Significance
Myeloproliferative neoplasms (MPN) are chronic hematopoietic disorders characterized by clonal proliferation of mature myeloid elements. A subset of MPNs transforms to acute myeloid leukemia (AML). The mechanisms and pathways that contribute to transformation from MPN to AML have not been well delineated. We have characterized the somatic mutational spectrum of post-MPN AML and demonstrate that somatic tumor protein 53 (TP53) mutations are common in JAK2V617F-mutant, post-MPN AML but not in chronic-phase MPN. We demonstrate that expression of JAK2V617F combined with Tp53 loss in a murine model leads to fully penetrant AML in vivo. We have characterized this model and used it to test therapeutic strategies. These data reveal novel insights into the pathogenesis of, and potential therapeutic strategies for, leukemic transformation.
Keywords: leukemia, genetics, myeloproliferative neoplasm, cancer biology, targeted therapy
Abstract
Patients with myeloproliferative neoplasms (MPNs) are at significant, cumulative risk of leukemic transformation to acute myeloid leukemia (AML), which is associated with adverse clinical outcome and resistance to standard AML therapies. We performed genomic profiling of post-MPN AML samples; these studies demonstrate somatic tumor protein 53 (TP53) mutations are common in JAK2V617F-mutant, post-MPN AML but not in chronic-phase MPN and lead to clonal dominance of JAK2V617F/TP53-mutant leukemic cells. Consistent with these data, expression of JAK2V617F combined with Tp53 loss led to fully penetrant AML in vivo. JAK2V617F-mutant, Tp53-deficient AML was characterized by an expanded megakaryocyte erythroid progenitor population that was able to propagate the disease in secondary recipients. In vitro studies revealed that post-MPN AML cells were sensitive to decitabine, the JAK1/2 inhibitor ruxolitinib, or the heat shock protein 90 inhibitor 8-(6-iodobenzo[d][1.3]dioxol-5-ylthio)-9-(3-(isopropylamino)propyl)-9H-purine-6-amine (PU-H71). Treatment with ruxolitinib or PU-H71 improved survival of mice engrafted with JAK2V617F-mutant, Tp53-deficient AML, demonstrating therapeutic efficacy for these targeted therapies and providing a rationale for testing these therapies in post-MPN AML.
Myeloproliferative neoplasms (MPNs) are hematopoietic disorders characterized by clonal proliferation of mature myeloid elements which manifest clinically as an excess of RBC [polycythemia vera (PV)], platelets [essential thrombocytosis (ET)], or WBC [primary myelofibrosis (PMF)] (1). Mutations in JAK2 have been identified in the majority of patients with PV, ET, and PMF (2–6), underscoring the importance of activated JAK–STAT signaling to the pathogenesis of chronic-phase MPN. Despite the increasing use of empiric and targeted therapies, a subset of MPN patients transform to secondary acute myeloid leukemia (AML). Leukemic transformation occurs in 1%, 4%, and 20% of patients over a 10-y period in ET, PV, and PMF, respectively (7). MPN patients who develop leukemic transformation have a dismal outcome, with a median survival of less than 6 mo (8). Advanced age (>60 y) and exposure to chemotherapy increase the risk of leukemic transformation; however, the mechanisms and pathways that contribute to transformation from MPN to AML have not been well delineated. Importantly, the use of standard AML therapies, including induction chemotherapy, has not been shown to improve outcome for patients with post-MPN AML (8, 9). These data indicate a need for new models and improved therapeutic approaches to improve outcomes for patients who have transformed from MPN to AML and to identify genetic lesions associated with leukemic transformation.
Genetic studies of paired samples before and after leukemic transformation have suggested there are at least two distinct routes for leukemic transformation. Some patients who present with a JAK2/MPL-positive MPN progress to JAK2/MPL-positive AML that is associated with the acquisition of additional genetic alterations (10–13). A second, more complex route to AML from MPN has been described in which a JAK2/MPL-positive MPN is followed by JAK2/MPL-negative AML (14, 15). Clonality studies using X-chromosome inactivation in informative females demonstrated that JAK2/MPL-positive MPN and JAK2/MPL-negative AML are clonally related, consistent with transformation of an antecedent, preJAK2/MPL-mutant clone which can progress to AML. As such, there is a need to elucidate better the mutations that govern the two divergent pathways that lead to leukemic transformation.
Recent candidate gene studies have begun to improve our understanding of the somatic mutations that contribute to transformation from MPN to AML. These data suggest that post-MPN AML has a somatic mutational spectrum that is distinct from that observed in de novo AML. JAK2V617F mutations are relatively rare in de novo AML (16), and AML patients with JAK2V617F mutations almost always have a history of an antecedent MPN (17). The most common mutations observed in de novo AML, including nucleophosmin (NPM1), DNMT3A, and FMS-like tyrosine kinase-3 (FLT3), are largely absent from post-MPN AML (10). SNP array analysis of post-MPN AML patients has identified recurrent gains and losses involving chromosomes 8, 12, 17, and 21, which contain MYC, ETV6, tumor protein 53 (TP53), and RUNX1, respectively (18, 19). Candidate gene studies have identified specific, recurrent point mutations that are more common in post-MPN AML than in de novo AML, including mutations in tet methylcytosine dioxygenase 2 (TET2), serine/arginine-rich splicing factor 2 (SRSF2), TP53, and isocitrate dehydrogenase 1/2 (IDH1/2) (10–13). These genetic data suggest post-MPN AML is molecularly distinct from de novo AML.
In this study we used high-throughput sequence analysis to characterize the somatic mutational spectrum of post-MPN AML and used these genetic data to develop a genetically accurate murine model of leukemic transformation. We demonstrate that loss of Tp53 in combination with expression of JAK2V617F results in the development of post-MPN AML. This leukemia is transplantable and is enriched for blasts expressing markers consistent with hematopoietic stem and progenitor cell populations, particularly megakaryocyte/erythroid progenitors (MEPs). In vitro and in vivo studies reveal therapeutic liabilities that can be used to inform the development of novel therapies for patients with in post-MPN AML. These data collectively reveal novel insights into the pathogenesis of leukemic transformation and demonstrate new potential therapeutic strategies for this disease.
Results
Genomic Analysis of Post-MPN AML.
Genomic profiling of post-MPN AML patient samples has demonstrated recurrent mutations in epigenetic modifiers, including TET2, additional sex combs like transcriptional regulator 1 (ASXL1), and IDH1/2 (11, 18, 20), as well as mutations in spliceosome complex members, most commonly SRSF2 (10). Acquisition of mutations in TP53 also has been suggested to be a frequent event at the time of transformation (12). Most recently, mutations in CALR have been identified in chronic-phase ET and MF patients and are exclusive of the JAK2V617F mutation (21, 22); however, the frequency of calreticulin (CALR) mutations in post-MPN AML has not been delineated. To characterize the somatic mutational spectrum of post-MPN AML better, we performed capture-based, next-generation sequencing of the entire coding sequence of 374 cancer-associated genes in a cohort of 33 patients with post-MPN AML (Table S1) using a newly developed clinical mutational platform for patients with hematologic malignancies. This platform also allows evaluation of 24 genes frequently rearranged using DNA-seq and 258 genes frequently rearranged using RNA-seq. We also performed genomic analysis of paired chronic-phase MPN samples from 13 patients for whom we had a paired chronic-phase MPN sample antecedent to transformation. We achieved a median coverage depth of 511× (range 405–645×), which allowed us to identify missense, nonsense, and frameshift mutations with quantitative data on variant allele frequency (VAF) as well as structural rearrangements and copy number alterations in post-MPN AML (Table S1).
