Significance
Strigolactones (SLs) are plant hormones that inhibit shoot branching and are parasitic and symbiotic signals toward root parasitic plants and arbuscular mycorrhizal fungi, respectively. Therefore, the manipulation of SL levels potentially improves the yield of crops. To achieve this goal, the biosynthesis pathway of SLs must be fully understood. SLs are biosynthesized from a precursor, named carlactone (CL), which is derived from carotenoid. However, no downstream pathway of CL has been elucidated. In this study, we show that CL is converted into a carboxylated metabolite, named carlactonoic acid, by Arabidopsis MAX1, the enzymatic function of which had been unknown, and that its methyl ester has the ability to interact with a SL receptor and suppress shoot branching in Arabidopsis.
Keywords: strigolactone, biosynthesis, cytochrome P450, Arabidopsis, rice
Abstract
Strigolactones (SLs) stimulate seed germination of root parasitic plants and induce hyphal branching of arbuscular mycorrhizal fungi in the rhizosphere. In addition, they have been classified as a new group of plant hormones essential for shoot branching inhibition. It has been demonstrated thus far that SLs are derived from carotenoid via a biosynthetic precursor carlactone (CL), which is produced by sequential reactions of DWARF27 (D27) enzyme and two carotenoid cleavage dioxygenases CCD7 and CCD8. We previously found an extreme accumulation of CL in the more axillary growth1 (max1) mutant of Arabidopsis, which exhibits increased lateral inflorescences due to SL deficiency, indicating that CL is a probable substrate for MAX1 (CYP711A1), a cytochrome P450 monooxygenase. To elucidate the enzymatic function of MAX1 in SL biosynthesis, we incubated CL with a recombinant MAX1 protein expressed in yeast microsomes. MAX1 catalyzed consecutive oxidations at C-19 of CL to convert the C-19 methyl group into carboxylic acid, 9-desmethyl-9-carboxy-CL [designated as carlactonoic acid (CLA)]. We also identified endogenous CLA and its methyl ester [methyl carlactonoate (MeCLA)] in Arabidopsis plants using LC-MS/MS. Although an exogenous application of either CLA or MeCLA suppressed the growth of lateral inflorescences of the max1 mutant, MeCLA, but not CLA, interacted with Arabidopsis thaliana DWARF14 (AtD14) protein, a putative SL receptor, as shown by differential scanning fluorimetry and hydrolysis activity tests. These results indicate that not only known SLs but also MeCLA are biologically active in inhibiting shoot branching in Arabidopsis.
Strigolactones (SLs) are allelochemicals, exuded from plant roots, that stimulate seed germination of root parasitic plants, Striga spp., Orobanche spp., and Phelipanche spp. (1). The hyphal branching of the biotrophic arbuscular mycorrhizal (AM) fungi is also induced by SLs in the vicinity of host roots to ensure symbiosis with host plants (2). SLs are not only host recognition signals in the rhizosphere but also play important roles in the SL-producing plants themselves. Since the mid-1990s, the existence of novel hormone-like signals involved in shoot branching inhibition of plants had been proposed following the isolation and analysis of mutants with increased shoot branching, ramosus (rms) of pea (Pisum sativum), decreased apical dominance (dad) of petunia (Petunia hybrida), more axillary growth (max) of Arabidopsis (Arabidopsis thaliana), and dwarf (d) and high tillering dwarf (htd) of rice (Oryza sativa) (3–6). Recently, these mutants have been identified as SL-deficient or -insensitive mutants, providing decisive evidence that SLs function as shoot branch-inhibiting hormones (7, 8). In addition, further characterization of these mutants has shown that SLs affect root growth and development, leaf shape and senescence, internode elongation, secondary growth, and drought and salinity stress responses (9–11).
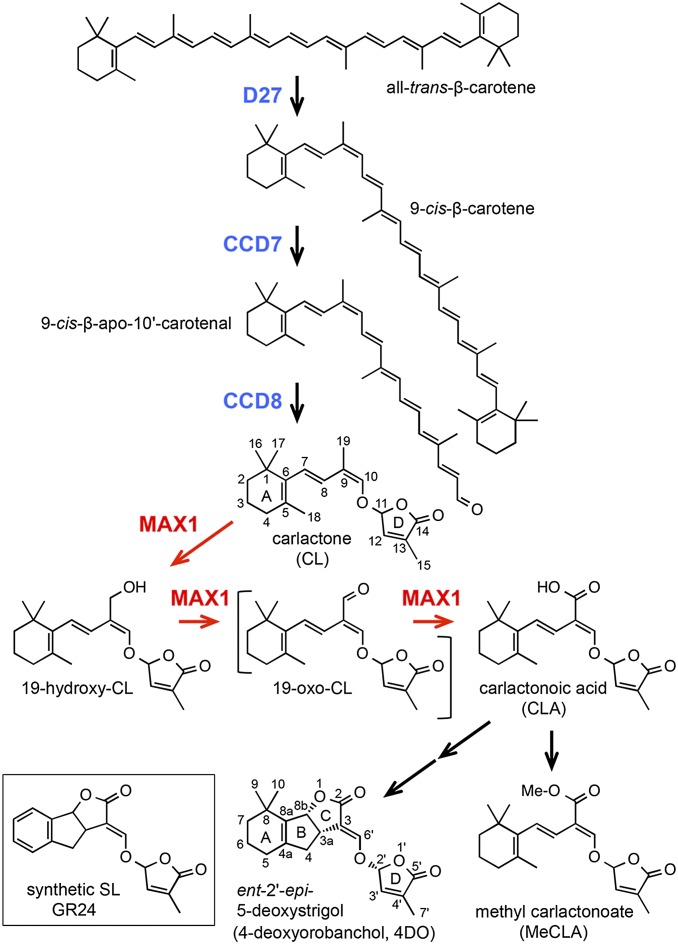
