Abstract
Background and Aims
Chromosomal evolution, including numerical and structural changes, is a major force in plant diversification and speciation. This study addresses genomic changes associated with the extensive chromosomal variation of the Mediterranean Prospero autumnale complex (Hyacinthaceae), which includes four diploid cytotypes each with a unique combination of chromosome number (x = 5, 6, 7), rDNA loci and genome size.
Methods
A new satellite repeat PaB6 has previously been identified, and monomers were reconstructed from next-generation sequencing (NGS) data of P. autumnale cytotype B6B6 (2n = 12). Monomers of all other Prospero cytotypes and species were sequenced to check for lineage-specific mutations. Copy number, restriction patterns and methylation levels of PaB6 were analysed using Southern blotting. PaB6 was localized on chromosomes using fluorescence in situ hybridization (FISH).
Key Results
The monomer of PaB6 is 249 bp long, contains several intact and truncated vertebrate-type telomeric repeats and is highly methylated. PaB6 is exceptional because of its high copy number and unprecedented variation among diploid cytotypes, ranging from 104 to 106 copies per 1C. PaB6 is always located in pericentromeric regions of several to all chromosomes. Additionally, two lineages of cytotype B7B7 (x = 7), possessing either a single or duplicated 5S rDNA locus, differ in PaB6 copy number; the ancestral condition of a single locus is associated with higher PaB6 copy numbers.
Conclusions
Although present in all Prospero species, PaB6 has undergone differential amplification only in chromosomally variable P. autumnale, particularly in cytotypes B6B6 and B5B5. These arose via independent chromosomal fusions from x = 7 to x = 6 and 5, respectively, accompanied by genome size increases. The copy numbers of satellite DNA PaB6 are among the highest in angiosperms, and changes of PaB6 are exceptionally dynamic in this group of closely related cytotypes of a single species. The evolution of the PaB6 copy numbers is discussed, and it is suggested that PaB6 represents a recent and highly dynamic system originating from a small pool of ancestral repeats.
Keywords: PaB6, Prospero autumnale, Hyacinthaceae, chromosomal evolution, copy number, differential amplification, fluorescence in situ hybridization (FISH), genome size, pericentric satellite DNA, next-generation sequencing
INTRODUCTION
Genomes of higher plants contain a spectrum of repetitive DNAs (Schmidt and Heslop-Harrison, 1998; Macas et al., 2002; Ugarković and Plohl, 2002; Hemleben et al., 2007). This repetitive fraction is predominantly composed of dispersed mobile genetic elements (DNA transposons, retroelements) and tandemly repeated satellite DNAs (Hemleben et al., 2007; Weiss-Schneeweiss and Schneeweiss, 2013). Satellite DNA is typically species or genus specific, consisting of long arrays of late-replicating, tandemly arranged, head-to-tail repeats (Charlesworth et al., 1994; Richard et al., 2008).
Satellite DNA is a non-coding fraction of the genome of limited transcriptional capacity, subject to methylation, histone modification and chromatin remodelling (Volkov et al., 2006; Hemleben et al., 2007). It is preferentially localized in heterochromatic pericentromeric and sub-telomeric chromosomal regions, but also occurs interstitially (Charlesworth et al., 1994; Hemleben et al., 2007). No general function has been ascribed to satellite DNA (Ugarković and Plohl, 2002; Hemleben et al., 2007), although biological roles have been suggested for its specific families – the maintenance of chromosome structure (Ferree and Prasad, 2012), recognition of homologous chromosomes during meiosis (Willard, 1998; Ferree and Prasad, 2012), regulation of gene expression (Pezer et al., 2012), and heterochromatin organization and centromere function (Csink and Henikoff, 1998; Ugarković and Plohl, 2002; Ugarković, 2005; Hemleben et al., 2007; Martins et al., 2008; Gong et al., 2012; Pezer et al., 2012).
Higher plant genomes have from a few to many families of satellite DNAs (Hemleben et al., 2007; Macas et al., 2007, 2011). Individual satellite DNA families in a genome differ in sequence and copy number. Thus, one or a few families are usually present in high copy number, while others have low numbers of repeats (Hemleben et al., 2007). It has been proposed that groups of related taxa share a common ‘library’ of satellite DNA families, each of which may follow its own evolutionary trajectory (Meštrovič et al., 1998). As species diverge, some satellite DNA families reduce in copy number, or even disappear, while others amplify, and new variants may arise (Meštrovič et al., 1998; Nijman and Lenstra, 2001; Pons et al., 2004). Newly arising variants of a satellite DNA can rapidly replace previous copies due to concerted evolution, which results in intraspecific sequence homogenization (Plohl, 2010). The efficiency of homogenization is satellite DNA specific and depends on initial copy number, genomic location, repeat length and mode of reproduction (Dover, 1982; Stephan and Cho, 1994; Plohl et al., 2008; Navajas-Pérez et al., 2009; Kuhn et al., 2010). All these changes may parallel, or even precede, species diversification (Elder and Turner, 1995; Koukalova et al., 2010; Raskina et al., 2011; Belyayev and Raskina, 2013). Plant satellite DNA families are often derived from fragments of standard components of the genome, such as 35S rDNA (Lim et al., 2004; Almeida et al., 2012), 5S rDNA (Vittorazzi et al., 2011) or transposable elements (Sharma et al., 2013). Their subsequent evolution involves various processes such as replication slippage, unequal crossing-over, gene conversion or extrachromosomal circular DNA (eccDNA) formation (Smith, 1976; Walsh, 1987; Charlesworth et al., 1994; Elder and Turner, 1995; Cohen et al., 2008; Navrátilová et al., 2008).
The genus Prospero (Hyacinthaceae) consists of two chromosomally and morphologically stable species, P. hanburyi, 2n = 14 and P. obtusifolium, 2n = 8, and a chromosomally variable species complex referred to as P. autumnale. Prospero autumnale consists of a spectacular, and unparalleled, array of genetically, chromosomally and phylogenetically well-defined, recently evolved, diploid cytotypes, and a large array of polyploid derivatives (Vaughan et al., 1997; Jang et al., 2013). This complex shows near homogeneity in its morphology, and provides an excellent system for comparative and evolutionary genomic studies. It is distributed across the whole Mediterranean basin (Speta, 1998; Jang et al., 2013). Four chromosomally distinct diploid lineages (cytotypes) have been described, each of which possesses a unique combination of basic chromosome number (x = 5, 6, 7), DNA content and localization of rDNAs (Vaughan et al., 1997; Jang et al., 2013). Two cytotypes based on x = 7 are referred to as B7B7, distributed across the whole Mediterranean basin, and AA, which has larger chromosomes and genome size and is confined to the western-most Mediterranean and the Atlantic coast of Morocco, Portugal and Spain. The other two diploid cytotypes – with 2n = 12 (B6B6) and 2n = 10 (B5B5) – originated from a putative ancestor with 2n = 14 via independent chromosome fusions. B6B6 is endemic to Crete while B5B5 is endemic to Libya. With the exception of the most recently evolved cytotype B5B5, all diploids hybridize and undergo polyploidization in nature to give auto- and allopolyploids. Amongst polyploids, tetraploid and hexaploid cytotypes are most common and widespread (Ainsworth et al., 1983; Vaughan et al., 1997).
Phylogenetic and evolutionary relationships of the three species of Prospero have recently been established, and the ancestral basic number for the P. autumnale complex was inferred to be x = 7 (Jang et al., 2013). Evolution of the cytotypes AA and B6B6 has been shown to be accompanied by independent genome size increases (Jang et al., 2013). Large heterochromatic blocks, however, have been detected only in cytotype B6B6 (Ebert et al., 1996).
Recent developments in high-throughput next-generation sequencing (NGS; Margulies et al., 2005) allow in-depth analyses of all components of any genome (Wicker et al., 2009; Deschamps and Campbell, 2010), and thus rapid identification of satellite DNAs (Macas et al., 2007; Torres et al., 2011; Heckmann et al., 2013). The current study involves comparative evolutionary analysis of a satellite PaB6 identified by NGS from cytotype B6B6. Specifically, the aims are to: (1) isolate, characterize, and determine the abundance and localization of PaB6 in the diploid species and cytotypes of Prospero, and their homoploid diploid hybrids; (2) assess intra- and interspecific variation of the reconstructed PaB6 monomer at all levels of its organization – its DNA sequence, chromosomal localization and genomic abundance; (3) analyse, in a phylogenetic context, the evolutionary trajectories of PaB6 in all six diploid cytotypes of P. autumnale and their diploid homoploid hybrids; and (4) discuss the dynamics of PaB6 evolution in the context of major chromosomal rearrangements in the genus.
