Significance
All organisms regulate biological processes in response to changes in their environment. Bacteria often achieve this control via two-component signal transduction pathways, which use histidine kinases to perceive environmental signals and relay this information to downstream effectors. Despite substantial efforts, key aspects of the mechanisms by which histidine kinases are activated by these signals remain poorly understood. In this paper, we present structural and functional data that shed light on the signaling strategy used by a monomeric histidine kinase. Our results demonstrate the versatility of histidine kinases by expanding the prevailing view that they must form dimers to function, while also highlighting conserved aspects of their signaling strategies.
Keywords: two-component system, cell signaling, histidine kinase, photosensory, regulation
Abstract
Although histidine kinases (HKs) are critical sensors of external stimuli in prokaryotes, the mechanisms by which their sensor domains control enzymatic activity remain unclear. Here, we report the full-length structure of a blue light-activated HK from Erythrobacter litoralis HTCC2594 (EL346) and the results of biochemical and biophysical studies that explain how it is activated by light. Contrary to the standard view that signaling occurs within HK dimers, EL346 functions as a monomer. Its structure reveals that the light–oxygen–voltage (LOV) sensor domain both controls kinase activity and prevents dimerization by binding one side of a dimerization/histidine phosphotransfer-like (DHpL) domain. The DHpL domain also contacts the catalytic/ATP-binding (CA) domain, keeping EL346 in an inhibited conformation in the dark. Upon light stimulation, interdomain interactions weaken to facilitate activation. Our data suggest that the LOV domain controls kinase activity by affecting the stability of the DHpL/CA interface, releasing the CA domain from an inhibited conformation upon photoactivation. We suggest parallels between EL346 and dimeric HKs, with sensor-induced movements in the DHp similarly remodeling the DHp/CA interface as part of activation.
Prokaryotes primarily respond to environmental cues with two-component signaling systems, minimally composed of a sensor histidine kinase (HK) and a response regulator (RR) (1). The HK autophosphorylates at a conserved histidine residue, a reaction that is regulated by stimuli specific to each HK. The resulting phosphoryl group is subsequently transferred to the downstream RR, modulating its activity. Most simply, HKs consist of a sensor domain and a kinase core composed of dimerization/histidine phosphotransfer (DHp) and catalytic/ATP-binding (CA) domains. Despite the biological importance of HKs to bacterial signaling and as potential antibiotic targets (2), the mechanisms by which sensors control autophosphorylation are not well known due to complications in obtaining high-resolution structures of full-length HKs. A key premise among most signaling models is that HKs must form dimers to function, consistent with the oligomerization state of HKs studied to date (3–6). Here, we present data that challenge this tenet with the full-length structure of EL346, a blue light-sensing monomeric HK from the HPK11 subfamily (7). Together with biochemical and biophysical analyses of signal-induced conformational changes, we demonstrate how conformational changes originating in light–oxygen–voltage (LOV) and Per-ARNT-Sim (PAS) domains can propagate to control the activity of an effector. Our findings highlight general principles governing HK regulation independently of oligomerization state.
Results
EL346 Is Light-Activated and Monomeric.
EL346 was initially described in a survey of bacterial HKs containing light–oxygen–voltage (LOV) domains, demonstrating that several of these proteins exhibited light-enhanced kinase activities as anticipated by the presence of the photosensory LOV domains (8, 9). We confirmed that EL346 undergoes the expected LOV photocycle with spontaneous postillumination reversion with a time constant of ∼55 min in the ATP-free state, which accelerates to 42 and 32 min upon binding to ATP and its nonhydrolyzable analog adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP), implying some overall conformational change upon nucleotide binding (Fig. S1A). In addition to undergoing LOV photochemical changes upon illumination, the lit state has higher net autophosphorylation activity, with a 1.6-fold enhancement in initial rate (Fig. 1A). Phosphotransfer to EL_LovR and EL_PhyR, two RR substrates we previously identified as EL346 targets (10), was also enhanced by blue light (5.6- and 1.6-fold increases in initial rates for EL_LovR and EL_PhyR, respectively) (Fig. S1 B and C).
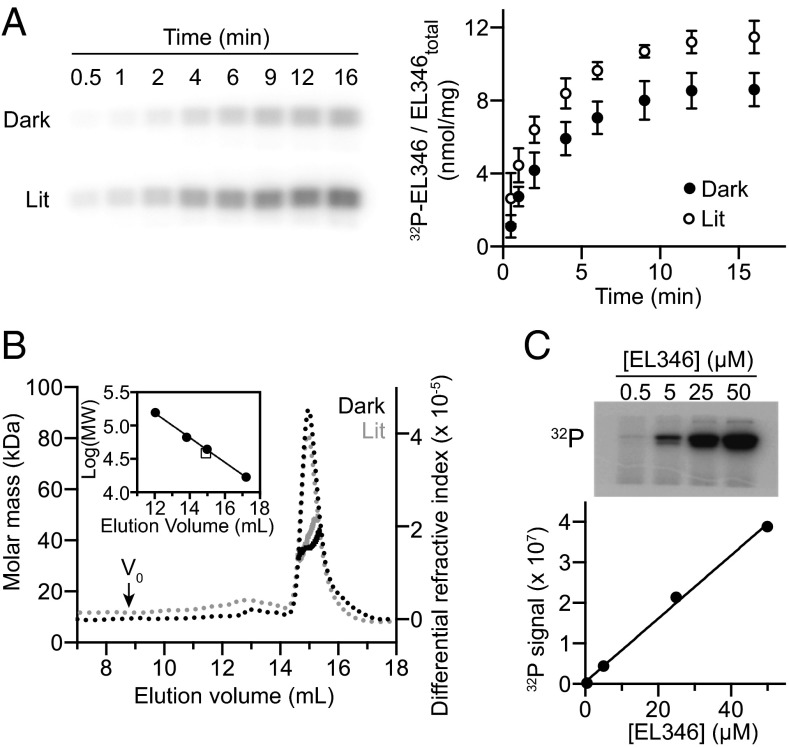
Fig. 1.