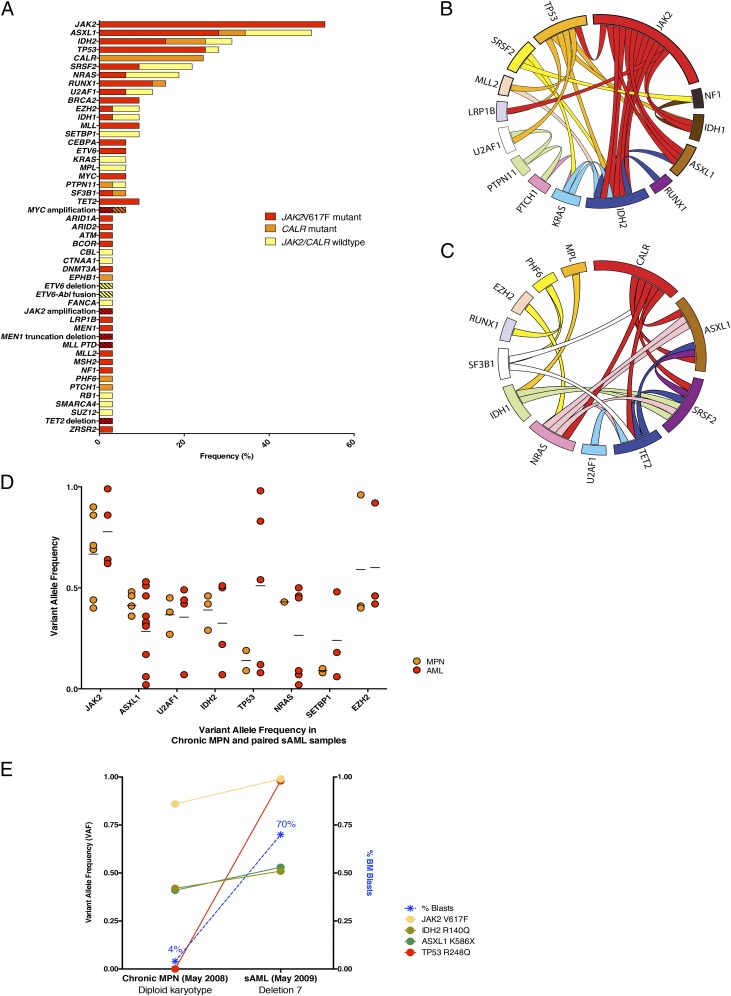
We identified frequent mutations in ASXL1 (47%), IDH2 (31%), SRSF2 (22%), and TP53 (27%), consistent with previous reports (Fig. 1A). We identified structural rearrangements in several cases (Table S1) that require further exploration in subsequent genomic and functional studies. Finally, we identified recurrent mutations not previously reported in post-MPN AML, including mutations in CALR (24.5%), MYC (7%), PTPN11 (7%), and SETBP1 (7%). Given that JAK2V617F mutations are not always retained at the time of leukemic transformation (14, 15), we analyzed the pattern of mutational co-occurrences in JAK2V617F-positive and -negative post-MPN AMLs (Fig. 1 B and C). We observed distinct mutational spectra in post-MPN AML samples segregated by JAK2V617F mutational status. In the JAK2V617F mutant subgroup, the most common co-occurring mutations were in TP53 (44%), ASXL1 (44%), and IDH2 (44%). In contrast, the most common mutations observed in JAK2-wild type post-MPN AML were in CALR (43%), ASXL1 (38%), and SRSF2 (29%). Of note, we did not identify any mutations in TP53 in JAK2-wild type post-MPN AML. Because the impact of CALR mutations in AML has not been evaluated previously, we examined whether there were differences in clinical outcome in secondary AMLs with JAK2 versus CALR mutations. However, no differences in survival were noted between patients with mutations in JAK2 or CALR or patients wild-type for both (Fig. S1A).
Fig. 1.
Genetic events in leukemic transformation of chronic-phase MPNs. (A) Frequency of mutations in post-MPN AML samples (n = 33). Hash marks indicate structural rearrangements (for scale reference, alterations were observed at a frequency of 3.1% beginning with ARID1A and extending to ZRSR2 in this representation). (B) Circos representation of co-occurring mutations in JAK2-mutant post-MPN AML. (C) Circos representation of co-occurring mutations in JAK2-wildtype post-MPN AML. (D) VAF of the most frequently mutated genes in post-MPN AML, in paired chronic-phase MPN and AML. (E) Representative analysis of VAF of mutations and bone marrow blasts occurring at chronic MPN phase and AML stage from a single patient.
Analysis of Mutational Heterogeneity Reveals Clonal Dominance of JAK2V617F/TP53 Mutant Cells in Post-MPN AML.
We next investigated mutant allele frequencies in paired chronic-phase MPN and post-MPN AML (Table S2) samples to investigate clonal dynamics during leukemic transformation. Although mutations in ASXL1 and NRAS were observed in a higher proportion of patients with leukemic transformation than in patients with chronic-phase MPNs (Fig. S1B), the VAF of these mutations remained constant after leukemic transformation, suggesting that these mutations do not lead to clonal dominance and to emergence of the AML clone (Fig. 1D). In the majority of cases analyzed, these mutations were present in less than 30% of reads, consistent with subclonal mutations that were not universal in the leukemic clone. In contrast, mutations in JAK2 and TP53 nearly always were present at high VAF in patients with post-MPN AML, as is consistent with their presence in the dominant AML clone (Fig. 1D). The VAF of TP53 was significantly greater in post-MPN AML samples than in MPN samples (7% versus 57%, respectively, P < 0.01) (Fig. S1C). Analysis of clonal architecture at the time of transformation demonstrated several distinct patterns of clonal evolution, including the emergence of distinct clones from the chronic-phase MPN, clonal progression of the chronic-phase MPN, and development of leukemia and chronic-phase MPN from a common precursor clone (Fig. S1D). We also analyzed the VAF of mutant disease alleles before and after leukemic transformation in patients with JAK2V617F mutations and TP53 mutations (Fig. 1E and Fig. S1E). In these individuals, the VAF of TP53 markedly increased at the time of leukemic transformation, coincident with the increase in bone marrow blasts, whereas concurrent mutations in ASXL1 and IDH2 did not increase consistently with transformation to AML. In the majority of cases, the TP53 mutant clone had a VAF >50% at the time of leukemic transformation, consistent with the loss of wild-type TP53 and with selection for homozygous TP53 inactivation. These data collectively indicate that JAK2 and TP53 mutations lead to clonal dominance of post-MPN AML cells and suggest that the combination of JAK2V617F mutations and TP53 mutations potently synergizes to induce leukemic transformation.
Loss of Tp53 Cooperates with JAK2V617F to Promote AML.
Given the human genetic data implicating JAK2V617F mutations and TP53 mutations in post-MPN AML, we sought to demonstrate if these two mutations could cooperate in vivo. We expressed JAK2V617F in whole bone marrow derived from Tp53-null (Tp53-KO) mice and from wild-type mice and transplanted these cells into lethally irradiated recipients. As controls, JAK2V617F retrovirus or empty vector [murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-GFP, MIGR1] was transduced into littermate wild-type C57BL/6 whole bone marrow, or MIGR1 was transduced into Tp53-KO C57BL/6 whole bone marrow, and was transplanted into lethally irradiated recipients (Fig. 2A). Recipients transplanted with Tp53-KO bone marrow transduced with JAK2V617F retrovirus demonstrated significantly reduced survival compared with controls (P < 0.01, log-rank test) (Fig. 2B), with all mice developing lethal disease within 110 d. These data demonstrate potent in vivo cooperativity between JAK2V617F mutations and Tp53 loss in vivo.