MATERIALS AND METHODS
Plant material and DNA isolation
Plants from collections of F. Speta, Linz, and J. S. Parker, Cambridge, were grown in the Botanical Garden of the University of Vienna. The plants studied and their collection details are listed in Supplementary Data Table S1. Due to the high levels of chromosomal variation in Prospero (Jang et al., 2013), every plant was karyotyped prior to analysis. Only ‘standard’ individuals without structural chromosomal variants were used.
Total genomic DNA was isolated from leaves, of several individuals each, of P. obtusifolium, P. hanburyi and the four diploid cytotypes of P. autumnale, including homoploid diploid hybrids (Supplementary Data Table S1) using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987; Jang et al., 2013).
Next-generation sequencing and clustering-based repeat identification
Sequencing of randomly sheared total genomic DNA of the cytotype B6B6 of P. autumnale was performed by the Center for Medical Research, Graz, Austria using a Roche/454 GS FLX instrument with Titanium reagents (Roche Diagnostics). Sequencing half a 70 × 75 picotitre plate yielded 555 480 reads of average length 350 bp. Quality-filtered reads (397 694 corresponding to 2·2 % coverage of the genome) were subjected to graph-based clustering analysis, as described by Novák et al. (2010), to identify groups of reads representing repetitive elements (H. Weiss-Schneeweiss et al., unpubl. res.). One hundred and ninety-five out of a total of 19 751 clusters, corresponding to the most abundant families of genomic repeats, were analysed for their similarity to known sequences using RepeatMasker Open-3·0 (http://www.repeatmasker.org) and BLAST (Altschul et al., 1990) searches against GenBank databases and a database of plant mobile element protein sequences (Novák et al., 2013). Graphical layouts of individual clusters were examined using the SeqGrapheR program (Novák et al., 2010).
Characterization of monomers of satellite repeats
Only one genomically abundant cluster (CL0009) was identified amongst all clusters as containing a potential satellite repeat. Structural features of the tandem repeat motif and its sub-repeats within the contigs of this cluster were further analysed with DOTTER (Sonnhammer and Durbin, 1995). Identification of the most conserved sequence variants and consensus monomer reconstruction of satellite repeat PaB6 were conducted using k-mer frequency analysis as described previously (Macas et al., 2010), using 25 bp long k-mers for final sequence reconstruction.
PCR amplification, cloning, sequencing and phylogenetic analysis of PaB6
The reconstructed consensus sequence of the monomer of PaB6 was used for the design of oligonucleotide primers (PaB6F, 5′-ACCCTAATCAGAACTGGCCT; PaB6R, 5′- TAGAGTTATTGGGATGTGTAC) facing outwards (Fig. 1A). These primers were used for amplification of PaB6 monomers from genomic DNA of diploid Prospero species and cytotypes and three outgroup species (all of family Hyacinthaceae; Supplementary Data Table S1). Polymerase chain reactions consisted of 1 × buffer (MBI Fermentas, St Leon-Rot, Germany), 2·5 mm MgCl2 (MBI Fermentas) 0·5 μm of each of the dNTPs (MBI Fermentas), 0·2 μM of each primer (Sigma Aldrich, Vienna, Austria) and 1 U of RedTaq polymerase (Sigma Aldrich). Amplification was performed on an ABI thermal cycler 9700 (Applied Biosystems, Foster City, CA, USA) with the initial 3 min at 94 °C followed by 25 cycles each of 45 s at 94 °C, 45 s at 55 °C and 40 s at 72 °C, and a final elongation step at 72 °C for 10 min. Amplified fragments were separated on a 1·5 % agarose gel, and PCR products corresponding to the length of the monomers of satellite DNA PaB6 were purified from the gel using Invisorb® Fragment clean up (Invitek, Berlin, Germany). DNA was cloned using the pGEM-T Easy vector system and JM109 competent cells (Promega, Madison, WI, USA) following the manufacturer's instructions. Five inserts per individual were amplified from plasmids using colony PCR with universal M13 primers whereby recombinant colonies were added directly into the PCR mix and inserts amplified using reagents and conditions described in Park et al. (2007). Amplification products were treated with exonuclease I (ExoI) and calf intestine alkaline phosphatase (CIAP) according to the manufacturer's protocol (MBI Fermentas), and amplicons were cycle sequenced using Big Dye terminator chemistry (Applied Biosystems) and run on a 48 capillary ABI 3730 DNA Analyzer (Applied Biosystems). The sequences of satellite DNA were manually aligned in BioEdit v.7.0.9 (Hall, 1999). Phylogenetic analyses were performed using Splits-Tree (version 4.11.3; Huson and Bryant, 2006). Sequenced clones are available from GenBank under accession nos KF897587–KF897652 (Supplementary Data Table S1). Gradient PCR was performed on a peqstar thermocycler (peqlab, Erlangen, Germany) to check for the presence of PaB6 in related genera. Primers, the PCR set-up and the PCR program used were the same as described above, except that the annealing temperatures ranged from 50 to 55 °C (Fig. 2, and not shown).
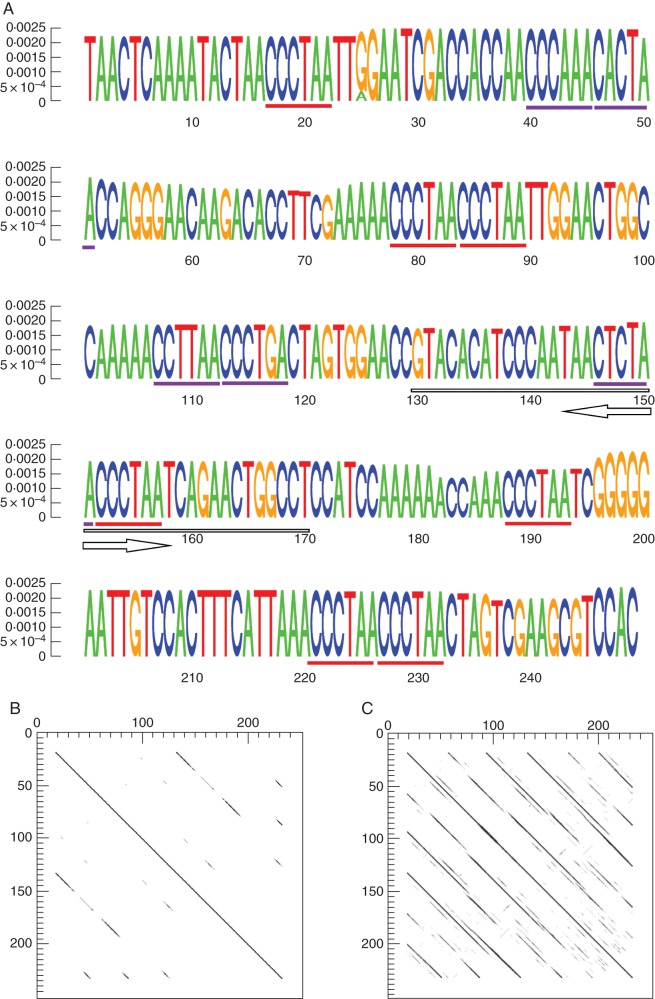
Fig. 1.
PaB6 monomer characterization. (A) Monomer sequence logo (Schneider and Stephens, 1990) with the height of the letters corresponding to k-mer frequencies. Arrows indicate the origin and direction of forward and reverse primers (underlined). Perfect telomeric sequences are underlined in red, and imperfect variants in violet. (B, C) Dot plots of the monomer sequence against itself with lower (B) and higher similarity stringency (C).
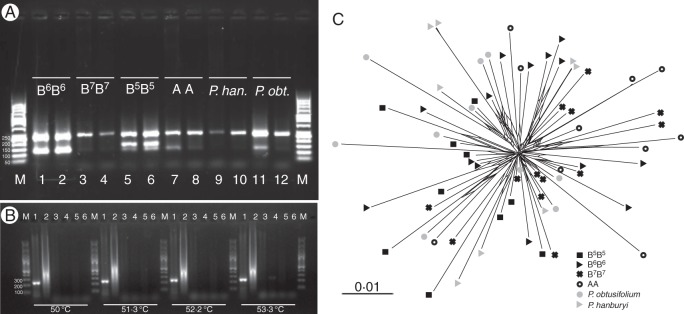
Fig. 2.
