Significance
V(D)J recombination occurs in lymphoid precursors to enable their maturation, but also induces DNA damage. Thus, it has been proposed that the activity of the tumor suppressor and gatekeeper protein p53 must be controlled during this process to prevent premature induction of apoptosis. In this study, we show that the transcription factor Miz-1 can exert such a function. Miz-1 activates expression of the ribosomal protein Rpl22, which in turn controls the translation of p53 specifically in lymphoid precursors. We propose that this Miz-1–Rpl22–p53 pathway prevents p53 from inducing cell death as a response to V(D)J recombination in lymphoid precursors from both the T-lineage and the B-lineage.
Keywords: Miz-1, p53, V(D)J recombination, Rpl22
Abstract
To be effective, the adaptive immune response requires a large repertoire of antigen receptors, which are generated through V(D)J recombination in lymphoid precursors. These precursors must be protected from DNA damage-induced cell death, however, because V(D)J recombination generates double-strand breaks and may activate p53. Here we show that the BTB/POZ domain protein Miz-1 restricts p53-dependent induction of apoptosis in both pro-B and DN3a pre-T cells that actively rearrange antigen receptor genes. Miz-1 exerts this function by directly activating the gene for ribosomal protein L22 (Rpl22), which binds to p53 mRNA and negatively regulates its translation. This mechanism limits p53 expression levels and thus contains its apoptosis-inducing functions in lymphocytes, precisely at differentiation stages in which V(D)J recombination occurs.
The development of T lymphocytes starts with early thymic progenitors (ETPs) that first enter the thymus after transiting through the bloodstream from the bone marrow. These progenitor cells differentiate through four CD4−CD8− double-negative stages of development (DN1–4) before becoming first CD4+CD8+ double-positive (DP) cells and then either CD4+ or CD8+ single positive T cells. The four DN subsets are differentiated based on their expression of the cell surface markers CD44 and CD25. Cells at the DN3 stage (CD44−CD25+) are fully committed to the T cell lineage and require signaling through cytokine receptors and Notch1 to survive (1). This DN3 population can be further subdivided into DN3a and DN3b based on their size and CD27 surface expression.
Importantly, DN3a cells (FSCloCD27lo) actively rearrange the genes encoding the T cell receptor β (TCRβ) chain through V(D)J recombination (1–3). Those DN3a cells that do not productively rearrange the TCRβ locus on both alleles are eliminated by apoptosis. In contrast, cells that productively rearrange their TCRβ chain genes are selected and become DN3b cells (FSChiCD27hi), which express a pre-T cell receptor (pre-TCR), a heterodimer between the TCRβ chain and a pTα chain. DN3b cells grow in size, owing in part to increased metabolic activity (4), and give rise to the (CD44−CD25−) DN4 subset, which in turn rapidly expands to produce CD4+CD8+ DP cells.
Because V(D)J recombination involves DNA double-strand breaks, it has been speculated that the activity of p53 as a DNA damage response factor must be contained and regulated during this process, to prevent DN3a cells from prematurely undergoing apoptosis (5, 6). V(D)J recombination occurs in DN3a cells when they are in G1, and a checkpoint might exist at this stage that specifically protects against DNA damage-induced p53-mediated apoptosis. Similar to pre-T cells, early B cell precursors in the bone marrow, so-called “pro-B” cells, begin to rearrange the genes encoding the Ig heavy chain, also through V(D)J recombination (7). Thus, a similar mechanism as for pre-T cells might also exist for pro-B cells at this stage to protect them from DNA damage-induced p53-mediated apoptosis (8). As for pre-T cells, pro-B cells that have productively rearranged the IgH locus become pre-B cells and express a pre-B cell receptor (pre-BCR) on their surface, which consists of the Ig heavy chain and two surrogate light chains (9). Those pro-B cells that have not productively rearranged the Ig heavy chain genes die by apoptosis, as is the case for DN3a cells that fail to productively rearrange their TCRβ chain genes, mainly because the pre-TCR– and pre-BCR–mediated proliferation and survival signals are lacking.
Myc-interacting zinc finger protein 1 (Miz-1) is an 87-kDa protein that was first described as an interaction partner for the proto-oncoprotein c-Myc (10). It contains 13 C2H2 type zinc finger domains at its C terminus and a BTB/POZ domain at its N terminus. This POZ domain is required for Miz-1 function, because it mediates the formation of homotetramers and interactions with various partner proteins (11). In addition, loss of this domain renders Miz-1 incapable of stably binding to chromatin (12). Among the validated Miz-1 target genes are those that encode the negative cell cycle regulators Cdkn2b (p15) (12, 13) and Cdkn1a (p21) (14, 15). Furthermore, a number of upstream factors regulate Miz-1 activity, including Bcl-6 (15, 16), which is itself a POZ domain transcription factor that binds directly to Miz-1, the DNA damage sensor protein TopBP1 (14), the E3 ubiquitin ligase HectH9 (17, 18), and the proto-oncoprotein Akt (10, 19). However, it also has been shown that recruitment of the c-Myc/Max complex by Miz-1 inhibits transcriptional activation, very likely by interfering with the formation of an activating Miz-1/p300 complex (12).
We have previously described a Miz-1–deficient mouse, in which the POZ domain of Miz-1 is conditionally deleted specifically in hematopoietic cells (Miz-1flox/flox × Vav-Cre; hereafter referred to as Miz-1ΔPOZ animals). This model has allowed us to identify a role of Miz-1 in the regulation of IL-7 receptor signaling in early steps of T cell and B cell development (20, 21). We also have demonstrated that Miz-1 regulates expression of p53 effector genes in DN3a cells undergoing pre-TCR selection to generate DN3b cells (22). Although Miz-1 does not affect V(D)J recombination, Miz-1–deficient DN3a cells die and exhibit increased transcriptional activation of a subset of p53 target genes known for inducing apoptosis (22).
In this study, we present evidence of a novel mechanism that controls p53 activity in pre-T and pro-B cells when V(D)J recombination occurs. Our results indicate that in these cells, Miz-1 activates transcription of the gene encoding ribosomal protein L22 (Rpl22), a component of the 60S ribosomal subunit. Because our data indicate that Rpl22 binds directly to p53 mRNA and negatively controls its translation, we propose that Miz-1 controls p53 protein expression levels via Rpl22 at stages of lymphoid development when p53 action must be contained.
Results
Ablation of Trp53 Restores the Development of Miz-1–Deficient αβ-Lineage Pre-T and Pro-B Cells.
