Significance
The prevailing dogma is that access to copper (Cu+) is restricted to the extent that protein–protein interactions mediate the routing of Cu+ from transporters in the plasma membrane to target cuproenzymes or transporters within subcellular compartments. The soluble proteins that distribute Cu+ are called metallochaperones. Although the chloroplast requires Cu+, a chaperone that delivers this essential cofactor has remained a missing link in the model for plastid Cu+ delivery. Using a comparative genomic approach and validating by biochemical characterization, we have discovered a missing chaperone. Uniquely, the previously unidentified chaperone family has evolved from the transporter to which it delivers Cu+. We also uncover an interaction between the thylakoid-localized transporter and the Cu+ chaperone for stromal Cu/Zn superoxide dismutase, which highlights the complexity of Cu+ distribution networks.
Keywords: Cu-transfer, inner envelope, metal transporter, Arabidopsis thaliana, Atx1
Abstract
Metallochaperones traffic copper (Cu+) from its point of entry at the plasma membrane to its destination. In plants, one destination is the chloroplast, which houses plastocyanin, a Cu-dependent electron transfer protein involved in photosynthesis. We present a previously unidentified Cu+ chaperone that evolved early in the plant lineage by an alternative-splicing event of the pre-mRNA encoding the chloroplast P-type ATPase in Arabidopsis 1 (PAA1). In several land plants, recent duplication events created a separate chaperone-encoding gene coincident with loss of alternative splicing. The plant-specific Cu+ chaperone delivers Cu+ with specificity for PAA1, which is flipped in the envelope relative to prototypical bacterial ATPases, compatible with a role in Cu+ import into the stroma and consistent with the canonical catalytic mechanism of these enzymes. The ubiquity of the chaperone suggests conservation of this Cu+-delivery mechanism and provides a unique snapshot into the evolution of a Cu+ distribution pathway. We also provide evidence for an interaction between PAA2, the Cu+-ATPase in thylakoids, and the Cu+-chaperone for Cu/Zn superoxide dismutase (CCS), uncovering a Cu+ network that has evolved to fine-tune Cu+ distribution.
The use of copper (Cu+) as a catalytic cofactor has necessitated the evolution of specific transport and delivery proteins. In eukaryotes, Cu+ trafficking pathways exist to shuttle Cu+ from the plasma membrane to cytosolic targets, such as Cu/Zn superoxide dismutase (SOD) (1, 2), or to intracellular compartments, such as vesicles of the trans-Golgi network (3, 4). In the latter case, Cu+ is often delivered to a transporter of the P1B subgroup of P-type ATPases (P1B-type ATPase), which catalyzes the ATP-dependent transport of cytosolic Cu+ across membranes of the secretory pathway (5). Central components of the Cu+ trafficking pathways are small, soluble proteins called Cu+ chaperones that bind Cu+ with high affinity (6–8) and deliver it through specific protein–protein interactions. In this way, Cu+ ions are directed to the correct apoproteins and compartments.
The prototypical Cu+ trafficking pathway involves Atx1-like Cu+ chaperones and P1B-type ATPases. Our understanding of how chaperones and Cu+-ATPases work together to distribute Cu+ is largely based on efflux pathways in bacteria, where these two proteins work in concert to transport Cu+ from the cytosol to the periplasm for detoxification and cuproenzyme metallation. Metal transfer is characterized by the specific and transient interaction of the Cu+-bound chaperone (Cu+-chaperone) with the ATPase and the unidirectional transfer of the ion. The crystal structure of Legionella pneumophila copper-translocating P-type ATPase (CopA) and biochemical analyses of Archaeoglobus fulgidus CopA support a model where the Cu+-chaperone interacts with an electropositive platform formed by a kink in the second transmembrane (TM) helix of the ATPase (8–10). The kink exposes three invariant residues (one Met and two Glu) that enable ligand exchange between the chaperone and the transporter (10). After ligand exchange, the Cu+ ions occupy the intramembrane metal-binding sites (11), and following the classic Albers–Post model, Cu+ export requires the metal-dependent catalytic phosphorylation of an invariant Asp (DKTGT) (5, 12, 13).
In addition to the electropositive platform, Atx1-like chaperones can interact with and deliver Cu+ to the N-terminal metal-binding domain (HMBD). Because this domain is not essential in vitro for activity (8, 10), one plausible role for the HMBD is in autoregulation of the ATPase; Cu+ delivery to this domain may be required before the chaperone can dock to the platform and deliver Cu+ to the intramembrane metal-binding sites (13). Interestingly, the HMBDs of the Cu+-ATPases and the Atx1-like chaperones are structurally similar with a ferredoxin-like fold and bind Cu+ through a conserved MxCxxC motif. Phylogenetic reconstruction suggests that the HMBDs and cognate Cu+ chaperones share a common origin that predates the prokaryotic-eukaryotic split (14). Nevertheless, although they are homologous, the chaperone, but not the HMBD, is able to transfer Cu+ directly to the intramembrane metal-binding sites (8).
Because of the presence of an extra Cu-using organelle, plants have a relatively complex Cu+ distribution network. In addition to the mitochondrion and trans-Golgi network, Cu+ must be delivered to and trafficked within the chloroplast to metallate plastocyanin (all green algae and land plants); Cu/Zn SOD (some green algae and land plants); and other lineage-specific enzymes, such as polyphenol oxidase in Populus tricorphora. Two P1B-type ATPases, P-type ATPase of Arabidopsis 1 (PAA1) and P-type ATPase of Arabidopsis 2 (PAA2) (15), have been localized to the chloroplast. A loss-of-function mutation in PAA1 from Arabidopsis thaliana causes a decrease in chloroplast Cu+ concentration that has a negative impact on the function of both stromal Cu/Zn SOD and plastocyanin, which binds Cu+ in the lumen, whereas mutations in PAA2 have little impact on total chloroplast Cu+ content but have a negative impact on plastocyanin activity (15). These phenotypes have led to the conclusion that PAA1 and PAA2 function in tandem; PAA1 imports cytosolic Cu+ into the stroma, and PAA2 imports Cu+ into the thylakoid lumen.
The Cu+ chaperones that must interact with PAA1 and PAA2 have remained unknown, leaving open the question of how Cu+ is routed to and within the chloroplast. The conservation of plastocyanin, PAA1, and PAA2 in green algae and land plants suggests that the Cu+ distribution route must have evolved early and specifically within the plant lineage. When we looked for homologs/paralogs of ATX1 that might serve this function, we noted that the ATX1 family had expanded in the land plants (16, 17), but these proteins seemed unlikely candidates for the missing link because of the absence of an ortholog in algae. The only Cu+ chaperone present in both the green alga Chlamydomonas reinhardtii and land plants is ATX1, which has a function analogous to ATOX1/Atx1 in mammals/yeast. The absence of an obvious Atx1-like chaperone candidate for the chloroplast suggests that a noncanonical chaperone must exist.
In this work, we describe a previously unidentified family of Cu+ chaperones that is conserved in C. reinhardtii and land plants. These chaperones have previously been overlooked because they are produced in most genomes as an alternative-splicing event of the PAA1 pre-mRNA. We provide evidence that in A. thaliana, this chaperone, which we have named plastid chaperone 1 (PCH1), delivers Cu+ with specificity to PAA1, highlighting a plant-specific adaptation not required for Cu+ delivery in cyanobacteria. We also uncover an interaction between CCS, the Cu+ chaperone for stromal Cu/Zn SOD, and PAA2.
Results
Comparative Genomic Analysis Reveals the Presence of a Plant-Specific Cu+ Chaperone.
Because Atx1-like chaperones share a common ancestor with the HMBDs of Cu+-ATPases, we were intrigued to find evidence that the pre-mRNA encoding the PAA1 ortholog (CTP2) in the unicellular, green alga C. reinhardtii is alternatively spliced, producing a small transcript that encodes solely the HMBD (we will refer to the short form as PCH1). The two transcripts, one encoding the full-length ATPase and the second encoding PCH1, share the first two exons (Fig. 1A). An exon that is skipped in the CTP2 transcript provides an alternative stop codon and alternative polyadenylation site for the PCH1 transcript (Fig. 1A).
Fig. 1.
