Abstract
Purpose
Reported prostate-specific antigen (PSA) values may differ substantially between assays with the Hybritech and World Health Organization (WHO) standardization. [-2]proPSA (p2PSA) and the Beckman Coulter prostate health index (phi) are newly approved serum markers, which are associated with prostate cancer risk and aggressiveness. Our objective was to study the influence of assay standardization on these markers.
Materials and Methods
PSA, % free PSA (%fPSA), and p2PSA were measured using the Hybritech calibration in 892 men undergoing prostate biopsy from a prospective multicenter study. Phi was calculated as: [p2PSA/ fPSA) × (square root of PSA)]. Performance characteristics of phi for prostate cancer detection were then determined using re-calculated WHO calibration PSA values.
Results
The median phi was significantly higher in men with prostate cancer compared to those with negative biopsies using the WHO values (47.4 vs 39.8, p<0.001). Phi offered improved discrimination of prostate cancer detection on biopsy (AUC 0.704) compared to %fPSA or total PSA using the WHO calibration.
Conclusions
Phi can be calculated using Hybritech or WHO standardized assays, and significantly improved the prediction of biopsy outcome over %fPSA or PSA alone.
Keywords: prostate cancer screening, proPSA, phi, prostate health index, assay
Introduction
Screening for prostate cancer (PCa) using serum blood tests for prostate specific antigen (PSA) is controversial because of the generally low specificity of the marker and possible over-diagnosis of indolent cancers.1 Although measurement of free PSA has been shown to improve specificity,2 it has not been widely adopted in clinical practice. More recently, the PSA isoform [-2]proPSA (p2PSA) was shown to be proportionally increased in prostate cancer compared to benign conditions.3 The p2PSA measurement may have greatest clinical utility when combined with PSA and free PSA in a mathematical formula called the Beckman Coulter Prostate Health Index (phi).4 Phi is calculated as: (p2PSA / fPSA) × (square root of PSA). In a recent multi-center trial, phi was shown to improve specificity compared to PSA and %fPSA and also was associated with prostate cancer aggressiveness.4 More recently, Sanda et al. showed that 59% of biopsies with insignificant cancer could have been avoided using a phi cutoff of 27 among men with PSA levels from 4-10 ng/ml (unpublished data). Both p2PSA and phi have recently been approved by the U.S. FDA.
A limitation of all PSA-based measurements is potential confounding from differences in assay standardization. Indeed, PSA values based upon the World Health Organization (WHO) standard[5] were reported to be approximately 20% lower than those obtained with Hybritech standardization in the same sample.5
The Hybritech PSA standard was used in many of the studies that defined the operating properties of PSA testing in clinical practice,6 as well as the initial studies demonstrating the usefulness of phi.4 Thus, we sought to evaluate the performance of various phi cutoffs based upon the WHO standardization in men from a large, prospective, multi-center clinical trial.
Materials and Methods
Study Population
From October 2003 to June 2009, 892 men aged ≥50 years from seven geographically diverse USA medical centers were enrolled in a study to evaluate the utility of p2PSA and phi. All men had PSA levels from 2-10 ng/ml and benign findings on digital rectal examination.
The study involved a multi-site, double-blind, case-control design and included about 97% prospectively enrolled subjects, as previously described.4 Approximately equal numbers of men with or without prostate cancer were enrolled at each institution with a total of 430 men with histologically-confirmed prostate cancer and 462 men without cancer on at least a 6 core biopsy (median 12, range 6-34, 98% with ≥10 core biopsy). Men were excluded if they had a prior history of prostate cancer or interventions such as transurethral resection of the prostate, were taking specific drugs known to affect PSA levels, or had a urinary tract infection at the time of the blood draw or biopsy.
Biomarker Measurement
All serum biomarkers were measured with the Access Hybritech p2PSA, PSA and fPSA assays on the Beckman Coulter Access 2 Immunoassay Analyzer. The p2PSA assay is a two-site immunoenzymatic sandwich assay using a monoclonal antibody capture bound to a paramagnetic particle and a second monoclonal antibody-alkaline phosphatase conjugate for analyte detection.7 Chemiluminescent substrate is added and the light generated by the reaction is measured by a luminometer. The amount of light produced is directly proportional to the concentration of p2PSA.