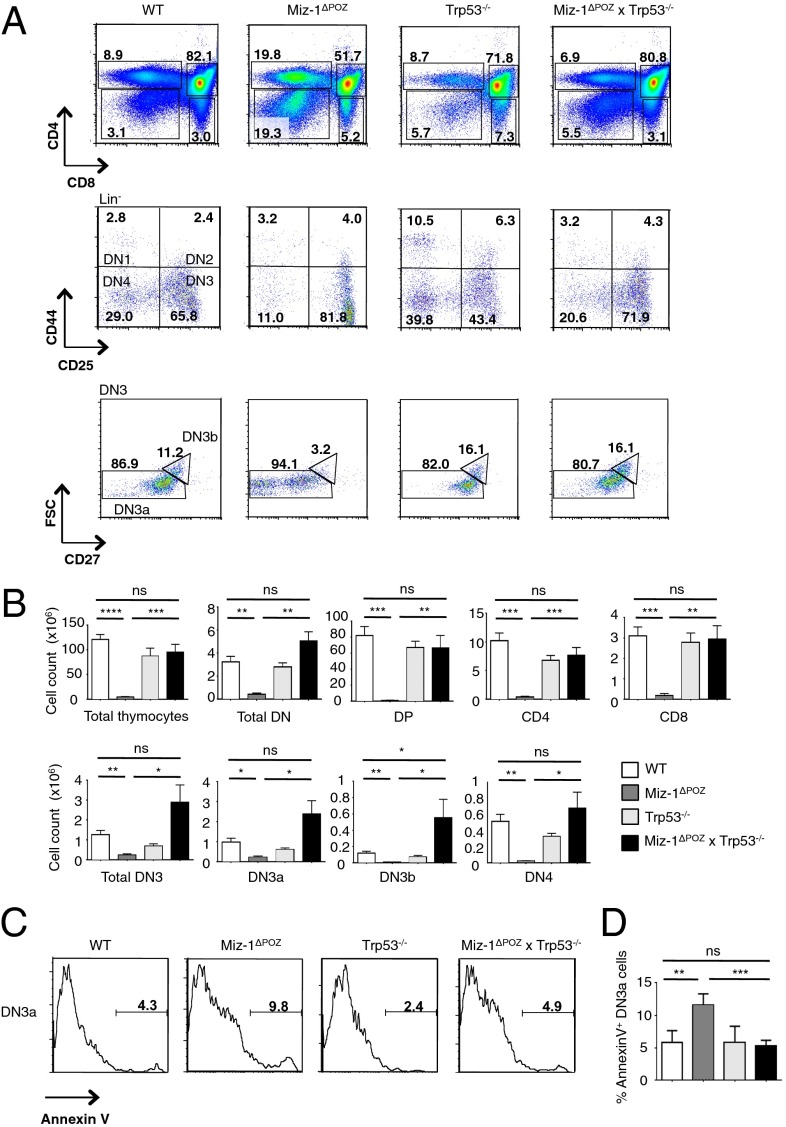
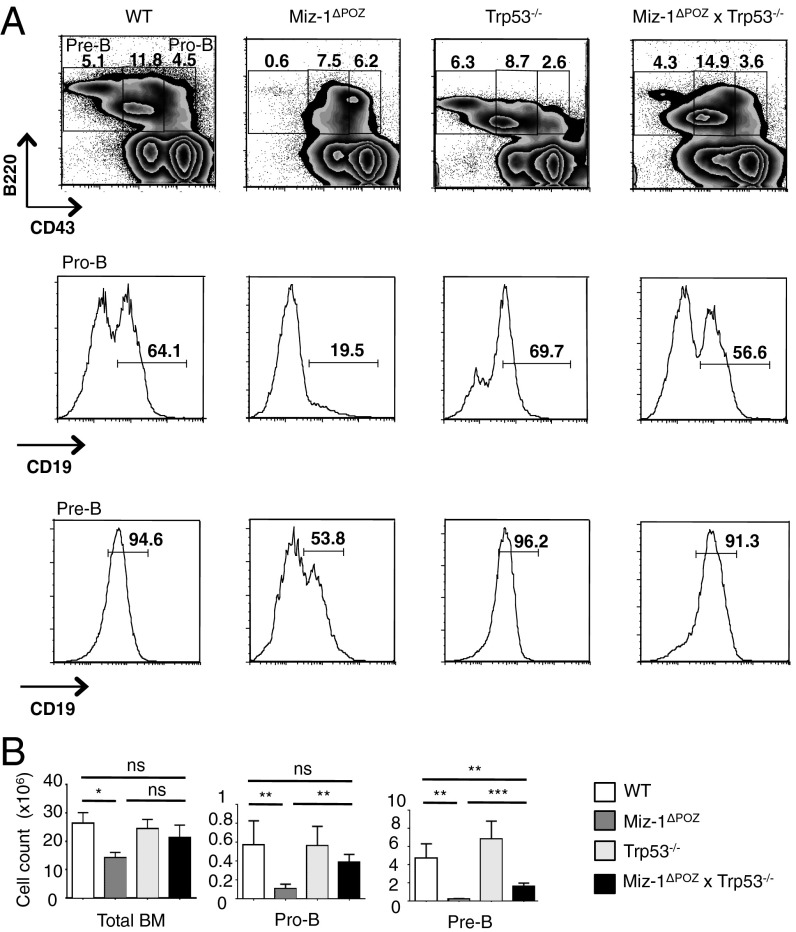
To determine whether the developmental block and increased apoptosis seen in Miz-1–deficient pre-T cells and pro-B cells are related to increased p53 signaling, we generated p53-deficient Miz-1ΔPOZ mice. We found significant increases in both the percentage and absolute number of DN3b and DN4 cells in Miz-1ΔPOZ × Trp53−/− mice, as well as almost completely restored thymic cellularity, compared with Miz-1ΔPOZ mice (Fig. 1 A and B). Deletion of p53 also partially restored the ability of Miz-1–deficient DN3 pre-T cells to transition to the DN4 stage in vitro on an OP9-DL4 stromal layer, whereas proliferation of these cells after DN4 remains impaired (Fig. S1A). Miz-1ΔPOZ DN3a pre-T cells have been shown to exhibit increased apoptosis and deficient progression through the cell cycle (22). The increase in apoptosis in Miz-1ΔPOZ DN3a pre-T cells is completely abolished in Miz-1ΔPOZ × Trp53−/− mice, as shown by annexin V staining (Fig. 1 C and D). Furthermore, ablation of p53 rescues the ability of Miz-1–deficient DN3 pre-T cells to progress through the cell cycle, as shown by both propidium iodide and BrdU stainings (Fig. S1 B and C). The development of Miz-1–deficient pro-B cells was rescued in Miz-1ΔPOZ × Trp53−/− mice as well, as indicated by the threefold increase in the percentage of pro-B cells expressing CD19 on the surface and the threefold increase in the absolute number of pre-B cells in the bone marrow (Fig. 2 A and B). These results suggest that the developmental defects in Miz-1ΔPOZ pre-T and pro-B cells are linked to both p53-mediated cell cycle arrest and apoptotic cell death.
Fig. 1.
Deletion of p53 restores pre-T cell development in Miz-1ΔPOZ mice. (A) FACS analysis of thymic subsets from WT, Miz-1ΔPOZ, Trp53−/−, and Miz-1ΔPOZ × Trp53−/− mice. FACS plots are a representative example of four independent experiments. (B) Absolute cell counts of thymocyte subsets from A. Data are averaged from four independent experiments and are presented as mean ± SD. (C) FACS analysis of annexin V staining on DN3 pre-T cells. FACS plots are a representative example of three independent experiments. (D) Quantification of percentage of annexin V+ cells from staining in C. Data are averaged from three independent experiments and are presented as mean ± SD.
Fig. 2.
Deletion of p53 restores early steps of B cell development in Miz-1ΔPOZ mice. (A) FACS analysis of B cell subsets from BM of WT, Miz-1ΔPOZ, Trp53−/−, and Miz-1ΔPOZ × Trp53−/− mice. FACS plots are a representative example of four independent experiments. (B) Absolute cell counts of B cell subsets from A. Data are averaged from four independent experiments and are presented as mean ± SD.
The Developmental Arrest in Miz-1–Deficient Pre-T Cells and Pro-B Cells Is Associated with Induction of Puma and Bax.
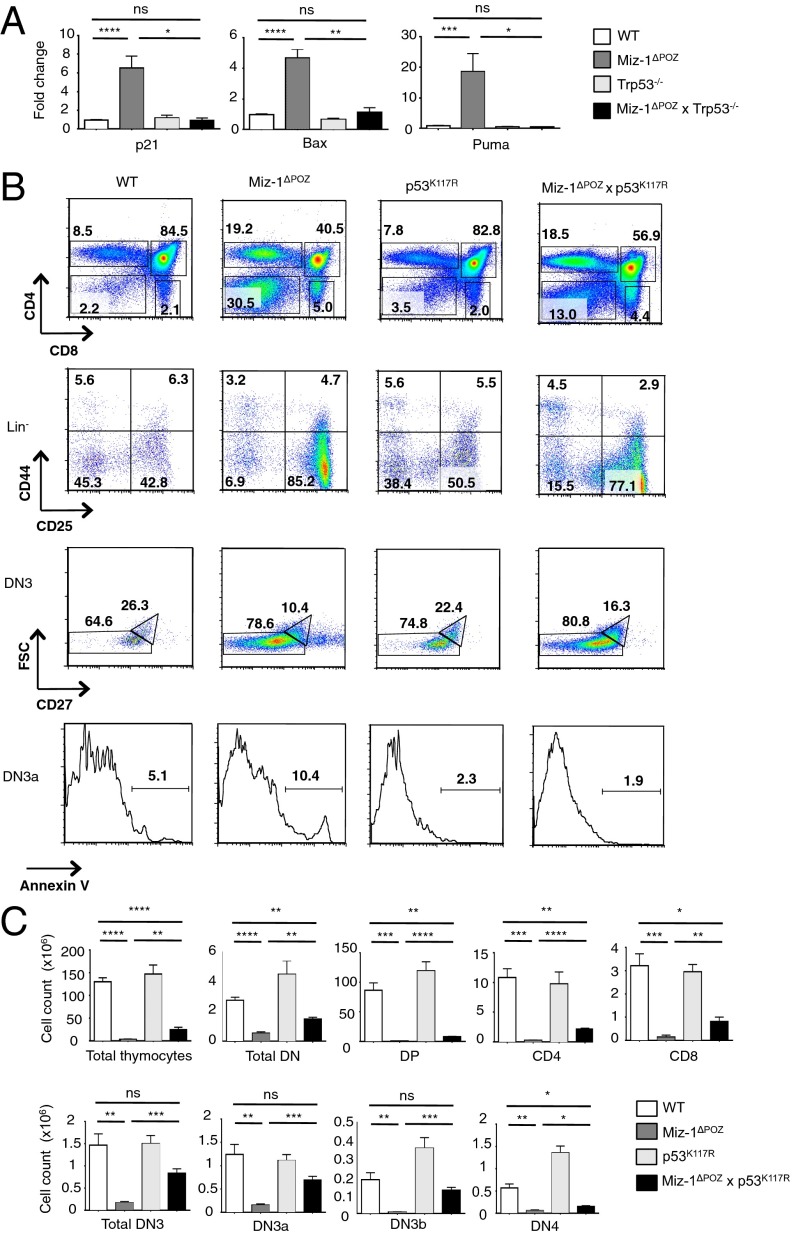
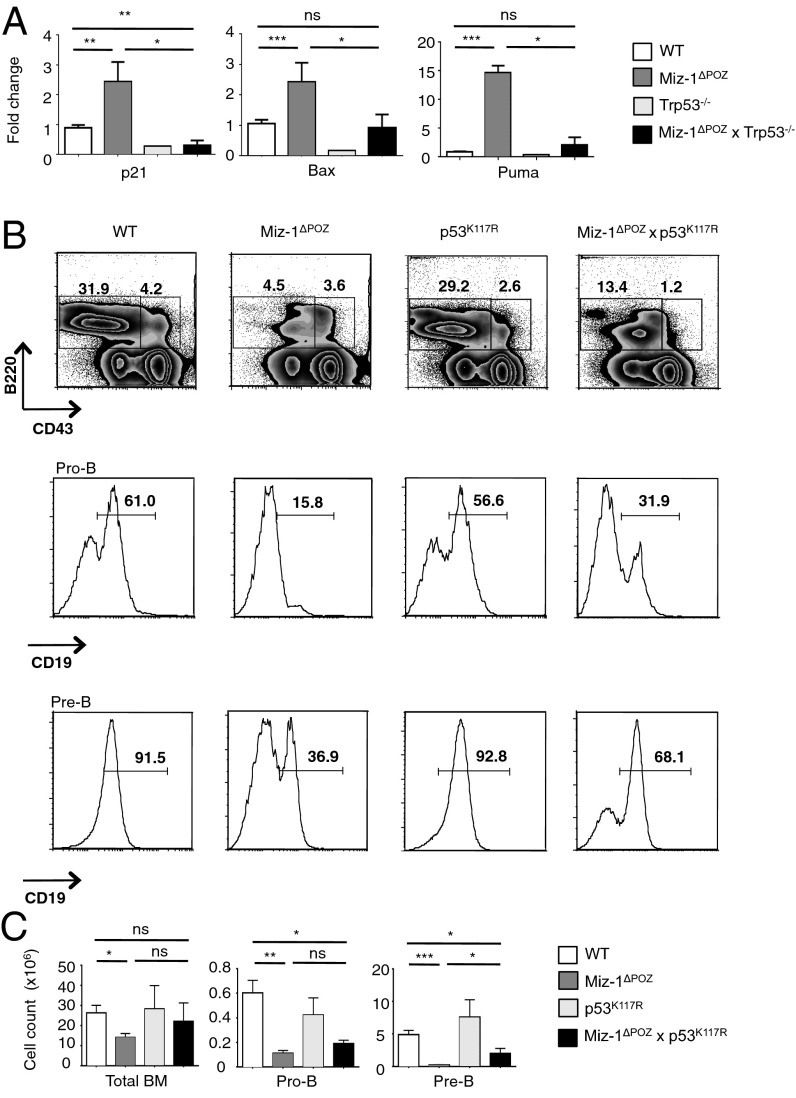
