Significance
Pod dehiscence is a critical step in the seed dispersal (shattering) of legume and crucifer crops and can cause significant yield losses. Upon drying, pod walls are dehisced by two factors: the reduction of pod-wall binding strength and the generation of dehiscing forces. Although the previously reported shattering-resistant mutants maintained binding strength, here, we show a gene regulating the dehiscing force. The gene, Pdh1, encodes a dirigent family protein, known to be involved in lignification, which increases dehiscing forces by promoting torsion of dried pod walls. The loss-of-function pdh1 gene has been widely used as a shattering-resistance gene in soybean breeding. This knowledge could be useful in improving other legume and crucifer crops, as well as soybean breeding.
Keywords: pod dehiscence, map-based cloning, QTL, dirigent protein, crop improvement
Abstract
Pod dehiscence (shattering) is essential for the propagation of wild plant species bearing seeds in pods but is a major cause of yield loss in legume and crucifer crops. Although natural genetic variation in pod dehiscence has been, and will be, useful for plant breeding, little is known about the molecular genetic basis of shattering resistance in crops. Therefore, we performed map-based cloning to unveil a major quantitative trait locus (QTL) controlling pod dehiscence in soybean. Fine mapping and complementation testing revealed that the QTL encodes a dirigent-like protein, designated as Pdh1. The gene for the shattering-resistant genotype, pdh1, was defective, having a premature stop codon. The functional gene, Pdh1, was highly expressed in the lignin-rich inner sclerenchyma of pod walls, especially at the stage of initiation in lignin deposition. Comparisons of near-isogenic lines indicated that Pdh1 promotes pod dehiscence by increasing the torsion of dried pod walls, which serves as a driving force for pod dehiscence under low humidity. A survey of soybean germplasm revealed that pdh1 was frequently detected in landraces from semiarid regions and has been extensively used for breeding in North America, the world’s leading soybean producer. These findings point to a new mechanism for pod dehiscence involving the dirigent protein family and suggest that pdh1 has played a crucial role in the global expansion of soybean cultivation. Furthermore, the orthologs of pdh1, or genes with the same role, will possibly be useful for crop improvement.
Seed dispersal is an essential process in many wild plants, providing their progeny with adequate space for growth and opportunities for survival under different environmental conditions. Some plant species use external vectors for seed dispersal, such as animals, wind and, water. Other plant species, such as Impatiens capensis (1), Impatiens glandulifera (2), and Cardamine hirsuta (3), have evolved reproductive organs that enable them to scatter seeds spontaneously after maturity. In domesticated crops, however, spontaneous seed dispersal, or seed shattering, causes significant yield losses (Fig. S1A). Shattering resistance, thus, has been preferentially selected during domestication as the single most important domestication trait (4).
In plant species having pod-fruit types, such as legumes and crucifers, pod dehiscence is the critical step in seed dispersal. In general, a pod forms abscission layers at the binding sites of its walls (valves) and accumulates the force to dehisce pod walls upon drying during and after maturation. When the dehiscing force exceeds the binding strength of the pod walls, the pod dehisces and seeds are dispersed. In Arabidopsis thaliana, several shattering-resistant mutants have been isolated, and the associated genes and mechanisms have been identified (5–10). In shattering-resistant (SR) mutants, the formation of abscission layers between valves and the replum is inhibited by two factors: defects in transcriptional factors regulating fruit patterning (6–9), or secondary cell wall formation (10), and the absence of polygalacturonase that degrades pectin, an adhesive polysaccharide binding the walls of adjacent cells (5). These shattering-resistance genes all increase the binding strength of abscission layers.
The cultivated soybean [Glycine max Merr. (L.)] is more resistant than the wild soybean (Glycine soja Sieb. et Zucc.) to shattering. Genetic analysis using a mapping population derived from a cross between these two species has not identified any quantitative trait loci (QTLs) with large effects on shattering, suggesting that multiple genes with minor effects contribute to shattering resistance in the cultivated species (11). A gene responsible for domestication, SHAT1-5 on chromosome 16, has very recently been identified (12). This gene, which is homologous to NST1/2 of A. thaliana, activates secondary cell-wall biosynthesis and promotes the thickening of fiber-cap cells in pod sutures (12), the dehiscence site in soybean pods. SHAT1-5 also enhances pod-wall binding strength.
Genetic variation in the degree of pod dehiscence is also present in cultivated soybean cultivars (13). Although shattering-susceptible (SS) cultivars are more shattering-resistant than wild soybeans, such cultivars are not suitable for harvesting under dry conditions. This problem with SS cultivars is particularly true in the case of mechanical harvesting (13), which is indispensable to large-scale cultivation. In contrast to results obtained from interspecific genetic analysis, a major QTL for pod dehiscence has been identified in the cultivated species on chromosome 16 (14–17). An anatomical analysis using near-isogenic lines (NILs) for this QTL, designated as qPDH1, has revealed no marked differences in suture morphology, including that of secondary cell-wall formation (18) (Fig. S1B). Furthermore, qPDH1 mapping has delimited this QTL to a 134-kb genomic region lacking candidate genes homologous to the Arabidopsis genes associated with pod dehiscence (19). These facts suggest the involvement of, at least, a novel gene and mechanism in the regulation of pod dehiscence associated with qPDH1.
Here, we report the map-based cloning of qPDH1. We demonstrate that qPDH1 encodes a dirigent-like protein that regulates dehiscing force, the torsion of pod walls under low humidity. We also provide evidence for the widespread distribution of the shattering-resistance allele at this locus in semiarid regions, including China and North America.
Results
Effects of qPDH1 on Pod Dehiscence and Pod-Wall Torsion Under Low Humidity.