Despite the fact that SLs play important roles in plant growth and development and in the rhizosphere, the biosynthesis pathway of SLs has not fully been elucidated. The natural SLs consist of a tricyclic lactone (ABC ring) connecting to a butenolide group (D ring) via an enol ether bridge. 5-Deoxystrigol (5DS) and ent-2′-epi-5-deoxystrigol [4-deoxyorobanchol (4DO); Fig. 1] are thought to be the precursors of other natural SLs, which have methyl group(s) on the A ring and hydroxyl or acetyloxyl group(s) on the A/B ring (1, 12). Because the mutations in the CCD7 (MAX3/RMS5/HTD1) and CCD8 (MAX4/RMS1/DAD1/D10) genes, both of which encode carotenoid cleavage dioxygenases, result in SL deficiency (7, 8), it has been thought that SLs are synthesized from carotenoids by these enzymes. Recently, it has been demonstrated that the Fe-containing protein D27 catalyzes the isomerization at C-9 of all-trans-β-carotene to produce 9-cis-β-carotene in vitro (13) (Fig. 1). The product 9-cis-β-carotene was a substrate for CCD7 to produce 9-cis-β-apo-10′-carotenal, and this cleavage product was subsequently catalyzed by CCD8 to produce an SL precursor named carlactone (CL) (13) (Fig. 1). More recently, we reported that CL was detected from rice and Arabidopsis, and exogenous CL was converted into SLs in rice, demonstrating that CL is an endogenous precursor for SLs (14). Because CL contains the A and D rings and the enol ether bridge but lacks the B and C rings, additional biosynthetic steps are needed for the conversion of CL to 5DS and 4DO in plants.
Fig. 1.
Proposed biosynthesis pathway for SL from carotenoid. The conversion from β-carotene to CL by D27, CCD7, and CCD8 enzymes has been confirmed previously by in vitro assay (13). The conversion from CL to CLA by MAX1 and the existence of CLA and MeCLA in Arabidopsis were shown in this study.
The most probable enzyme catalyzing these reactions is MAX1 (CYP711A1), a cytochrome P450 monooxygenase (5). In reciprocal grafting experiments of Arabidopsis, the hyperbranching phenotype in scions of the max4 (ccd8) mutant was rescued to WT shoot branching patterns when grafted to max1 rootstocks, whereas max4 rootstocks could not restore a WT shoot branching phenotype to max1 scions (5). These results suggested that MAX1 acts on a downstream pathway of CCD8 to produce a mobile signal for shoot branching inhibition. Recently, it was reported that CL could not rescue the max1 phenotype by exogenous application (15), and we found an extreme accumulation of CL in the max1 mutant (14). Hence, CL is the most probable candidate for the substrate of MAX1. In the present study, to elucidate the enzymatic function of MAX1 in SL biosynthesis, we performed in vitro conversion of CL using a recombinant MAX1 protein expressed in yeast microsomes. We then examined if CL is metabolized in a similar manner in vivo by detecting and identifying the CL metabolites in Arabidopsis and rice plants. In addition, to investigate the role of the CL derivatives for shoot branching inhibition, we examined their biological activities and interaction with Arabidopsis thaliana DWARF14 (AtD14), a putative SL receptor.
Results
MAX1 Oxidized CL at C-19.
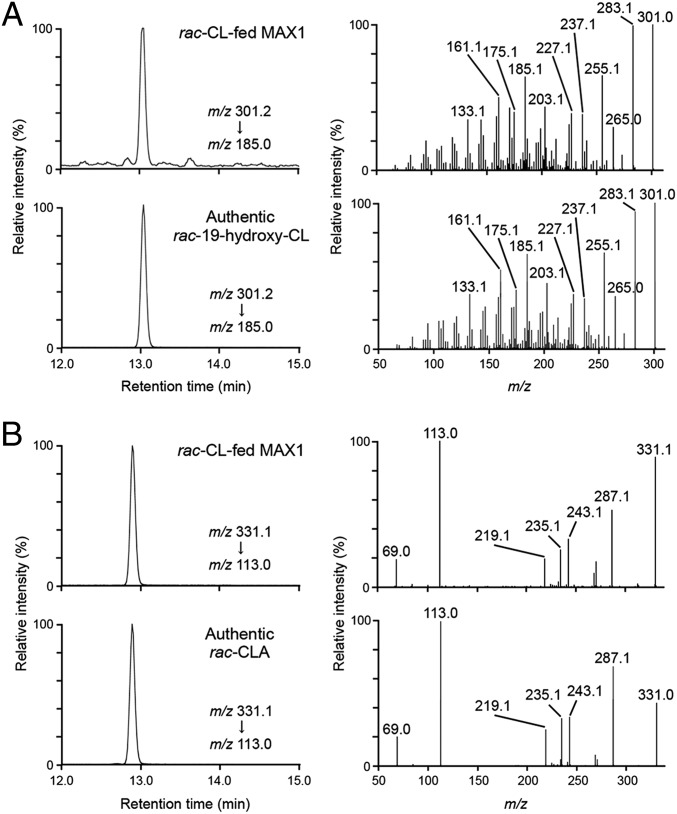
Arabidopsis MAX1 protein was expressed in yeast WAT11 strain that was generated to coexpress Arabidopsis NADPH-P450 reductase (ATR1) (16). Microsomes prepared from WAT11 expressing MAX1 (MAX1 microsomes) showed a P450-specific reduced carbon monoxide difference spectrum having an absorption peak at 450 nm, but control microsomes from cells transformed with an empty vector did not (Fig. S1), indicating that the recombinant MAX1 protein was an active P450 enzyme. The C-11 racemic (rac) CL as a candidate substrate of MAX1 enzyme was incubated with MAX1 and control microsomes. P450 enzymes are monooxygenases that catalyze extremely diverse reactions in biosynthetic and metabolic pathways in animals, plants, and microorganisms and generally mediate the insertion of oxygen atoms into substrates by the reductive activation of molecular oxygen (17). Because the oxidation of CL at C-19 position is needed to form the lactone of the C ring in the biosynthesis pathway of SL from CL (13), 19-hydroxy-CL was the most probable candidate for CL metabolites by MAX1 (Fig. 1). Therefore, we synthesized rac-19-hydroxy-CL (Fig. S2) and compared it with the metabolites of MAX1 using an electrospray ionization (ESI)-positive mode of LC-MS/MS for retention times and MS fragmentations. The transitions of the precursor ion [M+H–H2O]+ at m/z 301, generated by the loss of H2O from [M+H]+ at m/z 319, to product ions were detected in extracts from MAX1 microsomes incubated with rac-CL, as well as in the authentic rac-19-hydroxy-CL sample, but not in microsomes from the control, using selected reaction monitoring (SRM) (Fig. S3). The product ions and retention time of the metabolite were identical with those of the authentic rac-19-hydroxy-CL (Fig. 2A).