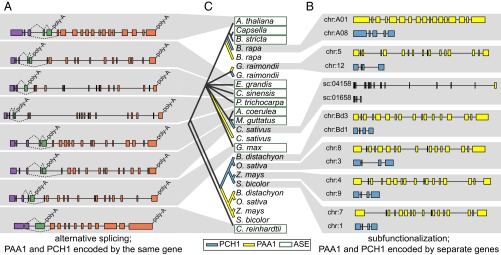
PCH1 in plants is encoded by either an alternative-splicing event (ASE) of the PAA1 pre-mRNA (A) or from a separate locus (B). In A, shared exons are colored purple, exons unique to PCH1 are colored green, and exons unique to PAA1 are colored orange. Splicing events are represented by broken lines linking exons. In B, gene models for PAA1 are colored yellow, and gene models for PCH1 are colored blue. Gray shading is used to connect each gene cartoon (A or B) with the relevant plant (C). In C, the phylogenetic history of PCH1 and the N-terminal HMBD of PAA1 were predicted using the neighbor-joining method. Branches representing PCH1 and the N-terminal HMBD of PAA1 that are produced from ASEs are collapsed, and the label is encased with a green box. In cases where the two proteins are encoded by separate genes, the branches representing PCH1 are colored blue and the branches representing the N-terminal HMBD of PAA1 are colored yellow. chr, chromosome; sc, scaffold.
We wondered whether this alternative-splicing event is unique to the PAA1 ortholog from C. reinhardtii or whether it occurred in other organisms as well. An exhaustive analysis of land plant genomes and ESTs revealed conservation of nearly identical alternative-splicing events in A. thaliana, Boechera stricta, Capsella rubella, Capsella grandiflora, Mimulus guttatus, Citrus sinensis, Eucalyptus grandis, Aquilegia coerulea, P. trichocarpa, and Glycine max (Fig. 1A). In another seven genomes, including Selaginella moellendorffii and Physcomitrella patens, two early diverging land plants, we found EST support for the short PCH1 transcript (minimally the last two 3′ exons, including the stop codon), but EST support for the unique exon junction of PAA1 is presently lacking (Fig. S1B). In each case, the short PCH1 transcript is predicted to encode a small soluble protein of 172–256 amino acids that preserves the N-terminal soluble portion of the corresponding ATPase, including the Cu+-binding motif MxCxxC (Fig. S1A).
We were further surprised to find that the absence of evidence for the alternative-splicing event in several plant genomes was often coincident with gene duplication and subfunctionalization. In Brassica rapa, Cucumis sativus, Gossypium raimondii, Brachypodium distachyon, Oryza sativa, Panicum virgatum, Sorghum bicolor, and Zea mays, PAA1 and PCH1 are encoded at separate loci (Fig. 1B). Phylogenetic analysis suggests that gene duplication, and subsequent subfunctionalization, has occurred independently in the Brassica, Cucumis, and Gossypium lineages, with closely related genera retaining the PAA1/PCH1 splicing event (Fig. 1C). This analysis also supports the existence of separate PAA1 and PCH1 genes in the common ancestor of grasses, which existed roughly 50 Mya (18). In G. max and P. trichocarpa, we found duplication of PAA1 with alternative splicing conserved at one locus, but EST support for alternative splicing at the second locus is lacking (Fig. S1D).
Because previous phylogenetic reconstruction had suggested that Atx1-like chaperones and the HMBDs of the Cu+-ATPases represent ancient lineages (14), the alternative-splicing event may represent a relatively recent and unique progenitor event. Indeed, we were unable to find such an event involving other plant Cu+-ATPases (Fig. S2). We did, however, find two distinct types of small soluble proteins that have evolved from the N-terminal HMBD of PAA1 (Fig. S3). The first type, PCH1, contains an N-terminal glycine-rich region after the putative signal peptide cleavage site characteristic of PAA1 (Fig. S4). The second type does not contain the polyglycine stretch and was not absolutely conserved as was found with PCH1. In grasses, phylogenetic analysis supports the conclusion that this second group arose from duplication of PCH1 and loss of the polyglycine stretch (Fig. S3). Multiple copies of this no-glycine stretch protein could be found in a single genome, but only one copy of PCH1 (either produced as an alternative-splicing event or from a separate gene) was ever found.
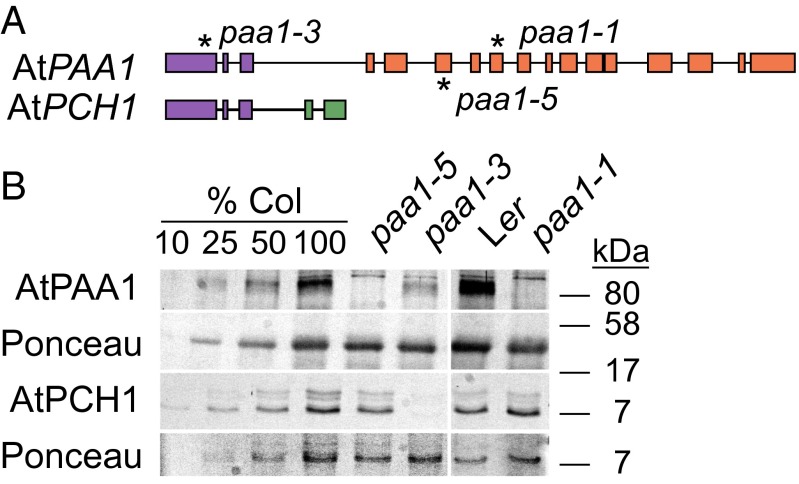
Short Alternative-Splice Form of A. thaliana PAA1 Is Translated.
Given the ubiquity and conservation of PCH1 as either an alternatively spliced transcript or a duplicated gene, we hypothesized that the short form of A. thaliana PAA1 (AtPAA1; gene model AT4G33520.1) is translated. Previous proteomic analysis of purified chloroplasts identified a peptide that corresponds to the N-terminal HMBD of AtPAA1 but with a native mass of 16 kDa and denatured mass of 12 kDa (19). Given its size and solubility, this protein is likely AtPCH1, but because this product could result from proteolysis of AtPAA1, we validated the presence of AtPCH1 using existing loss-of-function mutants (20). The mutants paa1-1 [background Landsberg erecta (Ler)] and paa1-5 [background Columbia-0 (Col-0)] contain point mutations (at positions 3,628 and 3,045, respectively) that should only affect AtPAA1, whereas paa1-3 (in Col-0) contains an in-frame deletion (Δ457–465) that should affect both AtPAA1 and AtPCH1 (Fig. 2A). Using an antibody raised against the HMBD of AtPAA1, we detected a protein with an apparent molecular mass of roughly 7 kDa in both the accessions Col-0 and Ler (Fig. 2B). As predicted, we did not detect AtPAA1 in either paa1-1 or paa1-5 but did detect the 7-kDa protein. In paa1-3 (Fig. 2B), AtPAA1 was detected but at a reduced level, whereas the 7-kDa protein was not. These data demonstrate unequivocally the presence of PCH1 in plants (independent of PAA1, and hence not a degradation product).
Fig. 2.
Alternative splicing produces two proteins from the AtPAA1 locus. (A) Schematic of the AtPAA1 and AtPCH1 transcripts. Asterisks mark the location of the paa1-1, paa1-3, and paa1-5 mutations. Exons are colored as in Fig. 1. (B) Immunoblot with antibody raised against the N-terminal HMBD of AtPAA1. Equal loading was confirmed with Ponceau staining of the membrane after transfer. Serial dilutions of Col-0 protein extract (%Col) were performed as indicated.
AtPCH1 Transfers Cu+ Specifically to AtPAA1.
Given the high similarity between PCH1 and documented Cu+ chaperones (Fig. S4), we hypothesized that PCH1 might be a Cu+ chaperone with function in delivering Cu+ either to PAA1 and/or PAA2. We used two biochemical assays to test whether PCH1 can interact with the ATPases in vitro by assessing its ability to (i) activate the ATPase activity and (ii) transfer Cu+. We used N-terminally truncated versions of PAA1 and PAA2 that lacked the N-terminal HMBDs (ΔN-AtPAA1 and ΔN-AtPAA2, respectively) to facilitate analysis of metal delivery to the intramembrane Cu+-binding sites without interference from metal transfer to the regulatory N-terminal HMBDs.