EL346 is a light-regulated, monomeric HK. (A) EL346 autokinase activity under dark and lit conditions. A representative autoradiogram is shown. Data points (dark and lit conditions with closed and open circles, respectively) represent the average ± 1 SD for n = 3 measurements. (B) Molecular weight of ATP-bound EL346 in the dark and lit states, as measured by SEC-MALS. (Inset) Correlations of elution volumes and log(MW) for EL346 and standards (158, 66.5, 44, and 17 kDa). (C) Concentration dependence of EL346 autokinase activity, showing a linear relationship between protein concentration and autokinase activity, consistent with EL346 not requiring dimerization to undergo autophosphorylation.
Sequence analyses predicted LOV, DHp (HisKA_2), and CA (HATPase_c) domains in EL346, with His142 inferred to be the phosphoacceptor histidine as predicted from the HPK11-type H-box sequence (7). We validated this assignment with data showing that an H142Q mutation eliminated autokinase activity (Fig. S1D). Despite the predicted DHp domain, which typically mediates tight dimerization between HKs, we observed that EL346 behaved as a monomer in solution using size exclusion chromatography with inline multiangle light scattering (SEC-MALS). These analyses established that ATP-bound EL346 had average solution molecular masses of 37.8 kDa and 41.8 kDa under dark and lit conditions, respectively, both consistent with a 38.6-kDa monomer (Fig. 1B). Similar values were obtained without nucleotide (Fig. S1E), indicating that EL346 is monomeric regardless of illumination or nucleotide state. These data imply that EL346 autophosphorylates in cis or, alternatively, transiently dimerizes to do so in trans. To address this key mechanistic point, we assayed autokinase activity with increasing concentrations of EL346 from 0.5 to 50 μM and found that activity increased linearly over this range (Fig. 1C). Coupling this observation with the monomeric state of EL346 in SEC-MALS experiments conducted at higher concentrations, our data indicate that autophosphorylation does not depend on the formation of a transient dimer, strongly implicating an in cis mechanism.
EL346 Adopts a Novel Arrangement of Conserved Sensor and Catalytic Domains.
To investigate the basis for light-dependent regulation of EL346 and its unexpected monomeric state, we determined this protein’s crystal structure. Crystals of full-length EL346 (EL346FL) grew in the presence of AMP-PNP and diffracted to a resolution of 2.92 Å. Because we were unsuccessful with attempts to solve the structure using LOV and HK homologs as molecular replacement models, we independently obtained coordinates for EL346 LOV (EL346LOV) and DHp/CA (EL346HK) constructs by molecular replacement and single-wavelength anomalous dispersion (SAD), respectively, and used these structures as molecular replacement models for the full-length structure (Table S1 and Fig. S2 A and B). EL346FL displays a previously unseen arrangement in which the LOV sensor and CA domains pack directly against two α-helices, α1 and α2, which resemble a single DHp domain from dimeric HKs (Fig. 2). Although crystals of EL346FL contained two molecules per asymmetric unit (Fig. S2C), the scarcity of contacts between the DHp-like (DHpL) domains from the two molecules is inconsistent with a solution-state dimer. Computational analyses of macromolecular interfaces using PISA (11) predict that EL346FL does not form stable higher order assemblies in solution, further supporting a monomeric solution state conformation. EL346FL’s DHpL does not dimerize to form a four-helix bundle as in other HK structures (6); instead, the sensor domain occludes the surface usually used for dimerization.
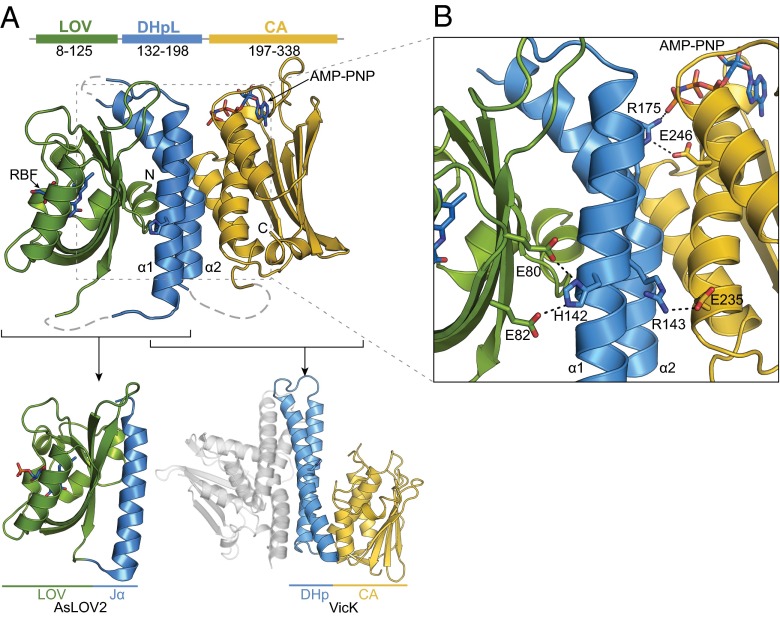
Fig. 2.
