Abstract
The CRISPR RNA-guided Cas9 nuclease gene-targeting system has been successfully used for genome editing in a variety of organisms. Here, we report the use of dual sgRNA-guided Cas9 nuclease to generate knockout mutants of protein coding genes, noncoding genes, and repetitive sequences in C. elegans. Co-injection of C. elegans with dual sgRNAs results in the removal of the interval between two sgRNAs and the loss-of-function phenotype of targeted genes. We sought to determine how large an interval can be eliminated and found that at least a 24 kb chromosome segment can be deleted using this dual sgRNA/Cas9 strategy. The deletion of large chromosome segments facilitates mutant screening by PCR and agarose electrophoresis. Thus, the use of the CRISPR/Cas9 system in combination with dual sgRNAs provides a powerful platform with which to easily generate gene knockout mutants in C. elegans. Our data also suggest that encoding multiple sgRNA sequences into a single CRISPR array to simultaneously edit several sites within the genome may cause the off-target deletion of chromosome sequences.
Recent research in targeted genome editing technology has made exciting progress in genome engineering, including those for zinc-finger nucleases (ZFNs)1,2,3, transcription activator-like effector nucleases (TALENs)4,5,6, and clustered regularly interspaced short palindromic repeats (CRISPR) RNA-guided Cas9 nucleases7,8,9,10,11,12,13,14,15,16,17,18,19,20. Both ZFNs and TALENs use sequence-specific DNA-binding modules linked to a non-specific DNA nuclease to create DNA lesions. The CRISPR/Cas9 system is a versatile RNA-guided genome editing technology that uses small guide RNA (sgRNA) to target and cleave DNA sequences16,21,22,23,24. sgRNA contains a hairpin that mimics the tracrRNA-crRNA complex and a short sequence complementary to the targeted DNA. When co-expressed with an artificial sgRNA targeting a cellular gene, the Cas9 endonuclease generates double-stranded breaks of DNA at the targeted locus. In addition, CRISPR/Cas9 can also precisely alter a target gene through homologous recombination (HR). Many selection schemes have been developed to allow for the HR-mediated insertion of large sequence tags to facilitate downstream mutant screening. CRISPR/Cas9 technology has recently been used for genome engineering in numerous organisms, providing a powerful tool for diverse biological applications11,25.
CRISPR/Cas9 technology has also been successfully applied to C. elegans5,26,27,28,29,30,31,32,33,34,35,36. This method typically produces small insertions and deletions (indels) that shift the open reading frame (ORF) of the targeted gene and result in premature termination of translation and loss-of-function phenotypes. Most, if not all, of the indels are sized less than 100 bp if only one sgRNA was injected. Recent work showed that large deletions can be induced by incorporating single-strand oligodeoxynucleotides together with dual sgRNAs in C. elegans37,38.
Despite this substantial progress, many remaining questions hinder the application of CRISPR/Cas9 technology. For example, different sgRNAs have substantially variable DNA cleavage efficiencies7,18,26. The discovery of small indels is time-consuming and laborious. Off-target effects result in unintended DNA cleavage, which requires multiple independent cell lines or the outcrossing of modified animals for gene analysis14,19,39. DNA cleavage by Cas9 at two genomic loci may induce inter- and intrachromosomal rearrangements in mammalian cells40.
Here, we report the use of dual sgRNA-guided Cas9 nuclease to generate gene knockout mutants in C. elegans. Co-injection of C. elegans with two sgRNAs results in the elimination of an interval up to 24 kb between two guide RNAs. Thus, the application of dual sgRNAs in the CRISPR/Cas9 system provides a powerful platform with which to generate gene knockout mutants. Our results suggest caution when incorporating multiple guide sequences into a single CRISPR experiment to simultaneously edit several sites within the genome.
Methods
Strains
Bristol strain N2 was used as the standard wild-type strain. The Hawaiian strain CB4856 was used for snp-SNP mapping. All strains were incubated on nematode growth medium (NGM) plates seeded with OP50 at 20°C41. GR1373:eri-1(mg366) and WM27: rde-1(ne219).
Construction of sgRNA expression plasmids
We manually searched for target sequences consisting of G(N)19NGG near the desired mutation sites22,30. The target sequences are listed in Supporting Information Table s2. We replaced the unc-119 target sequence in the pU6::unc-119 sgRNA vector30 with the desired target sequence using overlap extension PCR. The pU6::unc-119 sgRNA vector was diluted to 2 ng/µl and PCR amplified using the primers sgRNA F and sgRNA R to generate linear products. The PCR products were gel-purified and transformed into Trans10 Chemically Competent Cell (Transgene Biotech, Beijing) directly. We used PhantaTM super-fidelity DNA polymerase (Vazyme Biotech, Nanjing, Cat. No. P501-d1/d2/d3) in all PCR reactions. The primer sequences used for the construction of sgRNA expression plasmids are listed in Table s3.