The function of recombinant ΔN-AtPAA1 and ΔN-AtPAA2 was evaluated by measuring Cu+ dependence of their ATPase activities (independent of a Cu+-chaperone) and their Cu+-binding stoichiometries (Fig. 3 and Table 1). Both ΔN-AtPAA1 and ΔN-AtPAA2 displayed kinetic parameters compatible with proper folding and function (Fig. 3 and Table 1). Thus, both Cu+-ATPases interact in vitro with their substrate, independent of a chaperone, and can complete the catalytic cycle.
Fig. 3.
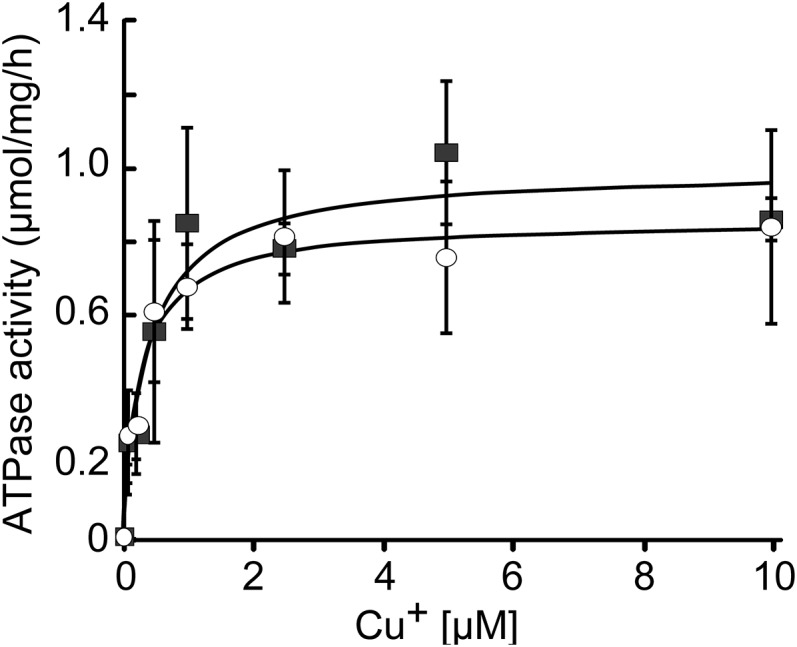
Cu+-dependent ATPase activity of ΔN-AtPAA1 (○) and ΔN-AtPAA2 (■). ATPase activity was measured in the presence of increasing concentrations of Cu+ in the absence of a chaperone.
Table 1.
Kinetic parameters of ATPase activity in the absence and presence of chaperones or N-terminal HMBD and Cu-binding stoichiometry of each protein
| Cu+ form | Vmax,* μmol/mg/h | K1/2,* μM | Cu+/protein stoichiometry† | |
| ΔN-AtPAA1 | Cu+ | 0.86 ± 0.04 | 0.27 ± 0.06 | 1.89 ± 0.15 |
| Cu+-AtPCH1 | 2.62 ± 0.25 | 1.81 ± 0.63 | ||
| Cu+-AtPAA1-HMBD | 1.61 ± 0.14 | 0.10 ± 0.06 | ||
| Cu+-AtPAA2-HMBD | −0.37 ± 0.27 | n.d. | ||
| Cu+-AtCCS | 0.07 ± 0.04 | n.d. | ||
| ΔN-AtPAA2 | Cu+ | 0.86 ± 0.06 | 0.38 ± 0.12 | 2.17 ± 0.046 |
| Cu+-AtPCH1 | 0.10 ± 0.03 | n.d. | ||
| Cu+-AtPAA1-HMBD | 0.10 ± 0.03 | n.d | ||
| Cu+-AtPAA2-HMBD | −0.03 ± 0.05 | n.d. | ||
| Cu+-AtCCS | 1.27 ± 0.04 | 0.79 ± 0.06 | ||
| AtPCH1 | Cu+ | 1.07 ± 0.08 | ||
| AtPAA1-HMBD | Cu+ | 0.91 ± 0.21 | ||
| AtPAA2-HMBD | Cu+ | 0.91 ± 0.12 | ||
| AtCCS | Cu+ | 1.10 ± 0.22 |
n.d., not determined.
Errors for Vmax and K1/2 are asymptotic SEs reported by the fitting software Kaleidagraph (Synergy).
Stoichiometry of Cu+ bound to ATPases or chaperones was estimated as moles Cu+/moles protein.
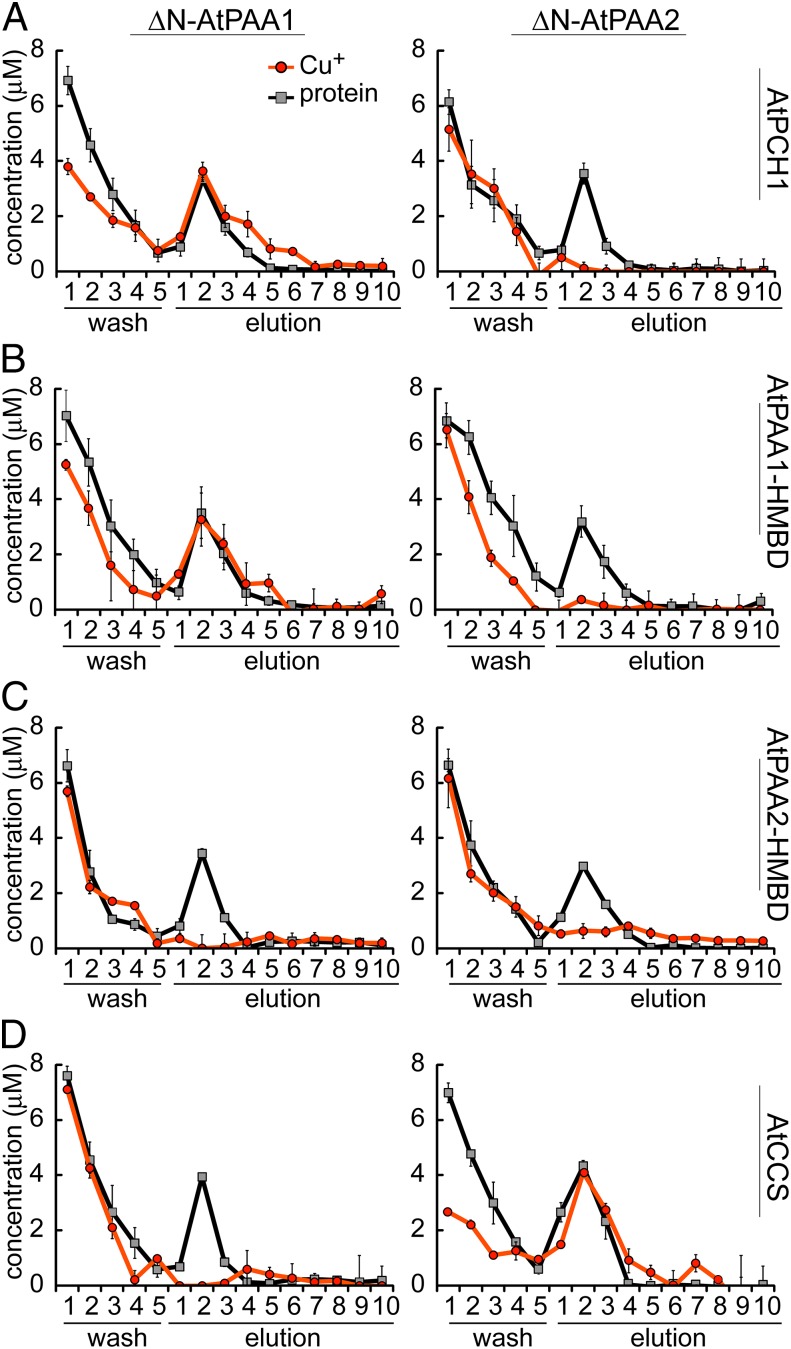
The ability of Cu+-AtPCH1 to interact with ΔN-AtPAA1 or ΔN-AtPAA2 was tested by measuring the activation of their ATPase activities by saturating concentrations (10 μM) of Cu+-AtPCH1 (Table 1). ΔN-AtPAA1 was activated by Cu+-AtPCH1 but ΔN-AtPAA2 was not, suggesting a specific interaction between Cu+-AtPCH1 and ΔN-AtPAA1. Moreover, as previously observed for other ATPases (21), the Vmax of ΔN-AtPAA1 in the presence of Cu+-AtPCH1 (2.62 ± 0.25 μmol⋅mg−1⋅h−1) was higher than the Vmax observed in the presence of free Cu+ (0.84 ± 0.26 25 μmol⋅mg−1⋅h−1) (Table 1).