Ablation of p53 in Miz-1ΔPOZ DN3 pre-T cells and pro-B cells restored the overexpression of p53 target genes Cdkn1a (p21), Bax, and Bbc3 (Puma) to WT levels (Figs. 3A and 4A). We had previously shown that deletion of Cdkn1a fails to rescue the transition from DN3a to DN3b (22), which excludes the possibility that p53-mediated cell cycle arrest through activation of Cdkn1a (p21) is responsible for this differentiation block. Thus, we attempted to restore pre-T cell development by preventing activation of the proapoptotic p53 targets Puma and Bax, using p53K117R knock-in mice in which p53 is specifically rendered unable to activate transcription of these genes (23–25). Lymphocyte development in Miz-1–deficient p53K117R (Miz-1ΔPOZ × p53K117R) mice is partially restored, as indicated by the decreased percentage of total DN cells compared with the Miz-1–deficient thymus (Fig. 3B). Furthermore, there was a fivefold decrease in the percentage of DN3a cells undergoing apoptosis compared with Miz-1ΔPOZ DN3a cells, as shown by annexin V staining (Fig. 3B). In addition, total thymic cellularity and absolute numbers of DN3b cells were increased in Miz-1ΔPOZ × p53K117R mice compared with Miz-1ΔPOZ mice (Fig. 3C). The development of pro-B cells was also partially restored in Miz-1ΔPOZ × p53K117R mice compared with Miz-1ΔPOZ mice, as indicated by the twofold increase in the percentage of pre-B cells expressing CD19 on the surface and the absolute number of pre-B cells in the bone marrow of Miz-1ΔPOZ × p53K117R mice (Fig. 4 B and C).
Fig. 3.
The developmental arrest in Miz-1–deficient thymocytes is associated with the overexpression of several p53 effectors. (A) Analysis of p53 target gene expression by RT-qPCR in sorted DN3 pre-T cells from WT, Miz-1ΔPOZ, Trp53−/−, and Miz-1ΔPOZ × Trp53−/− mice. Data represent average fold change over Gapdh from three independent experiments and are presented as mean ± SD. (B) FACS analysis of thymic subsets from WT, Miz-1ΔPOZ, p53K117R, and Miz-1ΔPOZ × p53K117R mice. FACS plots are a representative example of three independent experiments. (C) Absolute cell counts of thymocyte subsets from B. Data are averaged from three independent experiments and are presented as mean ± SD.
Fig. 4.
The developmental arrest in Miz-1–deficient pre-B cells is associated with several p53 effectors. (A) Analysis of p53 target gene expression by RT-qPCR in sorted pro-B cells from WT, Miz-1ΔPOZ, Trp53−/−, and Miz-1ΔPOZ × Trp53−/− mice. Data represent average fold change over Gapdh from two independent experiments and are presented as mean ± SD. (B) FACS analysis of B cell subsets from BM of WT, Miz-1ΔPOZ, p53K117R, and Miz-1ΔPOZ × p53K117R mice. FACS plots are a representative example of three independent experiments. (C) Absolute cell counts of B cell subsets from B. Data are averaged from three independent experiments and are presented as mean ± SD.
Miz-1–Deficient DN3 Cells Progress to DN4 in the Absence of V(D)J Recombination.
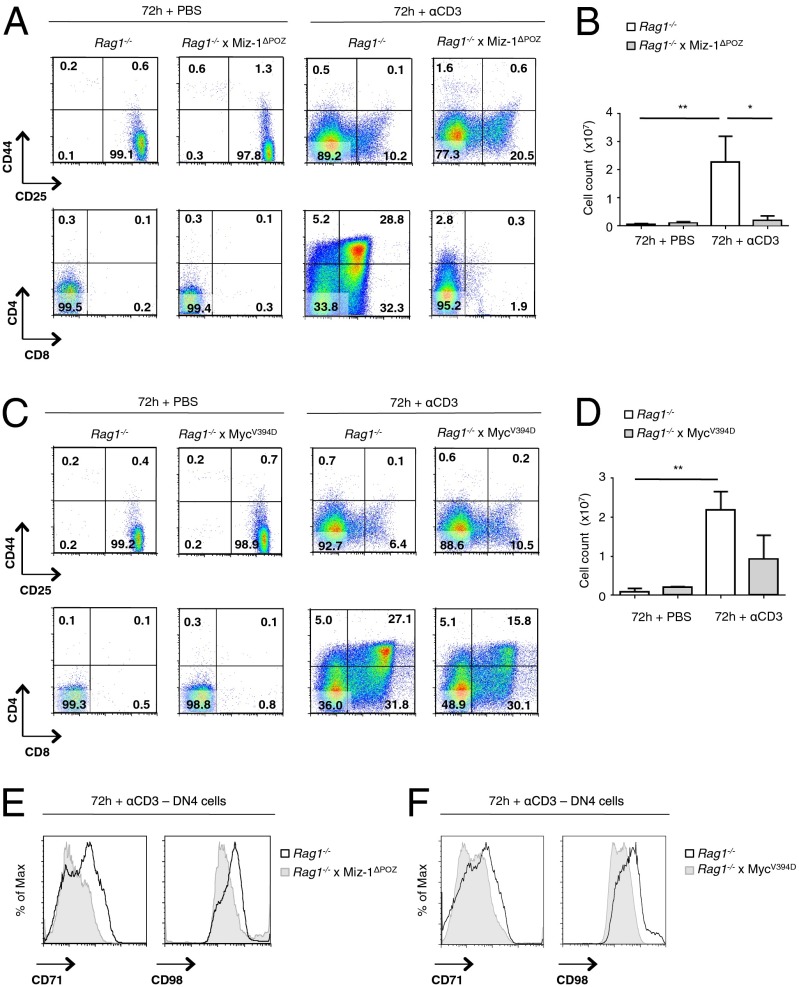
To determine whether V(D)J recombination-induced DNA damage might be responsible for the p53-mediated differentiation block of Miz-1–deficient pre-T cells at the DN3a stage, we crossed Miz-1ΔPOZ mice with Rag1−/− mice that are defective in rearranging TCR or Ig genes. We injected these Rag1−/− × Miz-1ΔPOZ mice with αCD3 antibodies to mimic pre-TCR signaling, and then followed the differentiation of DN3 cells into the DN4 and DP stages as described previously (26). Whereas αCD3 stimulation had no effect on Miz-1–deficient pre-T cells with a functional Rag1 gene (Fig. S2), the ablation of Rag1 in Miz-1ΔPOZ mice clearly allowed a full differentiation of Miz-1–deficient DN3 cells to DN4 cells (Fig. 5A); however, although Rag1−/− thymocytes were able to further differentiate into DP cells at 72 h after αCD3 injection, Rag1−/− × Miz-1ΔPOZ thymocytes were blocked at the DN4 stage (Fig. 5A) and were unable to proliferate to the same extent as Rag1−/− thymocytes (Fig. 5B).