Patterns of PCR amplification of PaB6 satellite DNA in Prospero and comparative phylogenetic analysis of the major monomer sequence. (A) PCR amplification products of PaB6 monomers [M, marker; 1–2, B6B6 (H166, H427); 3–4, B7B7 (H424, H428); 5–6, B5B5 (H582, H640); 7–8, AA (H541, H550); 9–10, P. hanburyi (H397, H115); 11–12, P. obtusifolium (H559; H563; Supplementary Data Table S1)]. (B) Gradient PCR amplification of PaB6 monomers in selected Prospero samples and outgroup taxa [M, marker; 1, P. obtusifolium H559; 2, P. autumnale B6B6 H166; 3, Othocallis siberica 2159/1; 4, Othocallis mischtschenkoana LI778; 5, Barnardia scilloides (JANG_1); 6, water as negative control] using annealing temperatures of 50–53·3 °C, as indicated. (C) Neighbour-net of PaB6 repeats cloned from diploid cytotypes of P. autumnale (AA, open circles; B5B5, black filled squares; B6B6, black filled triangles; B7B7, black crosses), P. obtusifolium (grey circles) and P. hanburyi (grey triangles).
Southern and slot blot hybridization
Abundance and restriction patterns of PaB6 monomers in selected individuals were analysed using the Southern blot technique. A 1 μg aliquot of total genomic DNA of each Prospero species and cytotype was digested with 0·7 μL of BstNI restriction endonuclease for 2 h at 37 °C. Digested DNA fragments were separated on a 1 % (w/v) agarose gel and transferred onto a positively charged nylon membrane, Hybond-XL, by the capillary flow method.
The probe used for hybridization was a 249 bp PCR product representing the PaB6 satellite of P. autumnale cytotype B6B6 (clone 4 of individual H195; GenBank accession no. KF897620). The probe was labelled either radioactively with 32P (DekaLabel kit, MBI Fermentas, Vilnius, Lithuania) or using a DIG-nick translation kit (Roche, Vienna, Austria). Radioactively labelled probe was hybridized to the membrane and washed under high-stringency conditions, as described in Matyášek et al. (2011). Hybridization bands were visualized with a PhosphorImager (Storm, Molecular Dynamics, Sunnyvale, CA, USA), and the data were processed in ImageQuant software (Molecular Dynamics).
Hybridization of digoxigenin-labelled probe (Dig Easy Hyb, Roche, Germany) to genomic DNA was carried out at 43 °C for 14 h, and it was then washed twice in 2 × SSC (saline-sodium citrate buffer) containing 0·1 % SDS (sodium dodecylsulphate) for 5 min at room temperature, and twice in 0·5 × SSC containing 0·1 % SDS for 15 min at 65 °C. Probe was detected with CSPD chemiluminescent substrate (Roche Applied Science, USA) using Dig Wash and Block Buffer Set (Roche Applied Science, Germany), and the hybridization signals were visualized on Fusion FX7 Advance (peqlab). Due to the lower sensitivity of chemiluminescent detection compared with radioactive systems, an additional hybridization experiment was performed with cytotypes B7B7, which had been shown to possess lower amounts of satellite DNA, using 1 μg and additionally also 3 μg of genomic DNA.
The copy number of PaB6 in all species and cytotypes was estimated using the slot blot technique. Briefly, the DNA concentration was estimated using Nanodrop 3300 (peqlab) with PicoGreen (Invitrogen) as DNA stain. Two or three dilutions of genomic DNA (100, 20 and 2 ng for B6B6 and B5B5 cytotypes; 2000, 200 and 20 ng for B7B7 and AA cytotypes; 2000 and 200 ng for P. hanburyi and P. obtusifolium), together with a series of dilutions of the unlabelled PaB6 insert corresponding to the monomer sequence, were denatured in 0·4 m NaOH and neutralized with 0·75 m NH4OAc. Samples were blotted onto a positively charged Nylon membrane (peqlab) using a vacuum slot blotter (VWR, Vienna, Austria). The probe and the hybridization conditions used were the same as described above for non-radioactive Southern hybridization. Copy number was estimated using Fusion FX7 Advance software (peqlab).
Methylation levels
The methylation level of PaB6 repeats in the B6 genome was assessed using a radioactive Southern blot (see above). The genomic DNA was digested with two restriction enzymes – BstNI (CCWGG) and ScrFI (CCNGG) – which recognize and cut nearly the same sequence, with ScrFI being sensitive to the inner C methylation.
Fluorescence in situ hybridization
Chromosomes were prepared by enzymatic digestion and squashing (Jang et al., 2013). Fluorescence in situ hybridization (FISH), probe labelling and detection were carried out according to the method of Jang et al. (2013).
The probes used for FISH were a monomer of satellite DNA PaB6 from the B6 genome in plasmid pGEM-T Easy and the genic region of 5S rDNA from Melampodium montanum (Asteraceae) in plasmid pGEM-T Easy, directly labelled with biotin or digoxigenin (Roche, Austria) by PCR (Jang et al., 2013). A 35S rDNA probe labelled with digoxigenin via nick translation (DIG-nick translation kit; Roche) was used in one experiment as a control for the PaB6 probe. Digoxigenin was detected with anti-digoxigenin conjugated with fluorescein isothiocyanate (FITC; 5 μg mL–1; Roche) and biotin with ExtrAvidin conjugated with Cy3 (2 μg mL–1; Sigma Aldrich), respectively.
Commercially available, directly Cy3-labelled, PNA (peptide nucleic acid) probe to vertebrate telomeric sequences (CCCTAA)3 was used as the third probe, as described in the manufacturer's protocol (Telomere PNA FISH Kit/Cy3; Dako, Denmark). For the directly labelled PNA probe, after stringent washes in 2 × SSC, 0·1 × SSC and 2 × SSC with 0·2 % Tween-20 at 42 °C, for 5 min each, preparations were mounted in antifade buffer Vectashield (Vector Laboratories, Peterborough, UK) containing 4′,6-diamidino-2-phenylindole (DAPI) counterstain (2 μg mL–1), and stored at 4 °C.
Preparations were analysed with an AxioImager M2 epifluorescent microscope (Carl Zeiss, Vienna, Austria); images were acquired with a CCD camera, and processed using AxioVision ver. 4.8 (Carl Zeiss) with only those functions that apply equally to all pixels. At least 30 well-spread metaphases and prometaphases were analysed in each individual.
RESULTS
Satellite DNA identification and characterization of the monomers
Clustering analysis of the shotgun Roche/454 reads of Prospero autumnale cytotype B6B6 (2n = 12) produced thousands of clusters differing in size, corresponding to the sequence composition and genomic abundance of the various genomic repeats. A set of 195 of the largest clusters, representing the most abundant repetitive elements with genome proportions exceeding 0·01 %, was searched for features typical of satellite repeats. Only one such cluster was identified, based on the shape of the cluster graph (Novák et al., 2010) and the presence of tandem repeats in assembled contigs (Fig. 1; Supplementary Data Fig. S1). This novel satellite has been designated as PaB6 – satellite DNA isolated from P. autumnale (Pa) cytotype B6B6 (B6). The number of reads in the cluster was 8461, or 1·8 % of the total, giving an estimate of the proportion of PaB6 in the genome. The consensus sequence reconstruction using 25 bp long k-mers (Macas et al., 2010) resulted in a monomer of 249 bp in length (Fig. 1A), with a GC content of 44 %. Detailed analysis, using the NGS dataset, revealed two large truncated sub-repeats which could have given rise to the present-day higher order monomer of 249 bp (Fig. 1B). Each of the two sub-repeats is typically composed of three even smaller secondary sub-repeats (Fig. 1C). The complex structure of this monomer is also indicated by the pattern of PaB6 amplification using PCR (see below).
The monomer of PaB6 contains seven intact vertebrate-type telomeric repeats (TTAGGG) dispersed amongst other sequences and in two instances forming dimers (Fig. 1A). Additionally, five imperfect telomeric-like repeats have been identified, and potentially other repeats degenerated to a higher degree (Fig. 1A). A pentanucleotide CAAAA, conserved in many satellites (Macas et al., 2002), occurred three times on the top strand. In addition, there were four A4 tracts important for DNA conformation and chromatin folding (Plohl et al., 2010).
Comparative sequence analysis of the monomers
The PCR amplification of the major type of the monomer, using primers designed for the reconstructed B6 genome monomer, resulted in products of the expected length in all four diploid cytotypes of P. autumnale and the two related species, P. hanburyi and P. obtusifolium. PCR with PaB6-specific primers yielded one strong band of approx. 250 bp, corresponding to the PaB6 monomer (Fig. 2A), a second band of approx. 120–130 bp and, occasionally, a third band of approx. 60–80 bp (Fig. 2B). The main bands, corresponding to the expected size of the monomer of PaB6, were isolated, cloned and sequenced from two or three individuals of each of the six taxa/cytotypes. Amplification of dimers or even longer fragments was not observed, or observed very rarely.
The outgroup taxa of the family Hyacinthaceae were subjected to the same PCR amplification protocol and primers. Representatives of the related genera Othocallis and Barnardia showed no bands after PCR amplification, regardless of the annealing temperature (Fig. 2B). Very faint, monomer-related bands, close to the limit of detection, were seen occasionally, without any consistent pattern regarding annealing temperature or taxon, and were regarded as contamination (Fig. 2B, and not shown).