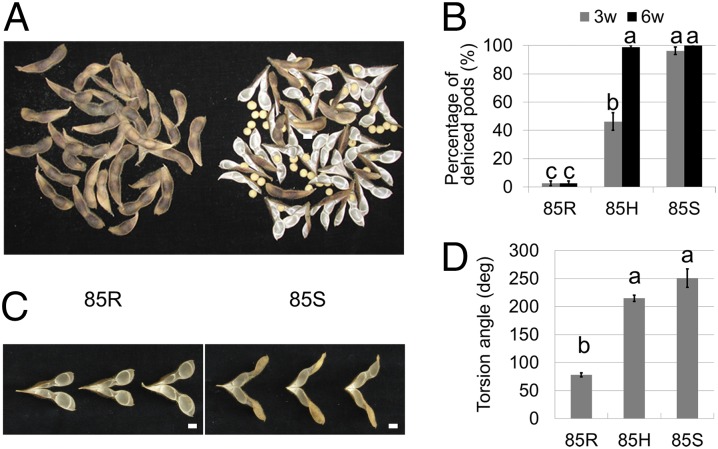
Fully mature pods of two NILs for qPDH1—line 85R carrying the shattering-resistance allele and line 85S carrying the shattering-susceptibility allele—showed contrasting degrees of pod dehiscence under low humidity (Fig. 1 A and B). The heterozygous genotype 85H displayed an intermediate level of pod dehiscence after 3 wk under dry conditions. After 6 wk, however, its level was almost identical to that of 85S (Fig. 1B). An analysis of variance (ANOVA) revealed significant differences among genotypes and between stages, and also a significant interaction effect of genotype × stage. These results indicate that the shattering-susceptibility allele at qPDH1 exhibits at least partial dominance.
Fig. 1.
Pod dehiscence and pod-wall torsion of near-isogenic cultivated soybean [Glycine max Merr. (L.)] lines for qPDH1 under low-humidity conditions. (Scale bars: 10 mm.) (A) Dried pods of the shattering-resistant (SR) line 85R (Left) and the shattering-susceptible (SS) line 85S (Right) at ambient humidity [∼40% relative humidity (RH)]. (B) Percentages of dehisced pods of 85R, 85H, and 85S after drying for 3 wk and 6 wk at 30% RH (mean ± SE; n = 8). The 85H indicates HC1-85H, the parental line of 85R and 85S with the heterozygous genotype at qPDH1. Different letters denote significant differences (P < 0.001). (C) Pod walls of 85R (Left) and 85S (Right) at 22% RH, dehisced after natural drying. (D) Torsion angles of dehisced pod walls of 85R, 85H, and 85S at 30% RH (mean ± SE; n = 6). Different letters denote significant differences (P < 0.001).
Because qPDH1 was unlikely to be associated with binding strength, we focused on the effect of this QTL on dehiscing force. Similar to mung bean (20) and bittercress (3), the dehiscing force in soybean is assumed to be associated with the coiling habit of pod walls upon drying. Under dry conditions, pod walls shrink and curl in a vertical plane perpendicular to the axis of fiber direction (21) (Fig. S1C), analogous to wood (22) and bittercress (3). Dehydration over the threshold leads to pod dehiscence when sutures dig inward (Fig. S1C). Because fiber and pod axes cross at an angle, this curling results in twisting, or spiral coiling, of pod walls after dehiscence (21). We accordingly measured the torsion of dried pod walls to compare dehiscing forces of NILs. At a low relative humidity, dehisced pod walls of 85R exhibited much lower degrees of torsion than those of 85S (Fig. 1 C and D), suggesting that the dehiscing force of 85R is weaker than that of 85S.
Map-Based Cloning of qPDH1.
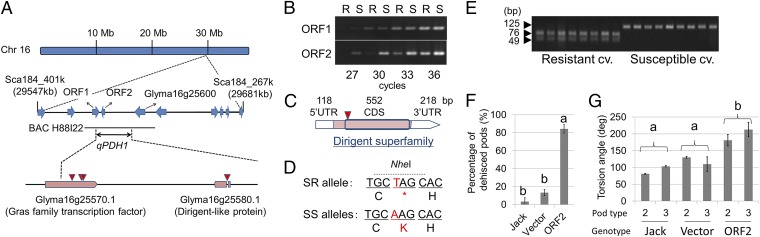
To fine-map qPDH1, we screened a large segregating population (∼2,500 plants) derived from HC1-85H, a line heterozygous only in the genomic region surrounding qPDH1. Seven plants showed recombination between the two DNA markers flanking the 134-kb genomic region. Progeny testing narrowed the candidate region to 20 kb (Fig. 2A and Fig. S2 A and B). An insertion/deletion (Indel) polymorphism in Glyma16g25600, located outside of the 20-kb region, has been speculated to be associated with shattering resistance (23). In lines in the present study, however, percentages of dehisced pods were not necessarily correlated with gap presence or absence (Fig. S2C), clearly indicating that this polymorphism is unrelated to qPDH1-associated shattering resistance.
Fig. 2.