Next, we tested whether Cu+ could be transferred from PCH1 to PAA1 or PAA2. We measured unidirectional Cu+ transfer under nonturnover conditions by incubating Cu+-loaded streptomycin (Strep)-tagged AtPCH1 with His-tagged versions of each ATPase (metal-free). The ATPases were recovered by metal affinity chromatography in a Ni2+ resin. Fractions eluting from the column were analyzed for protein and Cu+ content. We found that Cu+ coeluted with ΔN-AtPAA1 but not with ΔN-AtPAA2 (elution fractions 1–4; Fig. 4A), consistent with a specific interaction between AtPAA1 and Cu+-AtPCH1 as inferred from the ATPase activity assay (Table 1).
Fig. 4.
Interaction of ΔN-AtPAA1 and ΔN-AtPAA2 with soluble Cu+ chaperones. Cu+ (orange circles) and protein (gray squares) contents of the wash and elution fractions are shown. (A) Cu+ transfer assays between Cu+-AtPCH1 and ΔN-AtPAA1 or ΔN-AtPAA2. (B) Cu+ transfer assays between Cu+-AtPAA1-HMBD and ΔN-AtPAA1 or ΔN-AtPAA2. (C) Cu+ transfer assays between Cu+-AtPAA2-HMBD and ΔN-AtPAA1 or ΔN-AtPAA2. (D) Cu+ transfer assays between Cu+-AtCCS and ΔN-AtPAA1 or ΔN-AtPAA2.
Because they are produced by alternative-splicing events, AtPCH1 and the N-terminal HMBD of AtPAA1 are nearly identical except for 13 amino acids present uniquely at the C terminus of AtPCH1 (Fig. S5). In a previous work, we had noted that the N-terminal HMBD of A. fulgidus CopA was not able to deliver Cu+ to the intramembrane metal-binding sites of CopA (8). In that system, Cu+ transfer was specific for the separately encoded Cu+ chaperone, CopZ. This result raised the question of whether Cu+ transfer from the N-terminal HMBD to PAA1 might be a capability of other members of this subgroup of the P1B-ATPase family or whether the C-terminal 13 amino acids conferred this ability to PCH1. We expressed the soluble HMBD of AtPAA1 and AtPAA2 (by truncating the respective proteins just before the first TM helix) and used the purified proteins to test (i) activation of ATPase activity and (ii) Cu+ transfer in the absence of ATP. Unlike the situation with CopA, Cu+-AtPAA1-HMBD exhibited Cu+ chaperone activity in that it was able to activate the ATPase activity and transfer Cu+ to ΔN-AtPAA1 just like AtPCH1 did, indicating that this property was not conferred by the extra 13 amino acids at the C terminus of AtPCH1 (Fig. 4B and Table 1). On the other hand, Cu+-AtPAA2-HMBD did not activate either ΔN-AtPAA1 or ΔN-AtPAA2, and we could not detect Cu+ transfer from Cu+-AtPAA2-HMBD to either protein (Fig. 4C and Table 1), indicating that the Cu+ chaperone biochemical property of PCH1 and the N-terminal domain of PAA1 is unique.
AtCCS Delivers Cu+ to AtPAA2.
There is no evidence supporting the production of AtPAA2-HMBD as a stand-alone soluble protein, and Cu+ transfer was not detected from this recombinant protein to either ATPase. Therefore, to find a possible interacting partner for AtPAA2, we tested the interaction of the previously characterized chloroplast Cu+ chaperone for Cu/Zn SOD (AtCCS) (22) with ΔN-AtPAA1 and ΔN-AtPAA2. Although activation or Cu+ transfer was not detected with ΔN-AtPAA1, Cu+-AtCCS was able to activate and transfer Cu+ to ΔN-AtPAA2 (Fig. 4D and Table 1), revealing a possible interaction between AtPAA2 and AtCCS, and further highlighting the exclusive interaction between AtPAA1 and AtPCH1.
Orientation of PAA1 in the Inner Envelope.
All Cu+-ATPases in the plasma membrane of prokaryotes are considered to be orientated in the membrane with the N-terminal HMBD and ATP-binding domain facing the cytosol (5). By analogy with cyanobacteria, the closest free-living relatives of the plastid, it is predicted that the N-terminal HMBD and ATP-binding domain of PAA1 face the stroma. Given the proposed unidirectional transport of Cu+ away from the ATPase domains (23, 24), this orientation would imply that PAA1 exports Cu+ from the stroma toward the interenvelope space (IES). This function is at odds with loss-of-function phenotypes (20), which suggest AtPAA1 transports Cu+ into the stroma, and raises the question of which compartment it is that the PAA1/PCH1 interaction might occur.
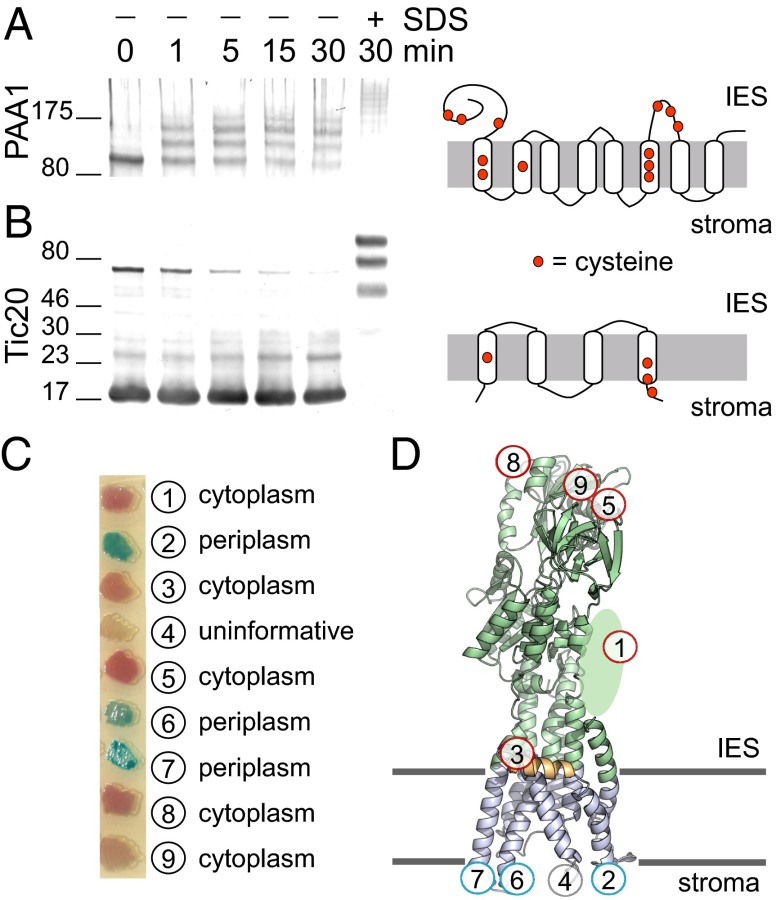
We therefore investigated the orientation of native PAA1 in right-side-out inner envelope vesicles from isolated Pisum sativum (pea) plastids. Based on the deduced structural model, PAA1 from P. sativum has six cysteines facing the same side of the membrane, whereas the remaining cysteines are occluded by the membrane (Fig. S6). Methoxypolyethylene glycol-maleimide (PEG-Mal) is a membrane-impermeable probe that reacts irreversibly with free thiols. If PAA1 faces the stroma (which becomes the vesicle lumen), the cysteines will be protected from PEGylation by the vesicle membrane. However, if PAA1 faces the IES, the cysteines will be free to react with the PEG-Mal. Three independent PEGylation events of pea PAA1 were readily detected (Fig. 5A) and consistent with the N-terminal HMBD and ATPase domains being located in the IES. Because three of the six accessible cysteines are present in the N-terminal HMBD used to raise antibodies, we cannot rule out the possibility that PEG-Mal modification interferes with antibody binding. However, if PAA1 were oriented in the opposite direction (facing the vesicle lumen), there would be no cysteines available for PEGylation. As a control for the orientation of the vesicles, PEGylation of Tic20, an inner envelope protein oriented with a cysteine facing the stroma (25, 26), was not detected unless SDS was added to solubilize the vesicles (Fig. 5B).