Sequence analysis of 66 cloned PaB6 monomers (Supplementary Data Table S1), representing monomers amplified from two or three individuals of each of the six diploid taxa, confirmed that they all carried PaB6 repeats. Fifty-one of these (83 %) were 249 bp long, with 12 shorter (18 %; 119, 175, 243, 247 and 248 bp) and three longer (4·5 %; 250 and 256 bp, the latter due to a TTAGGG insertion). High overall levels of sequence similarity amongst the amplified population of PaB6 monomers, both within (93–100 %) and between (92–100 %) the different diploid cytotypes of P. autumnale and two other Prospero species, were observed (Supplementary Data Table S2). Thus, the intercytotype sequence variation of repeats amplified with the reconstructed monomer primers was as equally low and random as that within cytotypes or between individuals. The variation was mostly due to single base pair indels or point mutations occurring at different positions along the monomer, and these were monomer specific (alignment available upon request).
Neighbour-net analyses of DNA sequences of all cloned inserts of PaB6 repeats from the six cytotypes corroborated the analyses of variation within the monomers, and did not reveal any cytotype-specific lineages (Fig. 2C). Instead, the repeats originating from different individuals were intermingled, regardless either of PaB6 overall copy number and abundance or of their phylogenetic relationship.
Copy number variation and genomic organization of PaB6
Copy numbers were estimated by quantitative chemiluminescent dot blot hybridization of labelled PaB6 as probe against known quantities of genomic DNAs of all three species and four cytotypes (Fig. 3A). Large differences in the genomic content of PaB6 between the four cytotypes of P. autumnale corroborated the results of Southern blot experiments and of FISH (Figs 3B–D and 4). The probe hybridized strongly to genomic DNA of cytotype B6B6 (Fig. 3A–C), moderately to cytotype B5B5 (Fig. 3A, C) and weakly to some individuals of cytotype B7B7 (Fig. 3A, B, D). A very weak signal was detected in genomic DNA of cytotype AA (Fig. 3A). The PaB6 probe hybridized only very weakly to DNAs of P. hanburyi and P. obtusifolium (Fig. 3A). The highest copy number was found in cytotype B6B6 with 1·8–2·1 × 106 copies per haploid genome (7–10 %), followed by cytotype B5B5 with 1·2–1·4 × 106 copies/1C (6–7 %). One accession of cytotype B7B7 had 2·1–2·5 × 104 copies/1C (approx. 0·13 %), while AA had 1·8–2·6 × 104 copies/1C (approx. 0·08 %; Table 1, Fig. 3A). A variant of the B7B7 cytotype, carrying a single 5S rDNA locus on chromosome 1 and stronger signals of PaB6 in all chromosomes, could not be analysed due to lack of appropriate quality plant material. Prospero obtusifolium and P. hanburyi possessed only very low amounts of PaB6, below the slot blot detection limit.
Fig. 3.
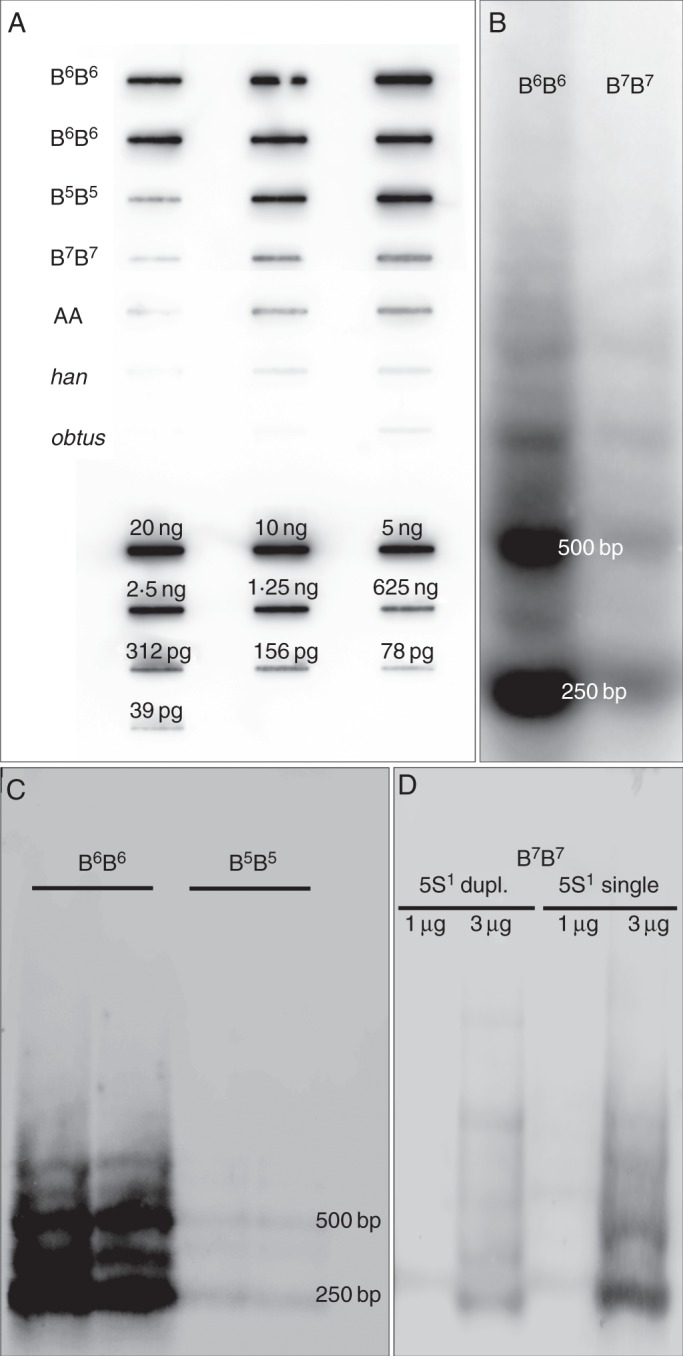
Copy number estimation of PaB6 using slot blotting (A) and analyses of genomic organization of PaB6 repeats in Prospero using (B–D) Southern blot hybridization. (A) Slot blot for PaB6 copy number determination; DNA amount: 2, 20, 100 ng (for B6B6 and B5B5); 20, 200 and 2000 ng for B7B7 and AA; 200 and 2000 ng for P. obtusifolium (obtus) and P. hanburyi (han) (Supplementary Data Table S1). (B) Radioactive detection of digoxigenin-labelled probe PaB6 hybridized to genomic DNA of cytotype B6B6 (H166) and B7B7 (H428). (C, D) Chemiluminescent detection of digoxigenin-labelled PaB6 probe in restricted genomic DNA: (C) B6B6 (H166, H468) and B5B5 (H637, H565); (D) B7B7 (H424, duplicated 5S1 rDNA) and B7B7 (H428, single 5S1 rDNA) each with 1 and 3 μg of DNA.
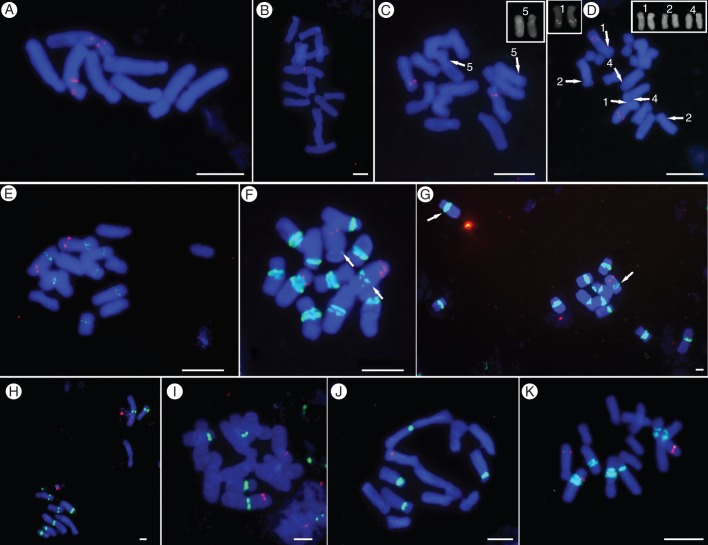
Fig. 4.