Samples were collected and processed within 8 hours, and subsequently stored frozen at ≤ -70°C. The PSA and fPSA assays were run using a single replicate, and the p2PSA assay was run in duplicate (first replicate used for data analysis) according to the testing protocol.
The PSA and free PSA assay results were initially generated using Hybritech calibration and were then re-calculated based on WHO calibrator assignments. Beckman Coulter has maintained the standardization of both assays as originally commercialized. The two standards are not the same, and PSA and fPSA values based on the WHO standards read approximately twenty percent lower compared to results based on the Hybritech standards. Mean calibration factors were used to convert PSA[8] and fPSA (unpublished data) results from the Hybritech to the WHO standard, consistent with published differences [6, 9-11]. There is no international standard for p2PSA, and calibration was done using a Hybritech standard. Phi was then calculated based upon both the Hybritech and WHO calibrations for PSA and fPSA.
Analysis
The primary objective of this study was to compare the performance characteristics of phi to %fPSA using the estimated WHO calibration values. Sensitivity and specificity were calculated for various WHO-based phi cutoffs, and receiver operating characteristic analysis was used to examine discrimination. The Wilcoxon rank-sum test was used to compare continuous variables between benign and cancer patients.
Additionally, we calculated the performance characteristics of phi for individual prostate cancer risk assessment using the estimated WHO calibration. An approximate 25% positive prostate cancer rate has been previously reported in a population of men with moderately elevated PSA levels > 4 ng/mL.8 Because the current study enrolled approximately 48.2% cancer subjects and 51.8% non-cancer subjects, bootstrapping was used to repetitively sample the study population to include 25% of cancer subjects for risk assessment. Each sampling consisted of 462 (75%) benign subjects and 154 (25%) cancer subjects. This random sampling process was repeated 1,000 times, and prostate cancer detection rates were then compared across phi ranges. In addition, we compared detection rates for Gleason ≥7 prostate cancer across phi ranges using the estimated WHO calibration.
Results
Demographics, PSA, fPSA, [-2]proPSA, %fPSA, and phi distributions are presented in Supplementary Table 1. The mean and median values for %fPSA and phi were different between the benign and cancer groups using both standardizations.
Supplementary Table 2 compares the phi distributions between the Hybritech and WHO calibrations using the cutoff values with 95% sensitivity. Overall, there were eight discordant pairs between the Hybritech and WHO calibrations. The observed relative negative, positive, and overall agreements were 0.9579, 0.9950, and 0.9910, respectively. Overall, the difference in phi was not statistically significant between the Hybritech and WHO calibrations for PSA and fPSA (p=1.000, McNemar's test).
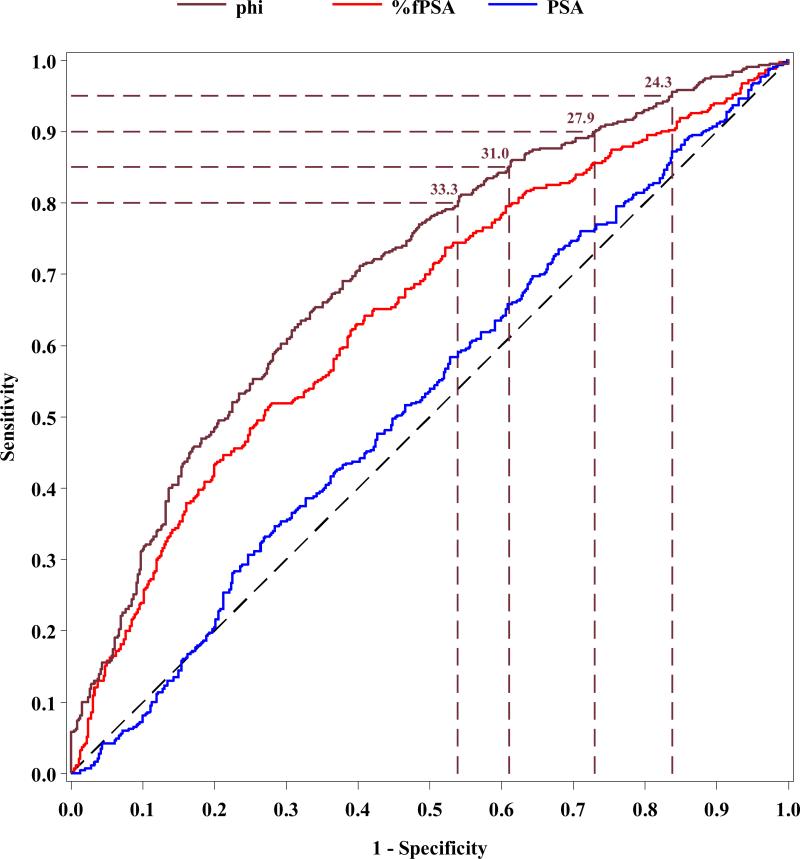
ROC analyses were then performed for phi compared to PSA and %fPSA using the WHO calibration (Figure 1). The AUC for phi (0.704) was significantly higher than for %fPSA (0.649, p=0.005) and total PSA (0.527, p<0.001) in men with PSA levels ranging from 1.6 – 7.8 ng/mL based on the WHO calibration (i.e., 2-10 ng/mL Hybritech calibration).