Map-based cloning of cultivated soybean [Glycine max Merr. (L.)] qPDH1. (A) Predicted ORFs in the previously determined candidate region of chromosome 16 (blue arrows) and predicted ORFs in the region delimited in the present study (pink boxes). BAC H88I22 indicates a BAC clone carrying the qPDH1 locus of a shattering-susceptible (SS) cultivar, Misuzudaizu. Red triangles indicate positions of single nucleotide polymorphisms (SNPs) between Hayahikari [shattering-resistant (SR)] and Toyomusume (SS). (B) Results from semiquantitative RT-PCR for transcripts of ORF1 and ORF2 in pod walls of near-isogenic lines for qPDH1. R and S indicate lines 85R and 85S, respectively. (C) cDNA structure of ORF2 of Toyomusume. (D) DNA and deduced amino acid sequences around the SNP. Red letters indicate the SNP and the resulting amino acid residue or termination signal. The SNP can be recognized by a restriction enzyme, Nhe I. (E) PCR–RFLP genotyping of SR and SS cultivars at the SNP using Nhe I. SR cultivars (leftmost lanes) and SS cultivars (rightmost lanes) are listed in SI Materials and Methods. In these cultivars, the presence of an SR or SS allele at qPDH1 on chromosome 16 was suggested in this study or previous studies (14, 15, 17, 48, 56). (F) Percentages of pod dehiscence of an SR cultivar, Jack, plants, which were nontransformed, transformed only with the vector, or transformed with ORF2 from Toyomusume, at 30% RH (mean ± SE; n = 3, 2, and 8). Different letters indicate significant differences (P < 0.001). Although nontransformed Jack plants were grown in a different growth chamber, pod dehiscence was simultaneously monitored in the same chamber. (G) Torsion angles of dehisced pod walls of two-seeded (bar 2) or three-seeded (bar 3) pods of nontransformed Jack plants, Jack plants transformed only with the vector, or Jack plants transformed with ORF2 from Toyomusume at 30% RH (mean ± SE; n = 3, 3, 2, 2, 8, and 6). Different letters indicate significant genotypic differences (P < 0.05), detected by two-way ANOVA.
Using the Phytozome soybean genome sequence database (24) (www.phytozome.net/soybean), all or part of ORFs ORF1 and ORF2 were predicted to be present in the 20-kb region. Because this database was constructed using an SR cultivar, Williams 82, an inserted fragment sufficiently large enough to contain other ORFs could be present in the SS genotypes. A bacterial artificial chromosome (BAC) clone containing both flanking sequences of the 20-kb region was accordingly isolated from a library constructed using the Japanese SS cultivar Misuzudaizu. Although we found Indel variations up to 29 nucleotides long as well as single-nucleotide polymorphisms (SNPs) in this clone, no large insertion sequence was detected in the region of interest. As in Williams 82, only ORF1 and ORF2 were predicted to be present in the genomic region of the SS cultivar.
Sequencing of these ORFs in the two HC1-85H parental cultivars, Hayahikari (SR) and Toyomusume (SS), revealed the presence of three SNPs in the ORF1 coding region, which were associated with one missense mutation and two synonymous substitutions, and one SNP in the predicted intron of ORF2 (Fig. 2A). The ORF1 missense mutation was not shared between the two SS cultivars, Toyomusume and Misuzudaizu (Fig. S3A), indicating no involvement of this SNP in shattering. In contrast, the genotype of the ORF2 SNP in Misuzudaizu was identical to that of Toyomusume. Additionally, expression analyses revealed differential transcript abundance between 85R and 85S for ORF2 but not ORF1 (Fig. 2B and Fig. S3 B and C).
To further characterize ORF2, we determined the full-length cDNA sequence of ORF2 of Toyomusume. ORF2 contained no introns and encoded a protein of 183 amino acids (Fig. 1C and Fig. S3D). Consequently, the SNP site in Hayahikari is responsible for a nonsense variant at the 31st amino acid residue (Fig. 2 C and D), indicating a defect in ORF2 in the SR genotype. The reduced transcript expression observed in 85R may be explained by nonsense-mediated mRNA decay resulting from the premature stop codon (25). This nonsense variant is commonly found in SR cultivars (Fig. 2E). These results strongly suggest that ORF2 is identical to qPDH1.
To confirm that ORF2 is qPDH1, we performed a complementation test. We introduced a 3.4-kb genomic fragment containing the 5′ and 3′ sequences of ORF2 from the SS cultivar Toyomusume into an SR cultivar, Jack (Fig. S4A). The sequence of the ORF2 of Jack was completely identical to that of Hayahikari. Moreover, a QTL analysis supported the hypothesis that Jack carried a shattering-resistance allele at qPDH1 (Fig. S4 B and C). Transformants with the SS allele from Toyomusume (Fig. S4 D and E) exhibited significantly higher degrees of pod dehiscence and torsion than control plants (Fig. 2 F and G). These results demonstrated that ORF2 is indeed qPDH1. We therefore designated the gene as Pod dehiscence 1 (Pdh1).
Predicted Amino Acid Sequence of Pdh1.
BLAST search analysis against the GenBank Conserved Domain Database v3.12 (www.ncbi.nlm.nih.gov/cdd/) indicated that the protein encoded by Pdh1 is a member of the dirigent (DIR) superfamily (Fig. 2C). This DIR superfamily contains a number of proteins whose expression levels are induced during plant-disease response and lignification. According to the classification of Ralph et al. (26), the Pdh1-encoded protein belongs to DIR protein group a, which includes DIR(-like) proteins from Forsythia × intermedia (27) and pea (28) (Fig. S5).
Pdh1 Gene Expression.
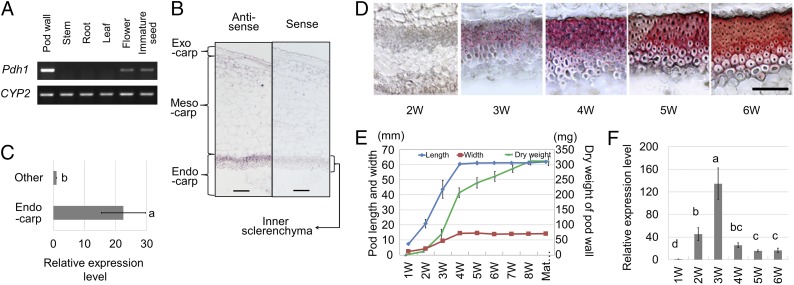
Analysis of Pdh1 expression in Toyomusume revealed an abundance of transcripts in pod walls, but only traces in flowers and immature seeds, and none in leaf, stem, or root tissues (Fig. 3A). In situ hybridization experiments indicated that the transcripts were abundant in the inner sclerenchyma of pod walls, the site of thick secondary cell-wall formation (Fig. 3B). Transcript levels in the pod-wall endocarp, which mainly consists of inner sclerenchyma tissue, were ∼22 times higher than those in the exocarp and mesocarp (Fig. 3C). Pdh1 expression levels also varied over the course of pod development. The highest level of expression occurred 3 wk after anthesis (Fig. 3F), when pod walls vigorously increased in dry weight and began to deposit lignin in the inner sclerenchyma (Fig. 3 D and E). These results indicate that the tissue-specific and developmental regulation of Pdh1 expression is similar to other DIR genes (29, 30).