Fig. 5.
Topology and orientation of PAA1. PEG-Mal labeling of P. sativum inner envelope vesicles in the presence or absence of 1% SDS was performed for the indicated time subsequent to detection of pea PAA1 (A) and Tic20 (B) by immunoblotting. The location of cysteines in each protein is shown to the right. (C) E. coli cells expressing various translational fusions between the PhoA-LacZα reporter cassette and AtPAA1 were plated on dual indicator plates, which contain chromogenic substrates. Hydrolysis of 5-bromo-4-chloro-3-indolyl phosphate by PhoA yields a blue color, whereas hydrolysis of 6-chloro-3-indolyl-β-d-galactoside by LacZ yields a red color, allowing assignment of the PhoA-LacZα reporter to either the periplasmic space or cytoplasm, respectively. (D) Location of each fusion construct from C is overlaid on the structural model of AtPAA1.
Membrane Topology of AtPAA1 in Escherichia coli.
Because interpretation of the PEG-Mal experiment is dependent on knowing the location of cysteines within the structure, we also confirmed the predicted topology of PAA1 with PhoA-LacZα sandwich fusions expressed in Escherichia coli. PhoA serves as a reporter for the periplasm; mature PhoA (alkaline phosphatase) contains two intramolecular disulfide bonds (27), which are formed within the oxidizing environment of the periplasm. LacZα serves as a reporter for the cytoplasm; LacZ (β-gal) activity requires association of the LacZα peptide with the cytoplasmic-localized LacZω peptide. Translational fusions within the N-terminal HMBD and ATPase domains of AtPAA1 displayed predominantly β-gal activity, whereas fusions in the intra-TM helix loops displayed predominantly alkaline phosphatase activity, with the exception of construct 4 in loop 3, which did not display activity of either reporter (Fig. 5C). These results support the structural model depicted in Fig. 5D.
Discussion
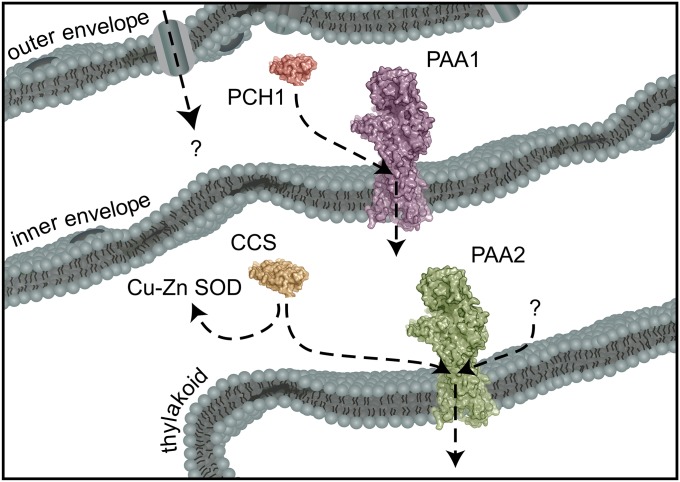
Ligand exchange and the transfer of Cu+ from soluble chaperones to transporters are fundamental to maintain Cu+ homeostasis (28–31). These interactions minimize the number of mismetallation events that would otherwise be caused by “free” Cu+ and provide the means to control the routing of Cu+ to distinct subcellular locations. Here, we have presented the identification and characterization of the Cu+ chaperone for PAA1, the chloroplast Cu+ importer. In contrast to A. fulgidus CopA, the N-terminal HMBD of PAA1 has evolved to transfer Cu+ to the intramembrane metal-binding sites, but only because this domain exists as a separate soluble protein within the plant lineage. Metallochaperones, such as PCH1, are the Cu+ handlers of the cell, and specific molecular recognition, such as we have described between PCH1 and PAA1, is a central component in Cu+ homeostasis that was previously missing in the model of plastid Cu+ delivery (Fig. 6).
Fig. 6.
Model of chloroplast Cu+ delivery. PAA1 faces away from the stroma, which places the chaperone docking platform and N-terminal HMBD in the IES. An interaction between CCS and PAA2 would occur in the stroma, but a ubiquitous Cu+ chaperone for PAA2 is still missing (represented by a question mark).
Delivery of Cu+ to Cu-Dependent Proteins Within the Plastid Required Plant-Specific Adaptations Compared with the Analogous Cu+ Transport Pathway Found in Cyanobacteria.
The chloroplast is the remnant of a cyanobacteria-like endosymbiont, and most of its metabolic and homoeostatic repertoire was inherited from this endosymbiont. Chloroplast Cu+ delivery, however, appears to have undergone several plant-specific modifications, which are likely a result of adapting to receiving Cu+ from an intracellular location rather than from the environment. Two Cu+-ATPases in cyanobacteria, CtaA, which is thought to reside in the plasma membrane, and PacS, which has been localized to the thylakoid membranes (32), are involved in supplying Cu+ to plastocyanin (33). The Cu+ chaperone Atx1 is proposed to function in transferring Cu+ between the two ATPases (34). In plants, neither pacS nor atx1 was retained, whereas ctaA was duplicated, giving rise to PAA1 and PAA2 (35). Reflecting their common origin, we found that ΔN-AtPAA2 displayed a Vmax comparable to the Vmax of CtaA, which is nearly an order of magnitude smaller than the Vmax of PacS (36). The relatively fast turnover rate of PacS is a characteristic required for Cu+ detoxification (21, 36), and because supply of Cu+ to the chloroplast is presumably restricted, there is no longer a need for a Cu+ detoxification pump. Thus, PacS function has been lost in the plant lineage.
The second plant-specific adaptation is the evolution of a new family of Cu+ chaperones that are derived from the N-terminal HMBD of PAA1. Because this domain uniquely exists as a separate soluble protein, a typical chaperone–ATPase interaction was able to evolve. Given the genetic evidence that PAA1 imports Cu+ into the stroma (15, 20) and the proposed transport mechanism of these ATPases (24), we propose that PCH1 delivers Cu+ to PAA1, which is oriented in the inner envelope membrane such that the chaperone platform faces the IES, which is a third plant-specific adaptation.
Previously, the polyglycine stretch following the transit peptide of AtPAA1 was proposed to target AtPAA1 to the inner envelope (15). Because the polyglycine stretch is conserved in all PAA1 orthologs and PCH1 orthologs, we suggest that the stretch may act as a conserved stop-transfer signal that keeps the N-terminal HMBD of PAA1 and PCH1 in the IES. In one model for the import of pre-Toc75, the polyglycine stretch serves to block translocation across the inner envelope (37, 38). Therefore, PAA1 may be inserted into the inner envelope on the IES side of the membrane, and PCH1 is retained in the IES. However, an outstanding question is where PCH1 binds Cu+. PCH1 may bind Cu+ in the cytosol before delivery to PAA1; this scenario is possible because small folded proteins can indeed be imported across the outer envelope (39). Such a mechanism was originally postulated for COX17, a soluble chaperone in the intermembrane space of the mitochondrion that delivers Cu+ for cytochrome c oxidase maturation (40). However, recent studies suggest that although the source of mitochondrial Cu+ remains unclear, metallation of cytochrome c oxidase appears to involve Cu+ transported from the mitochondrial matrix (41).
Conservation of PCH1 in the Green Lineage.
A comparative genomic analysis has enabled us to devise the evolutionary history of this new chaperone family. Conservation of alternative splicing that produces PCH1 in a Chlorophyte and in Embryophytes suggests that alternative splicing represents the ancestral state and that PCH1 evolved in several land plants through a rarely described mechanism: subfunctionalization of alternative-splice forms. At this point, we can only speculate as to why alternative splicing is maintained in some genomes and duplicated genes are found in other genomes, but given the absolute conservation of one or the other, PCH1 is an integral component of Cu+ homeostasis in plants.