Figure 1.
Receiver operating characteristic analysis comparing phi, %fPSA and PSA for prostate cancer detection in men with PSA levels of 1.6-7.8 ng/ml (WHO calibration)
Table 1 shows the sensitivity and specificity for various phi cutoffs using the Hybritech and estimated WHO calibration. For example, with the WHO calibration, at 95% sensitivity, the specificity for phi was 16.2% compared to 7.8% for %fPSA (p=0.008); while, at 90% and 80% sensitivity, phi had a specificity of 27.1% and 46.1%, respectively.
Table 1.
Sensitivity and Specificity of Prostate Cancer Cutoffs for Beckman Coulter phi in Men with Non-Suspicious DRE (Hybritech and WHO Calibration of PSA and fPSA)
| Hybritech Calibration | WHO Calibration | |||
|---|---|---|---|---|
| % Sensitivity | phi Cutoff | % Specificity | phi Cutoff | % Specificity |
| 99 | 17.2 | 5.2 | 19.7 | 5.8 |
| 98 | 18.4 | 8.4 | 20.7 | 7.8 |
| 95 | 21.3 | 16.0 | 24.3 | 16.2 |
| 90 | 24.1 | 26.2 | 27.9 | 27.1 |
| 85 | 27.2 | 39.0 | 31.0 | 39.0 |
| 80 | 29.3 | 45.2 | 33.3 | 46.1 |
| 75 | 31.1 | 52.6 | 35.5 | 52.6 |
| 70 | 33.4 | 60.0 | 38.4 | 60.4 |
| 65 | 35.0 | 65.2 | 40.0 | 66.2 |
| 60 | 37.5 | 70.3 | 42.8 | 70.8 |
| 55 | 39.1 | 74.2 | 44.8 | 74.7 |
| 50 | 42.2 | 79.0 | 47.4 | 78.1 |
| 45 | 44.3 | 82.7 | 50.5 | 83.3 |
| 40 | 46.7 | 85.7 | 53.2 | 86.1 |
| 35 | 49.3 | 87.4 | 55.5 | 86.8 |
| 30 | 52.6 | 90.7 | 60.0 | 90.3 |
| 25 | 55.9 | 91.8 | 62.9 | 91.3 |
| 20 | 61.9 | 93.7 | 70.0 | 93.3 |
| 15 | 67.6 | 95.2 | 77.3 | 95.7 |
| 10 | 78.1 | 97.6 | 89.5 | 97.8 |
| 5 | 104.2 | 100 | 118.3 | 100 |
Table 2 shows the probability and relative risks of detecting prostate cancer based upon the adjusted 25% proportion of cancer subjects. As seen, higher phi values, based on PSA and fPSA WHO calibrations, were associated with higher prostate cancer risk. Approximately 13% of the study population had a phi value > 62.0 and these men had a 4.4-fold increased risk of prostate cancer compared to the reference group (phi <29).
Table 2.