Localization of PaB6 in chromosomes of diploid Prospero species and cytotypes, and in three homoploid hybrids. The PaB6 loci are shown as green signals, and 5S rDNA in red. (A) P. obtusifolium (H563), (B) P. hanburyi (H115), (C–K) P. autumnale complex: (C) cytotype AA (H551, inset: chromosomes carrying PaB6 signals), (D, E) B7B7 with duplicated (D: H424, left inset, duplicated 5S rDNA signals; right inset, chromosomes carrying PaB6 signals) and single (E: H440) 5S rDNA locus in chromosome 1, (F, G) B6B6 with weak (F: H195) and strong (G: H427) signal of PaB6 in chromosome 2 (arrows), (H) B5B5 (H581), (I) AB5 (H567), (J) B5B7 (H633), (K) B6B7 (H518) diploid hybrid. Each individual has a unique ID (in parentheses, e.g. H563; see Supplementary Data Table S1). Scale bar = 5 μm.
Table 1.
Characterization of satellite PaB6 repeats in diploid species and cytotypes of the genus Prospero
| Taxon | 2n | Genome proportion % of PaB6 | Copy number/1C of PaB6 | Genome size (pg) per 1C* | Figure |
|---|---|---|---|---|---|
| P. autumnale | |||||
| Cytotype B5B5 | 10 | 6·3–7·4 | 1·25–1·37 × 106 | 4·86 ± 0·002 | 2A; 3A, C; 4H |
| Cytotype B6B6 | 12 | 7·16–10·71 | 1·76–2·06 × 106 | 6·27 ± 0·083 | 2A; 3A–C; 4F, G; 5A, B |
| Cytotype B7B7: single 5S1 rDNA | 14 | NA | NA | 4·23 ± 0·048 | 2A; 3B, D; 4E |
| Cytotype B7B7: duplicated 5S1 rDNA | 14 | 0·12–0·14 | 2·11–2·49 × 104 | 4·45 ± 0·023 | 2A; 3A, D; 4D |
| Cytotype AA | 14 | 0·06–0·08 | 1·75–2·56 × 104 | 7·85 ± 0·045 | 2A; 3A; 4C |
| P. hanburyi | 14 | ND | ND | 6·81 ± 0·017 | 2A; 3A; 4B |
| P. obtusifolium | 8 | ND | ND | 4·94 ± 0·039 | 2A; 3A; 4A |
NA, not analysed due to lack of material; ND, copy number could not be determined due to very low PaB6 contents.
Southern blot hybridization, using the satellite DNA single repeat (monomer) isolated from cytotype B6B6 as probe, was congruent in estimations of copy number of PaB6 repeats and also enabled analysis of their genomic organization. The hybridization pattern of PaB6 was typical of tandemly repeated DNAs, with the major 249 bp band and its multiples being most prominent in all samples. An additional, weaker, band about 375 bp in length, corresponding to an additional major sub-unit (Fig. 3B, C), has also been detected in all samples.
Methylation of PaB6 repeats was analysed in cytotype B6B6, after digestion with methylation-insensitive (BstNI) and methylation-sensitive (ScrFI) restriction enzymes with the same recognition site. The satellite DNA monomers were heavily methylated at CHG sites (Supplementary Data Fig. S3).
Chromosomal localization and organization of PaB6 repeats
PaB6 has been localized in all six cytotypes using FISH (Supplementary Data Table S3).
The variation in number and size of satellite DNA loci detected corresponded well to the Southern slot results. Thus P. obtusifolium (Fig. 4A) and P. hanburyi (Fig. 4B) had no PaB6 loci detectable by FISH due to very low copy numbers of PaB6 monomers (Figs 3A and 4A–B). Prospero autumnale diploids, in contrast, all exhibited hybridization signals using FISH, but were variable in numbers of sites and in signal strengths (Fig. 4C–K). PaB6 is predominantly located in pericentromeric regions of at least one, and sometimes all, chromosome pairs, and might, at least partly, span the centromeres (Supplementary Data Fig. S2).
In cytotype B6B6, major loci were present on all chromosomes of the complement (Fig. 4F, G). The pattern of satellite distribution was remarkably uniform between individuals and populations, and loci were of similar signal strength. Chromosome 1 showed the only polymorphism, with the locus size varying between homologues in some individuals (Fig. 4F, G).
In B5B5, PaB6 loci occurred on four of the five chromosome pairs (Fig. 4H) and were of similar signal strength. Chromosome 3 showed, at most, a very weak hybridization signal (Supplementary Data Table S3, and data not shown).
Cytotype AA had only a single locus of PaB6 – on chromosome 5 – but this was weak and barely detectable (Fig. 4C). The most variable PaB6 distribution was shown by cytotype B7B7. Some individuals possessed medium-sized signals in pericentromeric regions of all chromosomes (Fig. 4E; Supplementary Data Table S3), while others had much weaker signals limited to three chromosome pairs (Fig. 4D; Supplementary Data Table S3). These patterns correlated with a duplication polymorphism of 5S rDNA present on chromosome 1 (5S1; see also Fig. 6). Thus the five individuals with a single 5S1 rDNA locus showed moderate amplification of PaB6 on all chromosomes (Fig. 4E), while the six plants with a duplicated 5S locus carried weakly amplified PaB6 loci only on chromosomes 1, 2 and 4 (Fig. 4D). The number and localization of PaB6 satellite DNA loci in all cytotypes are shown in Fig. 6 and Supplementary Data Table S3.
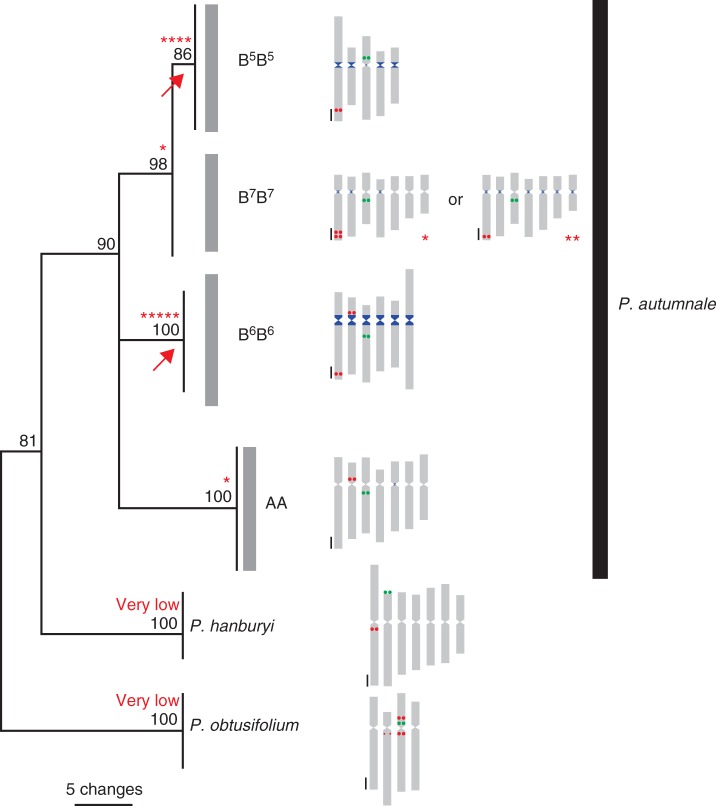
Fig. 6.
Model of evolution of PaB6 in diploid taxa of Prospero. Idiograms of all analysed species and cytotypes are mapped onto the ITS (internal transcribed spacer) tree (adapted from Jang et al., 2013). PaB6 satellite DNA is indicated as blue blocks, 5S rDNA as red circles and 35S rDNA as green circles. Asterisks indicate lineages which have experienced significant amplification of PaB6. Arrows mark amplification events accompanying fusions.
All six F1 diploid hybrids possessed perfectly additive numbers and strengths of PaB6 loci compared with their diploid parents. This was supported by Southern blot hybridization of a B6B7 hybrid, which also indicated additivity (not shown).
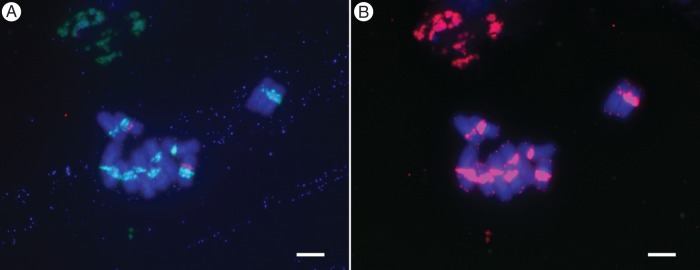
The PaB6 monomer contains seven perfect and a few imperfect vertebrate-type telomeric sequences (TTAGGG), typical of the monocot order Asparagales to which Prospero belongs. TTAGGG sequences were detected at chromosome ends (Fig. 5) but additionally co-localized with the PaB6 loci. Signal intensity in the pericentric chromosome regions using a telomeric DNA probe corresponded to signal strength and localization of the PaB6 probe itself (Fig. 5A, B).
Fig. 5.