Fig. 2.
Identification of 19-hydroxy-CL and CLA produced from CL by recombinant MAX1. rac-CL was incubated with MAX1 microsomes. The extracts of the microsomes and authentic standards were analyzed by LC-MS/MS [a triple quadrupole/linear ion trap instrument (QTRAP)]. SRM chromatograms (Left) and product ion spectra (Right) derived from the precursor ion [M+H–H2O]+ (m/z 301) of (A) 19-hydroxy-CL and [M–H]– (m/z 331) of (B) CLA are shown.
MAX1 Catalyzed Consecutive Three-Step Oxidations.
It has been known that carboxylations are often performed by consecutive three-step oxidative reactions catalyzed by P450 in biosynthesis pathways of plant secondary metabolites (18, 19). To investigate whether MAX1 catalyzes three-step oxidations to convert C-19 methyl group of CL into carboxylic acid (Fig. 1), we synthesized rac-9-desmethyl-9-carboxy-CL [designated as carlactonoic acid (CLA)] (Fig. S4A) and used it as a standard to trace the compound in extracts from MAX1 microsomes incubated with rac-CL. The transitions of a distinct ion at m/z 331 corresponding to the pseudomolecular ion [M–H]– to product ions of authentic rac-CLA was detected in extracts from the MAX1 microsomes but not in those from the control, by an ESI-negative mode of LC-MS/MS (Fig. S5A). The product ions and retention time of the metabolite were identical to those of the authentic rac-CLA (Fig. 2B). CLA was also detected when rac-19-hydroxy-CL was incubated as a substrate with MAX1 microsomes (Fig. S5B). However, the occurrence of 19-oxo-CL, a putative intermediate between 19-hydroxy-CL and CLA (Fig. 1), could not be confirmed because we could not synthesize authentic 19-oxo-CL due to its instability. No metabolites such as known SLs were detected when rac-CLA was incubated with the recombinant microsomes.
MAX1 Preferred 11R-CL to 11S-CL as a Substrate.
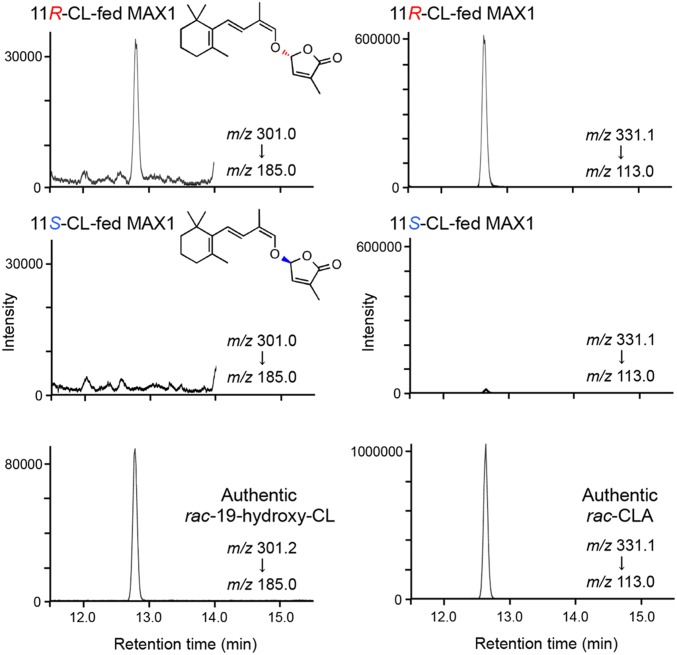
The apparent dissociation constant (Km) of rac-CL and rac-19-hydroxy-CL were 413 ± 65 and 960 ± 87 nM, respectively, when product CLA was quantified by LC-MS/MS using [1-13CH3]rac-CLA as an internal standard (Fig. S5C). The apparent catalytic constant (kcat) of rac-CL and rac-19-hydroxy-CL was 0.063 ± 0.003 and 0.114 ± 0.003 min−1, respectively (Fig. S5C). The C-11 of CL corresponds to the C-2′ of SL (Fig. 1). The configuration of C-11 in CL is thought to be important to determine the stereochemistry of SL because C-2′ configuration of all natural SL is (R) (1). Stereoisomers 11R-CL and 11S-CL (14) were incubated with MAX1 microsomes at 500 nM, and the products 19-hydroxy-CL and CLA were quantified using their [1-13CH3]-labeled rac-internal standards. 19-Hydroxy-CL and CLA (both are assumed to be 11R) were produced from 11R-CL at 0.101 ± 0.005 and 0.025 ± 0.002 min−1, respectively, whereas no 19-hydroxy-CL and a small amount of CLA (assumed to be 11S) was detected from 11S-CL (Fig. 3).