An estimated 700 My (42), four whole-genome duplications and one triplication event (43), separate C. reinhardtii from A. thaliana, yet PCH1 is formed by alternative splicing in both plants. One argument for the retention of alternative splicing is relatively rapid regulation of the two forms (because regulation occurs at the transcript-splicing level rather than at the expression level) (44). However, a survey of RNA-sequencing data in C. reinhardtii has not revealed a significant difference in the ratio of the two transcripts under various growth conditions, including different Cu2+ and Zn2+ supplementations. Another possibility is that alternative splicing is maintained such that expression of the two forms is identical (44). In prokaryotes, the Cu+ chaperone and Cu+-ATPase are frequently encoded by the same polycistronic mRNA (i.e., operon). In grasses and other plants where subfunctionalization has occurred, such tight control of transcript abundance may no longer be necessary. A third possibility is that retention of alternative splicing eliminates the possibility of sequence drift between PCH1 and the N-terminal HMBD of PAA1. The N-terminal HMBD is proposed to serve a regulatory function, and in addition to delivery of Cu+ to the intramembrane Cu+-binding sites, PCH1 may interact with the transporter HMBD, but such an interaction between the two HMBDs is presently unknown.
Interaction Between AtCCS and AtPAA2.
In this study, we unexpectedly observed an interaction between AtCCS and AtPAA2. Cu+ transfer from AtCCS to AtPAA2 is likely mediated through the AtCCS N-terminal Cu+-binding domain that is homologous to both the Atx1-like Cu+ chaperones and the N-terminal HMBD of the Cu+-ATPases. Genes encoding CCS and Cu/Zn SOD (CSD2) are not found in the C. reinhardtii genome, which would be expected if CCS were an integral component of the PCH1-PAA1-PAA2 Cu+ pathway, and the absence of a noticeable photosynthetic defect as a result of AtCCS disruption (45) supports the notion that AtCCS might not be critical for delivery of Cu+ to AtPAA2. However, an interaction between AtPAA2 and AtCCS may have evolved to fine-tune the distribution of Cu+ between Cu/Zn SOD (nonessential function) and plastocyanin (essential function). Interestingly, mutation of AtPAA2 was previously found to cause a significant increase in the amount of AtCSD2 abundance and activity, particularly at low concentrations of Cu+ (15). At the time, the increase in AtCSD2 activity was attributed to an increase in AtCSD2 transcript abundance in the mutant; however, in light of an interaction between AtCCS and AtPAA2, the absence of AtPAA2 may free-up AtCCS for Cu+ delivery to AtCSD2, leading, at least in part, to higher AtCSD2 activity.
Elucidating the Chloroplast Cu+ Network.
In summary, the data support two plausible mechanisms by which Cu+ gains access to the intramembrane metal-binding sites of the chloroplast Cu+-ATPases PAA1 and PAA2. The N-terminal HMBD of PAA1 is conserved throughout the plant lineage as an independent soluble protein, and in support of a chaperone role, the protein delivers Cu+ to the intramembrane metal-binding sites of AtPAA1. Such an interaction between AtPCH1 and AtPAA2 has not evolved, likely because the two proteins are not localized to the same compartment. Additionally, we did not find evidence that the N-terminal HMBD of PAA2 exists as an independent soluble chaperone, and Cu+ transfer experiments showed that this domain does not deliver Cu+ to the intramembrane metal-binding sites of either Cu+-ATPase. However, we did find that an interaction between AtPAA2 and AtCCS is possible, and such an interaction has not evolved between AtCCS and AtPAA1 (likely because AtPAA1 does not face the stroma). Clearly, given its absence in C. reinhardtii, CCS is not a conserved delivery route for PAA2. Instead, this interaction could have been selected as a way to ensure that PAA2, which supplies an essential cuproenzyme, can access Cu+ that would otherwise be solely channeled to AtCSD2, a nonessential cuproenzyme.
Methods
Bioinformatic Analyses.
Identification of plant-encoded proteins containing at least one heavy metal-associated (HMA) domain was performed by BLAST search (blastp with default parameters) of the Phytozome database (46). Reversed position specific (RPS)-BLAST of these sequences to the Conserved Domain Database (47) was used to identify the amino acid coordinates of each HMA domain. The protein similarity network (48) was constructed as described previously (49) from all-vs.-all blastp alignments of each HMA domain using an E-value cutoff of 2 × 10−8. Protein identification codes and amino acid coordinates of the HMA domains analyzed are available in Dataset S1. Identification of alternative-splicing events required at least one EST bridging the last shared exon and the unique exon of the short form and at least one EST bridging the last shared exon and the first unique exon of the long form. The phylogenetic tree of the N-terminal portions of AtPAA1 orthologs in plants and closely related soluble HMA domain-containing proteins was computed using the neighbor-joining method (50). Branches corresponding to partitions reproduced in less than 50% bootstrap (51) replicates (out of 500 replicates) were collapsed. All positions with less than 5% site coverage were eliminated. Evolutionary analyses were conducted in MEGA6 (52).
Cloning, Protein Expression, and Purification.
The coding region of AtPAA1 was amplified by PCR using PAA1_His_For and PAA1_His_Rev (amino acids 223–949) or PAA1HMBD_Strep_For and PAA1HMBD_Strep_Rev (amino acids 103–245) (Table S1) and the A. thaliana cDNA clone pda19661 obtained from the RIKEN BioResource Center (BRC). The full-length coding region of AtPAA2 was codon-optimized for expression in E. coli and synthesized (GenScript Corp.). For cloning of the ΔN-AtPAA2 construct (amino acids 160–883), primers PAA2_His_For and PAA2_His_Rev were used (Table S1). For AtPAA2-HMBD (amino acids 66–175), primers PAA2HMBD_Strep_For and PAA2HMBD_Strep_Rev were used (Table S1). The coding region of AtPCH1 (amino acids 103–237) was amplified using AtPCH1_Strep_For and AtPCH1_Strep_Rev (Table S1) and the cDNA clone pda04914 obtained from the RIKEN BRC. The coding region of AtCCS (amino acids 1–229) was amplified from HALO_SFI_24-E11 obtained from the Arabidopsis Biological Resource Center using the primers CCS_Strep_For and CCS_Strep_Rev (Table S1). The resulting products were digested with NdeI and SalI (or AseI and SalI for AtCCS) and ligated into pET21b vector engineered to contain a C-terminal tobacco etch virus protease recognition site followed by an in-frame His6-tag (for ATPase constructs) or an in-frame Strep-tag (for soluble chaperone constructs). The insert sequences of the resulting vectors were confirmed by Sanger sequencing. Plasmids were transformed into the E. coli BL21(DE3) strain, and heterologous expression was induced following the autoinducing media protocol (53). Purification of membrane and soluble proteins was performed as previously described (8, 54). Solubilized lipid/detergent micellar forms of ΔN-AtPAA1 and ΔN-AtPAA2 proteins were stored in 25 mM Hepes (pH 8.0), 50 mM NaCl, 10 mM ascorbic acid, 0.01% n-dodecyl-β-d-maltopyranoside (DDM), and 0.01% asolectin (buffer C) until use. Soluble chaperones were stored in buffer W containing 100 mM Tris (pH 8) and 150 mM NaCl. Protein concentrations were determined by Bradford assay using a BSA standard (55). Molar protein concentrations were estimated assuming that the proteins were pure, using Mr = 79,830 for ΔN-AtPAA1, Mr = 79,046 for ΔN-AtPAA2, Mr = 15,539 for AtPCH1, Mr = 16,753 for AtPAA1-HMBD, Mr = 14,203 for AtPAA2-HMBD, and Mr = 26,538 for AtCCS. To eliminate any bound metal, all purified proteins were treated with metal chelators as described previously (56). Briefly, the proteins were incubated for 45 min at room temperature with 0.5 mM EDTA and 0.5 mM tetrathiomolybdate in either buffer C (ATPases) or buffer W (chaperones). Chelators were removed by buffer exchange using either 30-kDa (ATPases) or 3-kDa (chaperones) cutoff Centricon (Millipore). The final purity of all protein preparations was at least 95%, as verified by SDS-PAGE followed by Coomassie Brilliant Blue staining and immunoblotting with either anti-His6 (Genscript) or anti–Strep-tag antibodies (IBA) (Fig. S7).
Cu+ Loading to Proteins.