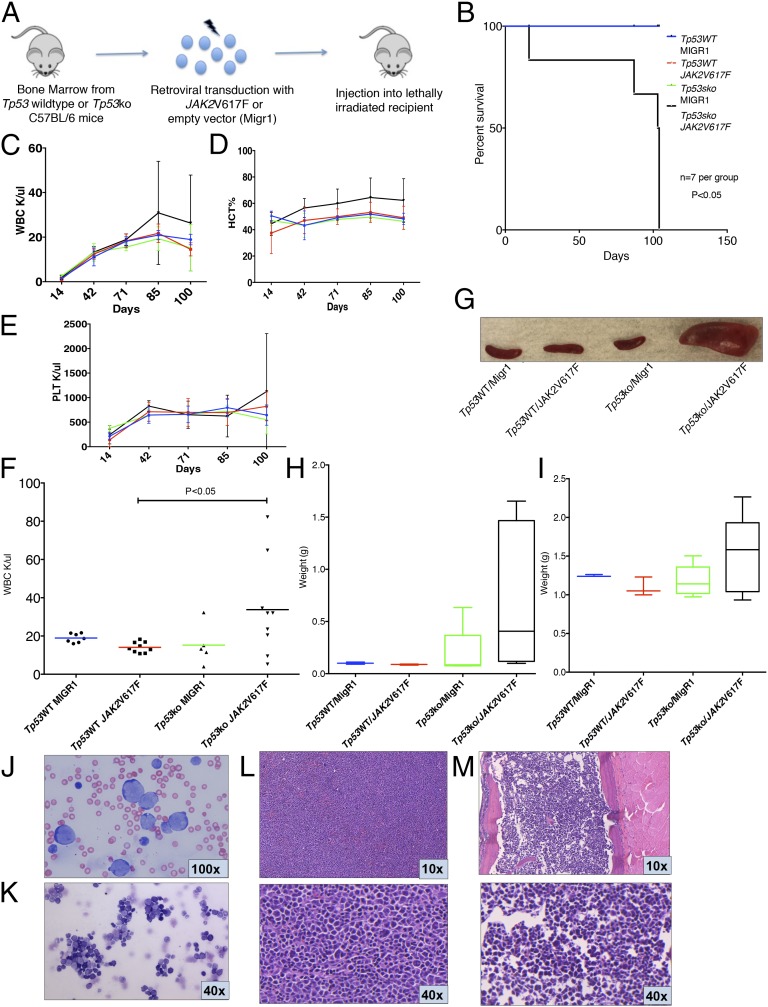
Fig. 2.
JAK2V617F collaborates with Tp53 loss to induce AML. (A) Bone marrow from Tp53-null (Tp53-KO) or wild-type C57BL/6 mice was harvested and transduced with either JAK2V617F- or MigR1-containing retrovirus. Transduced cells then were injected into lethally irradiated congenic recipients. (B) Survival of mice injected with Tp53-KO bone marrow transduced with JAK2V617F (Tp53-KO/JAK2V617F) was reduced significantly compared with control arms (P < 0.01, t test). (C–E) Trend of WBC (C), HCT (D), and PLT (E) in Tp53-KO/JAK2V617F mice compared with control arms from day 14 posttransplantation to day 100. (F) WBC count of recipients injected with Tp53-KO/JAK2V617F bone marrow was significantly greater than that of p53WT/JAK2V617F control (P < 0.05, t test) at day 100 posttransplantation, with a trend toward increase compared with other control arms. Blood counts displayed are derived from two independent experiments. (G) Representative spleens from killed animals. (H and I) Spleen (H) and liver (I) weights demonstrating increased organ weights in Tp53WT/JAK2V617F mice. (J and K) Representative peripheral blood smear (J) and bone marrow cytospin (K) from Tp53WT/JAK2V617F mice demonstrate increased numbers of intermediate to large blasts with round and irregular nuclei, high nuclear:cytoplasmic ratios, and finely stippled chromatin, consistent with acute leukemia; maturing myeloid and erythroid precursors are markedly reduced (Wright–Giemsa stain). (L and M) Representative spleen sections (L) and bone marrow (M) from Tp53WT/JAK2V617F mice demonstrating increased blasts consistent with acute leukemia (H&E stain).
Mice transplanted with JAK2V617F, Tp53-deficient cells were characterized by increasing WBC counts, hematocrit (HCT), and platelets (PLT) (Fig. 2 C–E; for WBC counts on day 100 posttransplantation, see Fig. 2F). A significant increase in HCT was observed in mice transplanted with JAK2V617F, Tp53-deficient cells (P < 0.05 for each comparison, t test) (Fig. S2A) compared with wild-type and single-mutant controls. We did not observe differences in PLT counts between mice transplanted with JAK2V617F, Tp53-deficient cells and controls (Fig. S2B). Tp53-KO/JAK2V617F mice were characterized by marked splenomegaly (Fig. 2 G and H) and hepatomegaly (Fig. 2I) because of the infiltration of AML cells. Likewise, cytologic examination of the peripheral blood and bone marrow of Tp53-KO/JAK2V617F mice showed expansion of blasts, consistent with transformation to AML (Fig. 2 J–M).
Leukemic Mice Demonstrate Enhanced Lineage-Negative, c-Kit–Positive, Sca-1–Positive and MEP Populations.
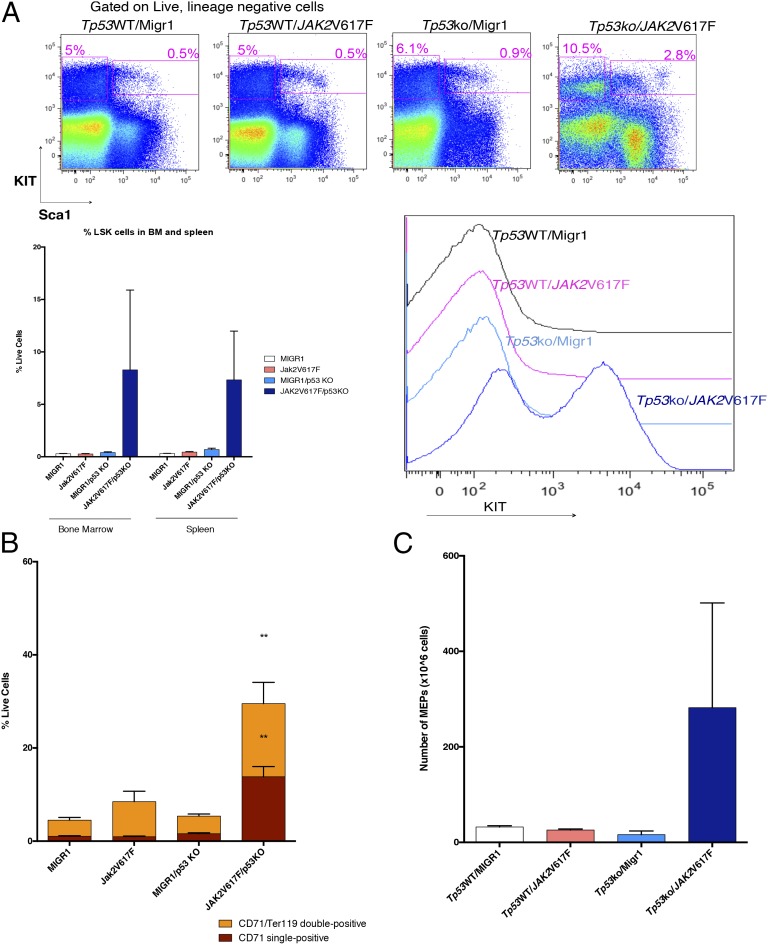
Analysis of the bone marrow and spleen demonstrated an increase in the percent of lineage-negative, c-Kit–positive, Sca-1–positive (LSK) cells in leukemic mice compared with controls (Fig. 3A), as is consistent with the known role of TP53 in regulating stem hematopoietic cell self-renewal (23). In addition, we noted expansion of the MEP (lineage-negative, c-Kit–positive, Sca-1–negative, FcγR-negative, CD34-negative) population in leukemic mice compared with control mice. Given that expression of JAK2V617F alone leads to PV in vivo (24), we also assessed the impact of concomitant JAK2V617F expression and Tp53 loss on erythroid differentiation compared with JAK2V617F expression alone. Tp53 wild-type mice transduced with JAK2V617F demonstrated a trend toward an increase in the CD71+/Ter119+ erythroid progenitor cells (Fig. 3B), as is consistent with expansion of differentiated erythroid progenitors. In contrast, mice expressing JAK2V617F in the setting of Tp53 loss were characterized by expansion of CD71+Ter119− cells (P < 0.05, t test) and CD71+/Ter119+ cells, as is consistent with erythroid expansion in vivo (Fig. 3B and Fig. S3A). Expansion of the MEP population was observed in both the spleen (Fig. 3C and Fig. S3C) and bone marrow (Fig. S3B) of Tp53-KO/JAK2V617F mice as compared with controls.