Full-length EL346 crystal structure highlights conserved features of sensor and catalytic domains. (A, Top and Middle) Domain architecture and crystal structure of EL346FL, with domains highlighted in green (LOV), blue (DHpL) and yellow (CA). The riboflavin cofactor (RBF), His142, and AMP-PNP are represented in sticks. (Bottom) Structural comparisons with AsLOV2 (PDB ID code 2V0U) (13) and residues 199–302 of VicK chain B, with chain A shown in gray (PDB ID code 4I5S) (19). (B) Detail of selected residues in the LOV/DHpL and DHpL/CA interfaces.
Our full-length structure provides insights into the EL346 photosensory process, which initiates within the N-terminal LOV domain with the formation of a reversible covalent bond between a flavin chromophore and a conserved cysteine residue. Although this domain has a standard mixed α/β fold surrounding the chromophore, we observed it binding riboflavin in lieu of more common FMN or FAD (12) in both the full-length and EL346LOV structures. Both FMN and riboflavin were detected in liquid chromatography-mass spectrometry (LC-MS) analyses of extracts of freshly purified protein samples (Fig. S3), suggesting that the EL346 might be more prone to bind riboflavin than other LOV domains, at least under overexpression conditions. A potential root of this preference is the presence of an alanine residue in the Fα helix (Ala72) instead of the conserved basic residue that often interacts directly with the FMN/FAD phosphoryl group.
Within the full-length structure, the LOV β-sheet forms a sizeable interface with the DHpL domain (>1,200 Å2) centered on a LOV hydrophobic patch that contacts the core of the DHpL alpha-hairpin (Fig. 3 A and B). Additional LOV/DHpL contacts include hydrogen bonds between LOV Glu80 and Glu82 and the phosphoacceptor His142 in the DHpL α1 helix (Fig. 2B). The EL346 LOV/DHpL α1 helix interaction is reminiscent of the arrangement seen in other LOV/helix pairs, including the well-studied AsLOV2 LOV/Jα helix interaction from the Avena sativa phototropin 1 LOV2 domain (Fig. 2A and Fig. S4A) (13, 14). In this latter case, illumination generates allosteric changes that dissociate the Jα helix to control phototropin kinase activity (14, 15), implying a comparable signaling mechanism in EL346. This parallel is reinforced by similar arrangements of the EL346 DHpL α2 and the AsLOV2 N-terminal A′α helix with their respective LOV domains, the latter of which undergoes light-induced conformational changes as well (16). Coupled with other structural similarities between LOV and PAS domains with extended elements (Fig. S4), multiple signs suggest a conserved signaling mode through the β-sheet in these domains. It is also worth noting that this LOV/DHpL interaction also seems to occlude the area known to contain the residues that mediate interaction between HK and RR (17, 18), suggesting that the LOV domain might also play a role in regulation at the level of phosphotransfer and/or phosphatase activity.
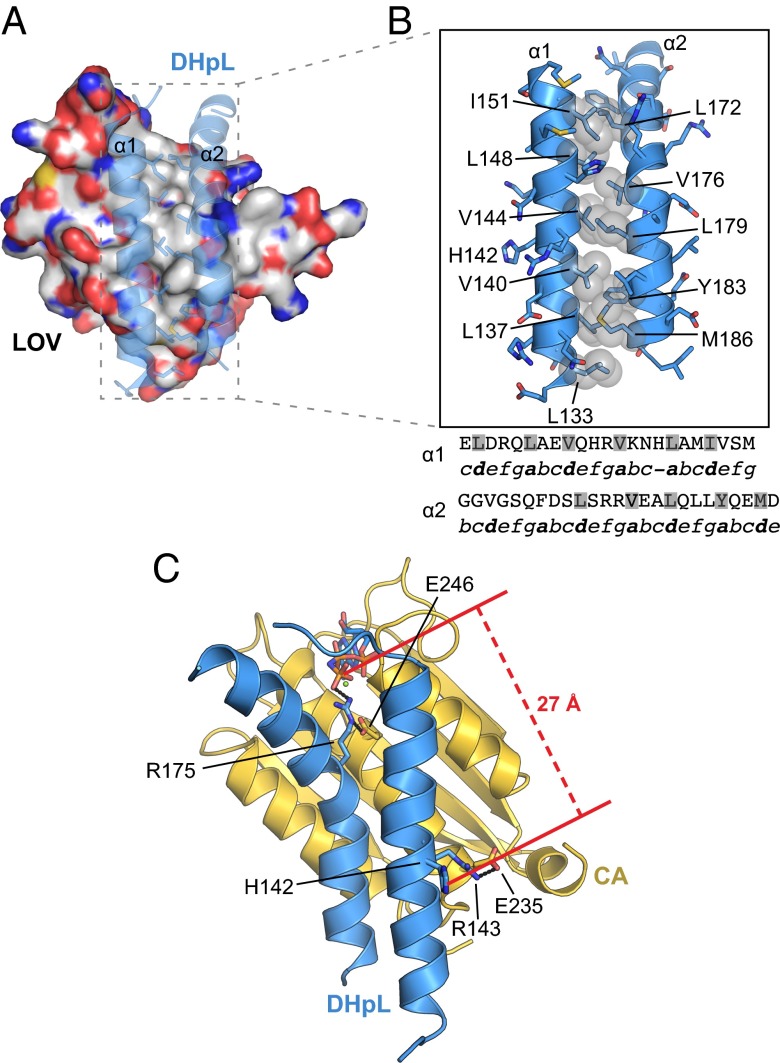
Fig. 3.