Fig. 5.
Mitogenic stimulation of Rag1−/−, Rag1−/− × Miz-1ΔPOZ, and Rag1−/− × MycV394D KI pre-T cells. (A) FACS analysis of Rag1−/− and Rag1−/− × Miz-1ΔPOZ mice at 72 h after injection with PBS or αCD3 antibodies. FACS plots are a representative example of three independent experiments. (B) Absolute cell counts of total thymocytes from A. Data are averaged from three independent experiments and are presented as mean ± SD. (C) FACS analysis of Rag1−/− and Rag1−/− × MycV394D KI mice at 72 h after injection with PBS or αCD3. FACS plots are a representative example of three independent experiments. (D) Absolute cell counts of total thymocytes from B. Data are averaged from three independent experiments and are presented as mean ± SD. (E) FACS analysis of CD71 and CD98 on Rag1−/− and Rag1−/− × Miz-1ΔPOZ thymocytes at 72 h after injection with αCD3. FACS plots are a representative example of three independent experiments. (F) FACS analysis of CD71 and CD98 on Rag1−/− and Rag1−/− × MycV394D thymocytes at 72 h after injection with αCD3. FACS plots are a representative example of two independent experiments.
Because c-Myc is a Miz-1 cofactor and has been identified as a mediator of the DN-to-DP transition, we tested whether the ability of c-Myc to bind to Miz-1 is critical for this function. For this test, we used MycV394D knock-in mice, in which c-Myc is unable to interact with Miz-1 owing to a mutation in its helix–loop–helix domain, but can still dimerize with Max (14, 27). As expected, stimulation of both Rag1−/− × MycV394D and Rag1−/− thymocytes with αCD3 led to full differentiation of DN3 cells to the DN4 stage (Fig. 5C). However, although both Rag1−/− and Rag1−/− × MycV394D thymocytes were able to differentiate into DP cells at 72 h after injection, this transition was less efficient in the Rag1−/− × MycV394D animals, as indicated by the lower poststimulation absolute cell counts and DP cell percentages in these mice (Fig. 5 C and D). Two c-Myc effectors, CD71 and CD98, which are highly expressed pre-TCR and post-TCR selection, were not up-regulated on the surface of αCD3-stimulated Rag1−/− × Miz-1ΔPOZ T cells (Fig. 5E). Furthermore, both CD71 and CD98 were less efficiently up-regulated on the surface of Rag1−/− × MycV394D T cells stimulated with αCD3 compared with Rag1−/−-stimulated T cells (Fig. 5F). These results indicate that in the absence of V(D)J recombination, Miz-1–deficient thymocytes regain the ability to pass through pre-TCR selection and give rise to DN4 cells, but still are unable to differentiate further to become DP thymocytes, because they likely require a functional Miz-1/c-Myc complex for this step.
Miz-1 Binds to the Promoter of Rpl22 and Regulates Its Expression in DN3 Pre-T cells and Pro-B Cells.
ChIP-seq experiments in P6D4 cells, a DN3 pre-T cell line, and 70Z/3 cells, a pre-B cell line, showed that Miz-1 does not bind to the promoters of p53 target genes, such as Cdkn1a, Bax, or Puma, in these cells, although they are deregulated in Miz-1ΔPOZ cells (Figs. S3A and S4A). P53 target genes are actively transcribed in these cells, however, as demonstrated by the presence of the histone activation marks in the ChIP-seq experiment and active transcription in the RNA-seq experiment performed in the same cells. The Vamp4 gene promoter contains a Miz-1–binding site and has been shown to be a bona fide Miz-1 target (28), and is used as a control. The data from the Miz-1 ChIP-seq experiments were confirmed by ChIP-quantitative PCR (qPCR) with P6D4 cells (Fig. S3B) and with sorted primary WT DN3 pre-T cells (Fig. S3C), as well as with 70Z/3 cells (Fig. S4B). These results suggest that the expression of the proapoptotic p53 target genes Puma and Bax, which are partially responsible for the developmental block in Miz-1ΔPOZ DN3 pre-T cells and pro-B cells (Figs. 3 and 4), is not directly regulated by Miz-1.
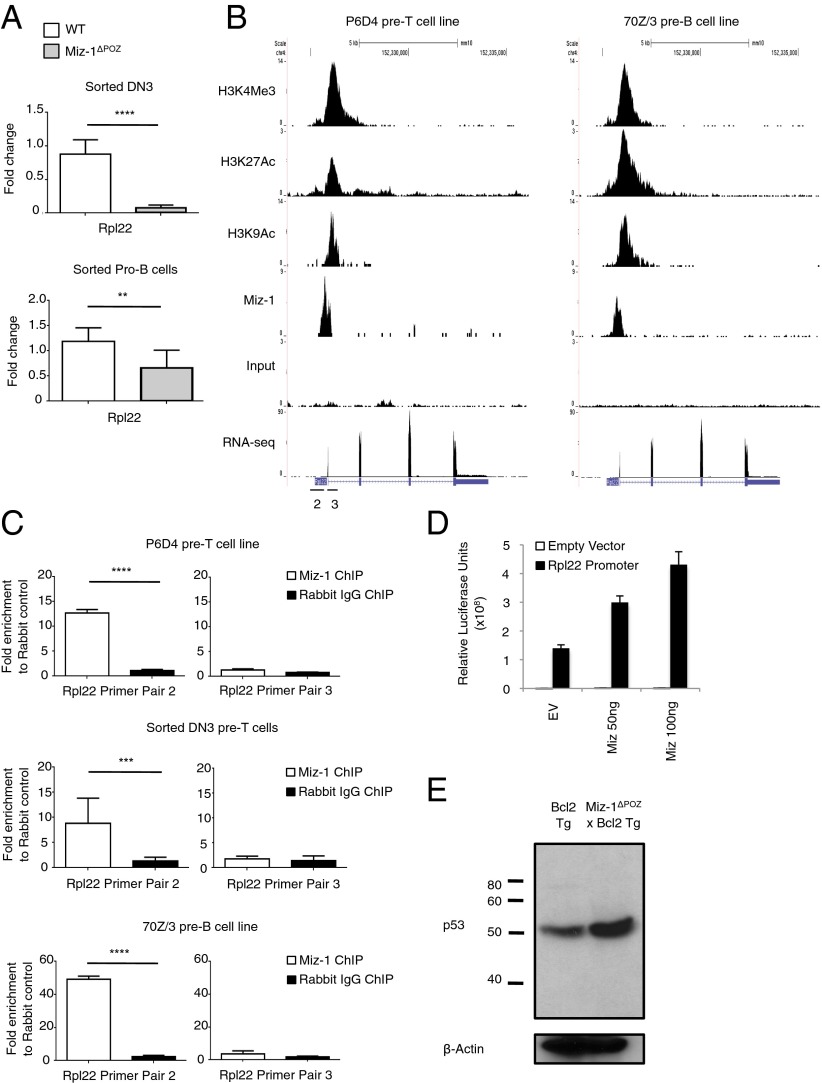
The analysis of a microarray dataset from Miz-1–deficient DN3 pre-T cells (22) identified Rpl22 as the most down-regulated gene in these cells compared with WT DN3 pre-T cell controls. Both DN3 pre-T cells and CD19+ pro-B cells were sorted from WT and Miz-1ΔPOZ littermates, and a significantly down-regulated Rpl22 mRNA expression level was confirmed in both cell types (Fig. 6A). Furthermore, ChIP-seq data from P6D4 and 70Z/3 cells indicated that Miz-1 occupies the promoter of Rpl22. A combination of the presence of active histone marks (ChIP-seq) and high levels of mRNA (RNA-seq) indicated that this gene is actively transcribed in both P6D4 pre-T cells and 70Z/3 pre-B cells (Fig. 6B). Miz-1 binding to the Rpl22 promoter was validated by ChIP-qPCR in P6D4 cells, sorted WT DN3 pre-T cells, and 70Z/3 cells (Fig. 6C) using different primer pairs (Fig. 6B, primer pairs 2 and 3), indicating that Rpl22 is a direct Miz-1 target gene.
Fig. 6.
