Significance
Transcription is affected, in part, by the spooling of promoter DNA in nucleosomes. EM analysis of single PHO5 gene molecules from budding yeast revealed a surprising degree of variation in the promoter nucleosome configuration between individual molecules. This variation could be explained on the theory of random transitioning between alternative configurations, suggesting that genes flicker between transcriptionally conducive and unconducive states. What is the origin of this random behavior? Here, we show that the nucleosomal promoter variation cannot be reduced to molecular variation in the gene’s intracellular surroundings, that is, it is intrinsically stochastic and thus, a cause of stochastic transcription rather than its consequence).
Keywords: gene expression noise, chromatin, transcription, PHO5
Abstract
Gene product molecule numbers fluctuate over time and between cells, confounding deterministic expectations. The molecular origins of this noise of gene expression remain unknown. Recent EM analysis of single PHO5 gene molecules of yeast indicated that promoter molecules stochastically assume alternative nucleosome configurations at steady state, including the fully nucleosomal and nucleosome-free configuration. Given that distinct configurations are unequally conducive to transcription, the nucleosomal variation of promoter molecules may constitute a source of gene expression noise. This notion, however, implies an untested conjecture, namely that the nucleosomal variation arises de novo or intrinsically (i.e., that it cannot be explained as the result of the promoter’s deterministic response to variation in its molecular surroundings). Here, we show—by microscopically analyzing the nucleosome configurations of two juxtaposed physically linked PHO5 promoter copies—that the configurational variation, indeed, is intrinsically stochastic and thus, a cause of gene expression noise rather than its effect.
Analysis of gene expression at the single-cell level has revealed that the number of gene product molecules varies, sometimes greatly, over time and between isogenic cells under identical conditions (1). Probabilistic theories have been invoked to explain this molecular variation or “noise,” because it seems to defy deterministic expectations. Gene expression noise may play crucial roles in stress response, signaling, and development (2), and noise measurements have been used to distinguish between competing mechanistic models of gene regulation (3). Probabilistic theories, furthermore, account for the observation that noninduced genes also exhibit some level of expression (background or “leaky” expression).
Where does the noise of gene expression come from? In part, variation in gene expression ensues, because the expression process responds deterministically to molecular variation in its intracellular environment. Variation in gene expression thus transferred from environmental or “extrinsic” variation is referred to as “extrinsic noise” (4). It carries information about the gene’s environment (5). In contradistinction, noise that is not explained by extrinsic variation is called “intrinsic noise” (4). It arises de novo because of inherently stochastic behavior or “intrinsic variation” within the gene expression process. Both extrinsic and intrinsic noise originate from intrinsic variation at last, because the intrinsic noise of one gene becomes extrinsic variation for other genes, which in turn, is transmitted as extrinsic expression noise. In short, intrinsic noise is generated or de novo variation, whereas extrinsic noise is transmitted or informational variation.
Noise can be experimentally decomposed into extrinsic and intrinsic components by observing the gene product molecules of two identical gene copies within the same cell (“conjugate reporter approach”), because extrinsic variation induces a correlation between the abundances of the molecules expressed from the two copies (4–9).
At the promoter level, intrinsic noise may arise, in other words, noise may be generated, by at least two mechanisms. First, local concentration fluctuations of surrounding bath molecules (e.g., transcription factors) are expected to generate (intrinsic) noise if the two conjugate reporter genes are spatially sufficiently far apart. (However, if the conjugate reporters are in close proximity, such fluctuations may be extrinsic variation and thus, would contribute to the extrinsic noise of expression). Second, conformational fluctuations of promoter DNA and nucleosomes may affect whether a chromatin remodeler can bind and displace a nucleosome or initiate its assembly, permitting or inhibiting transcription, respectively. Implicit in the latter proposition is the assumption of variation at the level of promoter chromatin structure.
Indeed, recent EM analysis showed that single-gene molecules exhibit different promoter nucleosome configurations under inducing conditions at steady state (10), consistent with earlier conclusions based on less direct observations (8, 11–13). The observed set of configurations for the induced PHO5 promoter in budding yeast encompassed every combinatorial possibility of occupying three nucleosome positions (23 = 8), including both the nucleosome-free and the fully nucleosomal configuration (the prevalent configuration under noninducing conditions). This finding suggested that individual PHO5 molecules at steady state pass, in some sequence, through alternative promoter nucleosome configurations; the structure of promoter chromatin is dynamic rather than static. The well-corroborated theory that nucleosomes are inhibitors to transcription (14) then predicts that transcription occurs in bursts [i.e., promoters alternate between (structural) states that are either conducive or unconducive to transcription]. [Specifically, it may be argued that configurations that lack a nucleosome in the central position, N-2, are the conducive configurations (10).]