Fig. 3.
Conversions of stereoisomers 11R-CL and 11S-CL by recombinant MAX1. 11R-CL and 11S-CL were incubated with MAX1 microsomes and the extracts were analyzed using LC-MS/MS (QTRAP). SRM chromatograms of products, 19-hydroxy-CL (Left) and CLA (Right), in MAX1 microsomes and authentic standards are shown.
Endogenous CLA Was Detected and Exogenous CL Was Converted into CLA in Arabidopsis.
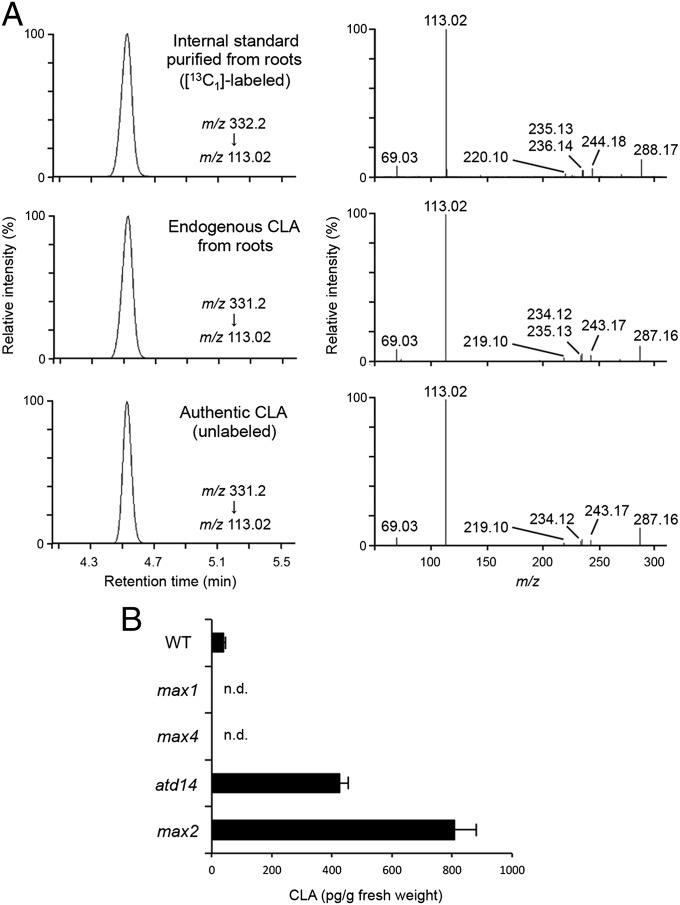
To confirm the existence of CLA in planta, we analyzed endogenous CLA in Arabidopsis plants. The CLA fraction was extracted from the roots of WT, the max1, max4, atd14, and max2 mutants of Arabidopsis grown hydroponically and analyzed by LC-MS/MS. The full-scan spectra and retention time of product ions confirmed the presence of endogenous CLA in the extracts of WT as well as those of the atd14 and max2 mutants that are defective in SL perception components (20) (Fig. 4A). In contrast, the endogenous contents of CLA were below the detection limit in the root extracts of the max1 and max4 mutants. The endogenous CLA was quantified using [1-13CH3]rac-CLA as an internal standard. The CLA content was 32.5 ± 5.2 pg/g fresh weight in WT and the content increased to ∼12- and 20-fold in the atd14 and max2 mutants, respectively (Fig. 4B). We could not detect known SLs in the tissues and exudates of Arabidopsis roots in these experiments. To further investigate whether CLA is also produced from CL in planta, the max4 mutant was grown hydroponically and incubated with [1-13CH3]11R-CL added into culture media. As a result, [13C1]-labeled CLA was detected in the max4 roots by LC-MS/MS analysis (Fig. S6A). However, when the max1max4 double mutant was used for the same feeding experiment, no [13C1]-labeled CLA was detected (Fig. S6A).
Fig. 4.
Endogenous analysis of CLA in Arabidopsis. Endogenous CLA was detected in WT, the max2 and atd14 mutants by LC-MS/MS [a quadrupole/time-of-flight instrument (QTOF)]. (A) Ion traces from the LC-MS/MS analysis (Left) and product ion spectra (Right) derived from respective precursor ions of [1-13CH3]-labeled (internal standard) and endogenous CLA extracted from max2 roots as a representative data and those of unlabeled authentic standard are shown. (B) Endogenous CLA was quantified in root extracts of WT, max1, max4, atd14, and max2 using [1-13CH3]CLA as an internal standard by LC-MS/MS (QTOF). Data are the means ± SD (n = 3). n.d., not detectable.
Endogenous CLA Was Detected and Exogenous CLA Was Converted into SLs in Rice.