Cu+ loading was performed by incubating each apo-chaperone (10 μM) in the presence of 20 μM CuSO4, 25 mM Hepes (pH 8.0), 150 mM NaCl, and the reducing agent 1 mM Tris(2-carboxyethyl)phosphine for 10 min at room temperature with gentle agitation. The unbound Cu+ was removed by washing in 30-kDa (ΔN-AtPAA1 and ΔN-AtPAA2) and 3-kDa (AtPCH1, AtPAA1-HMBD, AtPAA2-HMBD, and AtCCS) cutoff Centricons, and proteins were used immediately. The amount of Cu+ bound was verified by the bicinchoninic acid (BCA) method (57). CuSO4 solutions were used as standards.
ATPase Assays.
ATPase assays were performed at 37 °C in medium containing 50 mM Tris⋅Cl (pH 6.8), 3 mM MgCl2, 3 mM ATP, 0.01% asolectin, 0.01% DDM, 200 mM NaCl, 2.5 mM DTT, and 0.01 mg/mL purified protein. When activation by free Cu+ was tested, 20 mM cysteine was included in the buffer (8). Cu+ concentrations, either in the free form or bound to soluble chaperones, were varied to obtain curves reporting the Cu+ concentration for half-maximal ATPase activation (K1/2) and Vmax. ATPase activity was measured after 20 min of incubation. Released phosphate was determined according to Lanzetta et al. (58). ATPase activity measured in the absence of metal was subtracted from plotted values. Curves of ATPase activity vs. free Cu+ or Cu+-chaperone concentration were fit to v = (Vmax[donor⋅Cu+])/([donor⋅Cu+] + K1∕2).
Cu+ Transfer Assays.
Cu+ transfer from the various Strep-tagged chaperones to different His6-tagged ATPase constructs was performed as previously described (8, 10). Briefly, each Strep-tagged, Cu+-loaded chaperone was added independently in threefold molar excess to His-tagged ΔN-AtPAA1 or ΔN-AtPAA2 bound to Ni2+-nitrilotriacetic acid resin and incubated for 10 min at room temperature. Proteins were separated by washing with 25 mM Hepes (pH 8.0), 100 mM sucrose, 500 mM NaCl, 0.01% DDM, 0.01% asolectin, 10 mM ascorbic acid, and 20 mM imidazole, followed by elution with 25 mM Hepes (pH 8.0), 100 mM sucrose, 500 mM NaCl, 0.01% DDM, 0.01% asolectin, 10 mM ascorbic acid, and 300 mM imidazole. Cu+ content of the resulting fraction was determined by the BCA method (57). The absence of ΔN-AtPAA1 or ΔN-AtPAA2 in the wash fractions and AtPCH1, AtPAA1-HMBD, AtPAA2-HMBD, or AtCCS in the elution fractions was confirmed by immunoblotting with anti–His6-tag and anti–Strep-tag. Controls were performed where Cu+-loaded chaperones or ATPases were subjected to the same procedures individually, lacking a partner Cu+-exchanging protein.
Antibody Production and Immunoblot Analysis.
Polyclonal antiserum to the N-terminal metal-binding domain of AtPAA1 (amino acids 103–245) was produced in rabbits by injection of purified recombinant protein (Covance Research Products). Polyclonal antibodies were affinity-purified by coupling the recombinant protein to NHS-activated agarose (Thermo Scientific Pierce) according to the manufacturer’s protocol. A. thaliana plants, ecotypes Col-0 and Ler and the mutants paa1-1, paa1-3, and paa1-5 (20), were grown in Sunshine Mix no. 2 (Sun Gro Horticulture) with 16 h of light (110 μmol of photons per m−2⋅s−1) and 8 h of dark. Protein extracts were prepared from the aerial portions of 2-wk-old seedlings according to Martínez-García et al. (59). Protein was quantified by BCA protein assay (Pierce) using BSA as a standard. Equal protein amounts corresponding to 30 μg were separated by SDS/PAGE [7.5% (wt/vol monomer) (acrylamide/bis acrylamide, 37.5:1) for detection of AtPAA1 and 20% (wt/vol monomer) for detection of AtPCH1].
Isolation of Inner Envelope Vesicles and PEGylation Assay.
Right-side-out inner envelope vesicles from P. sativum chloroplast membranes were isolated from intact chloroplasts of 12-d-old plants as described previously (60). Inner envelope vesicles were incubated with 10 mM PEG-Mal (Laysan Bio, Inc.) in 100 mM Tris⋅HCl (pH 7.0) containing 1 mM EDTA at room temperature in the dark. The reaction was quenched by addition of 100 mM DTT and Laemmli sample buffer. Bis-Tris gels [7.5% (wt/vol monomer) for PAA1 and 10% (wt/vol monomer) for Tic20] were used with MES running buffer. PAA1 and Tic20 were detected by immunoblotting using anti–AtPAA1-HMBD and anti-Tic20 antibodies (25), respectively.
PhoA-LacZα Fusions.
The gene encoding the PhoA-LacZα reporter was amplified from pMA650 (61) using PhoALacZ_For and PhoALacZ_Rev. The 5′ region of AtPAA1 was amplified using PAA1_PhoLac_For and PAA1_PhoLac_1Rev for construct 1, PAA1_PhoLac_2Rev for construct 2, PAA1_PhoLac_3Rev for construct 3, PAA1_PhoLac_4Rev for construct 4, PAA1_PhoLac_5Rev for construct 5, PAA1_PhoLac_6Rev for construct 6, PAA1_PhoLac_7Rev for construct 7, PAA1_PhoLac_8Rev for construct 8, or PAA1_PhoLac_9Rev for construct 9. The 3′ region of AtPAA1 was amplified using PAA1_PhoLac_Rev and PAA1_PhoLac_1For for construct 1, PAA1_PhoLac_2For for construct 2, PAA1_PhoLac_3For for construct 3, PAA1_PhoLac_4For for construct 4, PAA1_PhoLac_5For for construct 5, PAA1_PhoLac_6For for construct 6, PAA1_PhoLac_7For for construct 7, PAA1_PhoLac_8For for construct 8, or PAA1_PhoLac_9For for construct 9. For each construct, a four-way ligation was performed between pBAD22 (digested with NcoI/NheI), the 5′ fragment of AtPAA1 (digested with NcoI/BamHI), the gene encoding the PhoA-LacZα reporter (digested with BamHI/XmaI), and the 3′ fragment of AtPAA1 (digested with AgeI/NheI). The sequence-verified constructs were transformed into E. coli NEBα cells (New England Biolabs) and plated on dual-indicator plates as described by Alexeyev and Winkler (61), except that 0.2% arabinose was used for induction instead of isopropyl β-d-1-thiogalactopyranoside.
Supplementary Material
Acknowledgments
We thank Profs. Krishna Nyogi and Marinus Pilon for providing the A. thaliana mutants, Profs. Marinus Pilon and Kentaro Inoue for insightful discussion, Simona Stübe for technical support, and Drs. Ian Blaby and Patrice Salomé for critical reading of the manuscript. This work was supported by NIH Grant GM42143 (to S.S.M.) and National Science Foundation Grant MCB-0743901 (to J.M.A.). Expression/purification of AtPAA1 N-terminal HMBD for use as an antigen was supported by the Institute of Genomics and Proteomics at the University of California, Los Angeles [funded by the Office of Science (Biological and Environmental Research), United States Department of Energy, through Cooperative Agreement DE-FC02-02ER63421]. C.E.B.-H. acknowledges support from an Individual Kirschstein National Research Service Award (GM100753).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421545111/-/DCSupplemental.