Fig. 3.
Tp53-KO/JAK2V617F mice are characterized by an expansion of hematopoietic stem and erythroid progenitor populations. (A) Immunophenotypic analysis demonstrating an increase in the frequency of LSK cells compared with control mice. (Upper) Representative FACS plots. (Lower Left) Quantification of bone marrow and spleen LSK populations. (Lower Right) Histogram of c-kit expression on live, nucleated cells in spleen. (B) The CD71/Ter119 double-positive population and the CD71 single-positive population are increased significantly in bone marrow of Tp53-KO/JAK2V617F mice (P < 0.05, t test). (C) Analysis of spleen cells demonstrates an increase in the MEP population in Tp53-KO/JAK2V617F mice compared with controls. Three mice per group were analyzed.
Tp53-KO/JAK2V617F Hematopoietic Cells Have Enhanced Self-Renewal and Serial Transplantability, Including in the MEP Compartment.
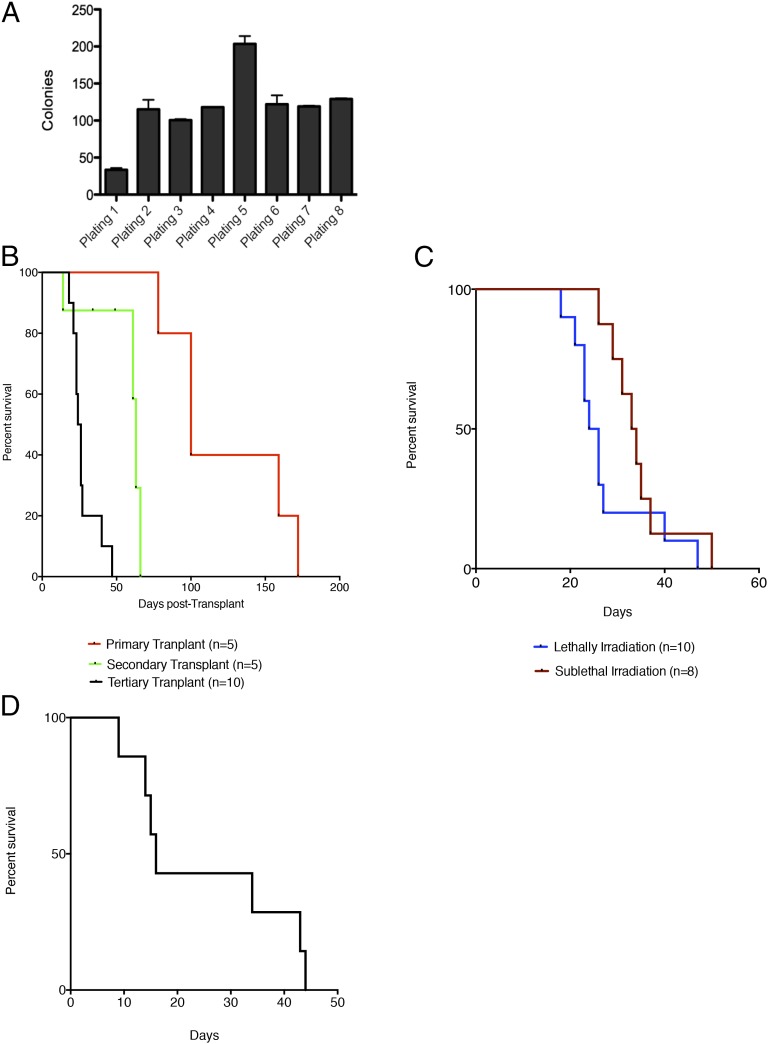
To assess for enhanced self-renewal (25), whole bone marrow cells from mice were serially passaged in methylcellulose containing myeloid/erythroid-promoting cytokines [IL3-, IL-6, cytokines stem cell factor (SCF), and erythropoietin (EPO)]. Bone marrow from Tp53-KO/JAK2V617F leukemic mice demonstrated increased serial replating in vitro, consistent with enhanced self-renewal capacity (Fig. 4A). We next investigated whether Tp53-KO/JAK2V617F cells showed increased self-renewal in vivo. Serial transplantation of Tp53-KO/JAK2V617F spleen cells induced disease in secondary and tertiary transplants. We observed significantly decreased latency with each serial transplant (P < 0.05 primary vs. secondary and primary vs. tertiary, log-rank test) (Fig. 4B), as is consistent with the progressive increase in self-renewal in vivo observed in other murine models of AML (26). We observed transplantable leukemia in sublethally and lethally irradiated recipients with no difference in latency of disease, indicating that the phenotype of Tp53-KO/JAK2V617F hematopoietic disease is consistent with an acute leukemia and not MPN (Fig. 4C). To assess for clonal evolution in primary and tertiary leukemias, we performed exome capture of leukemic cells from primary and tertiary transplant recipients compared with germline controls. We identified genomic alterations that were present in the tertiary transplanted leukemia but not in the germline or primary leukemia sample, including nonsense mutations, insertions, and deletions (Table S3). However, we did not identify any mutations in genes with a known role in human leukemogenesis.
Fig. 4.
Loss of Tp53 in the JAK2V617F setting confers increased self-renewal to committed progenitor populations. (A) Methylcellulose replating assay demonstrating enhanced self-renewal of Tp53-KO/JAK2V617F whole bone marrow. (B) Serial transplantation of whole spleen cells derived from leukemic Tp53-KO/JAK2V617F mice demonstrates significantly reduced latency with serial transplantation (P < 0.05 primary versus secondary, P < 0.05 secondary versus tertiary, log-rank test). (C) Transplantation of spleen cells from Tp53-KO/JAK2V617F leukemic mice results in the development of leukemia in both lethally and sublethally irradiated recipient mice. (D) Transplantation of the MEP population from Tp53-KO/JAK2V617F mice with leukemic phenotype into lethally irradiated recipients results in the development of leukemia in mice surviving longer than 20 d (n = 7).
