). The pH of the assay medium was controlled with a 20 mM MES/MOPS/Tris buffer.
Last, it is worth clarifying that the slight decline in the transport activity at high pH values is not associated with the deprotonation of D240, as it is also observed for D240N (Fig. 5). We speculate that this decline might be caused by the disruption of yet-unidentified interactions (elsewhere within the protein, or between the protein and the lipid membrane) required for the stabilization of the inward- and/or outward-facing conformations of the transporter.
Conclusions
The recent determination of the structure of a prokaryotic Na+/Ca2+ exchanger (NCX_Mj) at atomic resolution is a milestone in our understanding of this important family of membrane transporters, and will no doubt foster novel discoveries in this field. This notwithstanding, the crystallographic data are not by themselves conclusive with respect to the occupancy of putative binding sites for Na+ and/or Ca2+ within the transporter, the determination of which is key to establishing its exchange stoichiometry, its electrogenic properties, and the potential involvement of other ionic species in the transport mechanism. Such insights would be very important not only to facilitate a clear molecular interpretation of existing and future functional measurements for this particular exchanger, but also to permit its comparison with the eukaryotic NCXs characterized over the past decades.
Here, we have reported a systematic computer simulation study specifically designed to reveal the ion configuration that is most consistent with the crystallographic structure, and also the most probable from a thermodynamic standpoint. The computational results are conclusive in showing that this periplasmic-facing, occluded structure of NCX_Mj is loaded with three Na+ ions, a proton, and a water molecule. The three Na+ ions occupy the sites referred to as Sext, SCa, and Sint, while the water molecule resides in Smid. Interestingly, the proton binds to the side chain of D240, which is adjacent to Smid, implying that transport of Na+ is the electrogenic step (with net charge +1) in the Na+/Ca2+ exchange cycle.
Experimental assays of Na+/Ca2+ and Ca2+/Ca2+ exchange mediated by either wild-type NCX_Mj or D240 mutants confirm the notion that protonation of D240 is required for translocation of both Na+ and Ca2+, and hence, that D240 is not directly in contact with Na+ or Ca2+ at the Smid site during transport. The experiments do not rule out that D240 can be transiently deprotonated, or that Smid can be transiently occupied by an ion, but the resulting effect is an inhibition of the ion exchange cycle. Thus, these computational and experimental results support the notion that ion exchange stoichiometry in NCX_Mj is 3Na+:1Ca2+, and not 4:1 as proposed elsewhere.
That protonation of D240 is required for ion exchange in NCX_Mj also provides a simple explanation for the perplexing observation that the conserved residue at position 240 in known prokaryotic and eukaryotic Na+/Ca2+ exchangers is an asparagine, and not an aspartate. Indeed, the possibility of transient deprotonation of D240 in NCX_Mj may account, at least in part, for the much slower turnover rates of its ion exchange cycle (30), compared with other NCX orthologs. The substitution of aspartate for asparagine would thus provide a kinetic advantage, which may be physiologically beneficial for the rapid extrusion of cytosolic Ca2+ in excitable tissues (5, 31, 33).
Finally, it is pertinent to underscore that the conclusions of these study are not at odds with the initial finding that Ca2+ occupies the SCa site, although we would posit that Ca2+ binding to this site requires the displacement of Na+, and vice versa. The detection of anomalous scattering at this site is, after all, a clear indication of the presence of Ca2+. It should be noted that this observation implies that the protein crystal contains at least two populations of NCX_Mj molecules, one bound to Na+ and another to Ca2+. That is, it appears as if Ca2+, present during the protein purification, was not fully displaced from SCa upon exposure to the sodium-enriched crystallization buffer. Our simulations do indicate, however, that the configuration of the ion-binding region in the Ca2+-bound state is not accurately represented in the existing X-ray structure of NCX_Mj, and thus remains to be determined conclusively. This finding would not be inconsistent with the crystallographic data if the population of Ca2+-bound molecules in the NCX_Mj is minor, as the somewhat weak intensity of the anomalous scattering signal suggests. Further computational and experimental work is underway to determine the mode of Ca2+ binding and to rationalize how it differs from that of Cd2+ inhibition. Altogether, the computational and experimental insights presented here contribute to establishing the structural mechanism of ion recognition in NCX_Mj, and provide the basis for further mechanistic and functional studies.
Methods
MD Simulations of NCX_Mj.