Microinjection
DNA mixtures were microinjected into the gonads of young adult C. elegans. Plasmids for injection were prepared using a miniprep plasmid purification kit (Sangon, Shanghai, Cat. No. sk8192). For dual sgRNA experiments, we injected 50 ng/µl Cas9 expressing vector, 50 ng/µl sgRNA #1, 50 ng/µl sgRNA #2 (as indicated in the figures), and 5 ng/µl pCFJ90 vector. For single rde-12 sgRNA #2 experiments, we injected 50 ng/μl Cas9 expressing vector, 100 ng/μl rde-12 sgRNA #2, and 5 ng/μl pCFJ90 vector. After recovering from injection, each worm was placed onto an individual OP50 plate.
Screening for deletion mutants by PCR
Three days after injection, F1 animals expressing mCherry were transferred to individual NGM plates and allowed to produce F2 progeny for 2 to 3 days. F1 with corresponding F2 progeny were harvested and washed in M9 buffer, transferred to 50 µl lysis buffer (500 ug/ml Proteinase K, 100 mM NaCl, 50 mM Tris, 20 mM EDTA, and 1% SDS), and screened by PCR amplification with primers outside of the sgRNA-targeted regions. Mutants with large deletions were singled to NGM plates and further confirmed by PCR amplification and DNA sequencing. The primers used for PCR screening are listed in Table s4.
Imaging
Images were collected using Leica DM2500 and M165 FC microscopes.
RNAi
RNAi experiments were performed as described previously42. Bacteria expressing flr-1, nhr-23, mex-3, pos-1, lir-1, and unc-15 dsRNA were obtained from the Ahringer RNAi library and were sequenced to verify their identity43. The dpy-13 RNAi clone has been described previously42.
Genetic screening
To identify the factors specifically required for off-target RNAi silencing, we previously screened for the cellular factors required for the dpy-13 dsRNA-induced silencing of off-target RNAs but dispensable for canonical RNAi silencing42. Nine mutants were isolated from this genetic screening, three of which were identified by snp-SNP mapping and sequenced as missense alleles of rde-12(F58G11.2) (Fig. s1).
Quantitative RT-PCR
RNAs were isolated from embryos using a dounce homogenizer (pestle B) in TRIzol solution followed by DNase I digestion (Qiagen)42. cDNAs were generated from RNAs using the iScript cDNA Synthesis Kit (Bio-Rad) according to the vendor's protocol. qRT-PCR was performed using a MyIQ2 machine (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad). The primers for qRT-PCR analysis are listed in Table s5. eft-3 mRNA was used as an internal control for sample normalization. Data analysis was performed using a ΔΔCT approach.
Results
Using dual sgRNAs to knock out rde-12
We previously screened for factors required for RNAi silencing by chemical-induced mutagenesis42 and isolated three missense alleles of rde-12 (Fig. s1, Table s1). RDE-12 is an RNA helicase that engages targeted mRNA and Argonaute proteins to promote the synthesis of secondary siRNAs in C. elegans44,45. To further characterize rde-12, we sought to generate null alleles using CRISPR/Cas9 technology with dual sgRNAs.