The nucleosomal dynamics of promoter nucleosomes may be represented by a transition graph (8, 10, 11), with nodes for nucleosome configurations and directed edges for allowed transitions between them. Dynamical behavior is then described as the flow of probability mass between the nodes along the directed edges (stochastic process). At steady state, incoming and outgoing probability currents are balanced at each node, and probabilities are stable in time; the process is said to be stationary.
Remarkably, the microscopically observable structural variation between PHO5 promoter molecules at steady state could be explained by the assumption of a stationary process on a branched transition graph (10), that is, a graph with more than one outgoing edge per node (Fig. 1A). Transition graph branching implies that single-promoter molecules pass through a sequence of nucleosome configurations that is stochastic in the sense that the next configuration is not predetermined by the present configuration but may only be predicted statistically. Transcriptional bursting should then be stochastic as well. Indeed, eukaryotic gene expression generally conforms to the theoretical expectations of random transcriptional bursting (15–18).
Fig. 1.
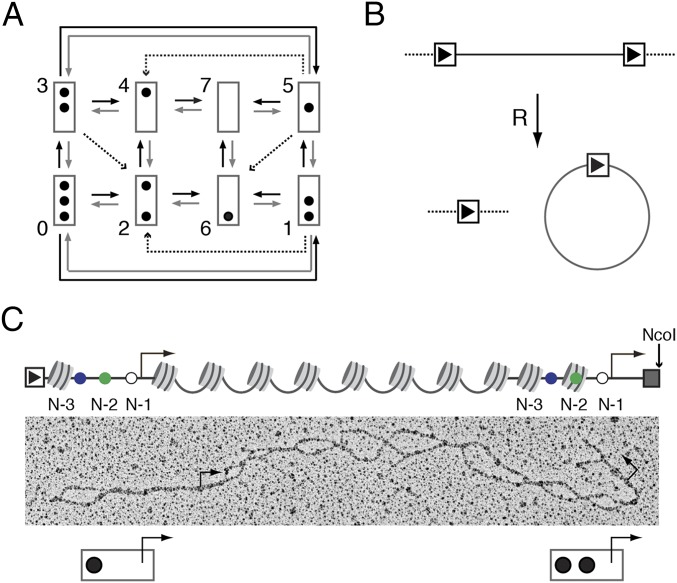
Transition graph, chromatin ring formation, and the double-promoter gene. (A) Transition graph for PHO5 promoter nucleosome dynamics (10). The promoter is represented by a box, and occupied nucleosome positions are represented by black dots. Position N-1 is at the top of the box, and position N-3 is at its bottom. Nucleosomes are assembled and disassembled one by one and slid out of but not into the central N-2 position. (B) Formation of a chromatin ring by R recombinase (R). (C) Linearized double-promoter ring. Dark gray dots indicate promoter nucleosome positions, and small dots indicate cis-regulatory sequences: UASp1 (blue), UASp2 (green), and the TATA box (white); the bent arrows mark the (mutated) transcription start sites. The rectangle with the arrowhead represents the recognition sequence for R, and the light gray box represents the lexA operator cluster.
Furthermore, the functional relationship between intrinsic noise and the mean of PHO5 expression has been explained on the conjecture that PHO5 expression is regulated by controlling the frequency of transcriptional bursting (8–10). That is, PHO5 transcription occurs in bursts, and bursting is intrinsically stochastic. Thus, if nucleosomal promoter variation provides a molecular basis for bursting, which has been suggested (10), then it must be intrinsic, that is, it cannot be reduced to extrinsic variation due to, for instance, unsynchronized cellular oscillations or fluctuations in the nuclear abundance of transcription factors or remodelers.
Here, we report the results of the first critical test, to our knowledge, of this conjecture. To this end, we applied the conjugate reporter approach to promoter molecules; specifically, we generated yeast strains that allowed us to isolate gene molecules bearing two copies of the PHO5 promoter. Thus linked, the two copies were assured to have shared the same intranuclear environment. If the structural variation arises from a deterministic response to extrinsic variation, then the two promoter copies will exhibit the same nucleosome configuration; knowledge of one copy’s nucleosome configuration entails knowledge of the other copy’s configuration, and the two copies will be stochastically dependent. However, if variation ensues purely from intrinsic variation, then knowledge of the state of one promoter copy contains no predictive information on the state of the other copy; the two copies will be stochastically independent.