To demonstrate that CLA is produced in other plant species, we analyzed endogenous CLA in Oryza sativa WT (cv. Shiokari). The LC-MS/MS analysis showed that CLA also exists in rice roots (Fig. S7A). Moreover, to investigate whether CLA is a biosynthetic precursor for SLs in planta, we examined the conversion from exogenous CLA into SLs using the d10-2 mutant (cv. Nipponbare), which is defective in CCD8 like the max4 mutant of Arabidopsis. The d10-2 mutant was grown hydroponically, and [1-13CH3]rac-CLA was added to culture media. Both [13C1]-labeled 4DO and orobanchol were detected in root exudates of d10-2 based on the comparison of the full-scan MS spectra and the retention time on LC with those of unlabeled authentic standards using LC-MS/MS analysis (Fig. S7B).
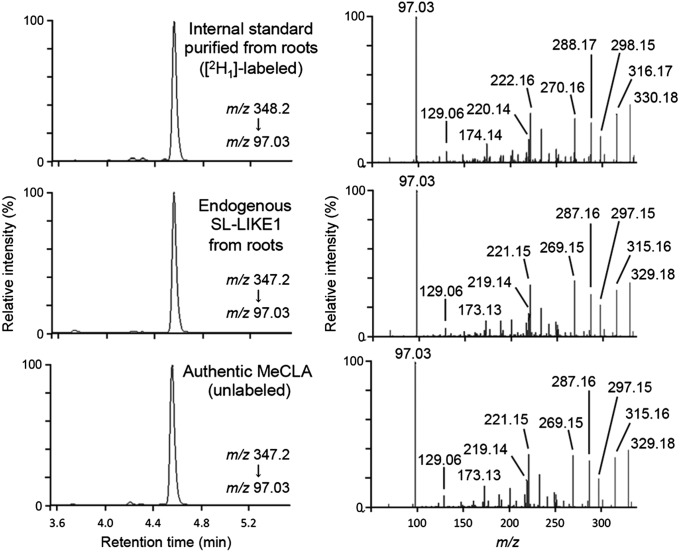
Identification of SL-LIKE1 as CLA Methyl Ester.
We reported the presence of an SL-like compound, SL-LIKE1, in root extracts of Arabidopsis, and that SL-LIKE1 was formed from exogenous CL in a MAX1-dependent manner (14). These results and the pseudomolecular ion of SL-LIKE1 ([M+H]+ at m/z 347) allowed us to predict that the chemical structure of SL-LIKE1 might be the methyl ester of CLA [methyl carlactonoate (MeCLA)]. To address this hypothesis, we synthesized MeCLA (Fig. S4B) and compared its chemical properties with SL-LIKE1 using LC-MS/MS. As a result, the product ions and retention time of MeCLA were identical with those of SL-LIKE1 extracted from roots of the Arabidopsis atd14 mutant (Fig. 5). Furthermore, the feeding of [1-13CH3]rac-CLA to the Arabidopsis max4 and max1max4 double mutants showed that MeCLA is formed from CLA in a MAX1-independent manner in planta (Fig. S6B).
Fig. 5.
Identification of endogenous MeCLA in Arabidopsis. Endogenous SL-LIKE1 was identified as MeCLA using LC-MS/MS (QTOF). Ion traces from the LC-MS/MS analysis (Left) and product ion spectra (Right) derived from respective precursor ions of [10-2H1]MeCLA (internal standard) and endogenous SL-LIKE1 extracted from roots of Arabidopsis atd14 mutant and those of unlabeled authentic MeCLA are shown.
CLA and MeCLA but Not CL and 19-hydroxy-CL Suppressed the Growth of Lateral Inflorescences in the max1 Mutant.
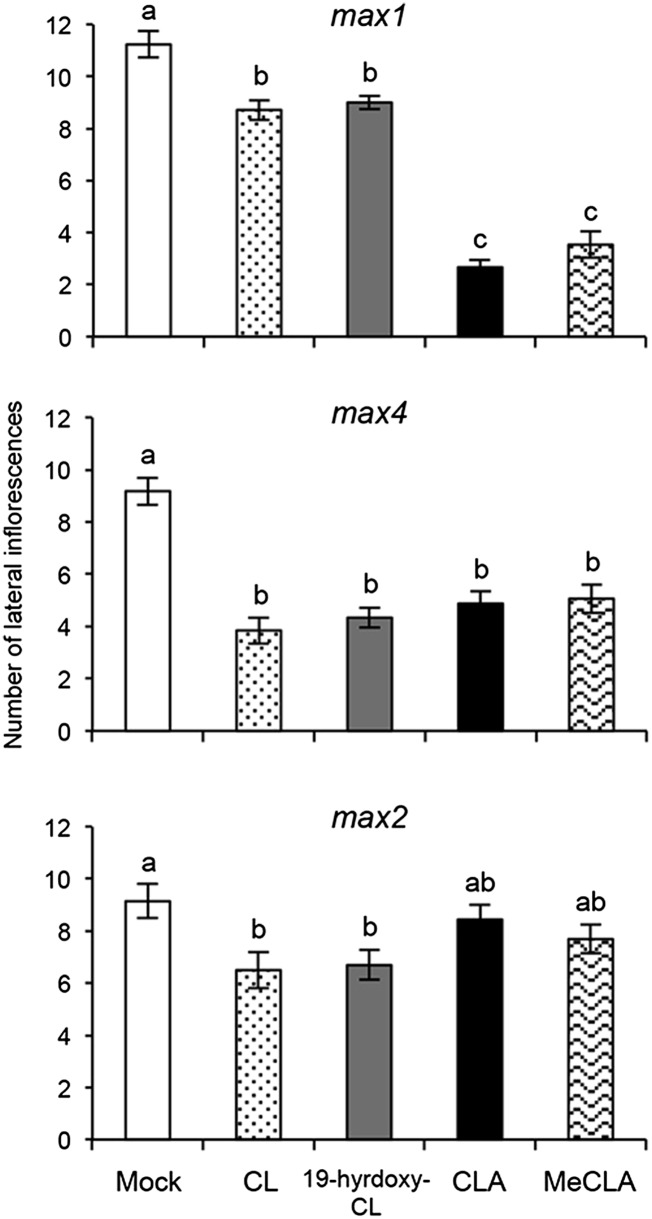
We examined inhibitory effects of CL, 19-hydroxy-CL, CLA, and MeCLA on the increased lateral inflorescence phenotype of the Arabidopsis max mutants. CLA and MeCLA dramatically reduced the number of lateral inflorescences at 10 μM compared with a mock treatment in the max1 mutant, whereas CL and 19-hydroxy-CL were significantly weaker inhibitors than CLA and MeCLA (Fig. 6). CLA showed a dose–response similar to that of the synthetic SL GR24 (Fig. S8A). In the case of the max4 mutant, CL, 19-hydroxy-CL, CLA, and MeCLA significantly inhibited the growth of lateral inflorescences (Fig. 6). The max2 mutant was less sensitive to CLA and MeCLA but slightly sensitive to CL and 19-hydroxy-CL (Fig. 6). Furthermore, we examined the germination stimulant activities of these CL derivatives on Orobanche minor seeds. CL, 19-hydroxy-CL, and CLA induced ∼70% germination at 10 μM, whereas ∼70% seeds were germinated by MeCLA and GR24 at 1 and 0.1 μM, respectively (Fig. S8B).