Given the expansion in the MEP compartment in Tp53-KO/JAK2V617F mice, we performed transplantation studies to determine if this population had leukemic-propagating potential in secondary recipients. MEPs were purified by FACS sorting and transplanted into lethally irradiated recipients with bone marrow support from congenic donors. All mice transplanted with Tp53-KO/JAk2V617F MEP cells developed a lethal AML, with morphologic and immunophenotypic features similar to those observed in primary Tp53-KO/JAK2V617F mice (Fig. 4D). These data demonstrate that the MEP population in Tp53-KO/JAK2V617F AML aberrantly exhibits self-renewal and can initiate AML in vivo.
JAK-STAT Inhibition Results in Antiproliferative Effects and Improves Survival in Tp53-KO/JAK2V617F Mice.
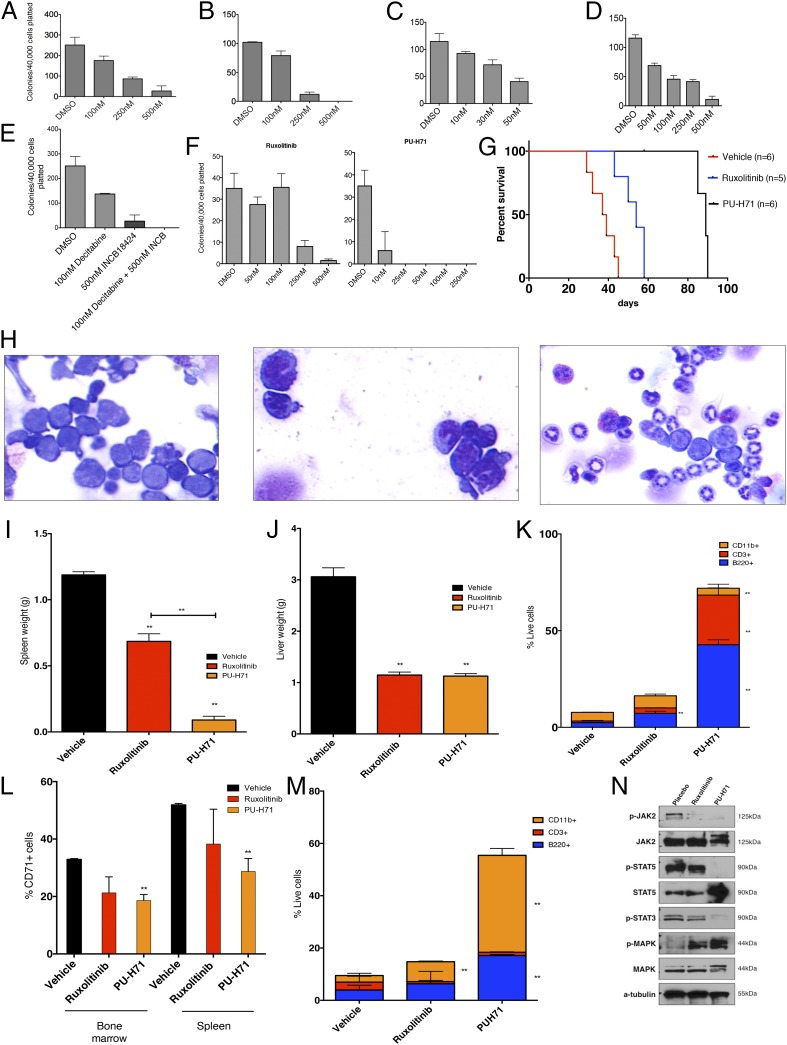
We next sought to use the Tp53-KO/JAK2V617F leukemic mouse model to test therapies in vitro and in vivo. We performed in vitro efficacy studies with a set of therapeutic agents using bone marrow cells from leukemic mice cultured in methylcellulose. Treatment with the JAK1/2 inhibitors ruxolitinib or CYT387 resulted in a concentration-dependent inhibition of colony formation (Fig. 5 A and B). We previously demonstrated that JAK2 is a client protein of heat-shock protein 90 (HSP90) and that treatment with the HSP90 inhibitor 8-(6-iodobenzo[d][1.3]dioxol-5-ylthio)-9-(3-(isopropylamino)propyl)-9H-purine-6-amine (PU-H71) results in efficacy in MPN patient samples and murine models (27). PU-H71 treatment resulted in concentration-dependent inhibition of colony formation at concentrations that are easily achieved in vivo (Fig. 5C). Recent early-phase clinical trials and case reports have suggested that hypomethylating agents show clinical activity in post-MPN AML (28, 29). Consistent with these data, decitabine exposure resulted in concentration-dependent inhibition of colony formation (Fig. 5D). More importantly, the combination of decitabine and ruxolitinib resulted in increased efficacy in suppressing colony formation of post-MPN AML cells (Fig. 5E). We next tested the ability of ruxolitinib or PU-H71 to inhibit colony formation in primary post-MPN AML blasts which had concurrent JAK2V617F and TP53 mutations. Exposure to either ruxolitinib or PU-H71 inhibited colony formation in a dose-dependent manner, consistent with murine in vitro observations (Fig. 5F). Notably, the PU-H71 doses required to inhibit colony formation are well below the 2–4 μM that can be achieved in vivo (27), as is consistent with the potent inhibition of post-MPN cell viability at feasible drug concentrations.
Fig. 5.
Pharmacologic inhibition of JAK2 as well as degradation of JAK2 alone and in combination with other therapies inhibits Tp53-KO/JAK2V617F leukemia. (A–E) The clonogenic capacity of Tp53-KO/JAK2V617F leukemic bone marrow cells in methylcellulose is inhibited on exposure to the JAK1/2 inhibitor ruxolitinib (A), the JAK1/2 inhibitor CTY387 (B), the Hsp90 inhibitor PU-H71 (C), decitabine (D), and a combination of decitabine and ruxolitinib (INCB18424) (E). For each condition, 40,000 spleen-derived cells were plated. (F) Clonogenic capacity of TP53 mutant/JAK2V617F human leukemic bone marrow cells in methylcellulose is inhibited on exposure to either ruxolitinib or PU-H71. (G) Treatment with either ruxolitinib or PU-H71 significantly prolongs survival (P < 0.01, log-rank test) of Tp53-KO/JAK2V617F leukemic mice relative to vehicle, and treatment with PU-H71 significantly prolongs survival compared with ruxolitinib (P < 0.01, log-rank test). PU-H71 was discontinued 1 wk after all ruxolitinib-treated mice were deceased. (H) Bone marrow cytospins demonstrating a predominance of blasts in vehicle-treated mice (Left), compared with increasing evidence of granulocytic maturation in mice treated with ruxolitinib (Center) and PU-H71 (Right). (I and J) Treatment with ruxolitinib or PU-H71 results in significant reductions in spleen (I; P < 0.05, P < 0.01, respectively) and liver (J; P < 0.01 for both treatments relative to vehicle) weights. PU-H71 significantly reduced spleen weight compared with ruxolitinib as well (P < 0.01, t test). (K) Treatment with ruxolitinib results in expansion of CD3+ cells (P < 0.05, t test), and treatment with PU-H71 results in expansion of CD3+ (P < 0.05, t test), CD11b++ (P < 0.01, t test), and B220+ (P < 0.01, t test) populations in spleens of treated mice. (L) The proportion of CD71+ cells is reduced in bone marrow of ruxolitinib- and PU-H71–treated mice (P < 0.05, t test, for PU-H71) and in spleen of ruxolitinib- and PU-H71–treated mice (P < 0.05, t test, for PU-H71). (M) Treatment with ruxolitinib or PU-H71 results in the expansion of CD11b+ (P < 0.05 for ruxolitinib, and P < 0.01 for PU-H71, t test) and B220+ (P < 0.01 for PU-H71, t test) cells in bone marrow of treated mice. (N) Western blot demonstrating reduction in phospho-JAK2 and phospho-STAT5 levels in mice treated with ruxolitinib or PU-H71 relative to placebo. **P < 0.05.