Results
To provide conjugated promoter copies, we inserted a second copy of the PHO5 promoter at the 3′ end of the endogenous gene followed by a cluster of binding sites for the bacterial DNA-binding protein LexA for affinity purification; to this end LexA was fused to a tandem affinity purification (TAP) tag. For short, we refer to this fusion protein as the “adapter molecule” (13, 19) (Fig. 1B). The TATA box sequence of both copies was replaced with an unrelated sequence to exclude possible effects of transcription on nucleosomes (13). [Promoter chromatin remodeling on PHO5 induction does not depend on the PHO5–TATA box (10, 13).] The construct was flanked with recombination sequences (RS) recognized by the R recombinase of Zygosaccharomyces rouxii, which allowed for its release from the chromosome in the form of a chromatin ring (10, 13, 19–21) (Fig. 1B). Rings were isolated as previously described (10) from cells grown under repressing conditions (i.e., high medium concentration of inorganic phosphate) and cells that expressed PHO5 constitutively because of the lack of Pho80, a repressor of the PHO signaling pathway (22). Constitutive activation of PHO5 in the pho80Δ mutant approached steady-state expression as closely as possible.
To map nucleosomes, purified dual-promoter chromatin rings were treated with psoralen, a UV-activated interstrand DNA cross-linker that selectively cross-links linker DNA between nucleosomes but not core particle DNA (23–25). The gene’s nucleosome configuration is, thus, engraved into the DNA. DNA segments that were previously occupied by nucleosomes become visible in the EM as single-stranded bubbles on chemical denaturation of the DNA (Fig. 1C). PHO5 promoter sequences were identified by restriction endonucleolytic cleavage of chromatin rings before denaturation with NcoI, which cut at a single site between the lexA operator cluster and the recognition sequence for the R recombinase (10). This site-specific cleavage positioned the two PHO5 promoter copies at opposite ends of the linearized molecule (Fig. 1C). The lexA operator cluster did not cross-link, supposedly because of protection by binding of adapter molecules and therefore, appeared as a fork of ssDNA at the 3′ end of every molecule (10). The DNA forks oriented each molecule and allowed for the assignment of cis-regulatory element positions by contour length measurements (10), including positions of the TATA box and the “upstream activating sequences” 1 and 2, UASp1 and UASp2 (Fig. 1C), each of which bears a binding site for the Pho4 activator.
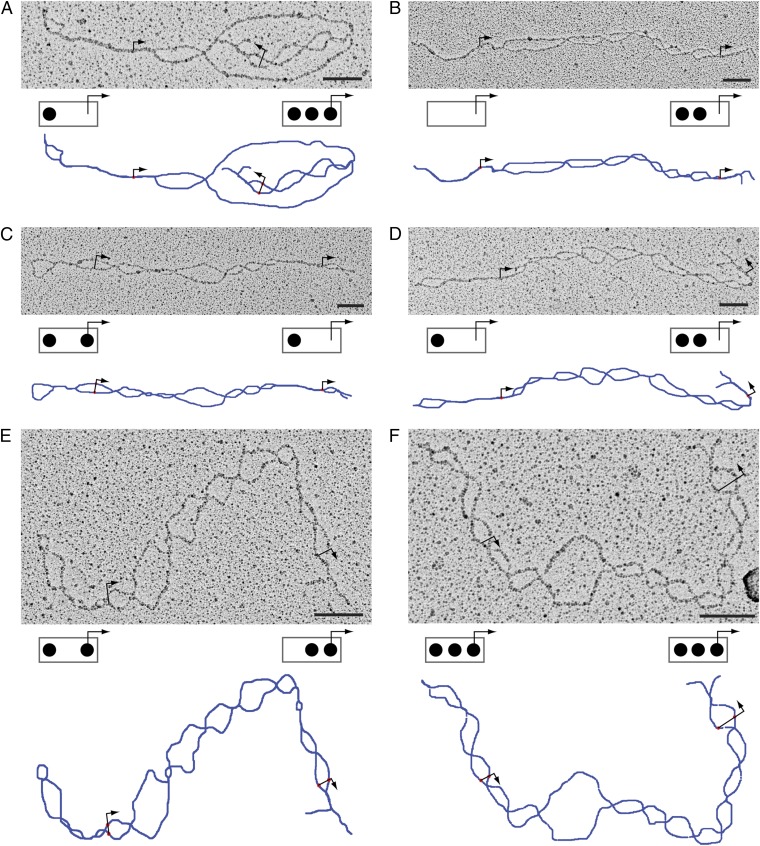
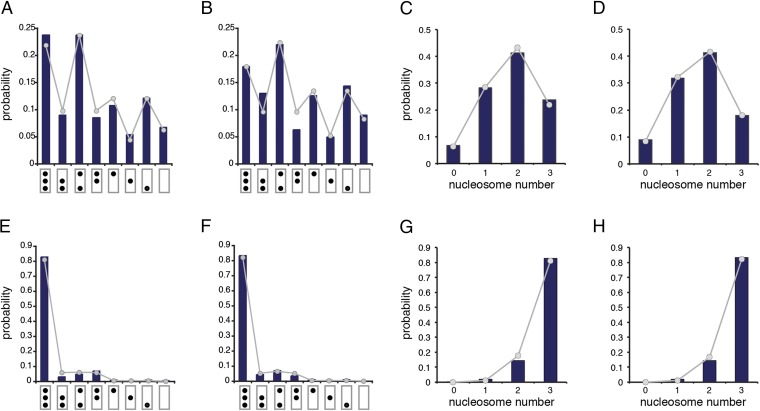
Fig. 2 shows examples of molecules isolated from induced cells (pho80Δ). All eight possible nucleosome configurations were observed at both promoter copies on molecules isolated from induced cells (Fig. 3 A and B). Consistent with earlier findings (10), probability mass shifted from configurations with more to those with fewer nucleosomes on PHO5 induction (compare Fig. 3 A–D with Fig. 3 E–H). The probability distributions (relative frequencies) of nucleosome configuration (Fig. 3 A, B, E, and F) and nucleosome number (Fig. 3 C, D, G, and H) of both promoter copies were similar to each other for both induced and noninduced cells (compare Fig. 3A with Fig. 3B, compare Fig. 3C with Fig. 3D, compare Fig. 3E with Fig. 3F, and compare Fig. 3G with Fig. 3H) and are well-explained by the assumption of a simple stationary process (Materials and Methods) on the transition graph in Fig. 1A (Fig. 3), consistent with earlier findings (10).