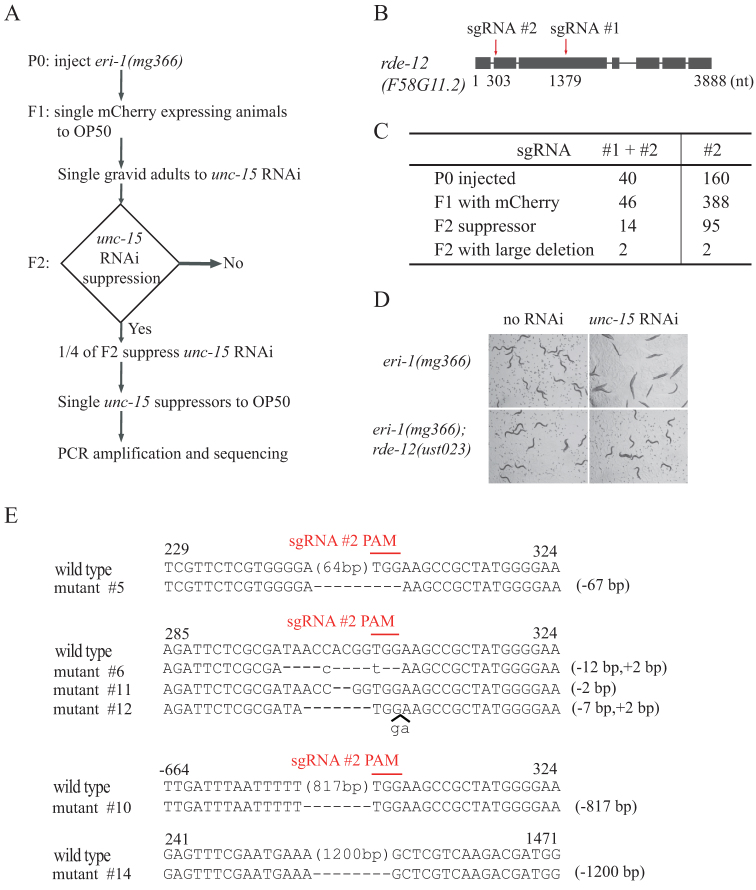
We co-injected sgRNAs targeting exons 2 and 3 of rde-12 and Cas9 and mCherry expression plasmids into eri-1(mg366) animals (Fig. 1). The mutation of eri-1 results in an enhanced RNAi (Eri) phenotype46. mCherry was used as a co-injection marker. From 40 injected animals, we obtained 46 fertile F1 animals expressing mCherry. F1 animals were first transferred to NGM plates and grow up to gravid adults, and then were singled to unc-15 RNAi plates. Whereas eri-1(mg366) control animals exhibited a paralyzed phenotype on unc-15 dsRNA, the mutation of rde-12 resulted in resistance to unc-15 RNAi, a phenotype termed RNAi defective (Rde) (Fig. 1D). Fourteen F1 animals produced unc-15 RNAi suppression progeny at the expected Mendelian frequency of approximately 25%, indicating that F1 animals were heterozygous for rde-12 loss-of-function mutations. Very few animals with an rde-12 mutation continued to express mCherry in subsequent generations, suggesting that the mCherry transgene expression was transient and present only in the F1 generation. The F2 suppressors were selected and further characterized by PCR and sequencing. We identified lesions in the rde-12 gene consistent with Cas9-directed cleavage (Fig. 1E). Four of the rde-12 mutants (#5, #6, #11, and #12) contained small indels that are likely directed by sgRNA #2 alone. Interestingly, we also obtained two mutants containing large deletions. One mutant (#10) eliminated 817 bases of genomic sequence, also likely guided by sgRNA #2 alone. Another mutant (#14) deleted 1,200 bases of genomic sequence, eliminating the entire interval between sgRNAs #1 and #2. This deletion was further confirmed by both PCR amplification and DNA sequencing (Figs. 1E, s2). The large deletion may reflect a simultaneous cutting directed by the two sgRNAs, whose targets are separated by 1,076 bases in this experiment. These data indicate that the deletion of large chromosomal segments can be induced by CRISPR/Cas9 technology with dual sgRNAs.
Figure 1. Dual sgRNA-guided deletion of the rde-12 gene.
(A) Schematic of the screen for CRISPR/Cas9 genome editing events. The dominant transformation marker mCherry was co-injected with Cas9 and sgRNA #1 and #2 expression plasmids. F1 animals with mCherry expression were grown on unc-15 RNAi, and phenotypes of F2 were scored. The RNAi suppressors were PCR amplified and sequenced. (B) Schematic of the rde-12 gene. Positions of sgRNA-guided cleavage sites are indicated. (C) Summary of microinjection experiments. (D) Sequence alignments of the rde-12 gene in wild-type and mutant animals. The PAM sequence is labeled in red and overlined. Dash indicates deletion. Lowercase indicates insertion. The numbers in parentheses within the sequence represent the number of bases not shown. The number of deleted (−) or inserted (+) bases is shown on the right of each indel. Numbers on the top of sequences indicate positions relative to the transcription start site.
We also noticed that 8 Rde mutants did not produce the expected PCR products. We speculated that dual sgRNAs may induce inversion or translocation at the targeted loci40. Alternatively, the deleted region may be larger than the PCR amplifiable region. These mutants were not followed in the present study.