MD simulations were carried out with NAMD, versions 2.7–2.9 (34), using the CHARMM27/CMAP force field for protein and lipids (35, 36) at constant temperature (298 K), pressure (1 atm), and membrane surface area (∼69 Å2 per lipid). Periodic boundary conditions were used in all directions. Electrostatic interactions were calculated using Particle Mesh Ewald with a real-space cutoff of 12 Å; the same cutoff distance was used for van der Waals interactions. The X-ray structure of the Na+/Ca2+ exchanger from Methanocaldococcus jannaschii [PDB entry 3V5U (22)] was initially processed using CHARMM (37), for an ion configuration chosen arbitrarily. DOWSER (38) was used to place water molecules in suitable binding sites inside the protein, and these were added to the water molecules revealed in the crystal structure. The protein–ion complex was then embedded in a hydrated palmitoyl-oleoyl-phosphatidyl-choline (POPC) membrane, which includes 208 POPC molecules and ∼14,790 water molecules, using GRIFFIN (39). To counterbalance the net charge of the protein–ion complex, which depends on the ion occupancy state considered, Cl− ions were added as needed. The simulation system thus amounts to a total of ∼77,300 atoms (Fig. 1C). The protein–membrane and protein–solvent interfaces were further optimized in two stages: first, a 25-ns MD simulation was carried out in which the motions of all nonhydrogen protein atoms (and ions) were individually restrained, while membrane and solvent molecules moved freely; this was followed by a 15-ns MD simulation in which the protein–ion complex could tumble freely within the membrane, but as a rigid body, owing to a strong RMSD-based restraint (including all nonhydrogen atoms) that preserves the conformation observed in the X-ray structure. After these preparatory simulations, each of the alternative ion occupancy states considered (Fig. 2A) were introduced and simulated independently. For each of the 11 ion configurations, the structure of the ion–protein complex was equilibrated in two additional stages: first, a 25-ns MD simulation was carried out with a conformational RMSD restraint relative to the X-ray structure that includes all Cα atoms in the protein and all nonhydrogen atoms in the residues flanking the putative ion-binding sites (Fig. 2B); this was followed by a 15-ns simulations in which only the RMSD of the Cα trace was restrained. After equilibration, MD trajectories of 100 ns were calculated for each the ion configurations without any restraints. These simulations were the basis for the distance violation analysis (see below) shown in Fig. 2B. For the three configurations that resulted in less than 10% violations, the simulations were extended to 200 ns and the violations recomputed. For these three systems, 200-ns MD simulations were also carried out using improved Lennard–Jones parameters for the cation−carboxylate interactions (SI Methods and Figs. S3 and S4).
Distance Violation Analysis.
For each of the ion occupancy states considered (Fig. 2A), the structure of the four putative ion-binding sites was characterized by monitoring 92 protein–protein distances during each of the MD simulations. The protein atoms that define this set of distances are all of the oxygen atoms within 3 Å of the putative ion-binding sites, except those in N81, D240, E213, and E54, for which the Cγ and Cδ atoms were included instead. Time averages and SDs were computed for all distances at the end of each simulation. It was considered that a distance violation occurs when the simulation average differs from the value in the X-ray structure by more than twice the calculated SD.
Free-Energy Calculations.
Free-Energy Perturbation (FEP) calculations were performed to assess the relative thermodynamic stability of selected ion configurations (Fig. 3A). These calculations were based on MD simulations analogous to those used in the distance violation analysis. The input coordinates of the ion–protein complex for each of the configurations considered were therefore extracted from the unrestrained simulations previously carried out, specifically through a clustering analysis based on the conformation of the ion-binding region (selecting the center of the most populated cluster in each case). The first set of free-energy calculations, in which configuration a is alchemically transformed into configuration c, and vice versa, comprised 50 intermediate simulations of ∼2 ns each, of which the first 500 ps were considered as equilibration and discarded. A soft-core van der Waals potential with a radius-shifting coefficient of 3 Å2 was used in this case to facilitate endpoint convergence. The starting points for each calculation, i.e., configuration a in the forward transformation and configuration c in the backward transformation, were obtained from the MD simulations carried out for the distance violation analysis, and are therefore uncorrelated. The second set of free-energy calculations, between configurations b and c, was carried out analogously, but used 25 intermediate simulations of ∼2 ns each and a soft-core van der Waals potential with a radius-shifting coefficient of 1 Å2. As noted in the thermodynamic cycles shown in Fig. 3A, these free-energy calculations involve transformations between protonated and deprotonated forms of aspartate, and between Na+ and water or K+, both in the context of the protein and in bulk solution. To avoid artifacts, these transformations were carried out in the same simulation box. The species transformed in solution (i.e., Na+, K+, and Ace-Asp-Nme) were kept at >16 Å away from each other and at >37 Å from the center of the membrane, to minimize artificial (i.e., not bulk-like) interactions among themselves and with the protein/lipid. To assess the force field dependence of the results, the calculated free-energy differences obtained with standard CHARMM27 were compared with values obtained with improved Lennard–Jones parameters for the cation−carboxylate interactions (SI Methods and Figs. S3 and S4). To do so, we calculated the free energy associated with the change in the relevant parameters for each ion configuration separately. The resulting values were then added/subtracted to the values computed with CHARMM27 as appropriate. These FEP transformations comprised 5–10 intermediate simulations of 1 ns each, of which the first 200 ps were considered as equilibration, and were also carried out in forward and backward directions. No soft-core van der Waals potential was necessary in this case.