Fig. 2.
Electron micrographs of denatured double-promoter PHO5 molecules. Micrographs A–F show examples of individual PHO5 molecules with two promoter copies. Schemes below the micrographs indicate the inferred promoter nucleosome configurations for the 5′ promoter (left box) and the 3′ promoter (right box). Bars denote 100 nm. Short linkers may fail to crosslink, in the body of the gene, for instance, where strings of bubbles tend to fuse (see panel A, for instance). Also shown below micrographs are manual tracings of the molecule (blue line). Bent arrows indicate the position of transcription start sites.
Fig. 3.
Marginal distributions. Relative frequencies of (A, B, E, and F) nucleosome configuration and (C, D, G, and H) nucleosome number are indicated by histogram bars (dark blue). Theoretical predictions (probabilities) on the assumption of a simple process with the transition graph in Fig. 1A are shown as dots connected by line segments for visual emphasis. (A) for 5′ promoter copy [i.e., the probability of configuration (C) i = 0,..., 7 given that the promoter is active (A)]. (B) for 3′ promoter copy. (C) for 5′ promoter copy [i.e., the probability of nucleosome number (N) i = 0,..., 3 given that the promoter is active (A)]. (D) for 3′ promoter copy. (E) for 5′ promoter [i.e., the probability of configuration i given that the promoter is repressed (R)]. (F) for 3′ promoter. (G) for 5′ promoter. (H) for 3′ promoter. In total, 223 and 233 molecules were analyzed for the induced (A) and noninduced (R) genes, respectively. Induction results in the translocation of the transcriptional activator Pho4 from the cytoplasm into the nucleus. Pho4 binds at UASp1 and UASp2 of the PHO5 promoter (Fig. 1C). With the kinetic parameter for nucleosome assembly set to 1 (on some timescale), maximum likelihood values for disassembly () and sliding () were (A and C) and , (B and D) and , (E and G) and , and (F and H) and .
More promoter nucleosomes, on average, were seen on molecules from both induced and noninduced cells compared with previous analyses (10)—∼0.3 nucleosomes for repressed and 0.5 nucleosomes for activated promoters—suggesting that the two promoter copies, which were juxtaposed on ring closure and separated by only 200 bp of DNA, compete for nucleosome disassembly activities. [Other possibilities, such as differences in growth conditions, are by no means excluded. However, topology assays with cells cultured under different conditions, viz., small culture flasks (topology) vs. fermenter (EM), consistently also indicated that fewer nucleosomes, on average, were removed from induced gene rings with two promoter copies than expected from previous analyses of rings with one promoter copy, whereas insertion of a spacer between both copies attenuated the apparent inhibitory effect of the copies on each other (Fig. S1).]