We examined the occurrence of small indels and large deletions by injecting only rde-12 sgRNA #2. We injected 160 P0, selected 388 F1 expressing mCherry, and recovered 95 F2 mutants that suppressed unc-15 RNAi (Fig. 1c). PCR screening of these 95 mutants identified 2 deletions larger than 500 bp. DNA sequencing confirmed the deletion of 826 bp and 517 bp, respectively (data not shown). We should point out that the occurrence of Cas9-induced small indels has been underestimated by the unc-15 RNAi, since many small indels may not change open reading frame or not result to strong loss-of-function alleles to suppress unc-15 RNAi. In contrast, the injection of dual sgRNAs favors the generation of strong loss-of-function alleles by creating large deletions.
Dual sgRNA-directed deletion of the linc-22 promoter
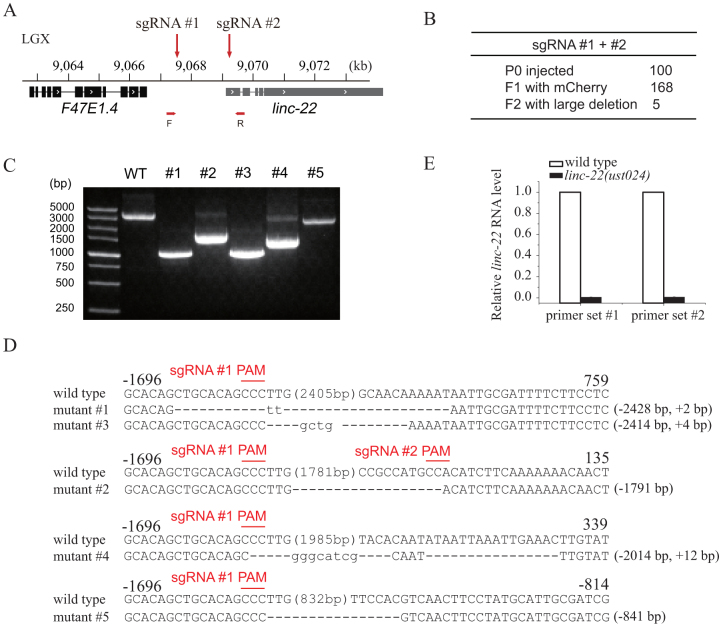
To further test this dual sgRNA strategy, we co-injected two sgRNAs targeting the promoter region of linc-22 into wild-type C. elegans (Fig. 2). linc-22 is a long intergenic non-coding RNA (lincRNA) that expresses abundant endogenous siRNAs42. We sought to knock down linc-22 expression by eliminating its promoter sequence to test whether the expression of linc-22 RNA is required for endo-siRNA production. We injected 100 animals and isolated 168 F1 animals expressing mCherry (Fig. 2B). F1 animals were singled to OP50 plates, laid F2 progenies, and were directly screened by PCR with primers flanking the sgRNA-targeted region. Five F2 animals contained large deletions in the linc-22 promoter region (Fig. 2C). The PCR products were subcloned and sequenced. Two mutants deleted approximately 2.4 kb of the promoter sequence, as expected (Fig. 2D). The expression level of linc-22 RNA was further examined by quantitative real-time PCR and confirmed the removal of the linc-22 promoter (Fig. 2E).
Figure 2. Dual sgRNA-guided deletion of the linc-22 promoter.
(A) Schematic of linc-22 gene. (B) Summary of microinjection experiments. F2 progenies were directly screened by PCR amplification. (C) PCR amplification of the targeted region in the deletion mutants. (D) Sequence alignments of wild-type and mutant animals. Dash indicates deletion. The numbers in parentheses within the sequence represent the number of bases not shown. The number of deleted (−) or inserted (+) bases is indicated on the right of each indel. (E) Quantitative real-time PCR detection of linc-22 expression. Total RNAs were isolated from embryos. eft-3 mRNA was used as an internal control for normalization. N = 3.