We next sought to pilot the efficacy of these therapeutic strategies in vivo. We used secondary recipients transplanted with Tp53-KO/JAK2V617F leukemic cells. After engraftment (14 d after transplantation), we randomized mice to treatment with vehicle (saline), ruxolitinib (administered daily at 60 mg/kg by oral gavage two times per day), or PU-H71 (administered at 75 mg/kg i.p. three times per week). Mice treated with ruxolitinib demonstrated significantly prolonged survival compared with mice treated with vehicle (P < 0.01, log rank test). Mice treated with PU-H71 also demonstrated significantly prolonged survival compared with mice treated with placebo and compared with mice treated with ruxolitinib (P < 0.01 for both comparisons, log-rank test) (Fig. 5G). Mice treated with ruxolitinib and PU-H71 demonstrated significant reductions in spleen (P < 0.05 and P < 0.01, respectively, t test) and liver (P < 0.01 for both comparisons, t test) weights at the time they were killed (Fig. 5 I and J). We observed significant reductions in WBC counts at day 42 with ruxolitinib and PU-H71 treatment compared with vehicle (P < 0.01 for both comparisons, t test) (see Fig. S5A) and a trend toward an increase in PLT count and hemoglobin in PU-H71–treated versus placebo- or ruxolitinib-treated mice. Examination of bone marrow cytospins revealed homogenous expansion of blasts in mice treated with vehicle. In contrast there was evidence of myeloid maturation and mature neutrophil production in mice treated with PU-H71 and, to a lesser extent, in mice treated with ruxolitinib (Fig. 5H). Flow cytometric analysis confirmed that PU-H71 therapy led to restored hematopoietic differentiation; PU-H71 therapy resulted in expansion of the CD11b+ (P < 0.05, t test), CD3+ (P < 0.05, t test), and B220+ (P < 0.01, t test) populations and in a reduction in the aberrantly expanded CD71+ population in the spleen compared with mice treated with vehicle (P < 0.05, t test) (Fig. 5 K and L). Similar results were observed in the bone marrow, with increases in the B220+ (P < 0.01, t test) and CD11b+ (P < 0.01, t test) populations and reduction of the CD71+ population (P < 0.05, t test) (Fig. 5M and Fig. S4B). We observed a significant, but less dramatic, effect of ruxolitinib on differentiation in vivo, including an increase in the proportion of CD11b+ (P < 0.05, t test) cells in the bone marrow and an increase in the CD3+ population in the spleen (P < 0.05, t test) compared with vehicle-treated mice (Fig. 5 K and M and Fig. S4C). Evaluation of the LSK populations (Fig. S5B) demonstrated a decrease in the total lineage-negative population and an increase in the proportion of LSK/myeloid progenitor in mice treated with either ruxolitinib or PU-H71.
Analysis of the JAK–STAT signaling pathway by Western blot in whole spleen cells from mice in all three arms demonstrated attenuated signaling with ruxolitinib and PU-H71, as is consistent with on-target JAK-STAT inhibition (Fig. 5N). Ruxolitinib led to a reduction in phosphorylated JAK2, STAT3, and STAT5 compared with vehicle-treated mice. In contrast, therapy with PU-H71 led to nearly complete inhibition of JAK2, STAT3, and STAT5 phosphorylation in post-MPN AML cells, as is consistent with greater pathway inhibition in vivo. These data demonstrate the therapeutic efficacy of PU-H71 and ruxolitinib in Tp53-KO/JAK2V617F AML mice, including inhibition of JAK–STAT signaling and restoration of hematopoietic differentiation with antileukemic therapy.
Discussion
Clinical studies have shown that transformation from chronic-phase MPNs to AML is associated with a dismal prognosis and with a poor response to conventional AML therapies. The spectrum of genomic alterations in post-MPN AML is strikingly distinct from de novo AML and previously has not been characterized to the same extent as de novo AML. Here we delineate the somatic mutational spectrum of post-MPN AML through high-throughput sequencing of a large set of genes with a known role in hematologic malignancies. This investigation allowed us to make several important observations. First mutations in CALR, JAK2, TP53, IDH2, and ASXL1 are frequent events in AML transformed from MPNs, whereas mutations in NPM1, cohesin complex members, FLT3, and CEBPA, which are common events in de novo AML, are rarely, if ever, observed in post-MPN AML. These data provide genetic evidence that post-MPN AML is a distinct disease from classical AML with a unique mutational spectrum and molecular pathogenesis and that lack of response to AML chemotherapy regimens reflects the divergent biology of post-MPN AML.
We also observed important differences in the mutational spectrum of JAK2V617F and JAK2-wild type in post-MPN AML. TP53 mutations are frequent, co-occurring events in patients with JAK2V617F mutations but not in patients with CALR mutations. Most importantly, patients with co-occurring JAK2V617F and TP53 mutations showed dominance of the JAK2/Tp53 mutant clone, consistent with potent cooperativity in vivo and with a dominant role in inducing transformation from JAK2V617F-positive MPN to JAK2V617F/Tp53-mutant AML. In contrast, mutations in RAS pathway members, SRSF2, and in ASXL1 were most common in JAK2-wild type post-MPN AML, and these mutations often were present as subclonal disease alleles at the time of transformation, suggesting they are not rate-limiting in the transformation from MPN to AML.
We used the observation that JAK2V617F and TP53 are common, clonally dominant lesions in post-MPN AML to investigate their ability to induce AML in vivo. Consistent with the human data, mice engrafted with Tp53-KO/JAK2V617F cells developed a fully penetrant, lethal AML that was transplantable into secondary and tertiary recipients. Of note, the leukemic clone appeared to arise from the aberrant MEP population, because this disease compartment was able to induce disease in secondary recipients. It is notable in this regard that acute erythroid leukemia and acute megakaryoblastic leukemia, which are rare immunophenotypic subtypes of de novo AML, often are observed in patients with post-MPN AML (8, 30). We hypothesize that post-MPN AML can arise through the acquisition of TP53 mutations by JAK2V617F-mutant MEP cells, which are expanded in chronic-phase disease but do not show increased self-renewal until the time of leukemic transformation. It will be important in subsequent studies to delineate if MEP cells are equally susceptible to leukemic transformation by other disease alleles enriched in post-MPN AML and to dissect the self-renewal mechanisms responsible for conferring leukemia-initiating potential in the MEP compartment.