Miz-1 regulates the expression of Rpl22 in DN3 pre-T cells and pro-B cells. (A) Rpl22 mRNA expression was assessed in sorted DN3 pre-T cells (Upper) and sorted pro-B cells (Lower) from WT and Miz-1ΔPOZ mice. Data are averaged from three independent experiments and are presented as mean ± SD. (B) ChIP-seq experiments for Miz-1 and histone activation marks from P6D4 murine pre-T cells (Right) and 70Z/3 murine pre-B cells (Left). Shown is the Rpl22 locus. Scale is in number of RPM. Primer pairs were designed in the promoter (2) or first exon (3) of Rpl22 to determine Miz-1 binding by ChIP. (C) ChIP-qPCR experiments to determine binding of Miz-1 to the promoter of Rpl22 in P6D4 murine pre-T cells (Top), sorted WT DN3 pre-T cells (Middle), and 70Z/3 murine pre-B cells (Bottom). Graphs show fold enrichment of anti–Miz-1 ChIP over rabbit IgG control ChIP. Data represent as average fold change ± SD from at least three independent experiments. (D) 293T cells were transfected with the human Rpl22 promoter fused to luciferase and pcDNA3.1 empty vector (EV) or pcDNA3.1 with human Miz-1 in varying concentrations. Data were normalized for transfection using β-galactosidase. Data are presented as average relative luciferase units ± SD and are representative of three independent experiments. (E) Whole protein extracts from total thymus of Bcl2 Tg or Miz-1ΔPOZ × Bcl2 Tg mice were evaluated for p53 expression by Western blot analysis. Data are representative of at least three independent experiments.
To confirm activation of the Rpl22 promoter by Miz-1, we cotransfected 293T cells with the human Rpl22 promoter fused to luciferase and increasing amounts of human Miz-1. We found that increasing amounts of Miz-1 led to increased activation of the Rpl22 promoter (Fig. 6D). This suggests that Miz-1 not only binds to the Rpl22 promoter in DN3 pre-T cells and pre-B cells, but also favors transcriptional activation of this gene.
We previously showed that overexpression of Bcl2 in Miz-1ΔPOZ mice (Miz-1ΔPOZ × Bcl2 Tg) rescues the apoptosis of Miz-1–deficient ETPs and partially rescues total thymic cellularity, but has no effect on the developmental block of Miz-1–deficient DN3 pre-T cells (21). Because Miz-1ΔPOZ × Bcl2 Tg mice have increased numbers of thymocytes compared with Miz-1ΔPOZ mice, we used them to test the effect of Miz-1 deficiency on the expression levels of p53 protein. Thymocyte extracts from Miz-1ΔPOZ × Bcl2 Tg mice showed increased p53 protein levels compared with Bcl2 Tg mice (Fig. 6E). This is consistent with a previous report that loss of Rpl22 correlates with an increased synthesis of p53 protein (29). Furthermore, this suggests that loss of Miz-1 leads to down-regulation of Rpl22, which increases the expression of p53 protein in DN3 pre-T cells.
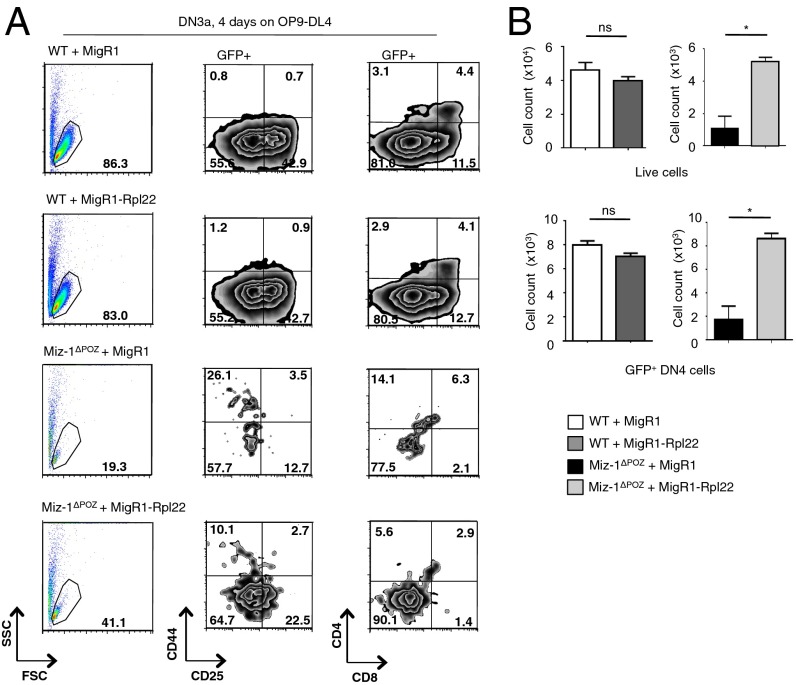
To determine whether overexpression of Rpl22 could rescue the phenotype of Miz-1ΔPOZ pre-T cells, we infected DN3a cells from WT and Miz-1ΔPOZ mice with a retrovirus expressing Rpl22. Overexpression of Rpl22 partially restored the ability of these cells to survive and differentiate into DN4 and cells in vitro on OP9-DL4 stromal cells after 4 d (Fig. 7A); however, although the absolute numbers of both live cells and GFP+ DN4 cells could be clearly restored on forced expression of Rpl22 (Fig. 7B), the cells were still unable to proliferate and differentiate past the DN4 stage into DP cells (Fig. 7A). Owing to the limited number of DN4 cells produced on the OP9-DL4 stromal layer in this experiment, it was not possible to measure the effect of Rpl22 overexpression on the levels of p53 protein; such an experiment will be necessary to further support direct links among Miz-1, Rpl22, and p53 protein levels.
Fig. 7.
Overexpression of Rpl22 in Miz-1–deficient DN3a cells partially restores survival and differentiation. (A) Flow cytometry analysis of sorted WT and Miz-1ΔPOZ DN3a cells infected with MigR1 or MigR1-Rpl22 after 4 d of coculture on OP9-DL4. Indicated graphs are gated on a GFP+ population. Data are representative of two independent experiments. (B) Absolute total live cell number (Upper) and total GFP+DN4 cell number (Lower) from OP9-DL4 experiments in A.
Rpl22 Binds to p53 mRNA to Regulate Translation.
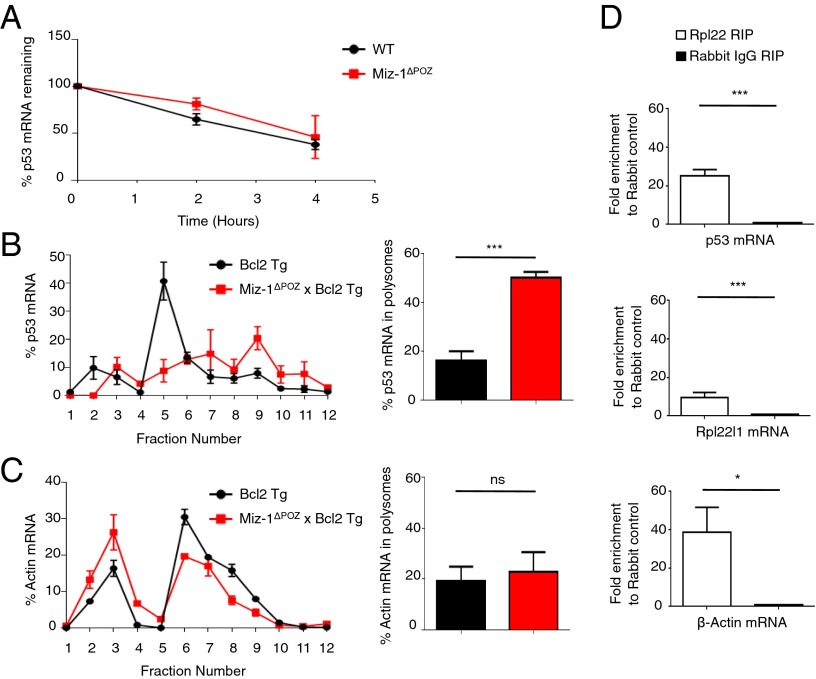
To test whether Miz-1 deficiency affects the stability of p53 mRNA, we sorted DN3 pre-T cells from WT and Miz-1ΔPOZ littermates and treated them with actinomycin D, an inhibitor of RNA polymerase II, over 4 h (Fig. 8A). The degradation of p53 mRNA over this time period is comparable in both WT and Miz-1–deficient cells, indicating that the increase in p53 protein is not related to increased stability of p53 mRNA. To determine whether loss of Miz-1 leads to increased incorporation of p53 mRNA into actively translating ribosomes, we again used Miz-1ΔPOZ × Bcl2 Tg mice, because the low number of thymocytes in Miz-1ΔPOZ animals limits experimentation. Thymocytes from Bcl2 Tg and Miz-1ΔPOZ × Bcl2 Tg mice were isolated, and lysates were sedimented through sucrose gradients. RNA from the fractionated gradients was extracted and analyzed by RT-qPCR for the presence of p53 mRNA. In Miz-1–deficient samples, the percentage of p53 mRNA in polysome fractions (fractions 8–12) was significantly increased compared with WT controls (Fig. 8B). Because the percentage of β-actin mRNA in polysomes is not significantly different in Miz-1–deficient and WT thymocytes (Fig. 8C), a global shift in translation in Miz-1–deficient cells can be ruled out, suggesting that Miz-1 is required to specifically regulate p53 protein levels by controlling the incorporation of p53 mRNA into actively translating ribosomes, likely via Rpl22.
Fig. 8.