Fig. 6.
Inhibitory effects of CL derivatives on increased lateral inflorescences of Arabidopsis max mutants. Each 10 μM solution of CL, 19-hydroxy-CL, CLA, and MeCLA (all racemic mixtures) was applied onto the basal region of the primary inflorescences of max1, max4, and max2 every second day for 2 wk, and then the number of lateral inflorescences from rosette leaves was counted. The 0.5% (vol/vol) acetonitrile solution without authentic sample was used as a mock treatment. Values are mean ± SEM (n = 15). The number of lateral inflorescence was 1.4 ± 0.3 in WT treated with the mock solution. Different letters indicate significant differences tested by the Holm-Sidak method under ANOVA (P < 0.05).
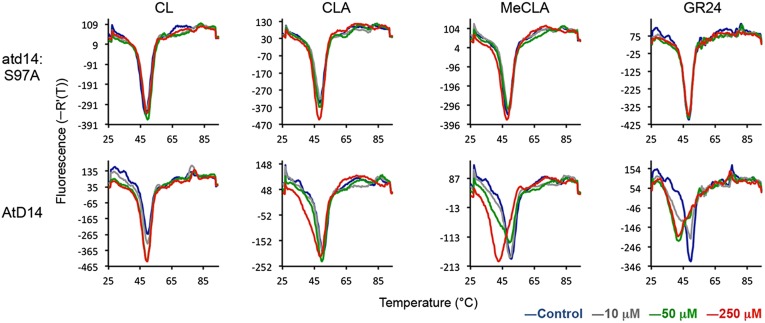
MeCLA but Not CLA Interacted with AtD14.
As described above, we determined the biological activity of CLA and MeCLA on the max1 mutant. However, it was still not clear whether they need further conversion(s) to be active hormones. To address this question, we tested the interaction of CL, CLA and MeCLA with AtD14 protein, a possible SL receptor of Arabidopsis (21), using differential scanning fluorimetry (DSF) method, which allows us to evaluate the interaction between a receptor protein and a ligand by detecting the melting temperature of the receptor protein. It has been reported that the melting point of DAD2, an ortholog of D14 in petunia, shifted to low temperature in the presence of GR24 (22). MeCLA, but not CL and CLA, could induce the shift of melting temperature of AtD14 protein as did GR24 (Fig. 7). In contrast, all substrates used did not affect the melting temperature of the mutant protein, atd14:S97A, which has a substitution change in the active site (Fig. 7). D14 family proteins belong to the α/β-fold hydrolase superfamily and have been reported to hydrolyze SLs such as GR24 (22–24). Therefore, we also tested the hydrolysis activity of AtD14 for CL, CLA, and MeCLA. GR24 was used as a positive control. After the incubation of AtD14 or atd14:S97A with each compound, we analyzed the remaining substrate and a common hydrolyzed product, hydroxymethylbutenolide (HMB; D ring), using LC-MS/MS (Fig. S9). No appreciable hydrolase activity of AtD14 was detected when CL and CLA were used as substrates, whereas MeCLA was hydrolyzed by AtD14 compared with results by atd14:S97A as well as GR24 (Fig. S9).
Fig. 7.
Biochemical analysis of the interaction between AtD14 and CL derivatives. DSF assays of AtD14 were carried out in the presence of CL, CLA, MeCLA, and GR24 (all racemic mixtures). The melting temperature curves of AtD14 are shown. atd14:S97A, the mutant protein having a substitution change in the active site.