Experimental Construct Preparation.
DNA encoding the wild-type NCX_Mj was amplified by PCR from a Methanocaldococcus jannaschii cDNA library (DSMZ) and ligated between the NcoI and BamHI restriction sites of a pET-28a plasmid as described elsewhere (22, 30). A C-terminal EGFP-fused NCX_Mj was made by ligating a SalI/XhoI-digested PCR-amplified EGFP-encoding fragment into the XhoI site of the pET28-NCX_Mj construct. Mutations were introduced by QuikChange mutagenesis (Stratagene) and confirmed by sequencing.
Preparation of E. coli-Derived Vesicles Containing Overexpressed NCX_Mj.
Expression vectors were transformed into E. coli BL21 (DE3) pLysS competent cells. The cells were grown, harvested, homogenized, and lysed as previously outlined (30). Cell lysates were loaded onto a three-step sucrose gradient (2.02, 1.4, and 0.7 M) for 15 h at 150,000 × g, 4 °C, following the protocol described elsewhere (30). Finally, the pelleted membrane vesicles were dissolved to homogeneity in 50 mM MOPS-Tris, pH 7.4, 250 mM sucrose, flash-frozen in liquid nitrogen, and stored in −80 °C until use. The total protein concentration in the vesicle preparations was determined using the Lowry protein assay. The expression levels of NCX_Mj were measured using a GFP-based assay (see below). To evaluate the orientation of NCX_Mj in the E. coli-derived cell membrane vesicles, digestion of a TEV-sensitive site attached to the C-terminal 6×His-tag was evaluated using an antibody against the 6×His-tag, as described previously (30). According to this test, ∼95% of NCX_Mj in the membrane vesicles (obtained by French-press disruption of the E. coli cells) adopts the right-side-out orientation, i.e., the periplasmic side of the protein faces the extravesicular space.
GFP-Based Assay for Evaluating the Expression Levels of NCX_Mj.
To determine the expression levels of NCX_Mj in the E. coli-derived membrane vesicles, relative to the total amount of protein, the concentration of GFP was estimated by its absorbance at 488 nm (e.c. = 56,000), and divided by the value evaluated by Lowry. The GFP absorbance signal at 488 nm was isolated from vesicle noise using a correction factor determined by measuring the nonexpressing vesicles. The correction factor was applied using a double measurement both at the GFP absorbance peak and outside of its spectrum (488 and 570 nm, respectively) using the following equation: A(488,GFP only) = A(488) − [A(570)/0.657]. According to this assay, the expression levels of NCX_Mj in the vesicular preparations of E. coli-derived cell membranes were in the range of 10–12% of the total membrane proteins (30).
45Ca2+-Uptake Assay in E. coli-Derived Vesicles.
Initial rates of Na+/Ca2+ and Ca2+/Ca2+ exchange were assayed by measuring 45Ca2+ uptake (t = 5 s) into the E. coli-derived vesicles containing overexpressed NCX_Mj, either in the wild type or mutagenized, as previously described (30). In standard assay conditions, vesicles loaded with either Na+ (160 mM) or Ca2+ (250 µM) were diluted 25- to 50-fold at 35 °C in an assay medium containing 20 mM MOPS/Tris, pH 6.5, 100 mM KCl, and 10–2,000 µM 45CaCl2. The 45Ca2+ uptake was quenched by cold EGTA buffer, and the quenched solutions were immediately filtrated on GF/C filters (Tamar Ltd.) as outlined before (30–32). E. coli-derived vesicles lacking NCX_Mj were used for blank assays, and the resulting nonspecific (background) signals were subtracted from the signals obtained with the vesicles containing wild-type or mutagenized NCX_Mj. GraFit 7.0 (Erithacus Software Ltd.) was used for data fitting at varying ionic concentrations. The kinetic parameters (Km and Vmax) were evaluated under conditions in which the concentration-dependent rates reach more than 85% saturation. Km and Vmax values were measured in three or more independent experiments; the data reported are mean ± SE values.