Because post-MPN AML patients do not commonly respond to standard induction chemotherapy used in the treatment of patients with de novo AML (8), there is a need for novel therapies for patients with this poor-prognosis myeloid neoplasm. Ruxolitinib has been approved for the treatment of myelofibrosis, and early-phase clinical trials reported significant, transient responses to ruxolitinib in post-MPN AML patients (31). Consistent with these data, we observed efficacy with ruxolitinib in our murine model of post-MPN AML; however, all mice progressed during therapy, suggesting a need to improve on ruxolitinib monotherapy in post-MPN AML. Notably, we observed in vitro efficacy with decitabine in our murine model, and we hypothesize that a combination of decitabine and ruxolitinib might show greater efficacy than either agent alone in this difficult-to-treat malignancy. Most importantly, treatment with the Hsp90 inhibitor PU-H71 showed increased efficacy in vivo, with increased inhibition of JAK-STAT signaling, reduced disease burden, and improved survival compared with ruxolitinib. Given that many other proteins expressed in AML cells are known clients of Hsp90, it is unclear if the effects demonstrated with Hsp90 inhibition are entirely attributable to attenuation of the JAK–STAT pathway or if inhibition of other Hsp90 client proteins contributes to the improved efficacy of PU-H71 in this context. Consistent with these data, we also observed restoration of normal hematopoietic differentiation with PU-H71 therapy in vivo, suggesting the possibility that PU-H71 impacts transcriptional regulation in post-MPN AML cells and that this contributes to the therapeutic efficacy of Hsp90 inhibition in this disease context. Most importantly, our data suggest novel therapeutic approaches for post-MPN AML which have the potential to improve outcomes for patients with this poor-prognosis hematologic malignancy.
Methods
Reagents.
PU-H71 was synthesized by the Chiosis laboratory as previously described (32). Stock 1-mM aliquots were prepared in DMSO, stored at −20 °C, and diluted in appropriate medium before use. For in vivo use, PU-H71 was formulated in 10 mM phosphate buffer at a pH of ∼6.4. The JAK1- and JAK2-specific inhibitor ruxolitinib was purchased from Chemitek. Decitabine was purchased from Sigma-Aldrich. Antibodies used for Western blotting included pSTAT5, phosphorylated and total JAK2, and STAT3 (all from Cell Signaling Technologies); STAT5 (Santa Cruz Biotechnology, Inc.); and actin (EMD Chemicals). The MSCV-mouse JAK2V617F–IRES-GFP (MSCV-mJAK2V617F-IRES-GFP) plasmid has been described previously (24).
Target Gene Sequencing.
DNA extracted from formalin-fixed paraffin-embedded tissue, blood, or bone marrow aspirates was quantified by PicoGreen (Life Technologies) or RiboGreen. Total RNA was converted to double-stranded cDNA using random hexamers, and DNA was sheared by sonication before generation of highly complex adaptor-ligated sequencing libraries which were enriched by solution hybrid capture using custom baitsets targeting 374 cancer-related genes, with tiled intron coverage of 24 genes frequently rearranged for DNA-seq. RNA was used for capture sequencing of 272 genes frequently rearranged in hematologic malignancies. Enriched libraries were sequenced to high depth; DNA-seq libraries averaged >590 median exon coverage, and RNA-seq libraries averaged >20 million total pairs (49 × 49 paired-end reads; Illumina HiSEq 2500). We achieved a mean coverage depth of 511× (range 405–645). DNA resequencing of all coding exons of CALR was performed as previously described (16, 17, 33).
Flow Cytometry.
Spleen and bone marrow cells were strained and washed in ice-cold PBS with 1% BSA. Cells were incubated with Fc block (BD Pharmingen) for 15 min, stained with monoclonal antibodies on ice for 20 min, washed again in ice-cold PBS with 1% BSA, and analyzed. Bone marrow or spleen mononuclear cells were stained with a lineage mixture comprised of antibodies targeting CD4, CD8, B220, NK1.1, Gr-1, CD11b, Ter119, and IL-7Rα. Cells also were stained with antibodies against c-Kit, Sca-1, FcγRII/III, and CD34. Cell populations were analyzed using a FACS-Fortessa (Becton Dickinson) and sorted with a FACSAria II instrument (Becton Dickinson). All antibodies were purchased from BD-Pharmingen or eBioscience. The following antibodies were used: c-Kit (2B8), Sca-1 (D7), Mac-1/CD11b (M1/70), Gr-1 (RB6-8C5), NK1.1 (PK136), Ter-119, IL7-Rα(A7R34), CD34 (RAM34), FcγRII/III (2.4G2), CD4 (RM4-5), CD4 (H129.19), CD8 (53-6.7), CD45.1 (A20), CD45.2 (104), CD150 (9D1), CD48 (HM48-1), B220 (RA3-6B2), BP-1 (6C3), CD24 (30-F1), CD43 (eBioR2/60), IgD (11-26), and IgM (eB121-15F9).
Murine Studies.
All animal studies were performed at Memorial Sloan-Kettering Cancer Center under an animal protocol approved by the Memorial Sloan-Kettering Cancer Center Instructional Animal Care and Utilization Committee. The JAK2V617F murine bone marrow transplant assay was performed as described previously (34). Briefly, bone marrow cells from 5-Flurouracil–treated (150 mg/kg) male donors were harvested and transduced with viral supernatant containing MSCV-mJAK2V617F-IRES-GFP, and 7.5 × 105 cells were injected into the lateral tail veins of lethally irradiated (2 × 4.5 Gy) C57BL/6 mice. Nonlethal bleeds were performed 14 d after transplantation and every 14–30 d thereafter to assess disease severity. For in vivo drug trials, mice were randomized to receive treatment with PU-H71 (75 mg/kg i.p., three times per week), ruxolitinib (60 mg/kg two times per day), or vehicle (10 mM phosphate buffer, pH ∼6.4), beginning 21 d after transplantation. All mice were treated until any one of several criteria for killing were met, including moribundity, more than 10% body weight loss, or palpable splenomegaly extending across the midline. Differential blood counts were assessed by submandibular bleeds before the trial, after 14 d of treatment/vehicle, and at study end points. For transplantation experiments, freshly dissected femurs and tibias were isolated from mice. Bone marrow was flushed with a 3-cm3 insulin syringe into PBS. The bone marrow was spun at 0.5 × g by centrifugation at 4 °C, and RBC were lysed in ammonium chloride-potassium bicarbonate lysis buffer for 5 min. After centrifugation, cells were resuspended in PBS, passed through a cell strainer, and counted. Cells were transplanted via tail-vein injection into lethally irradiated or sublethally irradiated mice. Animal care was in strict compliance with institutional guidelines established by the Memorial Sloan-Kettering Cancer Center, the Guide for the Care and Use of Laboratory Animals (35), and the Association for Assessment and Accreditation of Laboratory Animal Care International.
Colony-Forming Assays.
Whole bone marrow cells from mice were seeded at 40,000 cells per replicate in mouse recombinant IL3 (rmIL3), rmIL3, rmSCF, recombinant human EPO cytokine-supplemented methylcellulose medium (Methocult, M3434 and M4434; STEMCELL Technologies). Colonies propagated in culture were scored at day 7. Replating was performed using 20,000 cells per replicate.
Western Blot.
For Western blot analysis, spleen cells were harvested and were immediately centrifuged, washed in ice-cold PBS containing sodium orthovanadate, and collected in lysis buffer containing Protease Arrest (G-Biosciences), Phosphatase Inhibitor Mixture II (EMD Chemicals), 1 mM phenylmethylsulfonyl fluoride (MP Biomedicals), and 0.02 mM phenylarsine oxide. Protein was normalized using the Bio-Rad Bradford protein estimation and separated using 4–12% Bis-Tris electrophoresis gels (Invitrogen). Nitrocellulose membranes were blocked in Tris-buffered saline and Tween 20 with 5% (wt/vol) milk and incubated with appropriate dilutions of primary and secondary antibody.
