Bill Clinton was inaugurated as the 42nd president of the United Sates, Whitney Houston dominated the record charts, and the Noble Peace Prize was awarded to Mandela and de Klerk. Meanwhile, two groups quietly working in Texas and California would let curiosity lead them to uncover a mechanism of life essential for the evolution of multicellularity: the unfolded protein response (UPR). The year was 1993. It was a moment during which Peter Walter and Kazutoshi Mori (Fig. 1) would first describe a system that addressed one of the fundamental mysteries of the cell: how it could ensure that the millions of proteins it created and secreted were folded and functioning properly.
Fig. 1.
Kazutoshi Mori of University of Kyoto (A) and Peter Walter of University of California, San Francisco (B), winners of the Albert Lasker Basic Medical Research Award, 2014. (A) Image courtesy of the Albert and Mary Lasker Foundation, and (B) image courtesy of Elisabeth Fall.
The creation of a multicellular organism requires enormous coordination between the cells, tissues, and organs of the body, an achievement that depends on the secretion of proteins. The size of the human proteome is estimated at well over 20,000 proteins (1). Astoundingly, more than one third of these are targeted for a single subcellular compartment, the endoplasmic reticulum (ER), before being folded and transported to the cell’s membrane or secreted (2). Although many secreted proteins go on to form the essence of the extracellular structures that support the cell, they also represent an invaluable means by which the cell can convey information about its internal workings to other cells, making coordinated behavior between all of our tissues and organs possible. The sheer mass of proteins synthesized for this purpose can be astounding. For example, cells that are professional secretors, like liver cells, will synthesize more than 13 million secretory proteins per minute (3). It is easy to imagine how a failure in the systems responsible for their synthesis might be catastrophic.
How could the cell possibly handle the synthesis of such a large volume of proteins, and how could it ensure that they are folded into their correct form? The answers to this began with the observations of Christian Anfinsen, who first noticed that the protein RNase, once denatured, could refold on its own (4). The capacity for a protein to refold itself led Anfinsen to postulate that the 3D structure of a protein could be uncovered in its primary amino acid sequence, a discovery for which he was awarded a Nobel Prize in 1972 (5). However, the folding of proteins in vivo proved to be much more complex and required an orchestrated ensemble of helper proteins known as chaperones. The pioneering work of Hartl and Horwich, 2011 Lasker Awardees, revealed that the cell contains a cohort of proteins, chaperones, which help proteins find their native, active conformation (6). Among these chaperone proteins are the HSP70 class, which are the workhorses of the protein folding machinery. How the chaperones could be regulated to ensure the quality of the cellular proteome would require discoveries pieced together from disparate fields of research.
Additional clues as to mechanisms by which the cell monitors the quality of its proteins came in 1984, by an observation by Amy Lee of University of Southern California. She noticed that when cells grown in culture were starved of their normal food source, glucose, the synthesis of most proteins decreased, yet a few actually increased. The induced proteins were awarded the name of glucose regulated protein (GRP) and a number corresponding to their molecular weight (7).
For several more years, the ideas of GRPs and protein chaperones remained disconnected, until Hugh Pelham noticed that one of these GRPs that resided in the endoplasmic reticulum, GRP78, was oddly similar to the cytosolic chaperone HSP70 (8). Because of these observations, Pelham began speculating on the relationship between GRP proteins and heat shock proteins (HSPs) (9). Soon after, Mary Jane Gething and Joe Sambrook, working at University of Texas Southwestern Medical Center, found that GRP78 could bind to misfolded proteins (10). GRP78 not only looked like a chaperone, but it acted like one too. This was rather puzzling: why then would a protein whose levels are increased under energy restriction bind to misfolded proteins? In one classic experiment, Mary Jane Gething showed that the mere presence of a misfolded protein was enough to induce the production of GRP78 (11), again suggesting that GRP78 up-regulation was part of a concerted effort of the cell to combat protein misfolding stress. However, how could something as basic as the fold of a protein be detected and communicated to the nucleus to produce more chaperones?