Localization of telomeric PNA probe (TTAGGG) and satellite PaB6 in chromosomes of diploid Prospero autumnale cytotype B6B6 (H468): (A) PaB6 loci (green) and 35S rDNA locus (red); (B) telomeric PNA probe (red) localized to the same metaphase chromosomes spread as in (A). Scale bar = 5 μm.
DISCUSSION
Tandem repeats localize to heterochromatic segments in chromosomes (Hemleben et al., 2007). Prospero cytotypes differ in the amount and distribution of heterochromatin, both among and within cytotypes. So far, only the cytotypes AA, B7B7 and B6B6 have been analysed using C-banding (Ebert et al., 1996) and the only consistently detectable heterochromatic blocks co-localized with nucleolar organizer regions (NORs). However, cytotype B6B6 had a high amount of heterochromatin, detected as blocks (C-bands) in the pericentric regions of all chromosomes (Ebert et al., 1996). This was the rationale for selecting the B6 genome for repetitive DNA fraction analyses. Cytotype B7B7 was very variable in the number of heterochromatic blocks, but these were mainly dot-like and localized interstitially, except for slightly larger pericentric blocks which varied in size between individuals. Cytotype AA had only small interstitial heterochromatic blocks on six of the seven pairs. All of these pericentric heterochromatic blocks detected by Ebert et al. (1996) correspond to PaB6 signals. The additional, smaller and more polymorphic interstitial bands detected are most likely to be composed of other tandem repeat(s), some of which might be cytotype specific.
Satellite DNA repeats represent a substantial proportion of the genomes of many higher plants (e.g. VicTR-A/B in Vicia, Macas et al., 2000; FriSAT1 in Fritillaria, Ambrožová et al., 2011). The PaB6 repeat of Prospero is one of the most abundant satellites reported so far (Hemleben et al., 2007). It represents about 10 % of the genome in the B6B6 cytotype with 1·4 × 106 copies. In comparison, tandem repeat VicTR-A/B comprises about 1 % of the genome of most Vicia species with 106 copies (VicTR-A) but reaches 25 % of the genome with 1 × 106–5 × 106 copies (VicTR-B) in V. sativa (Macas et al., 2000), approaching the highest value reported in plants for the FokI element in V. faba (2·5 × 107 copies/1C; Kato et al., 1984). The 37–55 bp long PAF1 repeat in Picea abies occurs in 2·7 × 106 copies/1C (approx. 0·6 %; Sarri et al., 2008), while MCSAT in M. comosum has 9 × 105 copies representing 5 % of the genome (de la Herrán et al., 2001).
PaB6 is exceptional for its copy number variation between the closely related diploid cytotypes of one species complex. The satellite can clearly expand from a few hundred base pairs up to several hundred megabases in a relatively short evolutionary period. Such rapid changes should be reflected by genome size differences between Prospero cytotypes. The genome sizes of the derived cytotypes B5B5 and B6B6 are distinctly higher than those of cytotype B7B7, which has been inferred to be most similar to the ancestral karyotype (Jang et al., 2013; K. Emadzade et al., unpubl. res.). PaB6 amplification significantly contributes to these genome size increases and gives rise to heterochromatic blocks in B6B6. The correlation between genome size and PaB6 amount is particularly evident in the comparison of the youngest cytotype B5B5 and its close relative, and likely ancestor, B7B7 (Jang et al., 2013). The B5 genome is about 400 Mb (10 %) larger than the B7 genome, half of which can be attributed to PaB6 copy number increase (325 Mb in B5B5 vs. 7 Mb in B7B7). In contrast, the large size of the A genome is clearly not associated with the high copy number of PaB6.
Satellite DNA copy number can change relatively rapidly due to expansions and contractions of satellite arrays. Thus, the copy number of FRISAT1 in the genus Fritillaria varies within and between different subgenera (Ambrožová et al., 2011), and several genus-specific satellite DNAs differ in copy numbers between related Secale (Cuadrado and Jouve, 2002) and Nicotiana species (Lim et al., 2004). Such differences are also observed between varieties and cultivars of Phaseolus vulgaris and maize (Peacock et al., 1981; Ribeiro et al., 2011) indicating the highly dynamic character of satellite repeats. These changes may be accompanied by divergence of the monomer sequences during evolution, via accumulation and fixation of mutations in satellite families (Plohl et al., 2008). Interestingly, in Prospero, despite the dynamic changes in copy number, there is no indication of sequence divergence during lineage evolution.
In Barnardia and Othocallis (Fig. 2B), genera closely related to Prospero (Pfosser and Speta, 1999; Ali et al., 2012), no PaB6 monomers were detected, shown by a lack of amplification of PaB6 monomer-equivalent bands in PCR. Thus, PaB6 probably evolved during the emergence of the genus Prospero, and remained in low copy number as part of the library of repeats (Meštrovič et al., 1998) in the chromosomally stable species P. obtusifolium and P. hanburyi. PaB6 amplification, therefore, is specific to the chromosomally dynamic P. autumnale complex.
PaB6 dynamics can be assessed against the phylogeny of the genus (Jang et al., 2013). Prospero obtusifolium and P. hanburyi possess very few PaB6 monomers, and these can only be detected by PCR, because they are below the detection limit of all types of in situ hybridization. In contrast, the four diploid cytotypes of P. autumnale all possess PaB6 in amounts detectable by FISH and genomic DNA hybridization, although copy number varies substantially. PaB6 in B6B6 represents 8–10 % of the genome and 6–7 % in B5B5. Copy number estimation from NGS data, however, suggests that PaB6 represents about 1·8 % of the B6B6 genome, only a quarter of that from slot blot hybridization. This discrepancy is probably caused by PaB6 under-representation due to a bias affecting template preparation from satellite repeats during 454 sequencing (Macas et al., 2007; J. Macas et al., unpubl. res.).
In some plant and animal groups, patterns of copy number variation of a satellite DNA family in a group of closely related taxa carry a phylogenetic signal. However, similarity in copy number might result from independent satellite amplifications or contractions (Rosato et al., 2012). The two Prospero cytotypes whose genomes are enriched in PaB6 have reduced basic chromosome numbers of x = 6 and x = 5 derived from x = 7 via independent fusion events, so do not demonstrate a sister relationship (Jang et al., 2013; Fig. 6). Thus, the raised amounts of PaB6 in these two cytotypes could have resulted from independent amplifications, coinciding with fusions leading to basic number changes. This is particularly plausible for the phylogenetically young cytotype B5B5, which is nested within B7B7, a cytotype carrying relatively few copies of PaB6 (Jang et al., 2013; Fig. 6). However, high copy numbers in these two unrelated lineages might be a remnant of a common amplification event which was followed by differential loss. This hypothesis is more plausible for cytotype B6B6 than for B5B5. B6B6 clearly originated from x = 7, but does not strongly relate, phylogenetically or chromosomally, to any lineage of present-day B7B7, and may have arisen directly from the ancestral cytotype, or an as yet undiscovered B7 lineage, with high copy numbers of PaB6 (Jang et al., 2013). Thus, the lack of phylogenetic evidence of copy number of PaB6 in the ancestral karyotype of Prospero leaves the question open.
The presence of telomeric motifs in the PaB6 sequence is interesting with respect to the high karyotype instability within and between all P. autumnale cytotypes (Vaughan et al., 1997; Jang et al., 2013). The presence of interstitial telomeric repeats (ITRs) is often interpreted as a remnant of evolution by telomere–telomere chromosomal fusions. However, it may also result from rearrangements such as translocations or inversions (Uchida et al., 2002; Ruiz-Herrera et al., 2008; Rosato et al., 2012), particularly whole chromosomal arm inversions involving both the centromere and telomere (Presting et al., 1996). The occurrence of telomeric repeats within, or at the margins of, constitutive heterochromatin has been reported in vertebrates (Meyne et al., 1990) but is also known in plants (Presting et al., 1996; Uchida et al., 2002; Weiss-Schneeweiss et al., 2004; Mlinarec et al., 2009; Gong et al., 2012; He et al., 2013). It has been argued that these telomeric repeats can be an integral and long-established part of the satellite DNAs of constitutive heterochromatin (Slijepcevic et al., 1996; Garrido-Ramos et al., 1998; Metcalfe et al., 2004), originally inserted and amplified through DNA double strand breaks (DSBs) repaired by telomerase (Nergadze et al., 2004, 2007). The ITRs detected in Prospero are certainly an integral part of PaB6 interspersed amongst other sequence motifs. Their origin, however, cannot be unambiguously established.
Two mechanisms have been proposed for satellite DNA copy number change: unequal crossing-over with gene conversion (Liao, 1999; Eickbush and Eickbush, 2007), and amplification and homogenization of monomers by extrachromosomal circular DNA (eccDNA, ‘rolling circle’) molecules during recombination (Navrátilová et al., 2008; Cohen et al., 2010). They are not mutually exclusive and might operate in concert, resulting in mobility and homogenization of repetitive DNAs. Whether these mechanisms are also involved in expansion of PaB6 in Prospero remains unknown.