Fig. 3.
Cultivated soybean [Glycine max Merr. (L.)] Pdh1 expression patterns. (Scale bars: 100 μm.) (A) Results of RT-PCR targeting Pdh1 transcripts in several tissues. CYP2 corresponds to cyclophilin 2, used as a reference. (B) In situ hybridization for Pdh1 with an RNA probe in the antisense direction (Anti-sense) and with the probe in the sense direction (Sense) to detect nonspecific binding. (C) Relative expression levels of Pdh1 in the endocarp and the remaining portion of 5-wk-old pod walls detected by quantitative RT-PCR with reference to CYP2. (D) Cross-sections of inner parenchyma of 2- to 6-wk-old pod walls stained with phloroglucinol-HCl to reveal the degree of lignification. (Scale bar: 100 μm.) (E) Changes in pod length, pod width, and pod-wall dry weight during pod growth. (F) Relative expression levels of Pdh1 in 1- to 6-wk-old pod walls (means ± SE; n = 3). Different letters indicate significant differences (P < 0.05) between tissues (C) or among stages (F). 1W–8W correspond to the number of weeks after anthesis. Mat indicates maturity.
Distribution of the Pdh1 Genotype in Soybean.
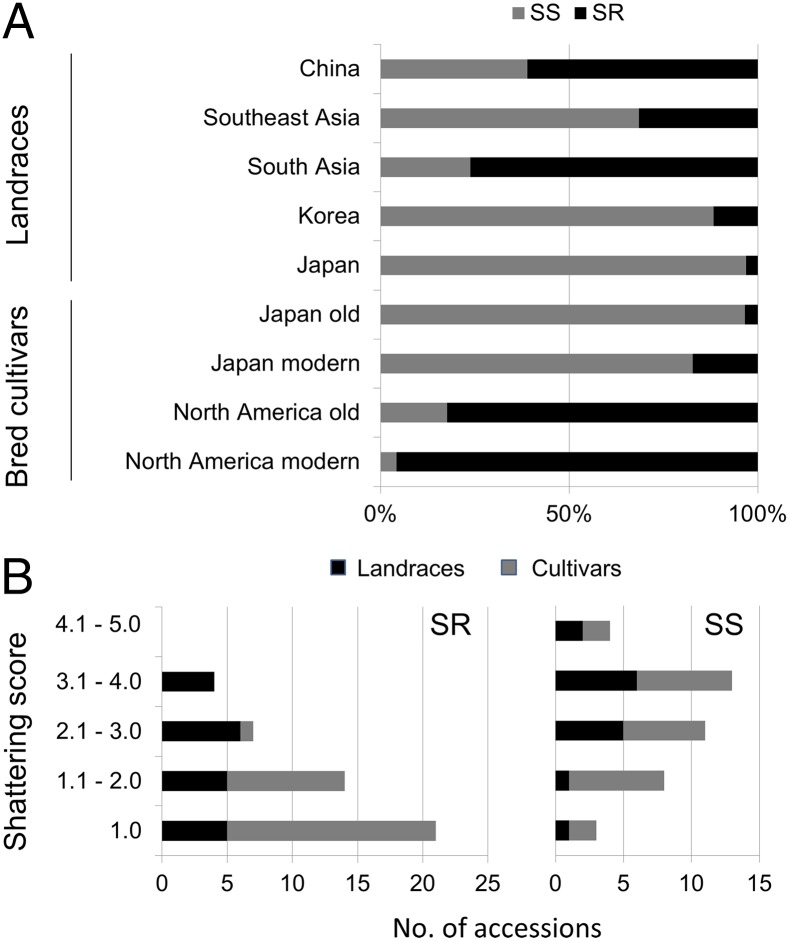
To assess the distribution of the shattering-resistance gene pdh1, we surveyed soybean germplasm in a mini-core collection established by Kaga et al. (31). This collection currently consists of 79 Japanese and 80 non-Japanese accessions, mostly landraces, and maximizes genetic diversity with its small population size (31). Sequencing of the Pdh1 coding region of the 159 active accessions in this collection revealed no other genotypes besides those of Hayahikari, Toyomusume, and Misuzudaizu, with the exception of a heterozygous genotype between Hayahikari and Toyomusume that is most likely the product of outcrossing (Table S1). Of the non-Japanese landraces, more than half of the Chinese landraces possessed the pdh1 allele (Fig. 4A). A few landraces from South Korea and Southeast Asia also contained pdh1 whereas a large proportion of South Asian accessions harbored this allele. We found only two accessions containing pdh1 among the Japanese landraces. We also examined accessions of old and modern soybean cultivars of North America and Japan for the SNP responsible for shattering resistance. Additionally, we tested East Asian landraces used in a previous study, which were collected mainly from China and considered to be the ancestors of North American cultivars (32). We found the SR genotype in 62% of the Chinese landraces and 20% of the Korean and Japanese landraces (Table S2), in agreement with the values estimated from the mini-core collection. The SR genotype was found in 80% of 17 old North American cultivars (Fig. 4) that have been reported to account for 86% of the collective parentage of North American soybean cultivars released between 1947 and 1988 (33). Among modern elite North American cultivars released between 1977 and 1990, only one possessed the SS genotype. In contrast, only one old Japanese cultivar carried the SR genotype. Although the SR genotype was somewhat more common in recently developed cultivars, more than 80% of modern Japanese cultivars still possessed the SS genotype. Only the SS genotype was detected in wild soybean G. soja accessions (Table S2).