Rpl22 regulates the translation of p53. (A) Sorted DN3 cells from WT or Miz-1ΔPOZ mice were treated with 5 μg/mL actinomycin D and harvested at the indicated time points. The percentage of p53 mRNA remaining at each time point was assessed by RT-qPCR and normalized to Gapdh. Data are averaged from three independent experiments and are presented as mean ± SD. (B) (Left) Total thymic extracts from Bcl2 Tg and Miz-1ΔPOZ × Bcl2 Tg mice were sedimented through a sucrose gradient and fractionated. qRT-PCR was performed to measure p53 mRNA in the fractions collected. Data are presented as the percentage of p53 mRNA in each fraction. The graph is representative of three independent experiments. (Right) Quantification of the percentage of p53 mRNA in polysomes (fractions 8–12) from left. Data are averaged from three independent experiments and are presented as mean ± SD. (C) (Left) Total thymic extracts from Bcl2 Tg and Miz-1ΔPOZ × Bcl2 Tg mice were sedimented on a sucrose gradient and fractionated. qRT-PCR was performed for β-actin on the fractions collected. Data are presented as the percentage of β-actin mRNA in each fraction. The graph is representative of three independent experiments. (Right) Quantification of percentage of β-actin mRNA in polysomes (fractions 8–12) from the left panel. Data are averaged from three independent experiments and are presented as mean ± SD. (D) RNA-IP of Rpl22 in P6D4 pre-T cells. The graph shows fold enrichment of anti-Rpl22 RIP over rabbit IgG control RIP. Rpl22l1 mRNA served as a positive control for Rpl22 RIP. β-actin mRNA is also enriched in anti-Rpl22 RIP. Data represent average fold change ± SD from three independent experiments.
To determine whether the increased translation of p53 is related to direct interaction of p53 mRNA with Rpl22 protein, we performed RNA immunoprecipitation (RIP) in P6D4 pre-T cells. Using this technique, we found a 25-fold increase in the amount of p53 mRNA bound by the Rpl22 protein compared with the rabbit IgG control (Fig. 8D). Rpl22 has been shown to bind the mRNA of its paralog, Rpl22l1 (30), which served as a positive control for the Rpl22 RIP. We found a 10-fold increase in the amount of Rpl22l1 mRNA bound by Rpl22 compared with the rabbit IgG control (Fig. 8D), confirming that both Rpl22l1 and p53 mRNA are bound by Rpl22. We also assessed the presence of β-actin mRNA in the RIP and found that although translation of this gene is not significantly different in Miz-1–deficient thymocytes compared with WT, it is bound by Rpl22.
Discussion
V(D)J recombination is necessary to rearrange TCR or Ig gene segments and to ensure generation of a large repertoire of antigen receptors. T and B lymphocytes, which carry one antigen-specific TCR or Ig at their cell surface, require such a repertoire to ensure recognition of a very large spectrum of antigens. The process of V(D)J recombination itself must be tightly regulated, however, and must be coordinated with DNA replication and mitosis to avoid genomic damage. This is achieved in part by linking V(D)J recombination to cell cycle progression by periodic phosphorylation and destruction of the Rag-2 recombinase that breaks and rejoins Ig or TCR gene segments, with the effect of allowing V(D)J recombination only in cells at G1 (31). In addition, however, it has been proposed that a DNA damage response pathway that may be initiated as a consequence of the double-strand breaks occurring during V(D)J recombination must be controlled to avoid the induction of p53-mediated apoptosis (5, 6). Here we provide evidence that the BTB/POZ domain transcription factor Miz-1 exerts such a function in both T- and B-lineage cells by restricting the expression of p53 through the inhibition of its translation via the ribosomal protein Rpl22 specifically in cells that undergo V(D)J recombination.
Miz-1–deficient pre-T cells express all of the necessary components to properly undergo pre-TCR selection (22), but are blocked at the DN3 stage and do not give rise to significant numbers of DN4 cells. In addition, DN3a cells lacking a functional Miz-1 show increased rates of apoptosis, and several experiments conducted during the present study suggest that this differentiation block and the increased cell death rate in DN3 pre-T cells is mediated by an overactive p53. First, as shown previously and confirmed here, various direct, known p53 target genes, including Cdkn1a, Bax, and Puma, are up-regulated in Miz-1–deficient cells. Second, deletion of p53 restores not only the numbers of DN3 pre-T cells, but also proper expression levels of Cdkn1a, Bax, and Puma and overall thymic cellularity, indicating full rescue of T cell differentiation. In addition, p53 deletion also partially restores the numbers of pre-B cells that otherwise do not develop from pro-B cells in Miz-1–deficient mice. Although there is a complete rescue of T cell differentiation in the thymus, loss of p53 expression only partially restores B cell development in Miz-1–deficient mice. This may be related to the fact that the previously reported defect in IL7 signaling has a stronger impact on B cell precursors than on T cell precursors (20). The fact that both pre-T cells and pro-B cells are affected by Miz-1 ablation and can regain their differentiation ability after concomitant p53 deletion precisely at stages in which V(D)J recombination occurs supports the hypothesis that this function of Miz-1 represents a general mechanism controlling potentially damaging consequences of V(D)J recombination regardless of cell lineage.
Although p53 affects both cell cycle progression and apoptosis, our data suggest that the proapoptotic p53 effectors Puma (Bbc3) and Bax are mainly responsible for the developmental block of Miz-1–deficient pre-T and pro-B cells. This suggestion is supported by our observation that Miz-1ΔPOZ mice that carry a p53K117R allele, in which p53 has specifically lost its ability to activate expression of both Puma and Bax, exhibited noticeable rescue of their pre-T and pro-B cell developmental defects. Although loss of the acetylation site at K117 in mouse p53 completely abolishes p53-mediated apoptosis, it has no effect on p53 translation or expression levels and does not affect p53-dependent cell cycle arrest or senescence (23–25). Moreover, total thymic cellularity is only partially restored in these mice, as is the percentage of DN3b pre-T cells. Furthermore, in the bone marrow B cell compartment, absolute numbers of pre-B cells are partially restored. This indicates that other functions of p53 also may be partially responsible for the pre-T cell and pro-B cell differentiation block in Miz-1–deficient mice.
Our findings are consistent with the hypothesis that Miz-1 is involved in regulating a p53-mediated response initiated by V(D)J-induced DNA damage in pre-T or pro-B cells. Further evidence supporting this contention is provided by our finding that Miz-1–deficient DN3 pre-T cells fully develop into DN4 cells in the absence of V(D)J recombination. Indeed, our experiments with Rag1−/− × Miz-1ΔPOZ mice showed a massive accumulation of DN4 cells on CD3 receptor stimulation, to the same extent as seen in Rag1−/− mice that express functional Miz-1; however, at this stage, the role of Miz-1 might not be limited to regulating the p53 pathway, because Miz-1ΔPOZ × Trp53−/− DN3 pre-T cells progress to DN4, but these DN4 cells are still less efficient than WT DN4 in their ability to proliferate and further differentiate to DP cells in vitro. In addition, although Rag1−/− × Miz-1ΔPOZ mice efficiently generate DN4 cells on αCD3 stimulation, they do not differentiate into DP cells.
Because the Miz-1 binding partner c-Myc is necessary for the transition of DN4 to DP (32), and because the c-Myc gene is activated by pre-TCR signaling, we reasoned that the interaction of c-Myc with Miz-1 might be required for an efficient DN4-to-DP transition. This idea is supported by data from Rag1−/− × MycV394D mice, which when stimulated with αCD3 antibodies were less efficient in producing DP cells than Rag1−/− mice with a functional c-Myc protein that still binds to Miz-1. In addition, CD71 and CD98, key metabolic genes up-regulated at the DN-to-DP transition and direct targets of c-Myc (33, 34), are not up-regulated on Rag1−/− × Miz-1ΔPOZ DN4 cells after mitogenic stimulation. Thus, we propose that, in contrast to the differentiation of DN3 cells to DN4, the DN4-to-DP transition represents a step requiring a functional c-Myc/Miz-1 complex. This is likely necessary for the activation of key metabolic pathways to support cell growth and differentiation of DN4 cells to generate a large number of DP thymocytes.
Although p53 mediates effectors involved in both cell cycle progression and regulation of apoptosis, it is unlikely that V(D)J recombination leads to p53-induced cell cycle arrest, given that the dissociation between DNA replication and V(D)J recombination is ensured by cell cycle stage-specific expression of Rag-2. We have found increased expression of Cdkn1a (p21), which can arrest cell cycle progression and is a p53 target gene, in Miz-1–deficient pre-T and pro-B cells; however, we previously showed that deletion of Cdkn1a in Miz-1–deficient cells did not rescue the defects caused by ablation of Miz-1, indicating that this part of the p53 pathway, although affected by Miz-1–dependent regulation of p53, is not responsible for the phenotype seen in Miz-1–deficient pre-T and pro-B cells. Thus, we propose that p53 can be activated to induce apoptosis in response to the DNA strand breaks that occur during V(D)J recombination, and that this activation of p53 is at least partially mediated through a mechanism involving Miz-1 and its effector, the ribosomal protein Rpl22. However, it is likely that additional mechanisms for activating p53 (e.g., through posttranslational mechanisms such as phosphorylation or mechanisms that do not involve p53) are also affected by Miz-1 and also play an important role at this critical point in pre-T cell and pro-B cell differentiation. For instance, it is possible that Rpl22 also acts on other proteins, not only via p53, and that forced expression of Rpl22 alone might not be sufficient to completely restore physiological p53 expression levels in Miz-1–deficient cells. Further experiments are needed to clarify the nature of these additional mechanisms and their link to Miz-1.