References
- 1.Culotta VC, et al. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272(38):23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 2.Pope CR, De Feo CJ, Unger VM. Cellular distribution of copper to superoxide dismutase involves scaffolding by membranes. Proc Natl Acad Sci USA. 2013;110(51):20491–20496. doi: 10.1073/pnas.1309820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamza I, Schaefer M, Klomp LW, Gitlin JD. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc Natl Acad Sci USA. 1999;96(23):13363–13368. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi Y, Heiny ME, Suzuki M, Gitlin JD. Biochemical characterization and intracellular localization of the Menkes disease protein. Proc Natl Acad Sci USA. 1996;93(24):14030–14035. doi: 10.1073/pnas.93.24.14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argüello JM, Eren E, González-Guerrero M. The structure and function of heavy metal transport P1B-ATPases. Biometals. 2007;20(3-4):233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 6.Banci L, et al. Affinity gradients drive copper to cellular destinations. Nature. 2010;465(7298):645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Singleton C, Le Brun NE. High Cu(I) and low proton affinities of the CXXC motif of Bacillus subtilis CopZ. Biochem J. 2008;413(3):459–465. doi: 10.1042/BJ20080467. [DOI] [PubMed] [Google Scholar]
- 8.González-Guerrero M, Argüello JM. Mechanism of Cu+-transporting ATPases: Soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc Natl Acad Sci USA. 2008;105(16):5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourdon P, et al. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475(7354):59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 10.Padilla-Benavides T, McCann CJ, Argüello JM. The mechanism of Cu+ transport ATPases: Interaction with CU+ chaperones and the role of transient metal-binding sites. J Biol Chem. 2013;288(1):69–78. doi: 10.1074/jbc.M112.420810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Guerrero M, Eren E, Rawat S, Stemmler TL, Argüello JM. Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. J Biol Chem. 2008;283(44):29753–29759. doi: 10.1074/jbc.M803248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan JH. Biochemistry of Na,K-ATPase. Annu Rev Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- 13.Mattle D, et al. On allosteric modulation of P-type Cu(+)-ATPases. J Mol Biol. 2013;425(13):2299–2308. doi: 10.1016/j.jmb.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Jordan IK, Natale DA, Koonin EV, Galperin MY. Independent evolution of heavy metal-associated domains in copper chaperones and copper-transporting atpases. J Mol Evol. 2001;53(6):622–633. doi: 10.1007/s002390010249. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell. 2005;17(4):1233–1251. doi: 10.1105/tpc.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barth O, Vogt S, Uhlemann R, Zschiesche W, Humbeck K. Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol Biol. 2009;69(1-2):213–226. doi: 10.1007/s11103-008-9419-0. [DOI] [PubMed] [Google Scholar]
- 17.Tehseen M, Cairns N, Sherson S, Cobbett CS. Metallochaperone-like genes in Arabidopsis thaliana. Metallomics. 2010;2(8):556–564. doi: 10.1039/c003484c. [DOI] [PubMed] [Google Scholar]
- 18.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101(26):9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peltier JB, et al. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol Cell Proteomics. 2006;5(1):114–133. doi: 10.1074/mcp.M500180-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Shikanai T, Müller-Moulé P, Munekage Y, Niyogi KK, Pilon M. PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell. 2003;15(6):1333–1346. doi: 10.1105/tpc.011817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Guerrero M, Raimunda D, Cheng X, Argüello JM. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol Microbiol. 2010;78(5):1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- 22.Abdel-Ghany SE, et al. AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Lett. 2005;579(11):2307–2312. doi: 10.1016/j.febslet.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 23.Argüello JM, Raimunda D, Padilla-Benavides T. Mechanisms of copper homeostasis in bacteria. Front Cell Infect Microbiol. 2013;3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson M, et al. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat Struct Mol Biol. 2014;21(1):43–48. doi: 10.1038/nsmb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovács-Bogdán E, Benz JP, Soll J, Bölter B. Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 2011;11:133. doi: 10.1186/1471-2229-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machettira AB, et al. The localization of Tic20 proteins in Arabidopsis thaliana is not restricted to the inner envelope membrane of chloroplasts. Plant Mol Biol. 2011;77(4-5):381–390. doi: 10.1007/s11103-011-9818-5. [DOI] [PubMed] [Google Scholar]
- 27.Schlesinger MJ, Barrett K. The reversible dissociation of the alkaline phosphatase of Escherichia coli. I. Formation and reactivation of subunits. J Biol Chem. 1965;240(11):4284–4292. [PubMed] [Google Scholar]
- 28.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem. 2000;275(33):25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 30.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109(10):4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Cellular copper distribution: A mechanistic systems biology approach. Cell Mol Life Sci. 2010;67(15):2563–2589. doi: 10.1007/s00018-010-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanamaru K, Kashiwagi S, Mizuno T. A copper-transporting P-type ATPase found in the thylakoid membrane of the cyanobacterium Synechococcus species PCC7942. Mol Microbiol. 1994;13(2):369–377. doi: 10.1111/j.1365-2958.1994.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 33.Tottey S, Rich PR, Rondet SA, Robinson NJ. Two Menkes-type atpases supply copper for photosynthesis in Synechocystis PCC 6803. J Biol Chem. 2001;276(23):19999–20004. doi: 10.1074/jbc.M011243200. [DOI] [PubMed] [Google Scholar]
- 34.Tottey S, et al. A copper metallochaperone for photosynthesis and respiration reveals metal-specific targets, interaction with an importer, and alternative sites for copper acquisition. J Biol Chem. 2002;277(7):5490–5497. doi: 10.1074/jbc.M105857200. [DOI] [PubMed] [Google Scholar]
- 35.Hanikenne M, Baurain D. Origin and evolution of metal P-type ATPases in Plantae (Archaeplastida) Front Plant Sci. 2013;4:544. doi: 10.3389/fpls.2013.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raimunda D, González-Guerrero M, Leeber BW, 3rd, Argüello JM. The transport mechanism of bacterial Cu+-ATPases: Distinct efflux rates adapted to different function. Biometals. 2011;24(3):467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldwin AJ, Inoue K. The most C-terminal tri-glycine segment within the polyglycine stretch of the pea Toc75 transit peptide plays a critical role for targeting the protein to the chloroplast outer envelope membrane. FEBS J. 2006;273(7):1547–1555. doi: 10.1111/j.1742-4658.2006.05175.x. [DOI] [PubMed] [Google Scholar]
- 38.Inoue K, Keegstra K. A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J. 2003;34(5):661–669. doi: 10.1046/j.1365-313x.2003.01755.x. [DOI] [PubMed] [Google Scholar]
- 39.Clark SA, Theg SM. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol Biol Cell. 1997;8(5):923–934. doi: 10.1091/mbc.8.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glerum DM, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem. 1996;271(24):14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- 41.Cobine PA, Pierrel F, Bestwick ML, Winge DR. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J Biol Chem. 2006;281(48):36552–36559. doi: 10.1074/jbc.M606839200. [DOI] [PubMed] [Google Scholar]
- 42.Becker B. Snow ball earth and the split of Streptophyta and Chlorophyta. Trends Plant Sci. 2013;18(4):180–183. doi: 10.1016/j.tplants.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 44.Copley RR. Evolutionary convergence of alternative splicing in ion channels. Trends Genet. 2004;20(4):171–176. doi: 10.1016/j.tig.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Cohu CM, et al. Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: Regulation and unexpected phenotypes in an Arabidopsis mutant. Mol Plant. 2009;2(6):1336–1350. doi: 10.1093/mp/ssp084. [DOI] [PubMed] [Google Scholar]
- 46.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchler-Bauer A, et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009;4(2):e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaby-Haas CE, Merchant SS. The ins and outs of algal metal transport. Biochim Biophys Acta. 2012;1823(9):1531–1552. doi: 10.1016/j.bbamcr.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 52.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Mandal AK, Cheung WD, Argüello JM. Characterization of a thermophilic P-type Ag+/Cu+-ATPase from the extremophile Archaeoglobus fulgidus. J Biol Chem. 2002;277(9):7201–7208. doi: 10.1074/jbc.M109964200. [DOI] [PubMed] [Google Scholar]
- 55.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 56.Raimunda D, Long JE, Sassetti CM, Argüello JM. Role in metal homeostasis of CtpD, a Co2+ transporting P(1B4)-ATPase of Mycobacterium smegmatis. Mol Microbiol. 2012;84(6):1139–1149. doi: 10.1111/j.1365-2958.2012.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenner AJ, Harris ED. A quantitative test for copper using bicinchoninic acid. Anal Biochem. 1995;226(1):80–84. doi: 10.1006/abio.1995.1194. [DOI] [PubMed] [Google Scholar]
- 58.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100(1):95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 59.Martínez-García JF, Monte E, Quail PH. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 1999;20(2):251–257. doi: 10.1046/j.1365-313x.1999.00579.x. [DOI] [PubMed] [Google Scholar]
- 60.Waegemann K, Soll J. Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol. 1995;50:255–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- 61.Alexeyev MF, Winkler HH. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J Mol Biol. 1999;285(4):1503–1513. doi: 10.1006/jmbi.1998.2412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.