DHpL inter- and intradomain interactions in the EL346FL structure. (A) DHpL and LOV interaction. The β-sheet side of the LOV domain is shown in surface representation, and the DHpL helices are presented in transparent ribbon representation with the side chains of heptad repeat residues as sticks. (B) DHpL heptad repeat. Hydrophobic residues that pack between helices α1 and α2 are indicated as gray spheres in the structure and shaded in gray in the sequences. Positions a to g in the heptad repeats are indicated in italics under the sequences. (C) DHpL/CA arrangement in the EL346FL structure showing the location of conserved residue pairs and distance between AMP-PNP and His142.
The DHpL domain contacts the CA domain through a 910-Å2 interface, keeping the nucleotide-binding site >27 Å away from His142 (Fig. 3C). Coupling this fact with similarity of the DHp/CA contacts observed in other inactive HK structures, such as the inactive protomers of VicK (19) (Fig. 2A) and EnvZchim (20), implies that we are observing an inactive conformation. Two pairs of highly conserved ionic interactions, Arg143-Glu235 and Arg175-Glu245 (Fig. 3C and Fig. S5A), suggest that EL346 homologs share a similar DHpL/CA arrangement. Additionally, Arg175 from the DHpL α2 helix forms a salt bridge with the AMP-PNP γ-phosphate oxygen (Fig. 2B). The observation that nucleotide binding accelerates photocycle kinetics in EL346 (Fig. S1A) suggests that this interaction has substantial effects that propagate back to the photosensory LOV domain itself.
Notably, the DHpL fold in the EL346HK construct (Fig. S5B) is quite different from the one in the EL346FL structure. Instead of forming a continuous helix, α1 in the EL346HK structure forms two shorter antiparallel helices that pack at an angle against α2. Helix α2 remains in the same position relative to the CA domain, and the Arg175/AMP-PNP interaction is preserved. In the EL346FL structure, there is a bend on helix α2. This bend is caused by the interruption of the heptad repeats in helix α1 by His147 in position d and the start of another repeat with Leu148 on position a (Fig. 3B). This arrangement forces α2 to bend at Val176 so that hydrophobic side chains on positions a and d can pack against the corresponding residues on α1. This bend is not present in the EL346HK structure, and the N terminus of α2 is partially unfolded. These observations suggest that the conformation of the DHpL domain is highly dependent upon interactions with the LOV domain.
Light-Sensing by the LOV Domain Induces Protein-Wide Conformational Changes and Alters the DHpL/CA Interface.
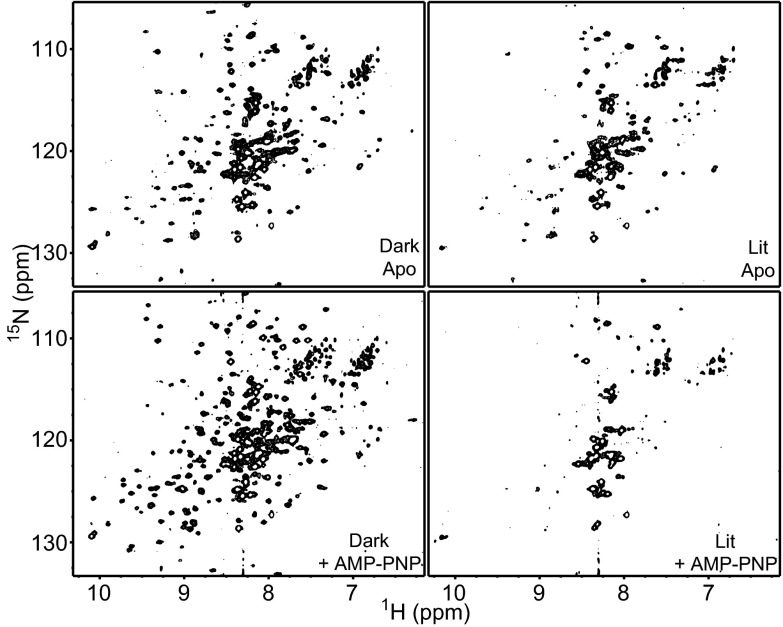
To characterize EL346 structural changes upon light activation, we used a combination of solution NMR and limited trypsin proteolysis. 15N/1H transverse relaxation-optimized spectroscopy (TROSY) spectra of dark-state EL346 demonstrated marked spectral improvement upon AMP-PNP addition, with the increasing numbers of peaks suggesting the stabilization of the structure (Fig. 4). This stabilization is consistent with nucleotide-mediated DHpL/CA interactions in our inhibited-state crystal structures, EL346FL and EL346HK (Fig. 2 and Fig. S2B). Further, we observed extensive spectral changes between dark and lit spectra (Fig. 4), especially with AMP-PNP bound, that revert to the dark state as illumination ceases (Fig. S6). These widespread changes in peak location and line width in the lit spectra are consistent with a high degree of structural disorder, indicating that the EL346 tertiary structure changes substantially in a light-dependent manner. The severe line broadening suggests that the folded domains have increased mobility in the lit state and that the protein exchanges among multiple conformations in the intermediate chemical shift exchange regime, leading to peak disappearance.