The regulation of p53 can occur at many levels. P53 expression is most commonly regulated at the posttranslational level, and ribosomal proteins have been shown to play a role in this regulation. Rpl5 (35), Rpl11 (36, 37), Rpl23 (38, 39), and Rpl26 (40) interact directly with the p53 E3 ubiquitin ligase Mdm2 and inhibit its activity, leading indirectly to enhanced stability of p53 under conditions of ribosomal biogenesis stress. Furthermore, Rpl26 has been shown to bind to both the 5′ and 3′ UTRs of p53 mRNA to increase its translation and induction after DNA damage, whereas nucleolin binds specifically to the p53 5′ UTR after DNA damage (41, 42). Our results indicate that, unlike other ribosomal proteins that have been shown to either activate p53 translation or act as Mdm2 antagonists under physiological conditions, Rpl22 directly suppresses p53 translation in a cell type- and developmental stage-specific manner, namely in cells undergoing V(D)J recombination. This finding agrees with previous reports that Rpl22-deficient mice exhibit no defects in global translation, but are defective specifically in regulation of p53 expression during pro-B cell and pre-T cell development (29, 30, 43). The data presented here strongly suggest that this regulation occurs through Miz-1, and that Miz-1 is a direct upstream regulator of the Rpl22 gene. Furthermore, the data presented here provide evidence for a previously unidentified regulation of p53 translation by a ribosomal protein, in that Rpl22 would be the only example of such a protein to repress the translation of p53 under physiological conditions, by decreasing its incorporation in actively translating ribosomes. In addition, the results presented here indicate that Rpl22 may bind numerous mRNA transcripts, including Rpl22l1 and β-actin, as part of its function in the ribosomal complex. However, based on our polysome fractionation experiment, the interaction between Rpl22 and p53 mRNA increases the incorporation of this mRNA specifically into polysomes and not other mRNA transcripts. How this specificity is achieved requires further investigation.
The expression of p53 must be tightly regulated to promote survival of cells that rearrange the TCRβ or IgH loci. These cells have to break and join DNA ends, a signal that normally initiates a p53-dependent DNA damage response. Our data support a function of Miz-1 that, via Rpl22, acts as a regulator of the translation of p53 in those cells at risk for elimination by p53-dependent cell death because they are undergoing V(D)J recombination.
Materials and Methods
Mice.
Mice had been bred on C57BL/6 background for at least 10 generations and were maintained in a specific-pathogen-free plus environment. Miz-1ΔPOZ mice have been described previously (20). Trp53−/− mice were purchased from Jackson Laboratory. p53K117R mice were provided by Dr. Wei Gu (23). The Institut de recherches cliniques de Montréal Animal Care Committee approved the animal protocols under which all animal experiments in this study were performed (protocol 2013–01), and all animal experimental procedures were performed in compliance with the guidelines of the Canadian Council of Animal Care (www.ccac.ca).
Flow Cytometry Antibodies and Cell Lines.
OP9-DL4/OP9 cultures and P6D4 SCID.adh murine thymic lymphoma and 70Z/3 pre-B cells were used, as described previously (20, 21). All antibodies were purchased from BD Bioscience except when indicated otherwise. DN thymic subsets were analyzed using CD25 (PC61.5; eBioscience) and CD44 (IM7) plus lineage marker-negative cells (Lin−) by staining thymocytes with the biotinylated antibodies against CD3ε (145-2C11), CD4 (RM 4–5), CD8α (53-6.7), CD45/B220 (RA3-6B2), Gr-1 (RB6-8C5), CD11b (Mac-1, M1/70), Ter-119 (Ly-76), NK1.1 (PK136; eBioscience), Pan-NK (DX5), and TCRγδ (GL3), followed by streptavidin-PerCPCy5.5 or PE. The same staining was performed with CD27 (LG.3A10) to differentiate between DN3a and DN3b subsets. Pre-B and pro-B cells were analyzed using B220 (RA3-6B2), CD43 (S7), and CD19 (1D3). Antibody incubations were performed at 4 °C for 20 min in PBS buffer. Cells were analyzed with a BD Biosciences LSR cell analyzer. Cell sorting was performed with a Cytomation MoFlo cell sorter.
Cell Cycle and Cell Death Analysis.
Cell cycle analysis was performed on sorted DN3 and DN4 cells. Cells were sorted directly into modified Krishan buffer (0.1% sodium citrate, 0.3% Nonidet P-40) containing 0.05 mg/mL propidium iodide (PI) and 0.02 mg/mL RNase and analyzed after 30 min of incubation on a BD FACSCalibur. For bromodeoxyuridine (BrdU) analysis, mice were injected i.p. with 100 mg/kg BrdU in PBS and killed after 16 h. Staining was performed using a FITC BrdU Kit (BD Pharmingen) according to the manufacturer’s instructions. Apoptosis rates were measured by annexin V staining (Annexin V-FITC Apoptosis Detection Kit; BD Pharmingen).
RNA Isolation and Real-Time PCR.
For RNA isolation, cells were FACS-sorted directly into RLT buffer with β-mercaptoethanol (RNEasy Micro Kit; Qiagen). RT-PCR was performed using SuperScript II (Invitrogen). Real-time PCR was performed in triplicate on the ViiA7 using SYBR Green reagent (Applied Biosystems) and indicated primers. The expression of the gene of interest was calculated relative to Gapdh mRNA (ΔCT) and is presented as fold induction relative to values obtained with the respective control (set as 1-fold). Primer sequences are provided in Table S1.
Polysome Fractionation.
Polysome fractionation was performed as described previously (44). In brief, single-cell suspensions of 10 × 106 cells from total thymus were treated with cycloheximide (100 μg/mL) for 15 min at 37 °C. Cells were washed in cold PBS containing 100 μg/mL cycloheximide and then lysed in ice-cold hypotonic lysis buffer (5 mM Tris⋅HCl pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 1× protease inhibitor mixture (complete Mini; Roche Diagnostics), and 400 U/mL RNaseIN (Promega)). The lysates were immediately adjusted to 100 μg/mL cycloheximide, 2 mM DTT, 0.5% Triton X-100, and 0.5% sodium deoxycholate. Cell extracts were centrifuged for 5 min at 20,817 × g, after which the supernatants were collected and loaded onto a 10–50% sucrose gradient. Gradients were placed in a Beckman SW40Ti rotor and centrifuged at 266,000 × g for 2 h at 4 °C. Fractions were collected (12 1-mL fractions) using a tube piercer system (Brandel) and a fraction collector. Samples were incubated with proteinase K, and RNA was extracted using phenol/chloroform. RT-PCR was performed using SuperScript II (Invitrogen). Real-time PCR was performed in triplicates on the ViiA7 using SYBR Green reagent (Applied Biosystems) and indicated primers.
RNA Immunoprecipitation.
Single-cell suspensions of 10 × 106 P6D4 cells were treated with cycloheximide (100 μg/mL) for 15 min at 37 °C. Cells were washed in cold PBS containing 100 μg/mL cycloheximide and then lysed in ice-cold hypotonic lysis buffer [5 mM Tris⋅HCl pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 1 mM DTT, 1% Triton X-100, 1% sodium deoxycholate, 100 μg/mL cycloheximide, 1× protease inhibitor mixture (Complete Mini; Roche Diagnostics), and 400 U/mL RNaseIN (Promega)] for 10 min. Cell extracts were centrifuged for 10 min at 20,817 × g, and the supernatants were collected and precleared with 10 μL of Pprotein A/G Dynabeads (Life Technologies) for 30 min. Immunoprecipitation was performed using 10 μg of anti-Rpl22 (H-106; Santa Cruz Biotechnology) or rabbit control IgG antibodies (Santa Cruz Biotechnology). Washes were performed using RIP wash buffer (50 mM Tris pH 7.5, 300 mM KCl, 12 mM MgCl2, 1% Triton X-100, 1 mM DTT, and 200 μg/mL cycloheximide). RNA was extracted using the Qiagen RNEasy Mini Kit, followed by RT-PCR and qPCR.
Immunoblot Analysis.
For immunoblot analysis, cells were lysed in RIPA buffer containing protease inhibitors (complete Mini; Roche Diagnostics) on ice for 20 min and then sonicated for 10 min in a water bath (Branson 5510). Immunoblotting was performed using anti-p53 (1C12; Cell Signaling), or anti-β-actin (AC-15; Sigma-Aldrich).
ChIP Sequencing and RNA Sequencing.