Discussion
We demonstrated that a recombinant Arabidopsis MAX1 protein produces 19-hydroxy-CL and CLA from CL (Fig. 2 and Figs. S3 and S5A) in vitro, indicating that MAX1 is an enzyme responsible for the oxidation at C-19 of CL and introduces a carboxyl group by consecutive oxidations (Fig. 1). 19-Hydroxy-CL is an intermediate produced by a single step oxidation at C-19 of CL because MAX1 produced CLA from 19-hydroxy-CL as a substrate (Fig. S5B). CLA appears to be a dominant end-product by Arabidopsis MAX1 because CLA was not further metabolized by the recombinant MAX1. We also identified CLA as an endogenous compound from Arabidopsis roots (Fig. 4A). CLA accumulated markedly in the atd14 and max2 mutants compared with WT (Fig. 4B). AtD14 is a putative SL receptor and interacts with an F-box protein, MAX2, which forms an SCF complex and ubiquitinates transcriptional regulators (25, 26). It is well known that the signal of plants hormones, such as gibberellins, represses the expression of own biosynthesis enzymes to maintain an optimum concentration of active hormone in plants (27). The accumulation of CLA seems to be due to the lack of a similar feedback loop from the signaling pathway in the atd14 and max2 mutants. Endogenous CLA was not detected in the roots of the max1 and max4 mutants (Fig. 4B), whereas [13C1]-labeled CLA was detected from [1-13CH3]11R-CL-fed max4 plants but not from [1-13CH3]11R-CL-fed max1max4 double mutant plants (Fig. S6A). These results indicate that MAX1 is responsible for the conversion of CL to CLA not only in the heterologously expressed system but also in planta. We also identified CLA from rice roots (Fig. S7A), suggesting that CLA is synthesized by monocots and dicots. One of the reasons for the existence of CLA in plants may be that it is a biosynthetic precursor for SLs. This is supported by the fact that [13C1]-labeled 4DO and orobanchol were detected from root exudates of [1-13CH3]rac-CLA-fed rice d10-2 plants (Fig. S7B). This result suggests that rice has enzyme(s) that can accept CLA as a substrate and catalyze the generation of the lactone C ring, the cyclization of the B ring, and hydroxylation at C-4. The Arabidopsis genome has one copy (CYP711A1/At2g26170) of the MAX1 gene, whereas five homologous genes for CYP711A1 exist on three distinct clades in the rice genome (CYP711A2/Os01g0700900, CYP711A3/Os01g0701400, CYP711A4/Os01g0701500, CYP711A5/Os02g0221900, and CYP711A6/Os06g0565100) (28, 29). Enzymatic and/or physiological functions of MAX1 homologs are predicted to be redundant in rice. Recently, simultaneous deletions of CYP711A2 and CYP711A3 have been found as a major quantitative trait locus (QTL) related to SL deficiency and increased shoot branching phenotype in the Indica rice cultivar, Bala, indicating that CYP711A2 and CYP711A3 are redundant and dominant enzymes responsible for SL synthesis in rice (30). However, it has been demonstrated that CYP711A5 and CYP711A6, as well as CYP711A2 and CYP711A3, could fully rescue the shoot branching phenotype of the Arabidopsis max1 mutant when expressed under the CaMV35S promoter (29, 30). In our study, endogenous CLA was identified in rice. Thus, rice CYP711As may have the same catalytic function as Arabidopsis MAX1. However, we could not exclude the possibility that some of them catalyze a reaction to produce compounds distinct from CLA that can suppress shoot branching in Arabidopsis.
In addition to endogenous CLA, we also identified its methyl ester, MeCLA, from Arabidopsis roots (Fig. 5). MeCLA turned out to be SL-LIKE1, an uncharacterized CL metabolite that we previously reported (14). Exogenous MeCLA, as well as CLA, significantly suppressed the increased lateral inflorescence of the max1 mutant compared with CL and 19-hydroxy-CL (Fig. 6). The structure–activity relationships of SLs in the germination stimulation of parasitic weed seeds are similar to that in the induction of hyphal branching of AM fungi. The C-D ring moiety, which is commonly found in all of the natural and synthetic SLs, is necessary, and the intact ABC ring is also preferable for these activities (1, 31). In the case of shoot branching inhibition of plants, the D ring is essential, whereas the ABC ring can be removed (32). CLA and MeCLA were 100- and 10-fold less active than GR24 in the germination stimulation of the root parasitic plant O. minor, respectively (Fig. S8B), whereas CLA and GR24 were equally active in the inhibition of growth of lateral inflorescences in the Arabidopsis max1 mutant (Fig. S8A). Intriguingly, MeCLA, as well as GR24, induced the shift of melting temperature of AtD14 protein and was hydrolyzed by AtD14, but not CL and CLA (Fig. 7 and Fig. S9). These results demonstrate that MeCLA, but not CL or CLA, can directly interact with AtD14 protein and induce the conformational change of the protein structure to transmit a signal as does GR24. Exogenous [1-13CH3]rac-CLA was converted into [13C1]-labeled MeCLA in an MAX1-independent manner in Arabidopsis plants (Fig. S6B). Therefore, MeCLA formed from CLA but not CLA itself may suppress the increased lateral inflorescences in max1 plants. Unlike rice plants (14), known SLs were not detected from the tissues and exudates of Arabidopsis roots in our experiments on a small scale, although Arabidopsis is reported to produce SLs (33). Our inability to detect SLs was probably due to the significantly low SL productivity of Arabidopsis compared with rice plants. These findings suggest that the formation of the BC rings appears to be low in Arabidopsis. Although further experiments are needed to clarify the structure–activity relationships of CL derivatives including MeCLA for shoot branching inhibition, our data, for the first time to our knowledge, provide a possibility that not only known SLs but also MeCLA can function as an active hormone for shoot branching inhibition in Arabidopsis.
Materials and Methods
Plant Materials.