Fig. 4.
Light induces widespread conformational changes in EL346 as revealed by solution NMR spectroscopy. 15N/1H TROSY spectra of EL346, showing the effects of illumination and AMP-PNP binding.
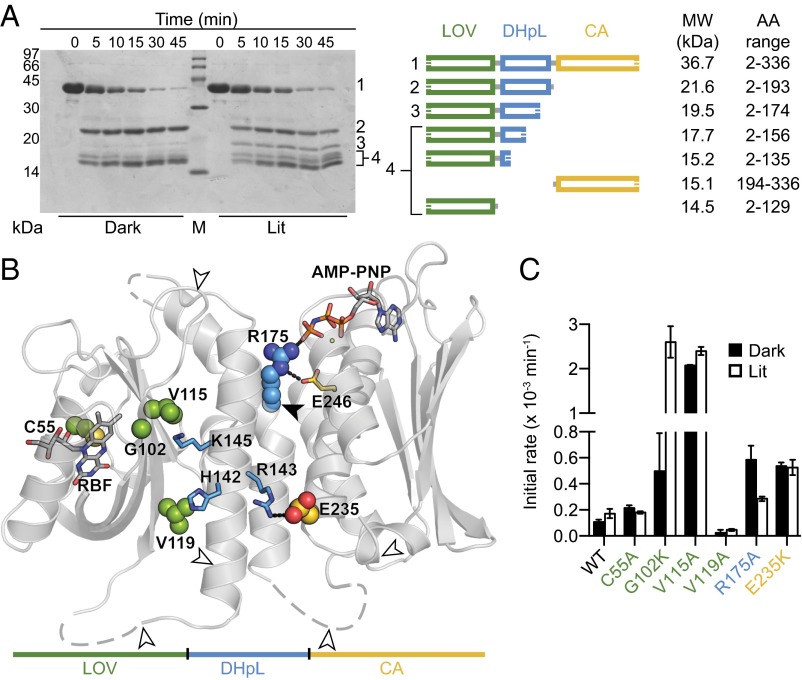
Coupled with the EL346FL crystal structure, these data suggest that light-induced changes in the LOV domain are propagated through the LOV/DHpL interface, altering the DHpL/CA interface and reorganizing the EL346 tertiary structure. To test this model, we used limited trypsinolysis of AMP-PNP–loaded EL346 under dark and lit conditions and identified the exact mass of the proteolytic fragments by mass spectrometry (Fig. 5A and Table S2). Most fragments were produced with comparable kinetics under both conditions; however, one fragment (band 3) was produced almost exclusively under lit conditions (Fig. 5A). This 19.5-kDa fragment (residues 2–174) resulted from cleavage in the middle of DHpL helix α2 at a site that is buried in the DHpL/CA interface in the dark-state crystal structure. This light-dependent cleavage is consistent with a disruption of this interface during activation, as implicated by reports suggesting that sensor domains control kinase activity by influencing DHp and CA domain orientations (19, 21, 22). Notably, controls with a C55A mutant that cannot form the photochemical protein/flavin adduct exhibited very low levels of the 2–174 fragment under lit conditions (Fig. S7A), confirming that DHpL/CA interface disruption is linked to signal propagation.
Fig. 5.
LOV domain photoactivation changes the DHpL/CA interface. (A) Limited trypsinolysis of EL346. (Left) SDS/PAGE of samples and molecular weight marker (M) reveals light-dependent changes in trypsinolysis pattern. (Right) Schematic of fragments corresponding to SDS/PAGE bands, using masses from electrospray ionization mass spectrometry (ESI‐MS) analysis. (B) Locations of residues selected for site-directed mutagenesis (spheres colored by domain assignment, shown on diagram representation of EL346). His142, riboflavin (RBF), and AMP-PNP are shown in sticks, as are additional residues that interact with those that were mutated. Arrowheads indicate light-independent (white) and light-dependent (black) cleavage sites observed by limited trypsinolysis. (C) Initial rate of autophosphorylation of mutants under lit and dark conditions. Assays were performed at least in duplicate, and their results are shown as the average ± 1 SD. A C55A mutant was used as a negative control for blue-light activation.
Mutations at the Domain Interfaces Affect Signaling and Autokinase Activity.