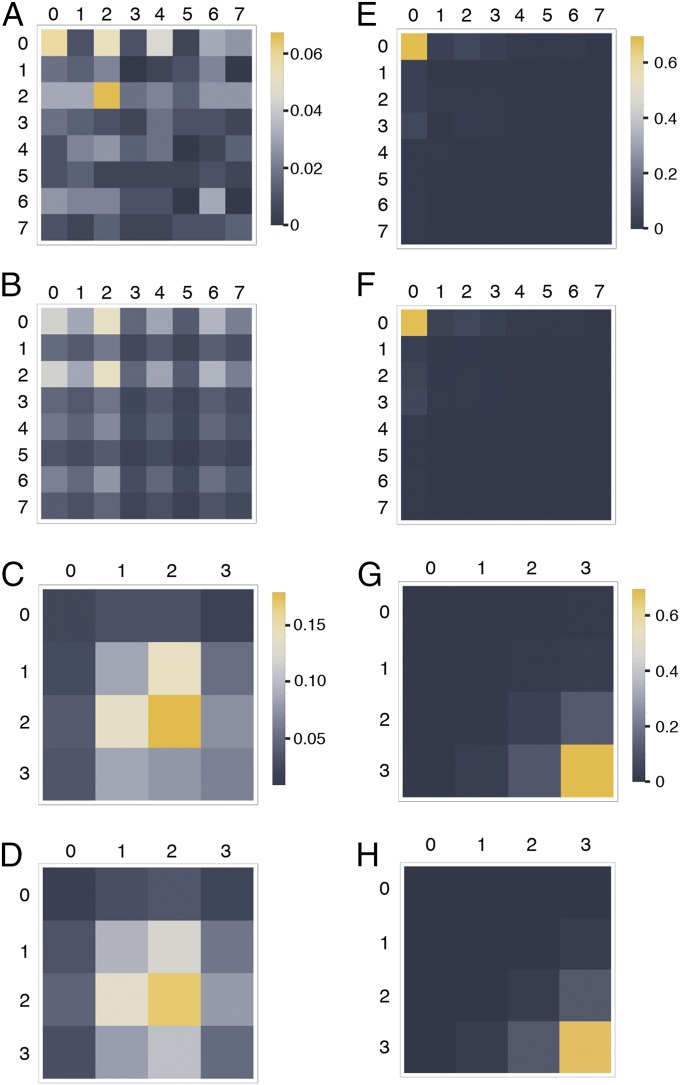
Among 223 molecules analyzed from induced cells, we observed 59 of 64 (8 × 8) possible combinations of 5′ and 3′ promoter nucleosome configurations (Fig. 4A). Predictions of joint probabilities similar to our experimental observations were obtained on the assumption that the structure of one promoter copy contains no information on the structure of the second copy (i.e., the nucleosome configurations of both copies are stochastically independent) (Fig. 4B). This assumption was supported by a χ2 statistic close to zero (, p ∼ 1) (Materials and Methods). Essentially, the same result was obtained for 233 molecules from noninduced cells (Fig. 4) (, p ∼ 1).
Fig. 4.
Joint probability distributions. (A) Observed joint probabilities [relative frequencies; ] for nucleosome configuration i at the 5′ promoter (rows) and nucleosome configuration j at the 3′ promoter (columns) of activated (A) gene molecules () (Fig. 1A). (B) Predicted joint distribution on the assumption of stochastic independence [i.e., ]. (C) Observed joint probabilities [] for the numbers of nucleosomes at the 5′ (rows) and 3′ (columns) promoters of induced gene molecules. (D) Joint probabilities [] predicted on the assumption of stochastic independence. (E–H) Equivalent to A–D, respectively, for repressed gene molecules; 223 and 233 molecules were analyzed for the induced and noninduced genes, respectively.
Because some of the 64 possible joint configurational promoter states were populated by fewer than five molecules in our dataset—and thus, fewer than desirable for application of the χ2 test—we regrouped our data according to nucleosome number, which reduced the number of joint promoter states to 16 (and hence, increased the number of molecules per state). Again, predictions of joint probabilities closely similar to experimental observations were obtained on the assumption of stochastic independence for molecules from both induced and noninduced cells (compare Fig. 4 C and D with Fig. 4 G and H). This assumption was consistent with correlation coefficients close to zero and further supported by vanishing χ2 statistics for gene molecules from both induced (correlation coefficient ρ of 0.025; , p ∼ 1) and noninduced (ρ = 0.0023; , p ∼ 1) cells.