Using dual sgRNAs to delete repetitive sequences
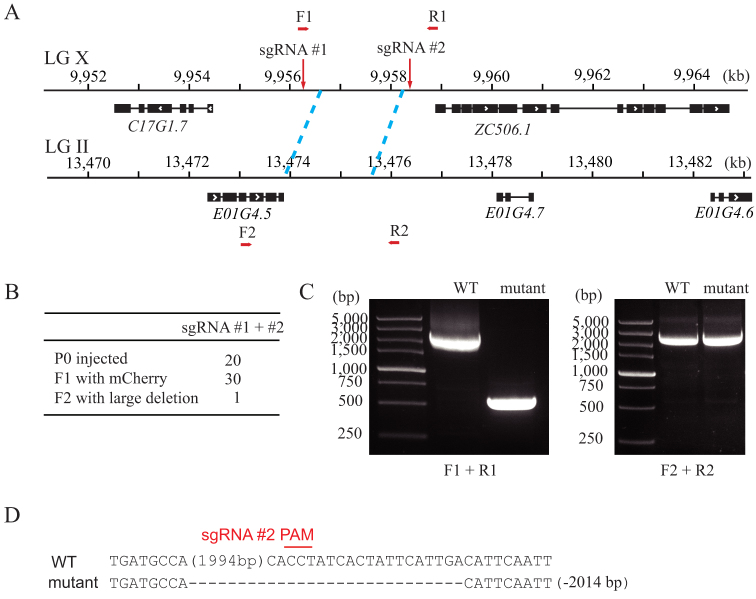
Repetitive sequences are widely present in the genome of C. elegans and may exhibit overlap or redundancy in gene function. Using a single guide RNA to direct the DNA cleavage of one gene can induce the off-targeted cleavage of another gene with highly sequence similarity. To study the function of one but not the other, we sought to use the dual sgRNA strategy to specifically knock out one of the repetitive gene pairs. e01g4.5 is a gene that generates the most abundant endogenous siRNAs42. The downstream sequence of E01G4.5 is highly similar to a region between c17g1.7 and zc506.1 on chromosome X, with 95.8% sequence identity for 1.75 kb (Fig. 3A). We chose sgRNA targeting sequences outside of the repetitive region on chromosome X. From 20 injected animals, we isolated 30 F1 animals expressing mCherry (Fig. 3B). These animals were directly screened by PCR with primers flanking sgRNA-targeted loci. We isolated one animal with an approximately 2 kb deletion at the region on chromosome X but an intact e01g4.5 region (Fig. 3C). The PCR product was subcloned and sequenced, confirming the deletion at the targeted region (Fig. 3D).
Figure 3. Dual sgRNA-guided deletion of a repetitive sequence.
(A) Schematic of gene structure. (B) Summary of the microinjection experiments. F2 progenies were directly screened by PCR amplification. (C) PCR amplification of the targeted region (left panel) and its homologous region (right panel). (D) Sequence alignment of wild-type and mutant animals. Dash indicates deletion. The number in parentheses represents the number of bases not shown. The number of deleted (−) bases is indicated on the right side.
Dual sgRNA-induced deletion of large chromosomal segments
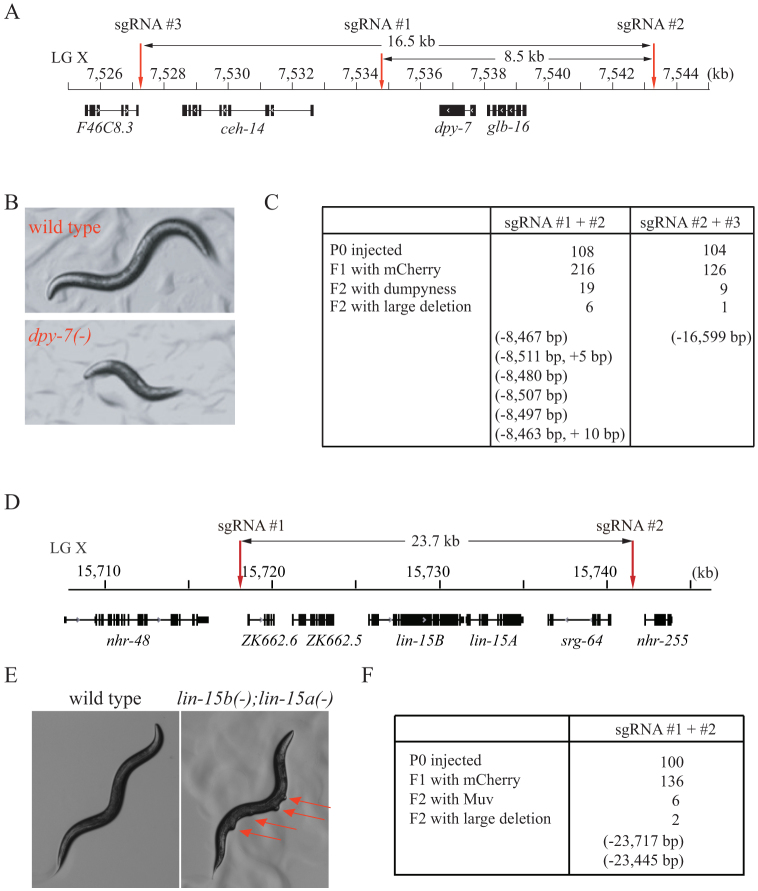
To further determine how large an interval can readily be eliminated using this method, we co-injected dual sgRNAs targeting distinct chromosome sites into wild-type C. elegans. To facilitate mutant selection, we targeted regions with visible genetic markers.