Fig. 4.
Genotypes for the single-nucleotide polymorphism (SNP) of Pdh1 and shattering degrees of accessions in soybean germplasms. (A) Proportion of Pdh1/pdh1 genotypes in various soybean germplasm pools: landraces from East, Southeast, and South Asian countries from a mini-core collection, old Japanese cultivars, modern Japanese cultivars, old North American cultivars, and modern North American cultivars. Old and modern North American cultivars correspond, respectively, to “ancestral” and “elite” North American cultivars in a previous study by Hyten et al. (32). Gray and black bars indicate percentages of accessions featuring SS and SR genotypes, respectively, at the SNP. The accessions used are listed in Tables S1 and S2. (B) Histograms of accessions with shattering scores, grouped by the shattering-susceptible (SS) or the shattering-resistant (SR) genotype at the Pdh1locus. The score increases with shattering degree as observed 2 wk after harvest (www.ars-grin.gov/). Accessions with “SHATLATE” values deposited in the GRIN database were analyzed.
To confirm the effect of the Pdh1 genotype on the shattering resistance of the accessions, we analyzed shattering scores deposited in the Germplasm Resources Information Network database (www.ars-grin.gov/npgs) (Table S2). The accessions with pdh1 had significantly lower shattering scores (P < 0.01) than those with Pdh1, even if only the landrace data were used (Fig. 4B). The genotypic effect of Pdh1/pdh1 was validated among Japanese modern cultivars as well (P < 0.01). When East Asian landraces with pdh1 were compared with old and modern North American cultivars with pdh1, the North American cultivars had significantly lower shattering scores, suggesting the use of additional shattering-resistance genes in breeding programs.
Discussion
The mechanisms and genes involved in pod dehiscence have been studied extensively in the model plant A. thaliana, with the importance of abscission-layer formation highlighted as a result. This accomplishment may be attributed to the relatively weak dehiscing force associated with pods of A. thaliana (3), as binding strength should play a more prominent role in such cases. In contrast, the pods of many other plant species have evolved to generate a strong dehiscing force (1–3) that should have a more important contribution than that found in A. thaliana. Various studies have investigated physical mechanisms, but genetic approaches have not been exploited. Our map-based cloning study using soybean has revealed important aspects of pod dehiscence, namely, the dehiscing force and the associated regulatory gene.
The identified gene, Pdh1, encodes a dirigent (DIR)-like protein. Davin et al. (34) first found the effect of a DIR protein on in vitro stereoselective bimolecular phenoxy radical coupling of a monolignol, (E)-coniferyl alcohol, which they noted in a woody plant, F. suspensa. The DIR protein led to the production of (+)-pinoresinol, a type of lignan, instead of a mixture of three racemic dimeric compounds (dilignols), including (±)-pinoresinol. The mediation of stereoselective coupling with DIR proteins has also been reported in moco cotton (35) and Arabidopsis (36, 37). Despite the presence of many genes encoding this protein family in vascular plants (26, 38), their only suggested role in planta was related to disease resistance (28, 39) until Hosmani et al. (40) reported the essential role of a dirigent domain-containing protein, ESB1, in the formation of lignin-based Casparian strips in roots. Our study has revealed a function of the DIR superfamily, the regulation of pod dehiscence.
The dehiscing force in C. hirsuta, or the pod-coiling habit, is considered to be generated by the second endocarp layer with its strongly asymmetrical cell-wall thickening (3). In the pod walls of this plant, the second endocarp layer has the thickest cell walls of any tissues. As seen in Fig. S1D, in soybean, the inner sclerenchyma should be the determinant of pod-wall shape because it possesses the highest cell density and the thickest cell walls of relevant plant tissues. Additionally, the difference in cell-wall thickness between the upper and lower layers also corroborates that the inner sclerenchyma plays a similar role to the second endocarp layer in C. hirsuta. The specific expression of Pdh1 in the inner sclerenchyma is consistent with the fact that Pdh1/pdh1 regulates the magnitude of dehiscing force. Taken together, Pdh1 may promote pod dehiscence by influencing the physical properties of the inner sclerenchyma. An alternative hypothesis, in which Pdh1 simply promotes dehydration of pod walls, can be excluded because no difference was observed in the pod-wall moisture content between Pdh1 genotypes (Fig. S6A).
The physical properties of dried pod walls are probably controlled by the high-molecular weight compounds constituting the cell walls. Although Pdh1 shows a high homology to DIR proteins belonging to group a, some of which have been reported to be involved in lignan biosynthesis, low-molecular weight compounds such as lignans are not likely able to influence physical properties. The most likely candidate of the cell-wall component associated with Pdh1 is lignin. Observed gene-expression patterns also suggest an association between Pdh1 and lignin deposition. Unlike a DIR gene reported in cotton (39), however, Pdh1 did not seem to significantly affect lignin content (Fig. S6B). Based on previous discussions (27, 29, 41), Pdh1 may regulate lignin primary structure. Alternatively, Pdh1 might affect the pattern of lignin deposition, being broadly similar to ESB1 (40), which also has little impact on lignin content (42).