Thus, while some degree of stochastic dependence or correlation cannot in principle be excluded (regardless of sample size), P values close to one (see Methods) and the very close agreement between prediction and measurement for pairs of nucleosome numbers (compare Fig. 4 C and D) suggested that deviations from the expectation of stochastic independence, such as missing joint states, e.g. (1,3) or (4,5), or higher than expected relative frequencies, such as states (0,0) and (2,2) (compare Fig. 4 A and B), may be attributable to small sample size.
Discussion
Based on the discovery of nucleosomal promoter variation and its explanation by a stochastic process on a branched transition graph, we previously conjectured that this variation is a source of gene expression noise rather than a reflection of extrinsic variation and a cause of transcriptional bursting rather than its symptom (10). This hypothesis was consistent with our findings, but was not critically tested by any previous experiments.
The conjugate reporter analysis of this study provides such a test and corroborating evidence, namely the stochastic independence of the nucleosome configurations of two physically linked promoter copies. Importantly, because knowledge grows by refutation and not corroboration of theories (26), our result falsifies many conceivable explanations of nucleosomal variation that previously could not be excluded (10).
Thus, the lack of stochastic dependence (and hence, correlation) between the two promoter copies refutes all theories that attribute the configurational variation of promoter nucleosomes to environmental variation. Excluded from further consideration are, for instance, all cyclical transition graphs that represent cellular oscillations, such as the cell cycle.
Also falsified are transition graphs with nodes that correspond to stable cellular states and hence, lack outgoing edges (“absorbing states”). In contradistinction to the dynamical theory espoused here, theories with absorbing states imply a steady state that is static. An example is the recent proposal by Small et al. (27), who suggested that gene molecules with a fully nucleosomal promoter represent a unique subpopulation of cells that failed to remodel the PHO5 promoter. If this assumption were true, then a second promoter copy within the same cell would always be fully nucleosomal as well. This prediction is refuted by our observations (Fig. 4).
The virtually complete lack of stochastic dependence between the two promoter copies is surprising, because it suggests an effectively uniform intranuclear background, whereas the existence of extrinsic gene expression noise indicates that the relevant molecular surroundings are not uniform but vary between cells and thus, supposedly, in time. Because the extrinsic component dominates PHO5 expression noise (9), the same may have been expected for the nucleosomal promoter variation.
Apparent temporal uniformity of the molecular surroundings may be explained by the assumption of fluctuations on different timescales. It is conceivable, for instance, that nucleosome removal requires a specific and thus, seldom sampled conformation of the nucleosome. If the abundance of available remodeler molecules fluctuates at a relatively short timescale, then such (extrinsic) fluctuations will remain without response most of the time; they are averaged out by slow intrinsic dynamics of the promoter, and the molecular surroundings appear uniform.
Our results indicate that the observed structural variation is intrinsic and not extrinsic variation. However, the question of what is the cause of this intrinsic variation remains open. Local (spatial) variation in molecular abundances—of transcription factors and remodelers, for instance—would be intrinsic if the two promoter copies are sufficiently far apart but extrinsic if sufficiently close to share the same local environment. Because the nucleosomal variation appears to be intrinsic entirely, it may be concluded that the two copies were, indeed, sufficiently far apart, despite their close proximity on the ring. However, the observed increase in promoter nucleosome occupancy for molecules with two promoter copies relative to molecules with one copy suggests competition between the copies and hence, a shared local environment. It might be argued that this notion is refuted by our observations, because competition should cause a negative correlation between the nucleosome numbers of both copies. However, this is not so necessarily. A separation of timescale between local abundance fluctuations and intrinsic promoter dynamics, as explained above, would effectively erase such a correlation. The two copies may, thus, compete and yet remain stochastically independent. As expected by the hypothesis of competition by proximity, insertion of a spacer between the copies appeared to allay promoter competition (Fig. S1). If this explanation is correct, then fluctuations other than abundance fluctuations of remodelers and transcription factors are at the origin of nucleosomal promoter variation (e.g., conformational fluctuations of promoter nucleosomes and DNA).
We conclude that nucleosomal promoter variation arises intrinsically and thus, is a source of gene expression noise—provided that nucleosomes matter to transcription—rather than its consequence. The variation, therefore, contributes to the intrinsic and not extrinsic noise of expression. The critical means for testing this conjecture has been the application of the conjugate reporter assay to gene molecules rather than mRNA and protein molecules. Because nucleosomes may form on any DNA sequence (albeit with different efficiencies), our theory should apply to eukaryotic genes in general.
Materials and Methods
Plasmids and Strains.