We chose an 8.5 kb and a 16.5 kb region on chromosome X (Fig. 4A). dpy-7 is a collagen gene inside these two regions and served as the selection marker. The mutation of dpy-7 leads to a dumpy phenotype with an animal size approximately half that of wild-type animals (Fig. 4B). We isolated 19 dumpy animals from 216 F1 animals expressing mCherry following the co-injection of sgRNAs #1 and #2. Six of these dumpy animals contained deletions of approximately 8.5 kb (Fig. 4C, s3). Following the co-injection of sgRNAs #2 and #3, we isolated one dumpy animal containing a 16,559 base deletion. Deletions in these mutants were confirmed by PCR amplification and sequencing (Fig. s3).
Figure 4. Dual sgRNAs can direct the deletion of large chromosome segments.
(A, D) Schematic of gene structures. (B) dpy-7(-) mutant exhibited a dumpy phenotype. (E) lin-15b/15a genes belong to syn-Muv gene families. Disruption of both lin-15b and lin-15a together results in a Muv phenotype. (C, F) Summary of the microinjection experiments. The dominant transformation marker mCherry was co-injected with Cas9 and sgRNA #1 and #2 or sgRNA #2 and #3 expression plasmids. F1 animals with mCherry expression were transferred to OP50 plates, and F2 animals were scored for dumpy (B) or Muv phenotypes (E). Mutant animals were further PCR amplified and sequenced.
We targeted another 23.7 kb region on chromosome X using the dual sgRNA strategy (Fig. 4D). Five genes are located in this region, including the lin-15b/15a operon. The disruption of lin-15b and lin-15a together results in a multi-vulva (Muv) phenotype (Fig. 4E). We isolated 6 Muv mutants from 136 F1 animals expressing mCherry (Fig. 4F). Two Muv mutants contained deletions of approximately 23.7 kb, which was confirmed by PCR amplification and sequencing (Fig. s4).
Some dumpy or Muv mutants did not produce the expected PCR products. These mutants may exhibit an inversion or translocation at the targeted loci40. Alternatively, the dual sgRNAs may have induced deletions larger than the PCR-amplifiable region. These mutants were not followed in the present study.
Discussion
The genetic manipulation of C. elegans is a crucial approach for the study of development and gene regulation. RNA interference and the creation of deletion mutants have both long been used to perturb gene function in C. elegans. The low efficacy and off-target effects of RNAi require null mutants that eliminate gene functions under many circumstances42,43,47. To generate knockout mutants in C. elegans, the random mutagen trimethylpsoralen (TMP) followed by UV irradiation was used to mutagenize a very large number of worms followed by PCR screening48,49,50. At a very low frequency, this strategy will generate a small deletion. The TMP/UV method is labor intensive and usually requires multiple rounds of sequential PCR screening to isolate the desired mutants. Alternatively, hopping out a Mos1 transposon from a chromosomal locus via the expression of a Mos1 transposase in the germline will create a double-strand break that results in DNA deletion. However, this method requires a strain bearing a Mos1 transposon inserted near the targeted site, which can also be problematic51,52,53. The CRISPR/Cas9 nuclease system has emerged as a new genome editing technology in C. elegans that provides an advantage compared to TMP/UV and Mos1 methods. In principle, this approach is limited only by the requirement of a PAM sequence. Here, we report a simple and efficient method to generate gene knockout mutants in C. elegans using the CRISPR/Cas9 system (summarized in Table 1). Specifically, we showed that the co-injection of two sgRNAs into C. elegans can induce a deletion of the chromosome sequence between the two sgRNA-targeted loci, which is a very useful technique for creating null alleles. In addition, single guide sgRNA-induced short indels are typically identified by PCR amplification followed by T7 or CEL nuclease digestion. We showed that large deletions can be easily identified by PCR and agarose electrophoresis, which is especially important for those mutants with no visible phenotypes. Alternatively, Paix et al. described a method to direct a 6 kb genome deletion by including single-strand oligodeoxynucleotides with dual sgRNAs during Cas9 injection, which may also facilitate the selection of mutants37. However, it is unknown whether this method is able to induce larger genome deletions up to tens of kilo base pairs.