Although copy number varies hugely between cytotypes within Prospero, the monomer sequence is conserved. This may indicate either relatively recent amplification of the monomer or efficient systems of sequence homogenization and gene flow between taxa (Hemleben et al., 2007). The geographically disjunct distributions of the cytotypes AA, B5B5 and B6B6, and consequent lack of gene flow between them, suggest that PaB6 represents a recent and highly dynamic system originating from a small pool of ancestral repeats (Mravinac et al., 2005; Plohl et al., 2010).
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Dr Franz Speta for his collection of Prospero. This work was supported by the Austrian Science Fund (P21440 to H.W.-S.); the Czech Science Foundation (P501/12/G090 to J.M.); and the Academy of Sciences of the Czech Republic (RVO:60077344 to J.M.).
LITERATURE CITED
- Ainsworth CC, Parker JS, Horton DM. Chromosome variation and evolution in Scilla autumnalis. In: Brandham PE, Bennett MD, editors. Kew Chromosome Conference II. London: Allen and Unwin; 1983. pp. 261–268. [Google Scholar]
- Ali SS, Yu Y, Pfosser M, Wetschnig W. Inferences of biogeographical histories within subfamily Hyacinthoideae using S-DIVA and Bayesian binary MCMC analysis implemented in RASP (Reconstruct Ancestral State in Phylogenies) Annals of Botany. 2012;109:95–107. doi: 10.1093/aob/mcr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida C, Fonseca A, dos Santos KGB, Pedrosa-Harand A. Contrasting evolution of a satellite DNA and its ancestral IGS rDNA in Phaseolus (Fabaceae) Genome. 2012;55:683–689. doi: 10.1139/g2012-059. [DOI] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ambrožová K, Mandáková T, Bures P, et al. Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Annals of Botany. 2011;107:255–268. doi: 10.1093/aob/mcq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyayev A, Raskina O. Chromosome evolution in marginal populations of Aegilops speltoides: causes and consequences. Annals of Botany. 2013;111:531–538. doi: 10.1093/aob/mct023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Cohen S, Houben A, Segal D. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. The Plant Journal. 2008;53:1027–1034. doi: 10.1111/j.1365-313X.2007.03394.x. [DOI] [PubMed] [Google Scholar]
- Cohen S, Agmon N, Sobol O, Segal D. Extrachromosomal chromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mobile DNA. 2010;1:11. doi: 10.1186/1759-8753-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Something from nothing: the evolution and utility of satellite repeats. Trends in Genetics. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Jouve N. Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. Journal of Heredity. 2002;93:339–345. doi: 10.1093/jhered/93.5.339. [DOI] [PubMed] [Google Scholar]
- Deschamps S, Campbell MA. Utilization of next-generation sequencing platforms in plant genomics and genetic variant discovery. Molecular Breeding. 2010;25:553–570. [Google Scholar]
- Dover GA. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Ebert I, Greilhuber J, Speta F. Chromosome banding and genome size differentiation in Prospero (Hyacinthaceae): diploids. Plant Systematics and Evolution. 1996;203:143–177. [Google Scholar]
- Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder JF, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Quarterly Review of Biology. 1995;70:297–323. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- Ferree PM, Prasad S. How can satellite DNA divergence cause reproductive isolation? Let us count the chromosomal ways. 2012. Genetics Research International Article ID 430136. [DOI] [PMC free article] [PubMed]
- Garrido-Ramos MA, de la Herran R, Ruiz Rejón C, Ruiz Rejón M. A satellite DNA of the Sparidae family (Pisces, Perciformes) associated with telomeric sequences. Cytogenetics and Cell Genetics. 1998;83:3–9. doi: 10.1159/000015151. [DOI] [PubMed] [Google Scholar]
- Gong Z, Wu Y, Koblízková A, et al. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. The Plant Cell. 2012;24:3559–3574. doi: 10.1105/tpc.112.100511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- He L, Jiang J, Liu J, et al. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Research. 2013;21:5–13. doi: 10.1007/s10577-012-9332-x. [DOI] [PubMed] [Google Scholar]
- Heckmann S, Macas J, Kumke K, et al. The holocentric species Luzula elegans shows interplay between centromere and large-scale genome organization. The Plant Journal. 2013;73:555–565. doi: 10.1111/tpj.12054. [DOI] [PubMed] [Google Scholar]
- Hemleben V, Kovařík A, Torres-Ruiz RA, Volkov RA, Beridze T. Plant highly repeated satellite DNA: molecular evolution, distribution and use for identification of hybrids. Systematics and Biodiversity. 2007;5:277–289. [Google Scholar]
- de la Herrán R, Robles F, Cuñado N, et al. A heterochromatic satellite DNA is highly amplified in a single chromosome of Muscari (Hyacinthaceae) Chromosoma. 2001;110:197–202. doi: 10.1007/s004120000115. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jang T-S, Emadzade K, Parker J, et al. Chromosomal diversification and karyotype evolution of diploids in the cytologically diverse genus Prospero (Hyacinthaceae) BMC Evolutionary Biology. 2013;13:136. doi: 10.1186/1471-2148-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Yakura K, Tanifuji S. Sequence analysis of Vicia faba repeated DNA, the FokI repeat element. Nucleic Acids Research. 1984;12:6415–6426. doi: 10.1093/nar/12.16.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukalova B, Moraes AP, Renny-Byfield S, Matyášek R, Leitch AR, Kovařík A. Fall and rise of satellite repeats in allopolyploids of Nicotiana over c. 5 million years. New Phytologist. 2010;186:148–160. doi: 10.1111/j.1469-8137.2009.03101.x. [DOI] [PubMed] [Google Scholar]
- Kuhn GCS, Schwarzacher T, Heslop-Harrison JS. The non-regular orbit: three satellite DNAs in Drosophila martensis (buzzatii complex, repleta group) followed three different evolutionary pathways. Molecular Genetics and Genomics. 2010;284:251–262. doi: 10.1007/s00438-010-0564-1. [DOI] [PubMed] [Google Scholar]
- Liao D. Concerted evolution: molecular mechanism and biological implications. American Journal of Human Genetics. 1999;64:24–30. doi: 10.1086/302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Skalicka K, Koukalova B. Dynamic changes in the distribution of a satellite homologous to intergenic 26–18S rDNA spacer in the evolution of Nicotiana. Genetics. 2004;166:1935–1946. doi: 10.1534/genetics.166.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Pozárková D, Navrátilová A, Nouzová M, Neumann P. Two new families of tandem repeats isolated from genus Vicia using genomic self-priming PCR. Molecular and General Genetics. 2000;263:741–751. doi: 10.1007/s004380000245. [DOI] [PubMed] [Google Scholar]
- Macas J, Mészáros T, Nouzová M. PlantSat: a specialized database for plant satellite repeats. Bioinformatics. 2002;18:28–35. doi: 10.1093/bioinformatics/18.1.28. [DOI] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navrátilová A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Neumann P, Novák P, Jiang J. Global sequence characterization of rice centromeric satellite based on oligomer frequency analysis in large-scale sequencing data. Bioinformatics. 2010;26:2101–2108. doi: 10.1093/bioinformatics/btq343. [DOI] [PubMed] [Google Scholar]
- Macas J, Kejnovský E, Neumann P, Novák P, Koblížková A, Vyskot B. Next generation sequencing-based analysis of repetitive DNA in the model dioecious plant Silene latifolia. PLoS One. 2011;6:e27335. doi: 10.1371/journal.pone.0027335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C, Baptista CS, Ienne S, Cerqueira GC, Bartholomeu DC, Zingales B. Genomic organization and transcription analysis of the 195-bp satellite DNA in Trypanosoma cruzi. Molecular and Biochemical Parasitology. 2008;160:60–64. doi: 10.1016/j.molbiopara.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Matyášek R, Fulnecek J, Leitch AR, Kovařík A. Analysis of two abundant, highly related satellites in the allotetraploid Nicotiana arentsii using double-strand conformation polymorphism analysis and sequencing. New Phytologist. 2011;192:747–759. doi: 10.1111/j.1469-8137.2011.03827.x. [DOI] [PubMed] [Google Scholar]
- Meštrovič N, Plohl M, Mravinac B, Ugarković D. Evolution of satellite DNAs from the genus Palorus – experimental evidence for the ‘library’ hypothesis. Molecular Biology and Evolution. 1998;15:1062–1068. doi: 10.1093/oxfordjournals.molbev.a026005. [DOI] [PubMed] [Google Scholar]
- Metcalfe CJ, Eldridge MDB, Johnston PG. Mapping the distribution of the telomeric sequence (T2AG3)n in the 2n = 14 ancestral marsupial complement and in the macropodines (Marsupialia: Macropodidae) by fluorescence in situ hybridization. Chromosome Research. 2004;12:405–414. doi: 10.1023/B:CHRO.0000034133.77878.88. [DOI] [PubMed] [Google Scholar]