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the WT. The T-DNA tagged mutants max1-4 (SAIL_25A05), max2-3 (SALK_092836), and max4-7 (SALK_082552) were provided by the Arabidopsis Biological Resource Center. The atd14-2 mutant was obtained from a TILLING project (14). The d10-2 mutant (cv. Nipponbare) and WT Shiokari of Japonica Oryza sativa were used in this study. Orobanche minor seeds were harvested from naturally grown plants parasitizing Trifolium pratense in Tochigi, Japan. Details of growth conditions are described in SI Materials and Methods.
Heterologous Expression of MAX1 in Yeast.
The full-length cDNA clone (pdx11320) of Arabidopsis MAX1 (At2g26170) was provided by RIKEN BRC. The coding region of MAX1 was ligated into pYeDP60 vector under a galactose-inducible GAL10-CYC1 promoter (16). The resulting plasmid and empty vector as a control were transformed into yeast strain WAT11 (16). Methods of microsomes preparation are described in SI Materials and Methods. Methods of LC-MS/MS analysis, chemical synthesis, and kinetics assay are described in SI Materials and Methods.
Endogenous Chemical Analysis and Exogenous Application Experiments.
Endogenous analysis and feeding of CL derivatives in Arabidopsis and rice plants were performed as described in SI Materials and Methods. The effects of CL derivatives on shoot branching inhibition of Arabidopsis and on germination stimulation of O. minor seeds were examined as described in SI Materials and Methods.
DSF and Hydrolase Activity Tests of AtD14 Protein.
The coding region of AtD14 (At3g03990) was cloned into modified pMALc5x vector (New England Biology). atd14:S97A was prepared using KOD-Plus-Mutagenesis Kit (TOYOBO). The recombinant proteins were expressed as a maltose-binding protein fusion in Escherichia coli and used for DSF and hydrolase activity tests as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ms. Akiko Takahashi for technical assistance. We also thank Dr. Junko Kyozuka (Tokyo University) for providing the rice d10-2 mutant and Dr. Denis Pompon (CNRS) for providing pYeDP60 vector and yeast strain WAT11. This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, in part by JSPS/MEXT KAKENHI (Grants 23380061 and 24114010), the Research Project of Utsunomiya University (UU-COE), and JST, CREST.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410801111/-/DCSupplemental.
References
- 1.Zwanenburg B, Pospísil T. Structure and activity of strigolactones: New plant hormones with a rich future. Mol Plant. 2013;6(1):38–62. doi: 10.1093/mp/sss141. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435(7043):824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 3.Johnson X, et al. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 2006;142(3):1014–1026. doi: 10.1104/pp.106.087676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snowden KC, et al. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell. 2005;17(3):746–759. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booker J, et al. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell. 2005;8(3):443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Arite T, et al. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51(6):1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 8.Umehara M, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455(7210):195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 9.Brewer PB, Koltai H, Beveridge CA. Diverse roles of strigolactones in plant development. Mol Plant. 2013;6(1):18–28. doi: 10.1093/mp/sss130. [DOI] [PubMed] [Google Scholar]
- 10.de Saint Germain A, et al. Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol. 2013;163(2):1012–1025. doi: 10.1104/pp.113.220541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha CV, et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci USA. 2014;111(2):851–856. doi: 10.1073/pnas.1322135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scaffidi A, et al. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 2014;165(3):1221–1232. doi: 10.1104/pp.114.240036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alder A, et al. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 14.Seto Y, et al. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc Natl Acad Sci USA. 2014;111(4):1640–1645. doi: 10.1073/pnas.1314805111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaffidi A, et al. Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 2013;76(1):1–9. doi: 10.1111/tpj.12265. [DOI] [PubMed] [Google Scholar]
- 16.Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 17.Nelson DR. Progress in tracing the evolutionary paths of cytochrome P450. Biochim Biophys Acta. 2011;1814(1):14–18. doi: 10.1016/j.bbapap.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Helliwell CA, Poole A, Peacock WJ, Dennis ES. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119(2):507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seki H, et al. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell. 2011;23(11):4112–4123. doi: 10.1105/tpc.110.082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett T, Leyser O. Strigolactone signalling: Standing on the shoulders of DWARFs. Curr Opin Plant Biol. 2014;22C:7–13. doi: 10.1016/j.pbi.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Waters MT, et al. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139(7):1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- 22.Hamiaux C, et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol. 2012;22(21):2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L-H, et al. Crystal structures of two phytohormone signal-transducing α/β hydrolases: Karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 2013;23(3):436–439. doi: 10.1038/cr.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura H, et al. Molecular mechanism of strigolactone perception by DWARF14. Nat Commun. 2013;4:2613. doi: 10.1038/ncomms3613. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L, et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504(7480):401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou F, et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504(7480):406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middleton AM, et al. Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc Natl Acad Sci USA. 2012;109(19):7571–7576. doi: 10.1073/pnas.1113666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4(1):59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Challis RJ, Hepworth J, Mouchel C, Waites R, Leyser O. A role for more axillary growth1 (MAX1) in evolutionary diversity in strigolactone signaling upstream of MAX2. Plant Physiol. 2013;161(4):1885–1902. doi: 10.1104/pp.112.211383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardoso C, et al. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proc Natl Acad Sci USA. 2014;111(6):2379–2384. doi: 10.1073/pnas.1317360111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akiyama K, Ogasawara S, Ito S, Hayashi H. Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 2010;51(7):1104–1117. doi: 10.1093/pcp/pcq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyer FD, et al. Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: Molecule design for shoot branching. Plant Physiol. 2012;159(4):1524–1544. doi: 10.1104/pp.112.195826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohlen W, et al. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011;155(2):974–987. doi: 10.1104/pp.110.164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.