Soybean is believed to have been domesticated from G. soja 3,000–5,000 y ago in China (33), with its cultivation subsequently spreading to surrounding Asian countries, including Japan (43). During domestication, soybean probably acquired several shattering-resistance genes, including SHAT1-5, to reach the minimum level of shattering resistance for a crop. The high Pdh1 frequency in landraces of Japan, Korea, and Southeast Asian countries suggests that the minimum shattering-resistance level was sufficient as long as manually harvested in a humid climate. However, more than half of the analyzed Chinese landraces were found to possess pdh1. A possible explanation is that cultivars containing pdh1 were developed later and were distributed throughout China by farmers, who found the shattering-resistance trait indispensable for cultivation in the relatively dry climate. This explanation is consistent with our observation of the high pdh1 allele frequency in the landraces of South Asia, considering the dry climate of this area, and that Chinese germplasm had a relatively late introduction to South Asia (31). Because no other major QTLs that condition pod dehiscence have been reported in cultivated soybean (44), pdh1 was likely distributed to large areas as an invaluable gene resource for shattering resistance. These data suggest a crucial role of pdh1 in the dissemination of soybean cultivation in semiarid areas.
Despite a relatively late introduction in the 18th century, North America has been the world’s leading soybean production area for more than 50 y (45). The recent marked increase in soybean acreage and production in North America is associated with the development of many SR cultivars (46). Our findings demonstrate that this achievement was made possible by the extensive use of pdh1 and the use of several additional minor shattering-resistance genes in breeding programs. In addition to the priority given in breeding programs to shattering resistance, the presence of pdh1 in most ancestral lines collected from East Asia likely facilitated its rapid North American distribution. Old North American cultivars with pdh1, such as S-100, Roanoke, and Tokyo (Table S2), also contributed in large part to the soybean’s genetic base in Brazil (47), currently the world’s other leading producer. We believe that the advent and extensive use of pdh1 in the breeding programs enabled soybean cultivation in semiarid areas and on large scales, leading to the current status of soybean as the most economically important legume.
Recently, high levels of shattering resistance have become necessary in moderately humid areas, such as Japan, due to modernized agricultural systems using mechanical harvesting (13). However, our results indicate that the presence of pdh1 is not common in modern Japanese cultivars. The near absence of pdh1 from Japanese landraces has obviously led to the low pdh1 frequency. The identification of the gene and the development of DNA markers will accelerate the use of pdh1 in breeding programs for areas with a low pdh1 frequency. For instance, pdh1 can be used to produce SR lines with various genetic backgrounds by the combined use of backcrossing and marker-assisted selection (48).
The prevention of pods from shattering is also a promising strategy for increasing the yields in some other crops, such as Brassica napus and Lotus corniculatus (49). To date, however, no agronomic achievement has been reported based on the use of the Arabidopsis shattering-resistance genes identified for fruit patterning. For instance, the introduction of the FRUITFULL gain-of-function gene, which dominantly suppresses the formation of abscission layers, to Brassica juncea made pods resistant to threshing (50). Although fine-tuning the INDEHISCENT gene expression has been proposed for crop improvement (51), the use of the orthologous gene for pdh1, or other genes regulating the dehiscing force, may provide promising alternatives because pdh1 has long proven its usefulness in agriculture.
In conclusion, our work provides insights into the mechanisms underlying pod dehiscence. Although the biochemical mechanisms remain to be shown, a biologically important function has been uncovered for the DIR protein family. We have also demonstrated the critical role of pdh1 in the worldwide expansion of soybean cultivation. Furthermore, the identified gene and the DNA marker developed in this study should be useful for the breeding of crop plants subject to pod dehiscence, including soybean, other legumes, and possibly crucifers.
Materials and Methods
Details about materials and methods are provided in SI Materials and Methods.
Plant Materials.
Soybean seeds were provided by the Hokkaido Research Organization (HRO), the National Agricultural Research Organization (NARO), the National Institute of Agrobiological Sciences (NIAS), the US Department of Agriculture, and soybean-breeding laboratories in Japan as described in Tables S1 and S2. A population of 2,535 self-pollinated progeny of HC1-85H were used for fine mapping of the qPDH1 locus. A set of F4:5 recombinant inbred lines was derived from a cross between Toyomusume and Jack for QTL analysis. Plants were grown in growth chambers or in the field.
Characterization of Pod Walls.
The pod dehiscence percentage was evaluated at low humidity [30% relative humidity (RH)] or by heat treatment. Pod-wall torsion angles were measured at low humidity with a newly developed instrument and image analysis system using two-seeded pods for 85R, 85H, and 85S plants or two- and three-seeded pods for Jack and transgenic plants. Cross-sections were stained with phloroglucinol-HCl before microscopic observation. The moisture content of each sample was determined based on fresh (or air-dried) and oven-dried weights. Lignin content was obtained from the sum of acid-insoluble (Klason) lignin and acid-soluble lignin contents.
BAC Screening.
BAC libraries constructed using total DNA of the SS soybean cultivar Misuzudaizu after partial digestion with BglII or HindIII were used for screening.
Genomic PCR, RT-PCR, Real-time PCR, 3′ and 5′ Rapid Amplification of cDNA Ends, in Situ hybridization, DNA Sequencing, and SNP Genotyping.
DNA and RNA isolation, PCR, in situ hybridization, and DNA sequencing were performed by commonly used methods. The PCR-restriction fragment length polymorphism (RFLP) method with the restriction enzyme Nhe I or “tetra-primer ARMS-PCR” (52) (Fig. S3E) was used for genotyping the soybean accessions for the SNP in Pdh1. Primer sequences are listed in Table S3.
Plant Transformation.
A biolistic transformation was performed using cultured cells derived from immature embryos of the cultivar Jack as described by Nishizawa et al. (53).
Bioinformatic Analyses.
The conserved domains search tool (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and MEGA6 program (54) were used for alignment and phylogenetic analysis of the Pdh1 protein, respectively. Windows Cartographer 2.5 (55) was used for QTL analysis.