Plasmid pM90.2 with two PHO5 promoter copies was derived from plasmid pM70.1 (13), which bears the PHO5 gene with a downstream lac operator cluster flanked with the sequence element for R recombination (RS). pM70.1 lacks the PHO5 TATA box, which had been replaced with an unrelated sequence of equal length (13). A segment of pM70.1 was cloned by PCR with Phusion (Finzyme) and primers P28 (5′-TTCTATTTACTGACCGAAAGTAGC-3′) and P66 (5′-GCCAGGGAAAGAGTAGTATGG-3′). The PCR product was cleaved with XhoI and EcoRV. The 649-bp fragment of this digestion, which encompassed the TATA-less PHO5 promoter and about 100 bp of the downstream sequence of the PHO5 ORF, was blunted with the Klenow fragment and inserted at the BsaBI site of pM70.1 at the 3′ end of the PHO5 gene. The two promoter copies are oriented head to tail on the resulting plasmid pM90.2 (Fig. 1B). The head-to-head orientation was not obtained, presumably because it was unstable in Escherichia coli (DH5α).
Yeast strains yM1.12 and yM17.3 (13), in which the PHO5 gene (including its promoter) was replaced with URA3, were transformed with pM90.2 after its digestion with NotI, which released a 4,034-bp fragment encompassing the PHO5 gene with two TATA-less promoter copies, the lexA operator cluster, two RS elements positioned at the 5′ end of the PHO5 promoter and the 3′ end of the lexA operator cluster (Fig. 1C), and ∼500 bp of upstream and downstream sequences for homologous recombination. Isolation of clones resistant to 5-fluoroorotic acid, where the ectopic URA3 gene was replaced with the double-promoter PHO5 construct (Fig. 1B), gave rise to strains yM192.1 (yM1.12 derivative; PHO80 WT) and yM193.4 (yM17.3 derivative; pho80Δ). Correct integration was tested by PCR. Strains yM192.1 and yM193.4 were transformed with plasmid pSH17 (21), generating strains yM192.1[pSH17] and yM193.4[pSH17].
Ring Purification, Psoralen Cross-Linking, and EM Analysis.
Chromatin rings were purified from 10 L yM192.1[pSH17] and yM193.4[pSH17], cultivated in a fermenter at 30 °C with constant air supply, and analyzed by psoralen cross-linking and EM as previously described (10). The adapter was constitutively expressed under the control of the weak TEF2 promoter (21). Expression of the R recombinase was controlled by the inducible GAL1 promoter (28). Excised chromatin rings were isolated by differential centrifugation and two steps of affinity chromatography as previously described (10). Nucleosome position N-1 was considered occupied if the TATA box (position), the transcription start site, or both were contained within a nucleosome-sized bubble. Similarly, position N-2 was considered occupied if UASp2 was positioned within a nucleosome-sized bubble, and nucleosome-sized bubbles upstream of UASp1 with midpoints no farther than 150 bp from UASp1 were scored as N-3 nucleosomes. Molecules were analyzed using custom-made programs.
Probabilistic Model.
The promoter nucleosome dynamics are described by the master equation of the process (8, 11):
where is the column vector with components () that are the probabilities of (nucleosome) configurations j = 0,…, 7; the matrix is the generator of the process, where is the rate constant for the probability current from node j into node i. At steady state,
A unique solution to the last equation (the stationary distribution p) always exists if the transition graph of the process is strongly connected (29) (i.e., if each node is connected to any other node through a string of one or more edges). The transition graph in Fig. 1A is strongly connected and has, therefore, a uniquely defined stationary distribution, which was determined by calculating the kernel of W for given using Mathematica (Wolfram).
The process was assumed to be simple (i.e., the rate constant only depended on the kind of transition). Three kinds were distinguished: nucleosome assembly, disassembly, and sliding transitions (10). The rate constant for assembly was set as equal to one (on some suitable timescale). The stochastic process model is, thus, limited to two degrees of freedom. Their numerical values were determined by maximizing the probability of the data given the transition graph in Fig. 1A (maximum likelihood) as previously described (10).
Calculations.
All calculations were performed with Mathematica 9 (Wolfram). Stochastic independence was tested by Pearson’s χ2 test [i.e., the χ2 statistic was calculated according to
where is the observed frequency of the configuration (nucleosome number) pair , N is the total number of gene molecules, and
is the expected frequency of given that the two promoter copies are stochastically independent]. The indicated P value (calculated from the χ2 distribution with 49 and 9 degrees of freedom for nucleosome configurations and nucleosome number, respectively) refers to the probability that a χ2 value identical to or larger than the measured value would be obtained by chance provided that the assumption of stochastic independence (null hypothesis) is correct.