To validate the signal-propagation model outlined above, we examined the functional and structural effects of point mutations designed to disrupt interdomain interfaces within EL346. We probed the LOV/DHpL interface with G102K, V115A, and V119A mutations, finding that autokinase levels increased above WT for G102K and V115A (Fig. 5 B and C). Notably, whereas G102K retained light responsiveness, the V115A mutation seemed to decouple sensor function by generating a near-constitutive enzyme. We suspect these differences may be due in part to the location of Val115 at the center of the LOV/DHpL interface whereas Gly102 is at its periphery. On the other hand, V119A’s drastically reduced activity and lack of 2–174 fragment after limited trypsinolysis (Fig. 5C and Fig. S7B) suggest that this mutant is strongly inhibited under dark and lit conditions. Contrarily, G102K and V115A showed a strong band for this fragment after limited trypsinolysis under both conditions (Fig. S7B). We conclude that, whereas V119A stabilizes the inhibited state, G102K and V115A destabilize it, resulting in high dark-state activity. Notably, residues Val115 and Val119 are located on the same Iβ-strand as Gln118, a conserved glutamine that changes its hydrogen bonding pattern with the flavin chromophore upon illumination (23–25), playing a key role in signaling (26, 27). Propagation of light-induced structural changes in Gln118 along the Iβ strand could contribute to the drastic impairment of EL346 regulation when the Val115 and Val119 sidechains were truncated.
We also tested R175A, E235K, R143A, and E246A mutations at the DHpL/CA interface (Fig. 5 B and C). R175A removes hydrogen bonds between the DHpL and AMP-PNP and Glu246 in the CA. Remarkably, this mutation reversed the signaling logic, doubling the initial phosphorylation rate in the dark compared with the light. Mutation E235K in the CA domain, which eliminates the ionic interaction with DHpL residue Arg143, exhibited constitutively high autokinase activity, suggesting that the Arg143–Glu235 interaction stabilizes the inhibited conformation observed in the crystal. These enhanced dark-state activities agree with the increased levels of 2–174 fragment in R175A and E235K limited trypsinolysis assays (Fig. S7C). R143A and E246A mutations diminished autokinase activity to levels below those measurable by our assays. Based on the recently reported structures of the autophosphorylating Michaelis complex (20, 28), we predict that Glu246 acts as the catalytic base whereas Arg143 helps position it through hydrogen bonding in the active state, making both residues essential for catalysis. Although we cannot precisely determine what specific changes are induced in the DHpL domain without detailed structural information on activated EL346, we can attempt to mimic the active site by superposing the individual DHpL and CA domains from the EL346FL structure onto the autophosphorylation-competent chain of the Enzchim structure (Fig. S8). In this model, EL346 Glu246 and Arg143 occupy the same location as the Enzchim catalytic base (Asp244) and helper residue (Asn343), respectively (20). The location of these residues is a conserved feature in HPK11-type HKs (7), strongly suggesting that the catalytic mechanism in the HPK11 family is similar to that proposed for other HK groups regardless of HK subclass or oligomerization state. Collectively, our results support the idea that interdomain interactions in EL346 hold the CA domain in an inhibited state; weakening them by mutations or light-dependent conformational changes leads to apparent activation.
Discussion
An open and fundamental question in two-component signaling is how HKs are activated by stimuli. Several studies have proposed mechanisms to explain how signals are propagated between the sensory and catalytic domains (19, 22, 29), typically suggesting stimulus-induced translation, rotation, or bending of helices within the intervening DHp four-helix bundle. However, it has been difficult to experimentally demonstrate a connection between signal reception by the sensor and the requisite conformational rearrangements between the DHp and CA domains. To address this issue, we investigated the inhibited-state structure of a full-length monomeric HK and the conformational changes that occur during its activation. The EL346 structure reveals a compact fold in which the DHpL domain, two helices analogous to the DHp in dimeric HKs, directly interacts with the LOV domain β-sheet, preventing dimerization. We note that the unusual monomeric nature of EL346 is not found in all LOV-HKs because several HKs with the same domain composition are established dimers (10, 22, 30). However, we anticipate that additional monomeric HKs will be found among other LOV-HK and PAS-HK proteins, given the tendency of both LOV and PAS domains to pack against helices in a manner analogous to EL346, facilitating competition between sensor:DHp (monomeric) and DHp:DHp (dimeric) interactions. We suspect additional determinants of monomer:dimer balance to be within the DHpL domain sequence; however, direct testing of this prospect was hindered by an inability to express and purify stable DHpL-only constructs for characterization.