Supplementary Material
Acknowledgments
We thank Prof. Youxing Jiang for helpful discussions of our simulation results. This work was funded in part by the Division of Intramural Research of the National Heart, Lung, and Blood Institute (National Institutes of Health, J.D.F.-G) and by the Israel Science Foundation Grant 825/14 (to D.K.). Financial support from the Fields Estate Foundation (to D.K.) is highly appreciated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415751111/-/DCSupplemental.
References
- 1.Noble D. The Initiation of the Heart Beat. Clarendon; Oxford: 1979. [Google Scholar]
- 2.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 3.Blaustein MP, Lederer WJ. Sodium/calcium exchange: Its physiological implications. Physiol Rev. 1999;79(3):763–854. doi: 10.1152/physrev.1999.79.3.763. [DOI] [PubMed] [Google Scholar]
- 4.Lytton J. Na+/Ca2+ exchangers: Three mammalian gene families control Ca2+ transport. Biochem J. 2007;406(3):365–382. doi: 10.1042/BJ20070619. [DOI] [PubMed] [Google Scholar]
- 5.Khananshvili D. The SLC8 gene family of sodium-calcium exchangers (NCX)—structure, function, and regulation in health and disease. Mol Aspects Med. 2013;34(2-3):220–235. doi: 10.1016/j.mam.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Philipson KD, Nicoll DA. Sodium-calcium exchange: A molecular perspective. Annu Rev Physiol. 2000;62:111–133. doi: 10.1146/annurev.physiol.62.1.111. [DOI] [PubMed] [Google Scholar]
- 7.Reeves JP, Hale CC. The stoichiometry of the cardiac sodium-calcium exchange system. J Biol Chem. 1984;259(12):7733–7739. [PubMed] [Google Scholar]
- 8.Rasgado-Flores H, Blaustein MP. Na/Ca exchange in barnacle muscle cells has a stoichiometry of 3 Na+/1 Ca2+ Am J Physiol. 1987;252(5 Pt 1):C499–C504. doi: 10.1152/ajpcell.1987.252.5.C499. [DOI] [PubMed] [Google Scholar]
- 9.Kang TM, Hilgemann DW. Multiple transport modes of the cardiac Na+/Ca2+ exchanger. Nature. 2004;427(6974):544–548. doi: 10.1038/nature02271. [DOI] [PubMed] [Google Scholar]
- 10.Hinata M, Kimura J. Forefront of Na+/Ca2+ exchanger studies: Stoichiometry of cardiac Na+/Ca2+ exchanger; 3:1 or 4:1? J Pharmacol Sci. 2004;96(1):15–18. doi: 10.1254/jphs.fmj04002x3. [DOI] [PubMed] [Google Scholar]
- 11.Dong H, Dunn J, Lytton J. Stoichiometry of the cardiac Na+/Ca2+ exchanger NCX1.1 measured in transfected HEK cells. Biophys J. 2002;82(4):1943–1952. doi: 10.1016/S0006-3495(02)75543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bers DM, Ginsburg KS. Na:Ca stoichiometry and cytosolic Ca-dependent activation of NCX in intact cardiomyocytes. Ann N Y Acad Sci. 2007;1099:326–338. doi: 10.1196/annals.1387.060. [DOI] [PubMed] [Google Scholar]
- 13.Khananshvili D. Distinction between the two basic mechanisms of cation transport in the cardiac Na+-Ca2+ exchange system. Biochemistry. 1990;29(10):2437–2442. doi: 10.1021/bi00462a001. [DOI] [PubMed] [Google Scholar]
- 14.Giladi M, Khananshvili D. Molecular determinants of allosteric regulation in NCX proteins. Adv Exp Med Biol. 2013;961:35–48. doi: 10.1007/978-1-4614-4756-6_4. [DOI] [PubMed] [Google Scholar]
- 15.Giladi M, Hiller R, Hirsch JA, Khananshvili D. Population shift underlies Ca2+-induced regulatory transitions in the sodium-calcium exchanger (NCX) J Biol Chem. 2013;288(32):23141–23149. doi: 10.1074/jbc.M113.471698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicoll DA, Hryshko LV, Matsuoka S, Frank JS, Philipson KD. Mutation of amino acid residues in the putative transmembrane segments of the cardiac sarcolemmal Na+-Ca2+ exchanger. J Biol Chem. 1996;271(23):13385–13391. doi: 10.1074/jbc.271.23.13385. [DOI] [PubMed] [Google Scholar]
- 17.Shigekawa M, Iwamoto T, Uehara A, Kita S. Probing ion binding sites in the Na+/Ca2+ exchanger. Ann N Y Acad Sci. 2002;976:19–30. doi: 10.1111/j.1749-6632.2002.tb04710.x. [DOI] [PubMed] [Google Scholar]