Supplementary Material
Acknowledgments
We thank the US Department of Agriculture Agricultural Research Service, the Hokkaido Research Organization, and Nagano Prefecture for providing soybean accessions; A. Kaga for providing total DNA of the National Institute of Agrobiological Sciences Japanese and world soybean core collections; R. Narita, S. Furuhata, R. Sugisawa, Y. Nakamoto, H. Kashihara, and Y. Yokota for technical assistance; and T. Suzuki, K. Oono, and M. Kuwahara for helpful discussions. This study was supported in part by Ministry of Agriculture, Forestry and Fisheries, Japan “Research and Development Projects for Application in Promoting New Policy of Agriculture Forestry and Fisheries” Grant 18038 (to H.F.) and “Genomics for Agricultural Innovation” Grant GMZ-1004 (to M.I.) and Japan Society for the Promotion of Science KAKENHI Grant 24580017 (to K.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database {DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank accession nos. AB823548 (Pdh1 full-length cDNA, cv. Toyomusume); AB826447 (3.4-kb Pdh1 genomic sequence of cv. Toyomusume transferred to cv. Jack); AB823549 (pdh1 sequence corresponding to coding sequence, cv. Hayahikari); AB823550 (Pdh1 coding sequence, cv. Misuzudaizu); AB823551 [ORF1 (Glyma16g25570.1), coding sequence, cv. Toyomusume]; AB823552 (ORF1 [Glyma16g25570.1], coding sequence, cv. Hayahikari); and AB823553 (partial sequence of BAC clone H88I22, cv. Misuzudaizu)}.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417282111/-/DCSupplemental.
References
- 1.Hayashi M, Feilich KL, Ellerby DJ. The mechanics of explosive seed dispersal in orange jewelweed (Impatiens capensis) J Exp Bot. 2009;60(7):2045–2053. doi: 10.1093/jxb/erp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deegan RD. Finessing the fracture energy barrier in ballistic seed dispersal. Proc Natl Acad Sci USA. 2012;109(14):5166–5169. doi: 10.1073/pnas.1119737109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaughn KC, Bowling AJ, Ruel KJ. The mechanism for explosive seed dispersal in Cardamine hirsuta (Brassicaceae) Am J Bot. 2011;98(8):1276–1285. doi: 10.3732/ajb.1000374. [DOI] [PubMed] [Google Scholar]
- 4.Fuller DQ. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann Bot (Lond) 2007;100(5):903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogawa M, Kay P, Wilson S, Swain SM. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell. 2009;21(1):216–233. doi: 10.1105/tpc.108.063768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrándiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289(5478):436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- 7.Liljegren SJ, et al. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404(6779):766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- 8.Rajani S, Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol. 2001;11(24):1914–1922. doi: 10.1016/s0960-9822(01)00593-0. [DOI] [PubMed] [Google Scholar]
- 9.Liljegren SJ, et al. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116(6):843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- 10.Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2008;56(5):768–778. doi: 10.1111/j.1365-313X.2008.03633.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, et al. QTL mapping of domestication-related traits in soybean (Glycine max) Ann Bot (Lond) 2007;100(5):1027–1038. doi: 10.1093/aob/mcm149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong Y, et al. Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun. 2014;5:3352. doi: 10.1038/ncomms4352. [DOI] [PubMed] [Google Scholar]
- 13.Tsuchiya T. Hysiological and genetic-analysis of pod shattering in soybeans. Jarq-Japan Agric Res Q. 1987;21(3):166–175. [Google Scholar]
- 14.Bailey MA, Mian MAR, Carter TE, Ashley DA, Boerma HR. Pod dehiscence of soybean: Identification of quantitative trait loci. J Hered. 1997;88(2):152–154. [Google Scholar]
- 15.Funatsuki H, et al. Simple sequence repeat markers linked to a major QTL controlling pod shattering in soybean. Plant Breed. 2006;125(2):195–197. [Google Scholar]
- 16.Kang ST, et al. Population-specific QTLs and their different epistatic interactions for pod dehiscence in soybean. Euphytica. 2009;166(1):15–24. [Google Scholar]
- 17.Yamada T, et al. A major QTL, qPDH1, is commonly involved in shattering resistance of soybean cultivars. Breed Sci. 2009;59(4):435–440. [Google Scholar]
- 18.Suzuki M, Fujino K, Funatsuki H. A major soybean QTL, qPDH1, controls pod dehiscence without marked morphological change. Plant Prod Sci. 2009;12(2):217–223. [Google Scholar]
- 19.Suzuki M, Fujino K, Nakamoto Y, Ishimoto M, Funatsuki H. Fine mapping and development of DNA markers for the qPDH1 locus associated with pod dehiscence in soybean. Mol Breed. 2010;25(3):407–418. [Google Scholar]
- 20.Isemura T, et al. Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata) PLoS ONE. 2012;7(8):e41304. doi: 10.1371/journal.pone.0041304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson JB, Lersten NR. 2004. Reproductive morphology. Soybeans: Improvement, Production, and Uses, Agronomy, eds Boema HR, Specht JE (Am Soc Agron-Crop Sci Soc Am-Soil Sci Soc Am, Madison, WI), No. 16, 3rd Ed, pp 59–95.