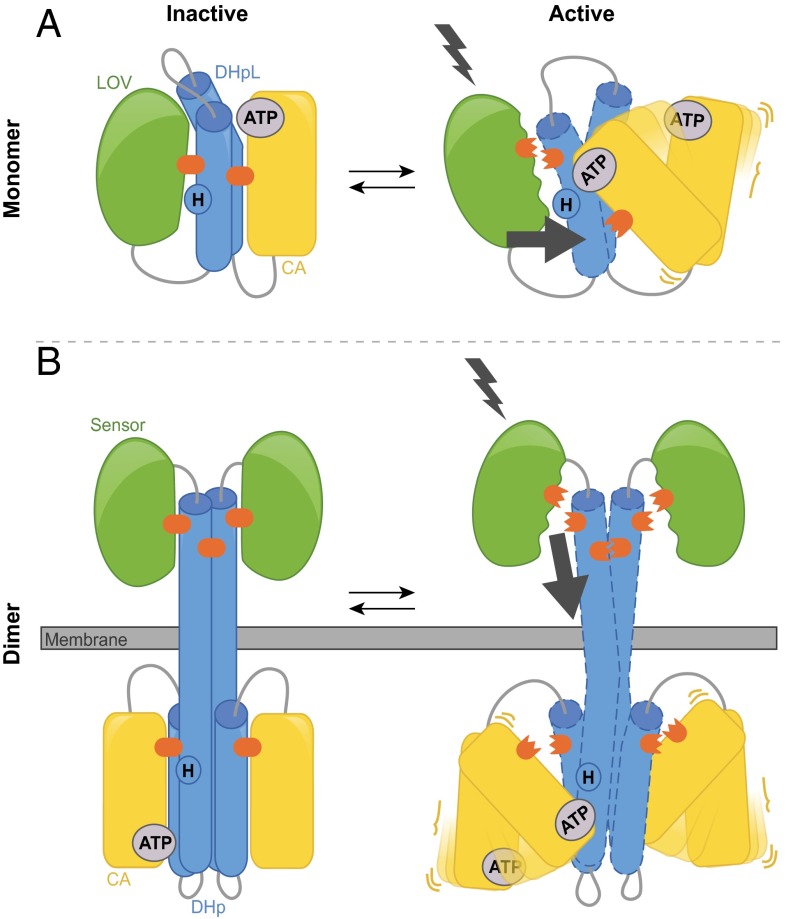
Our biochemical and biophysical data support an activation model for EL346 (Fig. 6A) in which light-induced conformational changes originating in the LOV domain propagate through the β-sheet to the DHpL domain via interactions between the two domains. This remodeling of interdomain interactions perturbs the DHpL helices—likely by some combination of motions as proposed for dimeric HKs (19, 21, 22, 29)—destabilizing the DHpL/CA interface observed in our crystal structures and breaking inhibitory contacts. This release of inhibitory contacts enables CA domain movement, allowing the ATP binding site and phosphoacceptor histidine to interact. Although our findings prove that signal propagation can be achieved in a monomeric context, they also provide insights into signaling within the much larger group of dimeric HK enzymes. The mechanism proposed for EL346 supports these overarching principles governing HK regulation, regardless of oligomeric state and cellular localization: (i) Changes in the sensor are propagated via movements of the DHp (or DHpL) helices and (ii) these movements result in structural remodeling of the interface with the CA, leading to changes in HK activity. Previous studies have shown that signaling relies on DHp movements (31) and that sensors often use helical connectors to affect DHp structure, dynamics, and flexibility at a distance (22, 30, 32, 33). Diverse experimental probes, ranging from limited proteolysis (30) to HDX-MS (31), have correlated changes in DHp structure and accessibility (caused by environmental change or mutation) to HK activity. Studies in other HK systems have found that altering interdomain interactions will impair signal transmission and even reverse its logic, as observed with the R175A mutation in EL346 (34, 35). Combining this evidence with our findings, we propose that DHp movements elicit the release of inhibitory interactions analogous to those observed to sequester the CA domain in EL346’s crystal structure (as evidenced by our limited trypsinolysis data), thus allowing the rearrangement of CA and DHp domains (Fig. 6B). This model is supported by several dimeric HK structures that show that the CA domain can adopt different arrangements with respect to the DHp domain, demonstrating the plasticity of this interface (28, 36). Although signaling relies on direct contact between the LOV β-sheet and the DHpL domain for EL346, we envision that, in dimeric HKs, it depends on sensors acting at a distance, modulating the movement of DHp helices from the two protein chains relative to each other.
Fig. 6.
Parallels between monomeric and dimeric HK activation models. (A) In the dark state, extensive interactions between domains (orange) hold the monomeric HK EL346 in an inhibited conformation. Photoactivation of the LOV domain alters LOV/DHpL interactions, leading to signal propagation (black arrow). These changes disturb critical DHpL/CA interactions, releasing the CA domain to move from its inhibited conformation to phosphorylate His142. (B) Analogous activation model for a membrane-bound dimeric HK. Here, sensors at a distance modulate DHp structure. Signal propagation through the DHp prompts the release of inhibitory interactions between DHp and CA.
Taken together, our results demonstrate that the sensor domains of some HKs can influence DHp/DHpL structure through direct binding as opposed to acting at a distance. We believe that additional examples of such interactions will become clear as further full-length HK structures become available, both in dimeric and monomeric contexts. More broadly, this model supports the potential for other protein and small-molecule factors to modulate HK activity by directly binding DHp domains. Such a model has been proposed for the Sda and KipI antikinases based on low-resolution structural data of their complexes with the DHp domain of the dimeric KinA kinase (37, 38). Our model provides an unanticipated line of support for this allosteric signaling mechanism, showing that the binding of proteins directly to the DHp/DHpL can affect the conformation and dynamics of this important element—possibly providing a route for cytosolic or membrane associated factors to influence transmembrane HKs (39). As such, we suggest that the mechanism acquired through evolution by EL346 and these antikinases will provide insights into regulatory processes that both inform our understanding of natural HK regulation while guiding the design of novel allosterically controlled recombinant proteins and new antibacterial therapies for research and therapeutic applications.