- 18.Doering AE, et al. Topology of a functionally important region of the cardiac Na+/Ca2+ exchanger. J Biol Chem. 1998;273(2):778–783. doi: 10.1074/jbc.273.2.778. [DOI] [PubMed] [Google Scholar]
- 19.Kang KJ, Kinjo TG, Szerencsei RT, Schnetkamp PP. Residues contributing to the Ca2+ and K+ binding pocket of the NCKX2 Na+/Ca2+-K+ exchanger. J Biol Chem. 2005;280(8):6823–6833. doi: 10.1074/jbc.M407933200. [DOI] [PubMed] [Google Scholar]
- 20.Kang KJ, Shibukawa Y, Szerencsei RT, Schnetkamp PP. Substitution of a single residue, Asp575, renders the NCKX2 K+-dependent Na+/Ca2+ exchanger independent of K+ J Biol Chem. 2005;280(8):6834–6839. doi: 10.1074/jbc.M412933200. [DOI] [PubMed] [Google Scholar]
- 21.Kamiya T, Maeshima M. Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J Biol Chem. 2004;279(1):812–819. doi: 10.1074/jbc.M309726200. [DOI] [PubMed] [Google Scholar]
- 22.Liao J, et al. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335(6069):686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 23.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 24.Forrest LR. Structural biology. (Pseudo-)symmetrical transport. Science. 2013;339(6118):399–401. doi: 10.1126/science.1228465. [DOI] [PubMed] [Google Scholar]
- 25.Besserer GM, Nicoll DA, Abramson J, Philipson KD. Characterization and purification of a Na+/Ca2+ exchanger from an archaebacterium. J Biol Chem. 2012;287(11):8652–8659. doi: 10.1074/jbc.M111.331280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nayal M, Di Cera E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J Mol Biol. 1996;256(2):228–234. doi: 10.1006/jmbi.1996.0081. [DOI] [PubMed] [Google Scholar]
- 27.Faraldo-Gómez JD, Forrest LR. Modeling and simulation of ion-coupled and ATP-driven membrane proteins. Curr Opin Struct Biol. 2011;21(2):173–179. doi: 10.1016/j.sbi.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Zdravkovic I, Zhao C, Lev B, Cuervo JE, Noskov SY. Atomistic models of ion and solute transport by the sodium-dependent secondary active transporters. Biochim Biophys Acta. 2012;1818(2):337–347. doi: 10.1016/j.bbamem.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 29.Waight AB, et al. Structural basis for alternating access of a eukaryotic calcium/proton exchanger. Nature. 2013;499(7456):107–110. doi: 10.1038/nature12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almagor L, et al. Functional asymmetry of bidirectional Ca2+-movements in an archaeal sodium-calcium exchanger (NCX_Mj) Cell Calcium. 2014;56(4):276–284. doi: 10.1016/j.ceca.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Khananshvili D, Weil-Maslansky E, Baazov D. Kinetics and mechanism: Modulation of ion transport in the cardiac sarcolemma sodium-calcium exchanger by protons, monovalent, ions, and temperature. Ann N Y Acad Sci. 1996;779:217–235. doi: 10.1111/j.1749-6632.1996.tb44789.x. [DOI] [PubMed] [Google Scholar]
- 32.Khananshvili D, Shaulov G, Weil-Maslansky E. Rate-limiting mechanisms of exchange reactions in the cardiac sarcolemma Na+-Ca2+ exchanger. Biochemistry. 1995;34(32):10290–10297. doi: 10.1021/bi00032a024. [DOI] [PubMed] [Google Scholar]
- 33.Baazov D, Wang X, Khananshvili D. Time-resolved monitoring of electrogenic Na+-Ca2+ exchange in the isolated cardiac sarcolemma vesicles by using a rapid-response fluorescent probe. Biochemistry. 1999;38(5):1435–1445. doi: 10.1021/bi981429u. [DOI] [PubMed] [Google Scholar]
- 34.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 36.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 37.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996;24(4):433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 39.Staritzbichler R, Anselmi C, Forrest LR, Faraldo-Gómez JD. GRIFFIN: A versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. J Chem Theory Comput. 2011;7(4):1167–1176. doi: 10.1021/ct100576m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.