- 22.Cockrell RT. A comparison of latewood pits, fibril orientation, and shrinkage of normal and compression wood of Giant Sequoia. Wood Sci Technol. 1974;8:197–206. [Google Scholar]
- 23.Gao M, Zhu H. Fine mapping of a major quantitative trait locus that regulates pod shattering in soybean. Mol Breed. 2013;32:485–491. [Google Scholar]
- 24.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 25.Mühlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779(9):538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Ralph SG, Jancsik S, Bohlmann J. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.) Phytochemistry. 2007;68(14):1975–1991. doi: 10.1016/j.phytochem.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 27.Gang DR, et al. Regiochemical control of monolignol radical coupling: A new paradigm for lignin and lignan biosynthesis. Chem Biol. 1999;6(3):143–151. doi: 10.1016/S1074-5521(99)89006-1. [DOI] [PubMed] [Google Scholar]
- 28.Wang YP, Nowak G, Culley D, Hadwiger LA, Fristensky B. Constitutive expression of pea defense gene DRR206 confers resistance to blackleg (Leptosphaeria maculans) disease in transgenic canola (Brassica napus) Mol Plant Microbe Interact. 1999;12:410–418. [Google Scholar]
- 29.Burlat V, Kwon M, Davin LB, Lewis NG. Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry. 2001;57(6):883–897. doi: 10.1016/s0031-9422(01)00117-0. [DOI] [PubMed] [Google Scholar]
- 30.Casu RE, et al. Identification of differentially expressed transcripts from maturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Mol Biol. 2004;54(4):503–517. doi: 10.1023/B:PLAN.0000038255.96128.41. [DOI] [PubMed] [Google Scholar]
- 31.Kaga A, et al. Evaluation of soybean germplasm conserved in NIAS genebank and development of mini core collections. Breed Sci. 2012;61(5):566–592. doi: 10.1270/jsbbs.61.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyten DL, et al. Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA. 2006;103(45):16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.TE C, et al. 2004. Genetic diversity in soybean. Soybeans: Improvement, Production, and Uses, Agronomy, eds Boema HR, Specht JE (Am Soc Agron-Crop Sci Soc Am-Soil Sci Soc Am, Madison, WI), No. 16, 3rd Ed, pp 303–416.
- 34.Davin LB, et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science. 1997;275(5298):362–366. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Stipanovic RD, Bell AA, Puckhaber LS, Magill CW. Stereoselective coupling of hemigossypol to form (+)-gossypol in moco cotton is mediated by a dirigent protein. Phytochemistry. 2008;69(18):3038–3042. doi: 10.1016/j.phytochem.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Pickel B, et al. An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew Chem Int Ed Engl. 2010;49(1):202–204. doi: 10.1002/anie.200904622. [DOI] [PubMed] [Google Scholar]
- 37.Kim KW, et al. Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra species. J Biol Chem. 2012;287(41):33957–33972. doi: 10.1074/jbc.M112.387423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ralph S, Park JY, Bohlmann J, Mansfield SD. Dirigent proteins in conifer defense: Gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.) Plant Mol Biol. 2006;60(1):21–40. doi: 10.1007/s11103-005-2226-y. [DOI] [PubMed] [Google Scholar]
- 39.Shi H, et al. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim Biophys Sin (Shanghai) 2012;44(7):555–564. doi: 10.1093/abbs/gms035. [DOI] [PubMed] [Google Scholar]
- 40.Hosmani PS, et al. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci USA. 2013;110(35):14498–14503. doi: 10.1073/pnas.1308412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davin LB, Lewis NG. Lignin primary structures and dirigent sites. Curr Opin Biotechnol. 2005;16(4):407–415. doi: 10.1016/j.copbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Baxter I, et al. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 2009;5(5):e1000492. doi: 10.1371/journal.pgen.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hymowitz T, Kaizuma N. Dissemination of soybeans (Glycine max): Seed protein electrophoresis profiles among Japanese cultivars. Econ Bot. 1979;33(3):311–319. [Google Scholar]
- 44.Funatsuki H, et al. Mapping and use of QTLs controlling pod dehiscence in soybean. Breed Sci. 2012;61(5):554–558. doi: 10.1270/jsbbs.61.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hymowitz T. On the domestication of the soybean. Econ Bot. 1970;24(4):408–421. [Google Scholar]
- 46.Caviness CE. Effects of relative humidity on pod dehiscence in soybeans. Crop Sci. 1965;5(6):511–513. [Google Scholar]
- 47.Wysmierski PT, Vello NA. The genetic base of Brazilian soybean cultivars: Evolution over time and breeding implications. Genet Mol Biol. 2013;36(4):547–555. doi: 10.1590/S1415-47572013005000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada T, et al. Production of soybean new line by back-crossing method and DNA marker assisted selection for shattering resistance and maturity loci. Bull NARO Inst Crop Sci. 2013;14:13–22. [Google Scholar]
- 49.Bennett EJ, Roberts JA, Wagstaff C. The role of the pod in seed development: Strategies for manipulating yield. New Phytol. 2011;190(4):838–853. doi: 10.1111/j.1469-8137.2011.03714.x. [DOI] [PubMed] [Google Scholar]
- 50.Østergaard L, Kempin SA, Bies D, Klee HJ, Yanofsky MF. Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol J. 2006;4(1):45–51. doi: 10.1111/j.1467-7652.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- 51.Girin T, et al. Brassicaceae INDEHISCENT genes specify valve margin cell fate and repress replum formation. Plant J. 2010;63(2):329–338. doi: 10.1111/j.1365-313X.2010.04244.x. [DOI] [PubMed] [Google Scholar]
- 52.Chiapparino E, Lee D, Donini P. Genotyping single nucleotide polymorphisms in barley by tetra-primer ARMS-PCR. Genome. 2004;47(2):414–420. doi: 10.1139/g03-130. [DOI] [PubMed] [Google Scholar]
- 53.Nishizawa K, Kita Y, Kitayama M, Ishimoto M. A red fluorescent protein, DsRed2, as a visual reporter for transient expression and stable transformation in soybean. Plant Cell Rep. 2006;25(12):1355–1361. doi: 10.1007/s00299-006-0210-x. [DOI] [PubMed] [Google Scholar]
- 54.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, Basten CJ, Zeng Z-B. Windows QTL Cartographer 2.5. North Carolina State University, Raleigh; NC: 2012. [Google Scholar]
- 56.Funatsuki H, et al. Confirmation of the location and the effects of a major QTL controlling pod dehiscence, qPDH1, in soybean. Breed Sci. 2008;58(1):63–69. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.