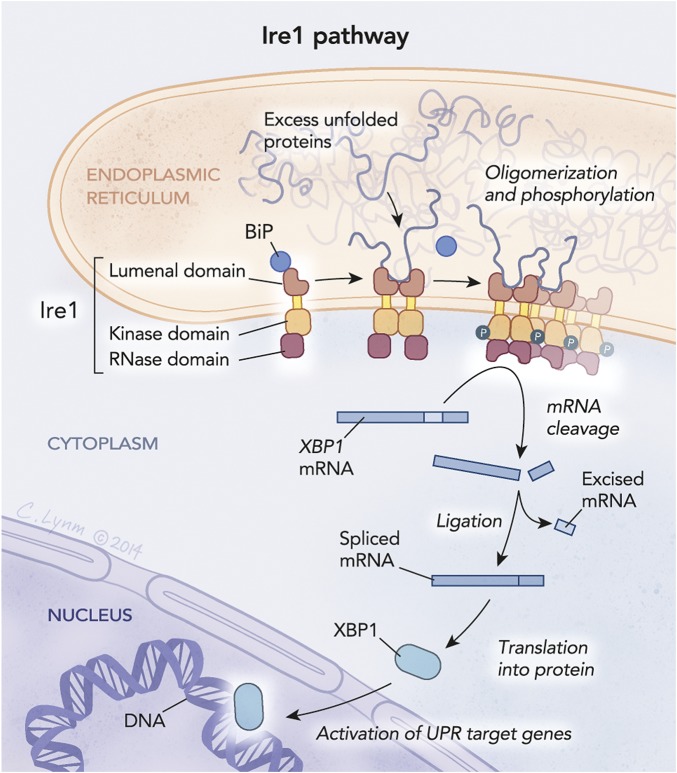
The answer to this came as, in 1989, Kazutoshi Mori left his secure position as an assistant professor at the Tokyo University to explore new areas of biology, starting a postdoctoral fellowship at UTSW working with Mary Jane Gething. Armed with a newly characterized GRP78 promoter and reporter genes, Walter and Mori independently set out to screen yeast mutants for their inability to turn on expression of GRP78. In 1993, both uncovered mutations in the gene encoding ire-1 (inositol requiring enzyme 1), laying the foundation for the discovery of the unfolded protein response of the ER (12, 13) diagrammed in Fig. 2. This discovery was a first and a giant step toward unveiling of an entire stress response network, capable of intricately and carefully regulating the folding and processing of the entire ER-targeted proteome.
Fig. 2.
The IRE-1 pathway is one of the pathways that make up the UPR. IRE-1 is a transmembrane protein that initiates UPR when excess unfolded proteins accumulate in the ER. When activated, IRE-1 splices mRNA located in the cytosol to produce the protein XPB1, a transcription factor that travels to the nucleus where it up-regulates UPR target genes that encode a variety of proteins that reduce the load of unfolded proteins in the ER. © Cassio Lynm.
Initially, the structure of IRE-1 seemed at odds with itself, as if it was a hybrid protein. Although it resided in the membrane of the ER, it had a kinase domain and an RNase domain, both of which lay in the cytoplasm. This structure suggested a capacity to translate information between these two subcellular locations. Because of its kinase domain, an early prevailing idea was that IRE-1 phosphorylated the major mediator for GRP78 induction. However, the only protein that IRE-1 phosphorylated was itself. The function of the RNase domain, in contrast, remained a mystery.
Mori and Walter continued their work, searching for downstream genetic targets of IRE-1. Three years later, Walter reported that, much like mutations in ire-1, mutations within a second gene—hac-1 (homologous to aft/CREB1)—also blocked induction of the UPR (14). Unlike ire-1, hac-1 encodes a transcription factor. Walter observed that, on protein folding stress in the ER, HAC1 protein levels increased even though its mRNA levels were relatively unchanged. Intriguingly, the hac-1 mRNA actually changed physically on ER stress: it became 26 nucleotides shorter (14). Suddenly, the RNase domain of IRE-1 had found a function. The selective removal of 26nts allowed efficient production of HAC1, allowing its nuclear entry and ability to bind to the GRP78 promoter to induce production of GRP78.