Materials and Methods
Full-length EL346, LOV, and DHpL/CA were expressed in Escherichia coli BL21 (DE3) cells and purified by affinity and size-exclusion chromatography. SEC-MALS analyses were performed with 50 μM EL346 on a Superdex 200 10/300 SEC column (GE Healthcare) as described previously (10). Crystals were grown at 20 °C by vapor diffusion. X-ray diffraction data were collected at beamline 19-ID at Argonne National Laboratory (Table S1). The structures of EL346LOV and EL346HK used as models in molecular replacement for the full-length structure were solved by molecular replacement and SeMet single-wavelength anomalous dispersion (SAD), respectively. NMR experiments were performed at 25 °C on a cryoprobe-equipped Varian Inova 800 MHz spectrometer. Lit-state NMR samples were exposed to blue-light pulses during data collection via a fiber optic. Kinase activity was assayed using γ-[32P]ATP and detected by autoradiography. Detailed procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Bogomolni, T. Swartz, G. Amarasinghe, M. Cobb, L. Rice, A. Stock, V. Ocasio, and other members of the K.H.G. laboratory for input; and Agilent Technologies for MS data on full-length EL346. This work was supported by NIH Grants R01 GM081875 and R01 GM106239, and Robert A. Welch Foundation Grant I-1424. The structures in this report are derived from work performed on beamline 19-ID at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research (Contract DE-AC02-06CH11357).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB), www.rcsb.org/pdb (PDB ID codes 4R3A, 4R38, and 4R39).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413983111/-/DCSupplemental.
References
- 1.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26(6):369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Matsushita M, Janda KD. Histidine kinases as targets for new antimicrobial agents. Bioorg Med Chem. 2002;10(4):855–867. doi: 10.1016/s0968-0896(01)00355-8. [DOI] [PubMed] [Google Scholar]
- 3.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96(1):131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 4.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24(24):4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surette MG, et al. Dimerization is required for the activity of the protein histidine kinase CheA that mediates signal transduction in bacterial chemotaxis. J Biol Chem. 1996;271(2):939–945. doi: 10.1074/jbc.271.2.939. [DOI] [PubMed] [Google Scholar]
- 6.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41(1962):139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 8.Swartz TE, et al. Blue-light-activated histidine kinases: Two-component sensors in bacteria. Science. 2007;317(5841):1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 9.Taylor BL, Zhulin IB. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63(2):479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correa F, Ko WH, Ocasio V, Bogomolni RA, Gardner KH. Blue light regulated two-component systems: Enzymatic and functional analyses of light-oxygen-voltage (LOV)-histidine kinases and downstream response regulators. Biochemistry. 2013;52(27):4656–4666. doi: 10.1021/bi400617y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9(10):713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46(49):14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 14.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301(5639):1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 15.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43(51):16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 16.Zayner JP, Antoniou C, Sosnick TR. The amino-terminal helix modulates light-activated conformational changes in AsLOV2. J Mol Biol. 2012;419(1–2):61–74. doi: 10.1016/j.jmb.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139(2):325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 18.Yamada S, et al. The signaling pathway in histidine kinase and the response regulator complex revealed by X-ray crystallography and solution scattering. J Mol Biol. 2006;362(1):123–139. doi: 10.1016/j.jmb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, et al. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 2013;11(2):e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casino P, Miguel-Romero L, Marina A. Visualizing autophosphorylation in histidine kinases. Nat Commun. 2014;5:3258. doi: 10.1038/ncomms4258. [DOI] [PubMed] [Google Scholar]
- 21.Dago AE, et al. Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc Natl Acad Sci USA. 2012;109(26):E1733–E1742. doi: 10.1073/pnas.1201301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diensthuber RP, Bommer M, Gleichmann T, Möglich A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure. 2013;21(7):1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14(5):1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaidya AT, Chen CH, Dunlap JC, Loros JJ, Crane BR. Structure of a light-activated LOV protein dimer that regulates transcription. Sci Signal. 2011;4(184):ra50. doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Möglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373(1):112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nash AI, Ko WH, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47(52):13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J Biol Chem. 2007;282(9):6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 28.Mechaly AE, Sassoon N, Betton J-M, Alzari PM. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 2014;12(1):e1001776. doi: 10.1371/journal.pbio.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17(2):190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell EB, McDonald CA, Palfey BA, Crosson S. An analysis of the solution structure and signaling mechanism of LovK, a sensor histidine kinase integrating light and redox signals. Biochemistry. 2010;49(31):6761–6770. doi: 10.1021/bi1006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31(11):2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takala H, et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509(7499):245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Möglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385(5):1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Etzkorn M, et al. Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat Struct Mol Biol. 2008;15(10):1031–1039. doi: 10.1038/nsmb.1493. [DOI] [PubMed] [Google Scholar]
- 35.Gleichmann T, Diensthuber RP, Möglich A. Charting the signal trajectory in a light-oxygen-voltage photoreceptor by random mutagenesis and covariance analysis. J Biol Chem. 2013;288(41):29345–29355. doi: 10.1074/jbc.M113.506139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albanesi D, et al. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci USA. 2009;106(38):16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacques DA, et al. Histidine kinase regulation by a cyclophilin-like inhibitor. J Mol Biol. 2008;384(2):422–435. doi: 10.1016/j.jmb.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Whitten AE, et al. The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J Mol Biol. 2007;368(2):407–420. doi: 10.1016/j.jmb.2007.01.064. [DOI] [PubMed] [Google Scholar]
- 39.Mascher T. Bacterial (intramembrane-sensing) histidine kinases: signal transfer rather than stimulus perception. Trends Microbiol. 2014;22(10):559–565. doi: 10.1016/j.tim.2014.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.