Histological Analyses.
Mice were killed, and tissue samples were fixed for 24 h in 4% (vol/vol) paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 μm and stained with H&E. Cytospins were performed by resuspending whole bone marrow or spleen cells (after ammonium-chloride-potassium lysis) in warm PBS. The cell suspension then was spun onto slides at 350 × g for 5 min. Slides were air-dried and stained using the Giemsa–Wright method. Images were acquired using a Zeiss Axio Observer A1 microscope (Zeiss).
Whole Exome Capture.
High-quality genomic DNA (2 μg) was captured by hybridization using the SureSelect XT Mouse All Exon kit (Agilent Technologies). Samples were prepared according to the manufacturer's instructions. PCR amplification of the libraries was carried out for eight cycles in the precapture step and for 10 cycles after capture. Samples were barcoded and run on a HiSEq 2000 instrument in a 75-bp/75-bp paired-end run, using the TruSeq SBS Kit v3 (Illumina). An average of 80 million paired reads was generated per sample, the duplication rate varied from 10 to 15%, and 90% of the targeted region was covered by at least 30×.
Statistics.
Data are displayed as mean ± SEM. Statistical significance between two groups was assessed using the t test to compare blood cell counts, organ weights, and stem and progenitor populations. Log-rank test was used for survival analysis. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Specialized Center of Research of the Leukemia and Lymphoma Society (to R.L.L. and S.V.) and National Cancer Institute (NCI) Grant 1R01CA151949-01 (to R.L.L.), support from the Susan and Peter Solomon Genomics Program at Memorial Sloan-Kettering Cancer Center, and a grant from the Geoffrey Beene Cancer Center (to R.R. and R.L.L.). G.C. is partly funded by NCI Grants R01 CA172546 and R01 CA155226. A.S.M is supported by National Institute of General Medical Sciences Grant T32GM007739 and National Cancer Institute Grant 5F30CA183497. R.L.L. is a Leukemia and Lymphoma Society Scholar, and O.A.-W. and R.R. are American Society of Hematology Scholars.
Footnotes
Conflict of interest statement: M.N., K.W., D.L., G.A.O., R.Y., P.J.S., and V.A.M., are employees of Foundation Medicine. R.R., O.A.-W., M.R.M.v.d.B., S.A.A., A.D., A.I., and R.L.L. have performed consulting for Foundation Medicine. Memorial Sloan-Kettering Cancer Center holds the intellectual rights to PU-H71. Samus Therapeutics, of which G.C. has partial ownership, has licensed PU-H71.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407792111/-/DCSupplemental.
References
- 1.Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355(23):2452–2466. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- 2.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434(7037):1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 3.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352(17):1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, et al. Cancer Genome Project Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365(9464):1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 5.Zhao R, et al. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem. 2005;280(24):22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7(4):387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Cervantes F, et al. Acute transformation in nonleukemic chronic myeloproliferative disorders: Actuarial probability and main characteristics in a series of 218 patients. Acta Haematol. 1991;85(3):124–127. doi: 10.1159/000204873. [DOI] [PubMed] [Google Scholar]
- 8.Mesa RA, et al. Leukemic transformation in myelofibrosis with myeloid metaplasia: A single-institution experience with 91 cases. Blood. 2005;105(3):973–977. doi: 10.1182/blood-2004-07-2864. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy JA, et al. Treatment outcomes following leukemic transformation in Philadelphia-negative myeloproliferative neoplasms. Blood. 2013;121(14):2725–2733. doi: 10.1182/blood-2012-10-464248. [DOI] [PubMed] [Google Scholar]
- 10.Zhang SJ, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119(19):4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Wahab O, et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70(2):447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harutyunyan A, Klampfl T, Cazzola M, Kralovics R. p53 lesions in leukemic transformation. N Engl J Med. 2011;364(5):488–490. doi: 10.1056/NEJMc1012718. [DOI] [PubMed] [Google Scholar]
- 13.Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362(4):369–370. doi: 10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- 14.Theocharides A, et al. Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood. 2007;110(1):375–379. doi: 10.1182/blood-2006-12-062125. [DOI] [PubMed] [Google Scholar]
- 15.Campbell PJ, et al. Mutation of JAK2 in the myeloproliferative disorders: Timing, clonality studies, cytogenetic associations, and role in leukemic transformation. Blood. 2006;108(10):3548–3555. doi: 10.1182/blood-2005-12-013748. [DOI] [PubMed] [Google Scholar]
- 16.Fröhling S, et al. Rare occurrence of the JAK2 V617F mutation in AML subtypes M5, M6, and M7. Blood. 2006;107(3):1242–1243. doi: 10.1182/blood-2005-09-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine RL, et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood. 2005;106(10):3377–3379. doi: 10.1182/blood-2005-05-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoennissen NH, et al. Prevalence and prognostic impact of allelic imbalances associated with leukemic transformation of Philadelphia chromosome-negative myeloproliferative neoplasms. Blood. 2010;115(14):2882–2890. doi: 10.1182/blood-2009-07-235119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klampfl T, et al. Genome integrity of myeloproliferative neoplasms in chronic phase and during disease progression. Blood. 2011;118(1):167–176. doi: 10.1182/blood-2011-01-331678. [DOI] [PubMed] [Google Scholar]
- 20.Beer PA, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115(14):2891–2900. doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 21.Klampfl T, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369(25):2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- 22.Nangalia J, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369(25):2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4(1):37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wernig G, et al. Expression of Jak2V617F causes a polycythemia vera-like disease with associated myelofibrosis in a murine bone marrow transplant model. Blood. 2006;107(11):4274–4281. doi: 10.1182/blood-2005-12-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huntly BJP, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6(6):587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 27.Marubayashi S, et al. HSP90 is a therapeutic target in JAK2-dependent myeloproliferative neoplasms in mice and humans. J Clin Invest. 2010;120(10):3578–3593. doi: 10.1172/JCI42442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascarenhas J, et al. Therapeutic options for patients with myelofibrosis in blast phase. Leuk Res. 2010;34(9):1246–1249. doi: 10.1016/j.leukres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Thepot S, et al. Groupe Francophone des Myelodysplasies (GFM) Treatment of progression of Philadelphia-negative myeloproliferative neoplasms to myelodysplastic syndrome or acute myeloid leukemia by azacitidine: A report on 54 cases on the behalf of the Groupe Francophone des Myelodysplasies (GFM) Blood. 2010;116(19):3735–3742. doi: 10.1182/blood-2010-03-274811. [DOI] [PubMed] [Google Scholar]
- 30.Abdulkarim K, et al. AML transformation in 56 patients with Ph- MPD in two well defined populations. Eur J Haematol. 2009;82(2):106–111. doi: 10.1111/j.1600-0609.2008.01163.x. [DOI] [PubMed] [Google Scholar]
- 31.Eghtedar A, et al. Phase 2 study of the JAK kinase inhibitor ruxolitinib in patients with refractory leukemias, including postmyeloproliferative neoplasm acute myeloid leukemia. Blood. 2012;119(20):4614–4618. doi: 10.1182/blood-2011-12-400051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulick K, et al. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7(11):818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koppikar P, et al. Efficacy of the JAK2 inhibitor INCB16562 in a murine model of MPLW515L-induced thrombocytosis and myelofibrosis. Blood. 2010;115(14):2919–2927. doi: 10.1182/blood-2009-04-218842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.