- Meyne J, Baker RJ, Hobart HH, et al. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;99:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- Mlinarec J, Chester M, Siljak-Yakovlev S, Papeš D, Leitch A, Besendorfer V. Molecular structure and chromosome distribution of three repetitive DNA families in Anemone hortensis L. (Ranunculaceae) Chromosome Research. 2009;17:331–346. doi: 10.1007/s10577-009-9025-2. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Ugarković D. Preservation and high sequence conservation of satellite DNAs suggest functional constraints. Journal of Molecular Evolution. 2005;61:542–550. doi: 10.1007/s00239-004-0342-y. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Quesada del Bosque ME, Garrido-Ramos MA. Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Molecular Genetics and Genomics. 2009;282:395–406. doi: 10.1007/s00438-009-0472-4. [DOI] [PubMed] [Google Scholar]
- Navrátilová A, Koblízková A, Macas J. Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biology. 2008;8:90. doi: 10.1186/1471-2229-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze SG, Rocchi M, Azzalin CM, Mondello C, Giulotto E. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Research. 2004;14:1704–1710. doi: 10.1101/gr.2778904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze SG, Santagostino M, Salzano A, Mondello C, Giulotto E. Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biology. 2007;8:R260. doi: 10.1186/gb-2007-8-12-r260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman IJ, Lenstra JA. Mutation and recombination in cattle satellite DNA: a feedback model for the evolution of satellite DNA repeats. Journal of Molecular Evolution. 2001;52:361–371. doi: 10.1007/s002390010166. [DOI] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next generation sequence reads. Bioinformatics. 2013;29:792–793. doi: 10.1093/bioinformatics/btt054. [DOI] [PubMed] [Google Scholar]
- Park JM, Schneeweiss GM, Weiss-Schneeweiss H. Diversity and evolution of Ty1-copia and Ty3-gypsy retroelements in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae) Gene. 2007;387:75–86. doi: 10.1016/j.gene.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Peacock WJ, Dennis ES, Rhoades MM, Pryor AJ. Highly repeated DNA sequence limited to knob heterochromatin in maize. Proceedings of the National Academy of Sciences, USA. 1981;78:4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezer Z, Brajković J, Feliciello I, Ugarkovć D. Satellite DNA-mediated effects on genome regulation. Genome Dynamics. 2012;7:153–169. doi: 10.1159/000337116. [DOI] [PubMed] [Google Scholar]
- Pfosser M, Speta F. Phylogenetics of Hyacinthaceae based on plastid DNA sequences. Annals of the Missouri Botanical Garden. 1999;86:852–875. [Google Scholar]
- Plohl M, Luchetti A, Meštrovič N, Mantovani B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero) chromatin. Gene. 2008;409:72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Plohl M. Those mysterious sequences of satellite DNAs. Periodicum Biologorum. 2010;112:403–410. [Google Scholar]
- Plohl M, Petrović V, Luchetti A, et al. Long-term conservation vs. high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity. 2010;104:543–551. doi: 10.1038/hdy.2009.141. [DOI] [PubMed] [Google Scholar]
- Pons J, Bruvo B, Petitpierre E, Plohl M, Ugarković D, Juan C. Complex structural features of satellite DNA sequences in the genus Pimelia (Coleoptera: Tenebrionidae): random differential amplification from a common ‘satellite DNA library. Heredity. 2004;92:418–427. doi: 10.1038/sj.hdy.6800436. [DOI] [PubMed] [Google Scholar]
- Presting GG, Frary A, Pillen K, Tanksley SD. Telomere-homologous sequences occur near the centromeres of many tomato chromosomes. Molecular and General Genetics. 1996;251:526–531. doi: 10.1007/BF02173641. [DOI] [PubMed] [Google Scholar]
- Raskina O, Brodsky L, Belyayev A. Tandem repeats on an eco-geographical scale: outcomes from the genome of Aegilops speltoides. Chromosome Research. 2011;19:607–623. doi: 10.1007/s10577-011-9220-9. [DOI] [PubMed] [Google Scholar]
- Ribeiro T, dos Santos KGB, Fonsêca A, Pedrosa-Harand A. Isolation and characterization of a new repetitive DNA family recently amplified in the Mesoamerican gene pool of the common bean (Phaseolus vulgaris L., Fabaceae) Genetica. 2011;139:1135–1142. doi: 10.1007/s10709-011-9615-8. [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiology and Molecular Biology Reviews. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato M, Galián JA, Rosselló JA. Amplification, contraction and genomic spread of a satellite DNA family (E180) in Medicago (Fabaceae) and allied genera. Annals of Botany. 2012;109:773–782. doi: 10.1093/aob/mcr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herra A, Nergadze SG, Santagostino M, Giulotto E. Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenetics and Genome Research. 2008;122:219–228. doi: 10.1159/000167807. [DOI] [PubMed] [Google Scholar]
- Sarri V, Minelli S, Panara F, et al. Characterization and chromosomal organization of satellite DNA sequences in Picea abies. Genome. 2008;51:705–713. doi: 10.1139/G08-048. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends in Plant Sciences. 1998;3:195–199. [Google Scholar]
- Schneider TD, Stephens R. Sequence logos: a new way to display consensus sequences. Nucleic Acids Research. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Wolfgruber TK, Presting GG. Tandem repeats derived from centromeric retrotransposons. BMC Genomics. 2013;14:142. doi: 10.1186/1471-2164-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slijepcevic P, Xiao Y, Dominguez I, Natarajan AT. Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma. 1996;104:596–604. doi: 10.1007/BF00352299. [DOI] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Speta F. Systematische Analyze der Gattung Scilla L. s. l. (Hyacinthaceae) Phyton. 1998;38:1–141. [Google Scholar]
- Stephan W, Cho S. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics. 1994;136:333–341. doi: 10.1093/genetics/136.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres GA, Gong Z, Iovene M, et al. Organization and evolution of subtelomeric satellite repeats in the potato genome. G3 (Bethesda)s. 2011;1:85–92. doi: 10.1534/g3.111.000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida W, Matsunaga S, Sugiyama R, Kawano S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes and Genetic Systems. 2002;77:63–67. doi: 10.1266/ggs.77.63. [DOI] [PubMed] [Google Scholar]
- Ugarković D. Functional elements residing within satellite DNAs. EMBO Reports. 2005;6:1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarković D, Plohl M. Variation in satellite DNA profiles – causes and effects. EMBO Journal. 2002;21:5955–5959. doi: 10.1093/emboj/cdf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan HE, Taylor S, Parker JS. The ten cytological races of the Scilla autumnalis species complex. Heredity. 1997;79:371–379. [Google Scholar]
- Vittorazzi SE, Lourenço LB, Del-Grande ML, Recco-Pimentel SM. Satellite DNA derived from 5S rDNA in Physalaemus cuvieri (Anura, Leiuperidae) Cytogenetic and Genome Research. 2011;134:101–107. doi: 10.1159/000325540. [DOI] [PubMed] [Google Scholar]
- Volkov RA, Komarova NY, Zentgraf U, Hemleben V. Molecular cell biology: epigenetic gene silencing in plants. Progress in Botany. 2006;67:101–133. [Google Scholar]
- Walsh JB. Persistence of tandem arrays: implications for satellite and simple-sequence DNAs. Genetics. 1987;115:553–567. doi: 10.1093/genetics/115.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Riha K, Jang CG, Puizina J, Scherthan H, Schweizer D. Chromosome termini of the monocot plant Othocallis siberica are maintained by telomerase, which specifically synthesises vertebrate-type telomere sequences. The Plant Journal. 2004;37:484–493. doi: 10.1046/j.1365-313x.2003.01974.x. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Schneeweiss GM. Karyotype diversity and evolutionary trends in angiosperms. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, editors. Plant genome diversity, Vol 2. Physical structure, behavior and evolution of plant genomes. Wien: Springer-Verlag; 2013. pp. 209–230. [Google Scholar]
- Wicker T, Taudien S, Houben A, et al. A whole genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. The Plant Journal. 2009;59:712–722. doi: 10.1111/j.1365-313X.2009.03911.x. [DOI] [PubMed] [Google Scholar]
- Willard HF. Centromeres: the missing link in the development of human artificial chromosomes. Current Opinion in Genetics and Development. 1998;8:219–225. doi: 10.1016/s0959-437x(98)80144-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.