Thus, it was discovered that the nuclease required to remove the 26nts from hac-1 resides within IRE-1. However, how does IRE-1 become activated? Through structural analysis, Robert Stroud and Peter Walter discovered that IRE-1 forms a dimer that can then oligomerize into a higher-order structure (15). The local high concentration of IRE-1 is thought to create large RNA processing center that can recognize not only hac-1, but also other RNAs to lighten the load placed on the ER. On the ER lumen side, the IRE-1 oligomers form a beautiful cleft to allow possible interactions with long hydrophobic stretches of proteins, common among misfolded proteins.
Meanwhile, researchers were working diligently to uncover more components of the elegant pathway that forms this coordinated response. GRP78 was only one of hundreds of targets of this response, which became known as the UPR. Soon, it became apparent that, in metazoans, IRE-1 was also not the only transmembrane receptor to sense unfolded proteins in the ER. Two other independent arms of the UPR, the transmembrane receptors PERK (discovered by David Ron and Ron Wek) and ATF6 (discovered by Mori), also play a major role to protect the ER proteome (16–18). Over the years, it became evident that the UPR had evolved to become more complex in multicellular organisms, a sign of its importance in distal communications between cells.
These initial discoveries by Mori and Walter led us to our current model of folding in the ER. To ensure that the proteins entering the secretory pathway have the capacity to become functional and fold into their proper 3D structures, a set of systems have evolved in the ER, which is the first stop for proteins that are targeted for secretion. There, cells institute an important quality check to monitor the integrity of the proteins found within it. If a protein adopts the shape of its native structure, it will glide past the surveillance system and continue toward secretion out of the cell. However, if a mutant or damaged protein is present in the ER, then the system recognizes it and shuffles it toward a fate of destruction.
In the absence of the ER surveillance system, faulty and damaged proteins could be secreted and convey misinformation to the neighboring cells, thus highlighting the importance of this system for cellular and organismal health. Damaged proteins are also at risk for forming aggregates, losing their functionality and physically disrupting the extracellular space. The function of the ER goes far beyond these initial functions, however. Its health relies unequivocally on the UPR. For these great discoveries, Peter Walter and Kazutoshi Mori are recognized by the Lasker foundation for their discovery of the UPR of the ER.
Footnotes
The author declares no conflict of interest.
References
- 1.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 2.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 3.Mullins C. The Biogenesis of Cellular Organelles. Kluwer Academic/Plenum; New York: 2005. [Google Scholar]
- 4.Anfinsen CB, Haber E. Studies on the reduction and re-formation of protein disulfide bonds. J Biol Chem. 1961;236:1361–1363. [PubMed] [Google Scholar]
- 5.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 6.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381(6583):571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 7.Lee AS, Bell J, Ting J. Biochemical characterization of the 94- and 78-kilodalton glucose-regulated proteins in hamster fibroblasts. J Biol Chem. 1984;259(7):4616–4621. [PubMed] [Google Scholar]
- 8.Munro S, Pelham HR. An Hsp70-like protein in the ER: Identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46(2):291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 9.Pelham HR. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986;46(7):959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- 10.Kozutsumi Y, et al. Identification of immunoglobulin heavy chain binding protein as glucose-regulated protein 78 on the basis of amino acid sequence, immunological cross-reactivity, and functional activity. J Cell Sci Suppl. 1989;11:115–137. doi: 10.1242/jcs.1989.supplement_11.10. [DOI] [PubMed] [Google Scholar]
- 11.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332(6163):462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 12.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73(6):1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 13.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74(4):743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 14.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87(3):391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 15.Korennykh AV, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457(7230):687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 17.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18(12):7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]