Probability of a) prostate cancer and (b) Gleason ≥7 disease in various ranges of Beckman Coulter phi (WHO calibrations)
| a) | |||
|---|---|---|---|
| phi Range (WHO calibration) | Probability of Cancer (95% Confidence Interval) | Relative Risk (95% Confidence Interval) | Percent of patients in phi range |
| 0-28.9 | 11.3% (6.8% - 16.0%) | 1.0 | 26.8% |
| 29.0-39.9 | 18.0% (13.4% - 22.8%) | 1.6 (1.0 - 2.9) | 31.4% |
| 40.0-61.9 | 34.0% (28.2% - 39.5%) | 3.0 (1.9 - 5.3) | 28.3% |
| 62+ | 49.6% (40.0% - 59.5%) | 4.4 (2.8 - 8.1) | 13.5% |
| b) | |||
|---|---|---|---|
| Gleason Score on Biopsy among Cancer Patients (N=429) | |||
| phi Range (WHO calibration) | Less than 7 n (%) | ≥7 n (%) | Risk Ratio (95% CI) |
| 0-28.9 | 40 (78.4) | 11 (21.6) | 1.0 |
| 29.0-39.9 | 68 (69.4) | 30 (30.6) | 1.4 (0.8, 2.6) |
| 40.0-61.9 | 117 (70.9) | 48 (29.1) | 1.3 (0.8, 2.4) |
| 62.0+ | 65 (56.5) | 50 (43.5) | 2.0 (1.1, 3.5) |
Note: One participant excluded with missing Gleason score.
Cochran-Armitage test for trend, p=0.003
Overall, the Gleason scores were ≤6, 7, and 8-10 in 290 (67.6%), 119 (27.7%), and 20 (4.7%) men, respectively. Table 2 shows the association between higher phi values using the WHO standardization and Gleason scores ≥7 (p-trend=0.003). For example, men with a phi value ≥62.0 had a 2.0-fold increased risk of Gleason ≥7 compared to the reference group (phi <29).
Discussion
PSA and fPSA concentrations based on calibration to the WHO standards (96/670 for PSA and 96/668 for fPSA) differ significantly from PSA and fPSA concentrations based on calibration to the original Hybritech standards.5 The original goal of using WHO calibration to standardize PSA and fPSA assays from different manufacturers has not been realized. This is most likely due to the heterogeneity of these molecules and the different antibodies used. In one study, comparison of 70 samples with six commercially available assays measured in the WHO PSA 96/670 reference preparation resulted in broad mean deviations ranging from 0.01 – 1.32 for PSA, and 0.05 – 0.49 ug/L for fPSA, with regression slopes varying from 0.99 to 1.22.9 This led to different results for patients classified according to commonly recommended PSA cutoffs for performing a biopsy.
Another study showed the potential confounding of PSA velocity due to assay standardization bias.10 For example, use of a WHO- then a Hybritech-standardized test in consecutive years could lead to pseudo-acceleration or vice versa.
The results of our study suggest that phi may be used with either the Hybritech or WHO calibrations. Clinical performance for the same population using the Hybritech calibrations for PSA and fPSA only were presented in a previous publication.4 Comparable phi results were obtained for both the Hybritech and WHO calibrations on ROC analysis and for assessing the probability of prostate cancer on biopsy. Phi specificity at sensitivities of 95%, 90%, and 80% between the two calibrations were 16.0% vs. 16.2%, 26.2% vs. 27.1%, and 45.2% vs. 46.1%, respectively. Phi AUCs were 0.703 vs. 0.704 for the Hybritech and WHO calibrations. Using the Hybritech calibration, the probability of PCa for phi <25 was 11.0% and for phi ≥ 55 was 52.1%. Using the WHO calibrations, the probability of PCa for phi < 29 was 11.3% and for phi ≥ 62 was 49.6%, comparable to those for the Hybritech calibrations. Indeed, we found comparable clinical utility for phi for prostate cancer detection using either calibration for PSA and fPSA, using adjusted cutoff values.
Furthermore, phi offered improved discrimination compared to %fPSA using the WHO calibration for the PSA and fPSA assays. Similar results were obtained by Jansen et al in a cohort of 5,865 men showing comparable results when using either the Hybritech or WHO calibrations for total PSA to determine the percent decrease in prostate biopsies (20% vs. 19%) and decrease in detected cancers (19% vs. 20%).5
Therefore, even with WHO standardized assays, consensus from a number of studies is that interchangeability of PSA and fPSA assays from different manufacturers remains inadequate.5, 9, 11 If the calibration is changed, the accepted laboratory practice is to establish a new baseline for patient management. Care should be taken to specify which assay and calibration was used to avoid misinterpretation of individual results and potentially erroneous clinical decisions about prostate cancer management.
Two main strengths of this study include the prospective design and the large sample size (892 total subjects) of men undergoing prostate biopsy with histologic confirmation. A potential limitation with this study is that subject enrollment was restricted to men with total PSA levels from 2 to 10 ng/ml, which may have affected performance characteristics. However, a recent publication by Guazzoni et al. showed that phi results (sensitivity, specificity, AUC, and probability of PCa) in a real-world setting support our findings.12
Conclusions
PSA and fPSA concentrations are dependent on the standard used to calibrate the assay. Both p2PSA and phi have recently been approved by the FDA. Although the phi calculation includes PSA and fPSA along with p2PSA, we found that phi had comparable performance characteristics using either the Hybritech or WHO standardization, and phi provided significantly improved discrimination of prostate cancer from benign disease in comparison to %fPSA.
Supplementary Material
REFERENCES
- 1.Barry MJ. Screening for prostate cancer--the controversy that refuses to die. N Engl J Med. 2009;360:1351. doi: 10.1056/NEJMe0901166. [DOI] [PubMed] [Google Scholar]
- 2.Catalona WJ, Partin AW, Slawin KM, et al. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. Jama. 1998;279:1542. doi: 10.1001/jama.279.19.1542. [DOI] [PubMed] [Google Scholar]
- 3.Mikolajczyk SD, Catalona WJ, Evans CL, et al. Proenzyme forms of prostate-specific antigen in serum improve the detection of prostate cancer. Clin Chem. 2004;50:1017. doi: 10.1373/clinchem.2003.026823. [DOI] [PubMed] [Google Scholar]
- 4.Catalona WJ, Partin AW, Sanda MG, Wei JT, Klee GG, Bangma CH, Slawin KM, Marks LS, Loeb S, Broyles DL, Shin SS, Crus AB, Chan DW, Sokoll LJ, Roberts WL, van Schaik RHN, Mizrahi IA. A Multicenter Study of [−2] Pro-Prostate Specific Antigen Combined With Prostate Specific Antigen and Free Prostate Specific Antigen for Prostate Cancer Detection in the 2.0 to 10.0 ng/ml Prostate Specific Antigen Range. J Urol. 2011;185:1650. doi: 10.1016/j.juro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen FH, Roobol M, Bangma CH, et al. Clinical impact of new prostate- specific antigen WHO standardization on biopsy rates and cancer detection. Clin Chem. 2008;54:1999. doi: 10.1373/clinchem.2007.102699. [DOI] [PubMed] [Google Scholar]
- 6.Catalona WJ, Ramos CG, Carvalhal GF, et al. Lowering PSA cutoffs to enhance detection of curable prostate cancer. Urology. 2000;55:791. doi: 10.1016/s0090-4295(99)00602-0. [DOI] [PubMed] [Google Scholar]
- 7.Sokoll LJ, Chan DW, Klee GG, et al. Multi-center analytical performance evaluation of the Access Hybritech(R) p2PSA immunoassay. Clinica chimica acta; international journal of clinical chemistry. 2012;413:1279. doi: 10.1016/j.cca.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Catalona WJ, Smith DS, Ratliff TL, et al. Measurement of prostate- specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991;324:1156. doi: 10.1056/NEJM199104253241702. [DOI] [PubMed] [Google Scholar]
- 9.Kort SA, Martens F, Vanpoucke H, et al. Comparison of 6 automated assays for total and free prostate-specific antigen with special reference to their reactivity toward the WHO 96/670 reference preparation. Clinical chemistry. 2006;52:1568. doi: 10.1373/clinchem.2006.069039. [DOI] [PubMed] [Google Scholar]
- 10.Loeb S, Chan DW, Sokoll L, et al. Prostate specific antigen assay standardization bias could affect clinical decision making. J Urol. 2008;180:1959. doi: 10.1016/j.juro.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 11.Stephan C, Klaas M, Muller C, et al. Interchangeability of measurements of total and free prostate-specific antigen in serum with 5 frequently used assay combinations: an update. Clinical chemistry. 2006;52:59. doi: 10.1373/clinchem.2005.059170. [DOI] [PubMed] [Google Scholar]
- 12.Guazzoni G, Nava L, Lazzeri M, et al. Prostate-specific antigen (PSA) isoform p2PSA significantly improves the prediction of prostate cancer at initial extended prostate biopsies in patients with total PSA between 2.0 and 10 ng/ml: results of a prospective study in a clinical setting. European urology. 2011;60:214. doi: 10.1016/j.eururo.2011.03.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.