The calculated correlation coefficient is Pearson's correlation coefficient, i.e.,
where X and Y are the random variables for the number of nucleosomes on the 5′ and 3′ promoter copies, respectively.
Supplementary Material
Acknowledgments
We thank Grant Hartzog for critical comments on the manuscript and National Science Foundation Grant 1243957 for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417527111/-/DCSupplemental.
References
- 1.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine JH, Lin Y, Elowitz MB. Functional roles of pulsing in genetic circuits. Science. 2013;342(6163):1193–1200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336(6078):183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99(20):12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowsher CG, Swain PS. Identifying sources of variation and the flow of information in biochemical networks. Proc Natl Acad Sci USA. 2012;109(20):E1320–E1328. doi: 10.1073/pnas.1119407109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 7.Hilfinger A, Paulsson J. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proc Natl Acad Sci USA. 2011;108(29):12167–12172. doi: 10.1073/pnas.1018832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao C, et al. Quantitative analysis of the transcription control mechanism. Mol Syst Biol. 2010;6:431. doi: 10.1038/msb.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304(5678):1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown CR, Mao C, Falkovskaia E, Jurica MS, Boeger H. Linking stochastic fluctuations in chromatin structure and gene expression. PLoS Biol. 2013;11(8):e1001621. doi: 10.1371/journal.pbio.1001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133(4):716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jessen WJ, Hoose SA, Kilgore JA, Kladde MP. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat Struct Mol Biol. 2006;13(3):256–263. doi: 10.1038/nsmb1062. [DOI] [PubMed] [Google Scholar]
- 13.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11(6):1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 14.Mao C, Brown CR, Griesenbeck J, Boeger H. Occlusion of regulatory sequences by promoter nucleosomes in vivo. PLoS ONE. 2011;6(3):e17521. doi: 10.1371/journal.pone.0017521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez A, Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342(6163):1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suter DM, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332(6028):472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 17.Larson DR, et al. Direct observation of frequency modulated transcription in single cells using light activation. eLife. 2013;2:e00750. doi: 10.7554/eLife.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senecal A, et al. Transcription factors modulate c-Fos transcriptional bursts. Cell Reports. 2014;8(1):75–83. doi: 10.1016/j.celrep.2014.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griesenbeck J, Boeger H, Strattan JS, Kornberg RD. Affinity purification of specific chromatin segments from chromosomal loci in yeast. Mol Cell Biol. 2003;23(24):9275–9282. doi: 10.1128/MCB.23.24.9275-9282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griesenbeck J, Boeger H, Strattan JS, Kornberg RD. Purification of defined chromosomal domains. Methods Enzymol. 2004;375:170–178. doi: 10.1016/s0076-6879(03)75011-3. [DOI] [PubMed] [Google Scholar]
- 21.Hamperl S, et al. Purification of specific chromatin domains from single-copy gene loci in Saccharomyces cerevisiae. Methods Mol Biol. 2014;1094:329–341. doi: 10.1007/978-1-62703-706-8_26. [DOI] [PubMed] [Google Scholar]
- 22.Lenburg ME, O’Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;21(10):383–387. [PubMed] [Google Scholar]
- 23.Cech T, Pardue ML. Cross-linking of DNA with trimethylpsoralen is a probe for chromatin structure. Cell. 1977;11(3):631–640. doi: 10.1016/0092-8674(77)90080-0. [DOI] [PubMed] [Google Scholar]
- 24.Cech T, Potter D, Pardue ML. Electron microscopy of DNA cross-linked with trimethylpsoralen: A probe for chromatin structure. Biochemistry. 1977;16(24):5313–5321. doi: 10.1021/bi00643a024. [DOI] [PubMed] [Google Scholar]
- 25.Sogo JM, Thoma F. Electron microscopy of chromatin. Methods Enzymol. 1989;170:142–165. doi: 10.1016/0076-6879(89)70045-8. [DOI] [PubMed] [Google Scholar]
- 26.Popper KR. Conjectures and Refutations. Routledge Classics; New York: 1963. [Google Scholar]
- 27.Small EC, Xi L, Wang JP, Widom J, Licht JD. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proc Natl Acad Sci USA. 2014;111(24):E2462–E2471. doi: 10.1073/pnas.1400517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansari A, Gartenberg MR. Persistence of an alternate chromatin structure at silenced loci in vitro. Proc Natl Acad Sci USA. 1999;96(2):343–348. doi: 10.1073/pnas.96.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirzaev I, Gunawardena J. Laplacian dynamics on general graphs. Bull Math Biol. 2013;75(11):2118–2149. doi: 10.1007/s11538-013-9884-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.