Table 1. Summary of dual sgRNA experiments.
| Targeted region (kb) | F1 with mCherry | F2 with deletion | Ratio (%) | |
|---|---|---|---|---|
| rde-12 | 1 | 46 | 2 | 4.3 |
| linc-22 promoter | 1.9 | 168 | 5 | 3.0 |
| e01g4.5 repeat sequence | 2.1 | 30 | 1 | 3.3 |
| dpy-7 region | 8.5 | 216 | 6 | 2.8 |
| 16.5 | 126 | 1 | 0.8 | |
| lin-15b/a region | 23.7 | 136 | 2 | 1.5 |
Homology-directed repair of double-stranded DNA breaks has been widely used for genome editing. If the donor molecule carries edits flanked by sequences homologous to the targeted locus, the edits will be integrated during the repair process. Both plasmid donors with long homology arms and single-strand oligodeoxynucleotides with short homology arms have been used to insert selection markers to targeted sequences in CRISPR/Cas9 technology29,31,35,36,37,38. In our dual sgRNA system, both methods can also be applied to insert selection markers to targeted sequences, which is beneficial to balance and maintain lethal mutants.
C. elegans expresses many long noncoding RNAs with unknown functions54. Many lincRNAs may serve as templates for endo-siRNA production or regulate the expression of other protein-coding genes. However, a lack of mutants hinders the mechanistic understanding of their function and regulation. Our work demonstrates an easy method with which to knock out lincRNAs using a dual sgRNA/Cas9 system that will facilitate the genetic study of these noncoding genes.
Encoding multiple guide sequences into a single CRISPR experiment to simultaneously edit several sites within the genome has been used to study gene function in mammalian cells8,15,55,56. In addition, the simultaneous use of multiple sgRNAs to target an individual gene has been reported to improve genome-editing efficiency8,20,57,58. Our data suggest that multiplexed guide RNAs may cause the off-target deletion of chromosome sequences in addition to gene editing at the targeted loci. Therefore, independent cell clones and rescue experiments are required to characterize gene function in mammalian cells in this set of experiments.
In summary, our work provides a novel platform with which to use CRISPR/Cas9 technology for gene knockout in C. elegans that has potential applications in other organisms.
Author Contributions
X.C. contributed to Figs. 2, 3, 4e, 4f, s4, and table s1. F.X. generated Figs. 4a, 4b, 4c, and s3. C.Z. generated plasmids for microinjection. J.J. generated Figs. 1c, 1d, 1e, and s2b. X.Z. contributed to Figs. 1b and s1. X.F. and S.G. designed the project and wrote the paper. All authors reviewed the manuscript.
Supplementary Material
supplementary info
Acknowledgments
We are grateful to Drs. Zhong Zhao, Ailong Ke, and Yupeng Yang and members of the Guang lab for their comments. We are grateful to the Caenorhabditis Genetics Center (CGC) for providing the strains. This work was supported by grants from the Chinese 973 Program (No. 2011CBA01100), the National Natural Science Foundation of China (Nos. 31171254 and 31371323), the Fundamental Research Funds for Central Universities (Nos. WK2060190018 and WK2070000034), and KJZD-EW-L01-2.
References
- Geurts A. M. et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325, 433, 10.1126/science.1172447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus M. H. & Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol 23, 967–973, 10.1038/nbt1125 (2005). [DOI] [PubMed] [Google Scholar]
- Wood A. J. et al. Targeted genome editing across species using ZFNs and TALENs. Science 333, 307, 10.1126/science.1207773 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell V. M. et al. In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118, 10.1038/nature11537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T. W. et al. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195, 331–348, 10.1534/genetics.113.155382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C. et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29, 143–148, 10.1038/nbt.1755 (2011). [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P. & Liu J. L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4, 220–228, 10.1016/j.celrep.2013.06.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823, 10.1126/science.1231143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23, 1229–1232, 10.1038/cr.2013.114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T. et al. Genome engineering of mammalian haploid embryonic stem cells using the Cas9/RNA system. PeerJ 1, e230, 10.7717/peerj.230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S. & Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278, 10.1016/j.cell.2014.05.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F. & Marraffini L. A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31, 233–239, 10.1038/nbt.2508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826, 10.1126/science.1232033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389, 10.1016/j.cell.2013.08.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87, 10.1126/science.1247005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H., Redding S., Jinek M., Greene E. C. & Doudna J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67, 10.1038/nature13011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918, 10.1016/j.cell.2013.04.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M. & Lander E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84, 10.1126/science.1246981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol, 10.1038/nbt.2889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A 110, 19012–19017, 10.1073/pnas.1318481110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997, 10.1126/science.1247997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H. & Doudna J. A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338, 10.1038/nature10886 (2012). [DOI] [PubMed] [Google Scholar]