Assays were performed on sorted primary DN3 cells, P6D4 SCID.adh cells, or 70Z/3 cellsas indicated. Cells were fixed with 1% formaldehyde. Cell lysis was performed using the following buffers: for total cell lysis, 5 mM Pipes pH 8, 85 mM KCl, 0.5% Nonidet P-40, 1× protease inhibitor mixture, and 1 mM PMSF; for nuclear lysis, 50 mM Tris, 10 mM EDTA, 1% SDS, 1× protease inhibitor mixture, and 1 mM PMSF. After sonication (Covaris E220 sonicator), immunoprecipitation was performed using Protein A/G Dynabeads (Life Technologies) and 10 μg of rabbit anti-Miz-1 (H-190; Santa Cruz Biotechnology), H3K4Me3 (Abcam), H3K27Ac (Abcam), H3K9Ac (Millipore), or rabbit control IgG antibody (Santa Cruz Biotechnology). The primers used in the ChIP-qPCR experiments are listed in Table S2.
For ChIP-Seq experiments, sequencing libraries were prepared from immunoprecipitated chromatin using an Illumina TruSeq DNA Kit in accordance with the manufacturer’s instructions, and sequenced using the TruSeq PE Clusterkit v3-cBot-HS on an Illumina HiSEq 2000 system. Sequencing reads were aligned to the mm10 genome using Bowtie2 v2.10 (45). Reads were processed and duplicates were removed using Samtools, and a genome coverage file was generated and scaled to reads per million reads (RPM) obtained for each sample using Bedtools (46, 47).
For RNA-Seq, a biological triplicate of sequencing libraries was prepared from RNA extracts using the Illumina TruSeq Stranded mRNA Kit according to the manufacturer’s instructions, and sequenced using the TruSeq PE Clusterkit v3-cBot-HS on an Illumina HiSEq 2000 system. Sequencing reads were aligned to the mm10 genome using Tophat v2.0.10. Reads were processed with Samtools and then mapped to Ensembl transcripts using HTSeq. Differential expression was tested using the DESeq R package (R Coding Team) (48). A genome coverage file was generated and scaled to RPM using Bedtools (46, 47) (GEO accession no. GSE57694).
Actinomycin D Treatment.
DN3 cells were FACS-sorted into serum-free OptiMEM medium (Gibco) and rested at 37 °C for 30 min. Cells were then treated with 5 μg/mL actinomycin D at 37 °C and lysed in RLT buffer with β-mercaptoethanol. RNA was extracted using the Qiagen RNEasy Mini Kit, followed by RT-PCR and qPCR.
Rpl22 Reporter Assay.
293T cells were transfected with 250 ng of human Rpl22 promoter fused to luciferase (Switchgear Genomics), β-galactosidase, and either pcDNA3.1 alone or with 0.5 or 1 μg of human Miz-1 in pcDNA3.1. Cells were lysed after 48 h and analyzed for Renilla luciferase expression and β-galactosidase expression for normalization, using LightSwitch Luciferase Assay Reagent (Switchgear Genomics) and ONPG, respectively.
Anti-CD3 Injections.
Four- to 6-wk-old Miz-1ΔPOZ × Rag1−/−, MycV394D × Rag1−/−, and Rag1−/− mice were injected i.p. with 50 μg of anti-CD3 (145-2C11) per mouse or with PBS alone (32). Mice were killed at the indicated times and analyzed by flow cytometry.
Retroviral Transfection.
Rpl22 was cloned into the MigR1 vector using the following primers: 5′-CGACTCGAGATGGCGCCTGTGAAAAAGCTTG-3′ and 5′-CGAGAATTCTTAATCCTCGTCTTCCTCCTCCTC-3′. MigR1 and MigR1-Rpl22 were generated using Phoenix Eco cells. DN3a cells were sorted and resuspended in viral supernatant in the presence of 8 μg/mL polybrene. Cells were centrifuged at 515 × g for 90 min. Media was changed at 4 h after infection, and cells were plated onto OP9-DL4 stromal cells with 5 ng/mL Flt3L and 1 ng/mL IL-7 for 4 d.
Statistical Analysis.
Quantitative data are presented as mean ± SD and were analyzed using one-way ANOVA or the two-tailed Student t test. A P value ≤0.05 was considered to indicate statistical significance (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Supplementary Material
Acknowledgments
We thank Mathieu Lapointe for technical assistance; Marie-Claude Lavallée, Mélanie St.-Germain, and Jade Dussureault for excellent animal care; Eric Massicotte and Julie Lord for FACS and cell sorting; and Odile Neyret for assistance with molecular biology. We also thank Martin Eilers and Elmar Wolf, Biozentrum, Universität Würzburg, and the other members of the Eilers laboratory for communicating results on Miz-1 promoter binding before publication. M.R. was supported by a fellowship from the Cole Foundation. J.R. and M.G. were supported by fellowships from the Canadian Institute for Health Research (CIHR). C.V. was supported by a fellowship from the Fonds recherche Québec-Santé. T.M. holds a Canada Research Chair (Tier 1) and a grant from the CIHR (MOP-115202) that supported this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.V.R. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE57694).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412107111/-/DCSupplemental.
References
- 1.Michie AM, Zúñiga-Pflücker JC. Regulation of thymocyte differentiation: Pre-TCR signals and beta-selection. Semin Immunol. 2002;14(5):311–323. doi: 10.1016/s1044-5323(02)00064-7. [DOI] [PubMed] [Google Scholar]
- 2.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2(5):309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg EV, Pant R. Origins of lymphocyte developmental programs: Transcription factor evidence. Semin Immunol. 2004;16(4):227–238. doi: 10.1016/j.smim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24(1):53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Guidos CJ, et al. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 1996;10(16):2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 6.Dujka ME, Puebla-Osorio N, Tavana O, Sang M, Zhu C. ATM and p53 are essential in the cell-cycle containment of DNA breaks during V(D)J recombination in vivo. Oncogene. 2010;29(7):957–965. doi: 10.1038/onc.2009.394. [DOI] [PubMed] [Google Scholar]
- 7.Melchers F, et al. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol Rev. 2000;175:33–46. [PubMed] [Google Scholar]
- 8.Lu L, Lejtenyi D, Osmond DG. Regulation of cell survival during B lymphopoiesis: suppressed apoptosis of pro-B cells in P53-deficient mouse bone marrow. Eur J Immunol. 1999;29(8):2484–2490. doi: 10.1002/(SICI)1521-4141(199908)29:08<2484::AID-IMMU2484>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173(5):1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peukert K, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16(18):5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stead MA, et al. A beta-sheet interaction interface directs the tetramerisation of the Miz-1 POZ domain. J Mol Biol. 2007;373(4):820–826. doi: 10.1016/j.jmb.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3(4):392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 13.Seoane J, et al. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3(4):400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 14.Herold S, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10(3):509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 15.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6(10):1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 16.Saito M, et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2009;106(27):11294–11299. doi: 10.1073/pnas.0903854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold S, et al. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J. 2008;27(21):2851–2861. doi: 10.1038/emboj.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123(3):409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Wanzel M, et al. Akt and 14-3-3eta regulate Miz1 to control cell-cycle arrest after DNA damage. Nat Cell Biol. 2005;7(1):30–41. doi: 10.1038/ncb1202. [DOI] [PubMed] [Google Scholar]
- 20.Kosan C, et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33(6):917–928. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 21.Saba I, Kosan C, Vassen L, Möröy T. IL-7R-dependent survival and differentiation of early T-lineage progenitors is regulated by the BTB/POZ domain transcription factor Miz-1. Blood. 2011;117(12):3370–3381. doi: 10.1182/blood-2010-09-310680. [DOI] [PubMed] [Google Scholar]
- 22.Saba I, Kosan C, Vassen L, Klein-Hitpass L, Möröy T. Miz-1 is required to coordinate the expression of TCRbeta and p53 effector genes at the pre-TCR “beta-selection” checkpoint. J Immunol. 2011;187(6):2982–2992. doi: 10.4049/jimmunol.1101451. [DOI] [PubMed] [Google Scholar]
- 23.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24(6):827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24(6):841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinkai Y, Alt FW. CD3 epsilon-mediated signals rescue the development of CD4+CD8+ thymocytes in RAG-2−/− mice in the absence of TCR beta chain expression. Int Immunol. 1994;6(7):995–1001. doi: 10.1093/intimm/6.7.995. [DOI] [PubMed] [Google Scholar]
- 27.Gebhardt A, et al. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol. 2006;172(1):139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf E, et al. Miz1 is required to maintain autophagic flux. Nat Commun. 2013;4:2535. doi: 10.1038/ncomms3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson SJ, et al. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26(6):759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 30.O’Leary MN, et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 2013;9(8):e1003708. doi: 10.1371/journal.pgen.1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin WC, Desiderio S. V(D)J recombination and the cell cycle. Immunol Today. 1995;16(6):279–289. doi: 10.1016/0167-5699(95)80182-0. [DOI] [PubMed] [Google Scholar]
- 32.Dose M, et al. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood. 2006;108(8):2669–2677. doi: 10.1182/blood-2006-02-005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Donnell KA, et al. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol. 2006;26(6):2373–2386. doi: 10.1128/MCB.26.6.2373-2386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14(11):7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23(23):8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai MS, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24(17):7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24(17):7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32(2):180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123(1):49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Kastan MB. 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 2010;24(19):2146–2156. doi: 10.1101/gad.1968910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadanlick JE, et al. Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. J Immunol. 2011;187(2):664–675. doi: 10.4049/jimmunol.1100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mamane Y, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2(2):e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R Coding Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.