- Jinek M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, 10.1126/science.1225829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P. & Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109, E2579–2586, 10.1073/pnas.1208507109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D. & Joung J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32, 347–355, 10.1038/nbt.2842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Fenk L. A. & de Bono M. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res 41, e193, 10.1093/nar/gkt805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I. & Sternberg P. W. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195, 1167–1171, 10.1534/genetics.113.155879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J. S. & Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195, 1177–1180, 10.1534/genetics.113.155853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J. & Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 10, 1028–1034, 10.1038/nmeth.2641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E. et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10, 741–743, 10.1038/nmeth.2532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y. B. et al. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195, 1181–1185, 10.1534/genetics.113.156075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I. & Grosshans H. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195, 1173–1176, 10.1534/genetics.113.155754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers S. & Boxem M. Engineering the Caenorhabditis elegans genome with CRISPR/Cas9. Methods, 10.1016/j.ymeth.2014.03.024 (2014). [DOI] [PubMed] [Google Scholar]
- Waaijers S. et al. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195, 1187–1191, 10.1534/genetics.113.156299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y. & Xue D. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res 24, 247–250, 10.1038/cr.2014.9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. et al. A Co-CRISPR Strategy for Efficient Genome Editing in Caenorhabditis elegans. Genetics, 10.1534/genetics.114.166389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A. et al. Scalable and Versatile Genome Editing Using Linear DNAs with Micro-Homology to Cas9 Sites in Caenorhabditis elegans. Genetics, 10.1534/genetics.114.170423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A. et al. Efficient Marker-Free Recovery of Custom Genetic Modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics, 10.1534/genetics.114.169730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B. et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11, 399–402, 10.1038/nmeth.2857 (2014). [DOI] [PubMed] [Google Scholar]
- Choi P. S. & Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun 5, 3728, 10.1038/ncomms4728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. Nuclear RNAi contributes to the silencing of off-target genes and repetitive sequences in Caenorhabditis elegans. Genetics 197, 121–132, 10.1534/genetics.113.159780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237, 10.1038/nature01278 (2003). [DOI] [PubMed] [Google Scholar]
- Shirayama M., Stanney W. 3rd, Gu W., Seth M. & Mello C. C. The Vasa Homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Curr Biol 24, 845–851, 10.1016/j.cub.2014.03.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. The DEAD box helicase RDE-12 promotes amplification of RNAi in cytoplasmic foci in C. elegans. Curr Biol 24, 832–838, 10.1016/j.cub.2014.01.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Wang D. & Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645–649, 10.1038/nature02302 (2004). [DOI] [PubMed] [Google Scholar]
- Feng X. & Guang S. Non-coding RNAs mediate the rearrangements of genomic DNA in ciliates. Sci China Life Sci 56, 937–943, 10.1007/s11427-013-4539-4 (2013). [DOI] [PubMed] [Google Scholar]
- Edgley M. et al. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res 30, e52 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Hazendonk E., Thijssen K. L. & Plasterk R. H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet 17, 119–121, 10.1038/ng0997-119 (1997). [DOI] [PubMed] [Google Scholar]
- Ahringer J. Reverse Genetics, The C. elegans Research Community, WormBook WormBook, doi/10.1895/wormbook.1.47.1, http://www.wormbook.org (Apr 6, 2006) (2006).
- Frokjaer-Jensen C. et al. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods 11, 529–534, 10.1038/nmeth.2889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Ailion M. & Jorgensen E. M. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods 9, 117–118, 10.1038/nmeth.1865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C. et al. Targeted gene deletions in C. elegans using transposon excision. Nat Methods 7, 451–453, 10.1038/nmeth.1454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J. W. & Bartel D. P. Long noncoding RNAs in C. elegans. Genome Res 22, 2529–2540, 10.1101/gr.140475.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver M. C. et al. Characterization of Genomic Deletion Efficiency Mediated by Clustered Regularly Interspaced Palindromic Repeats (CRISPR)/Cas9 Nuclease System in Mammalian Cells. J Biol Chem 289, 21312–21324, 10.1074/jbc.M114.564625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T., Nishikawa A., Kume S., Chayama K. & Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep 4, 5400, 10.1038/srep05400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. Dual sgRNAs facilitate CRISPR/Cas9-mediated mouse genome targeting. FEBS J 281, 1717–1725, 10.1111/febs.12735 (2014). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 46, 49–55, 10.1016/j.biocel.2013.10.010 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary info