Abstract
In order to ensure their physiological and cellular functions, plasma membrane (PM) proteins must be properly conveyed from their site of synthesis, i.e., the endoplasmic reticulum, to their final destination, the PM, through the secretory pathway. PM protein homeostasis also relies on recycling and/or degradation, two processes that are initiated by endocytosis. Vesicular membrane trafficking events to and from the PM have been shown to be altered when plant cells are exposed to mutualistic or pathogenic microbes. In this review, we will describe the fine-tune regulation of such alterations, and their consequence in PM protein activity. We will consider the formation of intracellular perimicrobial compartments, the PM protein trafficking machinery of the host, and the delivery or retrieval of signaling and transport proteins such as pattern-recognition receptors, producers of reactive oxygen species, and sugar transporters.
Keywords: plasma membrane, vesicular trafficking, plant–microbe interactions, signaling proteins, sugar transporters
INTRODUCTION
Plants co-exist with a vast diversity of mutualistic or pathogenic microbes such as bacteria, fungi, and oomycetes. Most of the land plants form a mutualistic relationship with symbiotic fungi such as mycorrhizal fungi, and legumes with rhizobia bacteria. Only a small fraction of plant–microbe encounters results in pathogenic relationship.
Indeed, the plants have developed intricate defense mechanisms, referred to as innate immunity, to identify and defend themselves against a wide range of pathogens (Dodds and Rathjen, 2010). The plasma membrane (PM) of plant cells possesses pathogen-recognition receptors (PRR) that recognize pathogen/microbe-associated molecular patterns (P/MAMP), formerly called elicitors, and initiate a pathogen-associated immunity (PTI; Boller and Felix, 2009). PTI, as a basal resistance, limits the invasion and propagation of pathogens. Adapted pathogens are able to suppress PTI by secreting virulence effectors that remain in the apoplast or penetrate the host cell cytoplasm, favoring pathogen survival, and mediating effector-triggered susceptibility. In turn, to confine and eliminate adapted pathogens, plants have evolved resistance (R) proteins that directly or indirectly recognize effectors and thwart pathogen attack. This second line of defense is named effector-triggered immunity (ETI) or R gene-mediated resistance (Win et al., 2012).
Many plant defense responses to infection are shared by PTI and ETI resistance such as protein phosphorylation, changes in ion fluxes, production of reactive oxygen species (ROS), synthesis of antimicrobial compounds, and pathogenesis-related proteins associated with cell wall reinforcement. ETI elicits more prolonged and robust immune responses than PTI, and is more often associated with the hypersensitive cell death that inhibits pathogen growth (Tsuda and Katagiri, 2010).
Accordingly, the plant cell PM contains integral or peripheral proteins that regulate plant biotic interactions by recognizing microbes, activating subsequent signaling cascades, and controlling cellular entrance and exit of molecules.
To reach the PM, proteins travel along the endomembrane system. They are first synthesized in the endoplasmic reticulum (ER) and then transported along the secretory pathway through the Golgi apparatus and the trans-Golgi network (TGN) to be delivered to the PM by exocytosis (Peer, 2011). Once at the PM, proteins remain there or are taken up by endocytosis, and either targeted to the lytic vacuole for degradation, or stored in endocytic compartments and recycled back to the cell surface when needed. The main and best-studied endocytic pathway in plant cells is the clathrin-dependent pathway (Chen et al., 2011). A membrane microdomain-associated endocytic pathway has also been described in plant that may involve specific microdomain proteins such as flotillins (Li et al., 2012).
Protein trafficking through the secretory and endocytic pathways relies on membrane-bound vesicles and a complex set of proteins involved in vesicle formation, transport, docking, and fusion with the respective target membrane (Bonifacino and Glick, 2004). By adjusting vesicle trafficking, plant cells can quickly respond to microbe confrontation. This topic has aroused considerable interest in recent years as shown by numerous reviews (Robatzek, 2007; Ivanov et al., 2010; Leborgne-Castel et al., 2010; Dörmann et al., 2014; Inada and Ueda, 2014; Teh and Hofius, 2014). Here, we review recent progress in our understanding of how both mutualistic and hostile plant–microbe encounters modulate membrane trafficking pathways to reshape the host PM. A special emphasis is placed on membrane trafficking regulators, and on integral PM proteins implicated in cell signaling and nutrient transport.
FORMATION OF PERIMICROBIAL COMPARTMENTS
Plants can establish intracellular associations with mutualistic as well as pathogenic microbes. The intracellular accommodation of microbes allows them to gain access to host nutrients. In a mutualistic relationship, the host cell also benefits from specific metabolites originating from the microbes. Microbes penetrate the cell wall and differentiate specialized intracellular feeding compartments by invagination and extension of the host cell PM to colonize the intracellular space (Figure 1). The perimicrobial compartment refers to the membrane of the host cell at the interface between the host cytoplasm and the microbe. Perimicrobial compartments are highly diverse in shape but their biogenesis relies on similar mechanisms, mainly an intricate balance between endocytosis and exocytosis (Ivanov et al., 2010).
FIGURE 1.
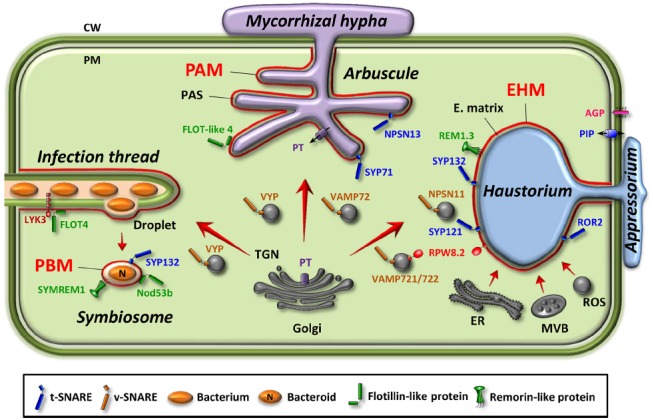
Perimicrobial membranes in three types of plant–microbe interactions. Location of proteins specifically targeted to or excluded from the perimicrobial membranes are indicated. Endomembrane compartments or vesicles involved are illustrated. Small GTPases and exocyst subunits are not represented. AGP, arabinogalactan protein; CW, cell wall; EHM, extrahaustorial membrane; E. matrix, extrahaustorial matrix; ER, endoplasmic reticulum; FLOT, flotillin; LYK3, lysM-family receptor-like kinase from Medicago; MVB, multivesicular body; N, fixed-nitrogen; Nod53b, nodule-specific 53-kDa protein from soybean; NPSN, novel plant SNARE, PAM, periarbuscular membrane; PAS, periarbuscular space; PBM, peribacteroid membrane; PIP, plasma membrane intrinsic protein (aquaporin); PM, plasma membrane; PT, phosphate transporter; REM, remorin; ROR2, required for mlo-specified resistance2 syntaxin; ROS, reactive oxygen species; RPW8.2, resistance powdery mildew8.2; SYMREM, symbiotic remorin1; SYP, syntaxin of plant; TGN, trans-Golgi network; VAMP, vesicle-associated membrane protein; VYP, vapyrin.
PERIBACTEROID MEMBRANE
Mutualistic association of legume plants with rhizobia culminates in the organogenesis of nitrogen-fixing root nodules. Rhizobia are first hosted in an intracellular infection thread (IT) that guides them through the root cell layers. The developing IT results from the local hydrolysis of the plant cell walls and invagination of the PM (Gage, 2004). When the IT reaches the nodule primordium, rhizobia are released from unwalled IT droplets into the plant cell cytoplasm by an endocytosis-like process to form the symbiosome (Figure 1). The symbiosome is an organelle-like compartment surrounded by the peribacteroid membrane (PBM) where rhizobia convert atmospheric nitrogen into ammonia. Numerous symbiosomes occupy each nodule cell; therefore an enormous amount of membrane material must be delivered to the PBM for symbiosome development and maintenance. Vesicle trafficking delivers new lipid and protein materials to the PBM, thus imparting a distinct biochemical identity to this membrane (Ivanov et al., 2010, 2012). In particular, remorin or flotillin proteins, two classes of membrane microdomain-associated proteins, localize to the PBM and are necessary for nodule organization and function (Haney and Long, 2010; Lefebvre et al., 2010; Haney et al., 2011). The actual function of these two classes of proteins remains unclear but they may play a role in IT initiation and elongation, and the entry of rhizobia.
PERIARBUSCULAR MEMBRANE
Mycorrhizal fungi form arbuscular mycorrhiza (AM) associated with plant roots. Intracellular fungal hyphae are enveloped by the periarbuscular membrane (PAM; Figure 1) that corresponds to an ∼10-fold expansion of the host cell PM (Alexander et al., 1989; Bonfante and Genre, 2010). The mechanisms leading to PAM biogenesis have been investigated, showing that pre-penetration responses and symbiotic interface construction are associated with extensive membrane dynamics as clathrin (Genre and Bonfante, 1999) and exocytosis markers accumulate at the site of PAM biogenesis (Genre et al., 2012). Moreover, gene expression reprogramming during AM symbiosis concerns, in addition to nutrient transporters, proteins involved in membrane dynamics, cell wall synthesis, and protein turnover (Guether et al., 2009; Harrison, 2012; Lota et al., 2013), thus suggesting an intense trafficking of cellular membrane components and cell wall precursors to the extending PAM. Recently, a qualitative proteome analysis of the Medicago root upon AM symbiosis evidenced flotillin-like proteins and proteins involved in vesicle trafficking supporting the importance of membrane trafficking events in mycorrhiza establishment and functioning (Abdallah et al., 2014). Although the PAM is continuous with the peripheral host PM, asymmetric distribution of transport proteins has been demonstrated. The prototypal protein of such asymmetric distribution is the phosphate transporter MtPT4 from Medicago that is found only in PAM without trace in the peripheral host PM in developed arbuscules (Harrison et al., 2002). Same focalized localization was observed for OsPT11 in rice (Kobae and Hata, 2010). MtPT4 polar localization results from a transient reorientation of secretion via the ER and TGN compartments (Pumplin et al., 2012).
EXTRAHAUSTORIAL MEMBRANE
During fungi and oomycete pathogen attack, the first responses of the plant cell are cell wall defensive appositions, called papillae, beneath the attempted pathogen penetration site deposited through the action of the secretory pathway and the cytoskeleton reorganization (Hückelhoven et al., 1999). Defense against penetrating pathogens also involves post-Golgi vesicles and multivesicular bodies (MVB)/late endosome for secretion of antimicrobial molecules at the host cell wall penetration site (An et al., 2006).
Adapted pathogens have evolved the ability to breach the host cell wall leading to the development of an infection structure, underneath the appressorium, from which a narrow hyphal strand grows and penetrates the plant cell wall (Koh et al., 2005). The penetration hypha then expands into the host cell cytoplasm and differentiates into a haustorium that allows the pathogen to extract nutrients and secretes effectors to suppress plant defense mechanisms (Figure 1). The haustorium is encased by a specialized membrane, the extrahaustorial membrane (EHM). Haustorium development is associated with extensive dynamics at the EHM and intense exocytosis of proteins and polysaccharides reaching the extrahaustorial matrix via the plant secretory pathway (Mendgen et al., 1995; Meyer et al., 2009). In addition, transmission electron microscopy has revealed swollen tubules and cisternae of plant ER in a very close proximity to EHM (Koh et al., 2005; Micali et al., 2011) that could allow a direct transfer of membrane components to EHM without involvement of the Golgi apparatus (Leckie et al., 1995; Figure 1). EHM is believed to originate from the remodeling of the host cell PM but an alternative hypothesis proposes that EHM is formed via de novo biogenesis (Koh et al., 2005). Many plant PM resident proteins, such as arabinogalactan protein (Micali et al., 2011) or aquaporins, are excluded from the EHM of fungi (Koh et al., 2005; O’Connell and Panstruga, 2006) and oomycetes (Caillaud et al., 2012; Lu et al., 2012). Some proteins, however, are specifically targeted to the mature EHM such as the resistance to powdery mildew8.2 (RPW8.2) in Arabidopsis thaliana (Micali et al., 2011; Wang et al., 2013), and the REM1.3 remorin in Nicotiana benthamiana that promotes susceptibility to Phytophthora infestans (Bozkurt et al., 2014) (Figure 1).
OPEN QUESTIONS
Differentiation of a perimicrobial compartment seems to rely heavily on polarized secretion (Ivanov et al., 2012; Pumplin et al., 2012). Is there an identity checkpoint along the secretory pathway that involves specific chaperones to retain some proteins and let go others such as the periarbuscular phosphate transporters MtPT4 and OsPT11, or the EHM RWP8.2 protein? Is polarized secretion also controlled at the target membrane? What is the role of the scaffolding proteins flotillin and remorin that are known to be associated with sterol- and sphingolipid-enriched microdomains? As focal exocytosis in animal cells depends upon the accumulation of N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs; see below) in PM microdomains (Puri and Roche, 2006; Tsai et al., 2007), it is tempting to speculate that, in plants, a similar phenomenon control the docking and fusion of vesicles loaded with perimicrobial membrane-specific proteins.
Another challenge will be to determine to what extent effectors secreted from mutualistic and pathogenic microbes manipulate the host secretory pathway and reorient secretion toward the perimicrobial membrane. The fact that the oomycete effector AVRblb2 colocalizes with REM1.3 in perihaustorial domains (Bozkurt et al., 2014) suggests that pathogens exploit the lateral organization of the host membrane to accommodate infection structures.
REGULATORS OF VESICLE TRAFFICKING
The transport of membrane components from one compartment to another within the endomembrane system is achieved through the continuous budding, movement and fusion of vesicles. The specificity of vesicular transport is in part ensured by small GTPases of the Rab, ARF (ADP-ribosylation factor; Nielsen et al., 2008), and ROP (Rho of plants)/RAC (Ras-related C3 botulinum toxin substrate; Yalovsky et al., 2008) subfamilies that cycle between an active GTP-bound state associated with membranes and an inactive GDP-bound state present in the cytosol (Rutherford and Moore, 2002). Rab and ARF GTPases work with soluble SNARE proteins to promote membrane fusion (Uemura et al., 2004; Bassham and Blatt, 2008). It is the pairing of complementary SNAREs associated with the vesicle and the target membrane that overcomes the repulsive forces between the two lipid bilayers and drives their fusion. In addition to small GTPases and SNAREs, vesicle tethering factors are necessary for the initial contact and subsequent fusion of the transport vesicle to its target membrane (Yu and Hughson, 2010). One of them is the exocyst, a protein complex composed of eight subunits (SEC3, SEC5, SEC6, SEC8, SEC10, SEC15, EXO70, and EXO84) in Arabidopsis (Zárský et al., 2013) that mediates the docking of exocytic vesicles to the PM in relation with the cytoskeleton (Synek et al., 2014) before SNARE complex formation (Fendrych et al., 2013).
As developed below, all three types of intracellular trafficking regulators have been found to be implicated in plant–microbe interactions.
SMALL GTPases
From the early stages of plant–rhizobial interaction, a major reprogramming of plant genes associated with the initiation of vesicle trafficking is induced that includes Rab and ARF genes and leads to IT development (Peleg-Grossman et al., 2007).
Rab GTPases that are involved in post-Golgi secretion to the PM appear to play a role in plant defense (Speth et al., 2009) and to regulate autophagy and immunity-associated hypersensitive response (Kwon et al., 2013). ARF GTPases have been implicated in preinvasive penetration resistance against powdery mildew in barley (Böhlenius et al., 2010).
ROP/RAC GTPases that control polarized cell growth have been implicated in directing exocytic vesicles via the cytoskeleton to sites of fast membrane expansion during interaction of fungal pathogens with barley (Opalski et al., 2005; Pathuri et al., 2008), Arabidopsis (Poraty-Gavra et al., 2013), and rice (Chen et al., 2010a), or mycorrhizal and oomycete root colonization in barley (Kiirika et al., 2012). A role for ROP GTPases in defense-related endocytic processes can also be suspected from the demonstration of the involvement of clathrin-mediated endocytosis in plant defense (Leborgne-Castel et al., 2010), and ROP GTPases regulation of clathrin-mediated endocytosis in Arabidopsis roots (Chen et al., 2012).
Small GTPases are activated and deactivated through interaction with guanine nucleotide exchange factors (GEFs) that catalyze the exchange of bound GDP with cytosolic GTP, and GTPase activating proteins (GAPs) that enhance GTP hydrolysis (Cherfils and Zeghouf, 2013).
Inhibition of host vesicle-mediated trafficking by targeting activators of GTPase proteins is one strategy of microbes to hinder plant defenses. Indeed, one well-known fungal toxin is brefeldin A, a potent inhibitor of ER-to-Golgi protein transport that targets ARF-GEF proteins (Anders and Jürgens, 2008). The bacterial effector HopM1 from Pseudomonas syringae destabilizes the Arabidopsis ARF-GEF AtMIN7 through the host 26S proteasome that leads to suppression of immunity (Tanaka et al., 2009; Nomura et al., 2011). Interestingly, both the effector and its target have been localized in the TGN suggesting a role for this compartment in defense. A candidate effector from Blumeria graminis was identified as an interactor of a barley ARF-GAP, suggesting interference with defense-associated host vesicle trafficking. A mutation in the orthologous gene in Arabidopsis allows higher entry of powdery mildew agent, Erysiphe pisi, thus confirming that this protein is required for defense against non-adapted fungal pathogens (Schmidt et al., 2014). Interestingly, the same mutant exhibits elevated resistance to the adapted downy mildew pathogen Hyaloperonospora arabidopsidis, thus signifying that a single ARF-GAP may function antagonistically in defense against adapted and non-adapted pathogens.
SNARE PROTEINS
PTI-dependent resistance against pathogenic fungi is characterized by the secretion of antimicrobial components into the plant apoplast, a process that requires the activity of several SNARE proteins. SYP121/PEN1 (for PENETRATION), which is the most studied PM-resident SNARE, plays a pivotal role in cell wall-based resistance against the powdery mildew agent, Blumeria graminis, in Arabidopsis (Collins et al., 2003; Assaad et al., 2004; Zhang et al., 2007; Pajonk et al., 2008), barley (Bhat et al., 2005), and grapevine (Feechan et al., 2013). The potato homolog of SYP121, StSYR1, also participates in the formation of callose-containing papillae for secretory defense responses of potato against the oomycete Phytophthora infestans (Eschen-Lippold et al., 2012). A loss-of-function mutation in the SYP121/PEN1 gene was recently shown to decrease hypersensitive cell death triggered by oomycete and bacterial effectors (Johansson et al., 2014) suggesting that this SNARE is also involved in ETI response.
SYP121/PEN1 forms a ternary SNARE complex with the adaptor protein SNAP33 and the sequence-related vesicle-associated membrane proteins VAMP721/722 at the PM that is necessary for pre-invasive immune responses in barley and Arabidopsis (Kwon et al., 2008). The R-protein RPW8.2, which is specifically targeted to the EHM, is transiently located on VAMP721/722 vesicles before incorporation into the EHM of mature haustoria (Figure 1). This reveals that VAMP721/722 vesicles are not only engaged in pre-invasive defense at the cell periphery but also in post-invasive defense to carry the resistance protein RPW8.2 at the EHM (Kim et al., 2014).
Moreover, the PM SNARE SYP132 has been identified to contribute to defense against bacterial pathogens in N. benthamiana, notably by participating in the secretion of antimicrobial proteins to fight the extracellular infection (Kalde et al., 2007). SYP132 also interacts with VAMP721/722 in the constitutive secretion pathway during Arabidopsis growth. In untreated plants, VAMP721/722 barely accumulate at the PM as they move continuously to and from the PM, and are rapidly degraded by the 26S proteasome (Yun et al., 2013). Upon application of flg22, the bacterium-derived active peptide of flagellin, which induces immune response as well as growth inhibition in plants, VAMP721/722 appears to be protected from proteosomal degradation (Yun et al., 2013). This regulation suggests that growth inhibition during MAMP responses is induced by the reorganization of the secretory pathway for defense. This could open new perspectives to reduce defense-associated fitness costs in crop plants (Sup Yun et al., 2013).
TaNPSN11, a wheat SNARE belonging to the plant-specific novel plant SNARE (NPSN) group (Sanderfoot et al., 2000), has been recently associated with defense against pathogens (Wang et al., 2014). TaNPSN11 is present in vesicular structures near cell membranes (Figure 1) in contact with fungal hyphae when wheat leaves are challenged with the stripe rust agent Puccinia striiformis f. sp. tritici (Wang et al., 2014). TaNPSN11 interacts with TaSYP132 in the TGN and PM, and when downregulated, confers a reduced resistance to an avirulent race of the fungal pathogen. TaNPSN11 thus appears to be involved in vesicle-mediated resistance to stripe rust. NPSN11 was initially described as a component of the fusion machinery involved in cell plate formation in Arabidopsis (Zheng et al., 2002; El Kasmi et al., 2013). The work of Wang et al. (2014) suggests that this plant specific SNARE may have a dual role during cytokinesis and plant-pathogen interactions.
Finally, a recent interactome analysis of SNAREs, using A. thaliana plants expressing GFP-tagged SYP/SNAREs, has identified several disease resistance proteins of the TIR-NBS class as potential interactants (Fujiwara et al., 2014), a result that argues for a role for SNAREs in ETI.
The involvement of PM SNARE proteins is also established for plant-mutualist interactions. Numerous membrane proteins are proposed to be targeted to the symbiosome membrane by SNAREs via the Golgi apparatus (Catalano et al., 2004). Notably, MtSYP132 from Medicago is localized to the IT and infection droplet membranes indicating that this SNARE might have a dual role in IT growth and symbiosome formation (Figure 1; Catalano et al., 2007). In Lotus japonicus, SYP71, which is expressed in vascular tissue, has been shown to play a pivotal role in symbiotic nitrogen fixation in nodules as Ljsyp71 mutant presents nodulation defects (Hakoyama et al., 2012).
Pre-penetration apparatus formation during AM infection is less documented but probably involves the main components of the exocytic machinery, with a major participation of the Golgi apparatus, such as proteins of the VAMP72 family as revealed by both transmission electron microscopy and in vivo GFP imaging (Figure 1; Genre et al., 2012). This data is supported by the recent identification of target-SNAREs (NSPNS13 and SYP71) and vesicle-SNAREs from VAMP72 protein family that is required for arbuscule development (Ivanov et al., 2012; Abdallah et al., 2014). Moreover, a vesicle-associated membrane protein, vapyrin, which contains several ankyrin protein–protein interaction repeat motifs, was shown to be involved in cellular remodeling during AM symbiosis (Pumplin et al., 2010), and also appears to be essential for the rhizobial infection in Medicago truncatula (Murray et al., 2011; Figure 1). The role of vapyrin in membrane trafficking is still questioned. The observation that vapyrin-GFP localizes to mobile puncta in mycorrhizal roots suggests that vapyrin may be associated with vesicles in exocytosis-driven polar tip growth in response to external signals such as Nod or Myc factors (Feddermann et al., 2010; Pumplin et al., 2010).
EXOCYST COMPLEX
Several recent findings accredit the role of the exocyst complex as a new regulator of plant–microbe interactions. Expression of genes encoding exocyst subunits, including EXO70B2, H1, H2, and H7, is highly responsive to different pathogens or elicitor treatment such as flg22 and elf18, the active peptide of the prokaryotic elongation factor EF-Tu, in Arabidopsis (Pečenková et al., 2011). Likewise, cryptogein, a defense elicitor from the oomycete Phytophthora cryptogea, triggers a 10-fold up-regulation of EXOH4 in tobacco cells (Bouhidel, unpublished data). Moreover, accumulation of the exocyst subunit EXO84b was revealed at the site of PAM biogenesis in AM (Genre et al., 2012).
Interestingly, Arabidopsis exo70B2 and exo70H1 mutant lines display a higher susceptibility to Pseudomonas syringae pv. maculicola infection, and exo70B2 mutants present an increase in the proportion of abnormal papillae after infection with Blumeria graminis f. sp hordei (Pečenková et al., 2011). Exo70B2 is also identified as a target of the plant U-box-type ubiquitin ligase 22 (PUB22), which is a negative regulator of PAMP-triggered responses, and it is supposed that PUB22-mediated ubiquitination and degradation of Exo70B2 contributes to the attenuation of PAMP-induced signaling (Stegmann et al., 2012).
In addition, RIN4, a well-known regulator of bacterial-triggered ETI as a guardee interacting with multiple plant R-proteins, has been shown to interact with components of the exocyst complex, raising the hypothesis that RIN4 is involved in defense-associated vesicle trafficking (Afzal et al., 2013).
OPEN QUESTIONS
Compelling progress has been made in recent years in our understanding of the role of membrane trafficking in plant–microbe interactions but we still do have a fragmentary picture of the processes and molecular actors that are involved in the regulation of vesicle trafficking. One should ask whether there is some unity in the mechanisms of vesicular transport. Some studies have already helped to put together some pieces of the puzzle. v-SNAREs of the VAMP72 family have been identified as common symbiotic regulators in exocytotic vesicle trafficking (Ivanov et al., 2012) and also appear to be involved in the exocytotic pathway that leads to EHM biogenesis (Kwon et al., 2008). A ternary SNARE complex between SYP121/PEN1, SNAP33, and VAMP721/722 at the PM has been identified that mediates the pre-invasion resistance against non-adapted pathogens (Kwon et al., 2008). Is there a defense- or a symbiosis-related “trafficking code,” similar to the well-known genetic and histone codes, that associates SNAREs, exocyst subunits and small GTPases to guide the intracellular journey of exocytic and endocytic vesicles during plant–microbe interactions? The identification of interactants of already known regulators may pave the way for a better understanding of the molecular mechanisms that control such membrane trafficking events.
TRAFFICKING OF INTEGRAL PM PROTEINS
Now that small GTPases, SNAREs, and exocyst subunits are established as important trafficking regulators in the plant cell responses to microbes, it is important to have an overview of the subcellular trafficking of mutualism- and defense-associated PM proteins. We describe hereafter selected examples of spatial regulation involving major actors in the responses of plants to microbes such as PRRs, respiratory burst oxygen species homologs (RBOHs), and sugar transporters.
PATTERN RECOGNITION RECEPTORS
The plant cell PM contains a plethora of receptors (PRRs) that sense changes in the biotic environment and trigger specific signaling cascades and physiological outputs inside the cell. PRRs do this by recognizing evolutionarily conserved MAMPs/elicitors or molecules released from damaged plant cells. PRRs are divided into two classes: leucine-rich repeat receptor-like proteins (LRR-RLPs) and receptor-like kinases (LRR-RLKs). Increasing evidence supports the claim that PRRs are part of multi-protein complexes associating regulatory proteins (Monaghan and Zipfel, 2012).
Control of ER exit
The ER is the first compartment of the secretory pathway. It ensures the quality-control process (ER-QC) for folding and maturation of client secretory proteins during and after their biosynthesis (Vitale and Boston, 2008). Many ER proteins such as chaperones involved in protein folding or components of the N-glycosylation pathway, which permit a correct trimming of oligosaccharides on nascent proteins, are major actors in the ER-quality control.
ER-QC dysfunction leads to folding defect of PRRs that affect their quality, accumulation, and/or complex formation at the PM necessary for signaling competency, and consequently for plant resistance to pathogens. The prototypal PRR that has been shown to require such control is EFR, the receptor of EF-Tu in Arabidopsis. A recent review suggests that, owing to the diversity of ER-proteins engaged, the anterograde trafficking of PRR may be a potential point of regulation to generate appropriate defense responses (Tintor and Saijo, 2014).
During symbiosis in M. truncatula, the trafficking of the lysin motif receptor-like kinase NFP, which is required for perception of Rhizobium Nod factors and establishment of symbiosis, is highly sensitive to regulation in the ER. Notably, NFP biological function depends upon the formation of disulfide bridges mediated by the ER protein disulfide isomerases in the receptor extracellular region (Lefebvre et al., 2012).
In addition to the ER-QC components, ER proteins of the reticulon-like protein family (RTNBs), which take part of ER network structuration (Nziengui and Schoefs, 2009), have been shown to interact with the FLAGELIN SENSING 2 receptor (FLS2; Lee et al., 2011). RTNB mutations affect both FLS2 and EFR signaling (Lee et al., 2011; Popescu, 2012) suggesting a role for RTNBs in PRR exit from the ER and trafficking along the secretory pathway.
Recently, ACD6 (ACCELERATED CELL DEATH6), a multipass membrane protein with an ankyrin domain, has been involved in the formation of a complex with FLS2 at the ER to promote FLS2 targeting to the PM and immune response under the signaling defense hormone salicylic acid SA (Zhang et al., 2014).
Endocytosis
Environmental changes could alter the abundance of plant PM proteins by the process of endocytosis (Leborgne-Castel and Luu, 2009). Once removed from the PM, integral proteins are either targeted to the vacuole for degradation or recycled back to the PM. Ubiquitination is used as a sorting signal that directs membrane proteins for degradation. Once internalized, ubiquitinated proteins are delivered via the TGN/early endosome to the MVB/late endosome, where they are recognized by the endosomal sorting complex (ESCRT), and sorted into internal vesicles that are subsequently discharged into the lytic vacuole when the MVB/late endosome fuses with the tonoplast.
Internalization of cell surface receptors, via the clathrin-mediated pathway or any other endocytic pathway, would serve to terminate signaling through degradation, sustain signaling through recycling, or relay signaling inside the cell through the formation of signaling endosomes (Geldner and Robatzek, 2008).
Evidence of MAMP-inducing PRR endocytosis comes from localization studies with the flagellin receptor FLS2 (Robatzek et al., 2006). Following flg22 elicitation, GFP-tagged FLS2 is targeted to a compartment with properties intermediate between the TGN/early endosome and the MVB/late endosome in Arabidopsis (Beck et al., 2012) and N. benthamiana (Choi et al., 2013). Detailed analysis of FLS2 location revealed its presence in the MVB lumen in association with components of the ESCRT-I complex, VPS37-1, and VPS28-2 (Spallek et al., 2013). Interestingly, VPS37-1 and VPS28-2 are required for flg22-induced stomatal closure, but not for a range of other flg22-induced defense responses (i.e., ROS production, kinase activation or callose deposition), linking late endocytic trafficking of FLS2 specifically with defense-associated stomatal closure. These data are consistent with the notion that FLS2 activates separate signaling branches from its targeting to various endomembrane compartments.
Moreover, while FLS2 constitutively recycles in a brefeldin A-sensitive manner, flg22-activated receptors traffic via endosomes insensitive to brefeldin A (Beck et al., 2012). This suggests the existence of two distinct endocytic trafficking routes that depend upon FLS2 activation status and involve distinct subgroups of RabA/11 GTPases (Beck et al., 2012; Choi et al., 2013). In addition, the traffic of de novo-synthesized FLS2 to the PM also involves a distinct RabA/11 subgroup demonstrating that a complex regulatory system exists to properly locate FLS2 (Choi et al., 2013).
Salicylic acid treatment up-regulates the expression of clathrin proteins in maize and Arabidopsis (Pajerowska-Mukhtar et al., 2012; Zeng et al., 2013). Knowing the role of SA in plant immunity, it is tempting to speculate that the expression of these coat proteins is a potential component of plant immunity. Indeed, the clathrin endocytic pathway is activated by the Phytophthora cryptogea elicitor cryptogein in tobacco (Leborgne-Castel et al., 2008) and the Trichoderma viride ethylene-inducing xylanase (EIX) in tomato (Bar and Avni, 2009). In addition, activation of endocytosis may contribute to downstream elicitor signaling (Ron and Avni, 2004; Sharfman et al., 2011; Adam et al., 2012). The EIX receptor LeEIX contains a clathrin endocytic motif, that is also found in some cell surface receptors from fungi (Kawchuk et al., 2001; Fritz-Laylin et al., 2005) or bacteria (Zipfel et al., 2006). By contrast, FLS2 does not contain such motif, but harbors ubiquitination motifs that serve as signals for endocytosis and sorting to the MVBs (Robatzek et al., 2006). The flagellin-triggered endocytosis of FLS2 is altered but not blocked by tyrphostin A23 an inhibitor of the clathrin-mediated endocytosis pathway (Spallek et al., 2013), and is insensitive to SA, which inhibits clathrin-mediated endocytosis of other PM proteins (Du et al., 2013). Therefore, the endocytic pathway that target FLS2 remains to be identified.
RESPIRATORY BURST OXIDASE HOMOLOGS
A rapid and transient production of ROS, the so-called “oxidative burst,” is a hallmark of successful recognition of plant pathogens (Lamb and Dixon, 1997). ROS are known to have antimicrobial activity, to drive cell wall reinforcement, and to act as second messengers in defense-related signaling pathways (Levine et al., 1994). Early ROS production is predominantly apoplastic and notably dependent upon PM-resident NADPH oxidases, also called RBOHs. Plants synthesize several RBOH isoforms with isoform D playing the most prevalent role during biotic stress (Torres et al., 1998; Marino et al., 2011). Owing to their pathosystem-dependent effects on hypersensitive cell death and disease resistance, RBOH-mediated ROS are now conceptualized as mediating agents in signaling pathways with opposite effects on plant defense reactions (Torres et al., 2005; Torres, 2010; Marino et al., 2011). In addition to their role in plant defense, RBOHs have also been described as a major source of ROS required for the establishment of rhizobial root nodules and mycorrhizal symbiosis (Montiel et al., 2012; Puppo et al., 2013; Arthikala et al., 2014).
PM distribution
Proteomic studies have shown that plant RBOHs are present in detergent-insoluble membrane fractions of the PM (Mongrand et al., 2004; Morel et al., 2006; Fujiwara et al., 2009; Stanislas et al., 2009). This suggests that they could be associated in vivo with sterol- and sphingolipid-rich PM microdomains also known as lipid rafts, like their animal counterparts (Jin et al., 2011). In line with this localization is the non-uniform distribution of several RBOHs within the PM of different cell types (Takeda et al., 2008; Liu et al., 2009; Lee et al., 2013; Noirot et al., 2014), and the discretely distributed RBOHD-dependent H2O2 patches observed along the PM of elicited tobacco cells (Lherminier et al., 2009).
Interestingly, RBOHD was recently found to form a complex with either of the PM immune receptors EFR or FLS2, and the receptor-like cytoplasmic kinase BIK1 (Kadota et al., 2014; Li et al., 2014). In response to MAMP elicitation, RBOHD is phosphorylated by BIK1, a post-translational modification required for its function in immunity against bacterial pathogens (Kadota et al., 2014; Li et al., 2014). It is tempting to speculate that the RHBOD-FLS2/EFR-BIK1 complex is part of a lipid raft-associated signaling platform since both RBOHD and FLS2 have already been found in detergent-insoluble membrane fractions (Mongrand et al., 2004; Keinath et al., 2010).
Subcellular dynamics
Reactive oxygen species species have also been detected in vesicles that migrate toward the site of infection. H2O2-containing vesicles accumulate at pathogen-challenge sites in the barley-powdery mildew interaction (Hückelhoven et al., 1999) and their incidence is influenced by mutations in the SNARE protein ROR2 (Collins et al., 2003; Figure 1). In tobacco cells, RBOHD has been found in two distinct intracellular compartments, one of them being the Golgi apparatus (Noirot et al., 2014). The Golgi-localized RBOHD pool is relocalized to the PM after stimulation with the oomycete elicitor cryptogein but prior to any transcriptional upregulation (Noirot et al., 2014). Internal reservoirs thus provide to the PM a supply of fresh enzymes to rapidly replace the PM-resident pool of RBOHD that is turned over following elicitor-induced activation, and to restore its signaling capacities. Recently, the PM dynamics of a GFP-tagged version of RBOHD were analyzed in Arabidopsis using variable-angle total internal reflection fluorescence microscopy (Hao et al., 2014). This study revealed that clathrin- and microdomain-dependent endocytic pathways cooperatively regulate the constitutive internalization of RBOHD. Furthermore, elicitation with flg22 was shown to increase the mobility and clustering of RBOHD, a potential prerequisite for its activation and subsequent internalization by endocytosis (Hao et al., 2014). The mechanism of endocytic turnover of RBOHD following cryptogein or flagellin elicitation remains, however, to be identified.
SUGAR TRANSPORTERS
Obtaining metabolic resources such as sugars from their hosts is a key mechanism for symbiotic and pathogenic microorganisms to survive and reproduce. Access to nutrients depends upon microorganism’s lifestyle. For instance, mutualistic microbes or biotrophic pathogens will either grow in the apoplastic space and/or invade living cells of their hosts to get nutrients from the cytoplasm. There is some evidence suggesting that microbes manipulate the transport machinery of the host PM to increase the efflux of sugar and, that, in return, the host plant attempts to restrict the availability of apoplastic sugars to halt microbe proliferation and disease progression (Ruan, 2014).
The pioneering work of Chen et al. (2010b) has provided the first experimental evidence that bacterial pathogens manipulate the plant sugar transport system to fulfill its nutritional needs. Chen et al. (2010b) identified a new family of sugar uniporters called SWEETs that facilitate sugar efflux into the apoplast. They further showed that Xanthomonas oryzae pv. oryzae (Xoo) upregulates the expression of two SWEET genes in rice, a plant response that results from the direct binding of TAL (transcriptional activator-like) effectors to the SWEET promoters (Chen et al., 2010b). Since then, a third SWEET gene was shown to be induced after Xoo infection (Liu et al., 2011).
Silencing of the N. benthamiana squalene synthase gene, a key enzyme in the phytosterol biosynthesis pathway, results in compromised basal and non-host resistances, events that are correlated with an enhanced efflux of nutrients, mainly sugars, in the apoplastic space (Wang et al., 2012). In this study, it was suggested that alteration of specific PM sterols affects permeability and fluidity of the PM as previously demonstrated in artificial membranes (Schuler et al., 1991). One might imagine that the activity and/or abundance of sugar transporters at the PM are modified by the alteration of PM sterols and fluidity as in animal cells for transmembrane proteins (Bhor and Sivakami, 2003; Levitan et al., 2010), leading to bacterial proliferation.
Depriving microorganisms of the sugars they need by changing sugar fluxes toward host cells appears as a potential defense mechanism in the battle against pathogens. Active resorption of hexoses has been observed in elicited pine suspension cells (Azevedo et al., 2006). Expression of the monosaccharide H+ symporter STP4 is rapidly induced in elicited Arabidopsis suspension cells as well as in plants exposed to fungal pathogens (Truernit et al., 1996). Recently, the sugar transporter STP13 was shown to be induced in Arabidopsis leaves challenged with Botrytis cinerea (Lemonnier et al., 2014). Phenotypic analysis of a knockout mutant and plants constitutively-expressing STP13 revealed that this sugar transporter participates in host intracellular sugar uptake as a basal resistance mechanism against B. cinerea (Lemonnier et al., 2014).
Degradation of sugar efflux carriers after internalization by endocytosis is another potential mechanism to reduce apoplastic sugar concentration. Circumstantial evidence suggests that endocytic degradation of sugar carriers could operate in the defense against pathogens. On the one hand, targeting and cycling of the potato sucrose transporter 1 (SUT1) at the PM are controlled by endocytosis during plant development (Liesche et al., 2010; Krügel et al., 2012). On the other hand, glucose efflux is inhibited after cryptogein elicitation of tobacco cells (Bourque et al., 2002). The mechanism of inhibition is not known but as cryptogein stimulates endocytosis (Leborgne-Castel et al., 2008), it is tempting to speculate that such event may modify the abundance and/or the activity of glucose transporters in such a way that glucose uptake is inhibited.
In AM, the symbiotic fungus offers nutrients, such as phosphorus and nitrogen, to the plant and get, in return, carbohydrates produced by photosynthesis (Harrison, 1999). The tomato SUT2, which is expressed in colonized cells, ensures an essential function for the mycorrhizal symbiosis; the control of sucrose transport back to the plant cytosol to balance the needs of the host and its symbiont (Bitterlich et al., 2014). A recent search for SUT2-interacting proteins has revealed proteins either associated to detergent-insoluble membranes or localized in intracellular vesicles (Bitterlich et al., 2014). This result suggests that SUT2 may be recycled or degraded via a microdomain-associated endocytic pathway. A similar mechanism of internalization has been evidenced in a yeast expression system for the plant sucrose transporter SUT1 (Liesche et al., 2010). The detailed SUT2 targeting pathway to the PAM and its regulation, where active nutrient exchange occurs, remains to be fully investigated.
Interestingly, an RNA-seq transcriptomic approach has reported, in addition to host transporter genes such as SUTs and SWEET1, a highly significant upregulation of remorin genes in ectomycorrhizal interaction (Tarkka et al., 2013). It may suggest that modification of membrane properties by remorins could enhance PM targeting and/or activity of transporters for exporting sugars into the plant apoplast to support fungus growth.
OPEN QUESTIONS
Interestingly, all PM proteins discussed above, and whose intracellular trafficking is modulated upon exposure to microorganisms, have been found in detergent-insoluble membranes or to co-localize with microdomain-associated proteins. Membrane microdomains are thought to temporally and spatially organize proteins and lipids into dynamic signaling complexes (Cacas et al., 2012). Such a functional link between PRR signaling and relocation into membrane microdomains has been suggested for the flagellin receptor FLS2 from proteomic studies (Keinath et al., 2010) but remains to be demonstrated in vivo. Alternatively, protein clustering in microdomains could promote internalization by endocytosis and thus extents signaling intracellularly or ensures its termination. Future studies will have to characterize the dynamics of association of proteins with microdomains at the PM but also in intracellular compartments to figure out the precise role of lateral compartmentalization of membranes in pathogenic or mutualistic interactions.
As endosomal signaling is now an established process in the defense reactions against pathogens, another open question is whether signaling endosomes are genuine targets for effectors produced by adapted pathogens. Recently, the HopW1 effector has been shown to increase the growth of Pseudomonas syringae on Arabidopsis plants through the disruption of the actin cytoskeleton and the inhibition of endocytosis (Kang et al., 2014). We may hypothesize that disruption of actin-dependent endocytosis reduces endosomal signaling (i.e., from PRRs) and thus leads to plant infection. Identification of the intracellular targets of microbial secretory effectomes might shed some lights on this issue.
CONCLUSION
This review highlights the dynamics of the PM as a key process for a better molecular dialog between the plant cell and its biotic environment.
Intracellular colonizers (mutualists or pathogens) trigger a massive remodeling of the host cell PM that leads to the formation, and maintenance, of a perimicrobial membrane with a unique identity. Perimicrobial membranes are characterized by the presence of microdomain-associated proteins of the remorin and flotillin families. These proteins are believed to play a role as molecular scaffolds for the building of signaling platforms. Future should reveal whether they are involved in the polarized delivery of specific PM proteins and/or the endocytic retrieval of others.
Recent studies have identified vesicle trafficking regulators as modulators of the plant response to pathogenic or mutualistic microorganisms. A challenge that lies ahead will be to identify commonalities and differences in the vesicle pathways implicated in the different types of interactions. It will also be important to determine to what extent pathogen effectors affect the accumulation of PM proteins for subversion of plant defense. Deciphering new mechanisms of pathogenicity as well as novel aspects of plant immunity would benefit from the identification of the whole repertoire of protein trafficking components that are targeted by pathogen effectors. The identification of new R-genes should help to design novel strategies for manipulating crop plants toward resistance to ravaging pathogens.
Conflict of Interest Statement
The Guest Associate Editor Daniel Wipf declares that, despite being affiliated to the same institution as the authors, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGMENTS
We thank the French Ministère de l’Enseignement Supérieur et de la Recherche (MESR) and the Institut National de la Recherche Agronomique (INRA) for funding.
Glossary
Abbreviations
- AM
arbuscular mycorrhiza
- ARF
ADP-ribosylation factor
- EFR
elongation factor-TU receptor
- EHM
extrahaustorial membrane
- ER
endoplasmic reticulum
- ETI
effector-triggered immunity
- flg22
22-amino acid sequence of the flagellin
- FLS2
FLAGELIN SENSING 2 receptor
- IT
infection thread
- MAMP
microbe-associated molecular pattern
- MVB
multivesicular bodies
- PAM
periarbuscular membrane
- PAMP
pathogen-associated molecular pattern
- PBM
peribacteroid membrane
- PM
plasma membrane
- PRR
pattern-recognition receptor
- PTI
PAMP-triggered immunity
- RAC
Ras-related C3 botulinum toxin substrate
- RBOH
respiratory burst oxidative homolog
- ROS
reactive oxygen species
- SA
salicylic acid
- SNARE
N-ethylmaleimide-sensitive factor attachment protein receptor
- TGN
trans-Golgi network.
REFERENCES
- Abdallah C., Valot B., Guillier C., Mounier A., Balliau T., Zivy M., et al. (2014). The membrane proteome of Medicago truncatula roots displays qualitative and quantitative changes in response to arbuscular mycorrhizal symbiosis. J. Proteom. 108, 354–368. 10.1016/j.jprot.2014.05.028 [DOI] [PubMed] [Google Scholar]
- Adam T., Bouhidel K., Der C., Robert F., Najid A., Simon-Plas F., et al. (2012). Constitutive expression of clathrin hub hinders elicitor-induced clathrin-mediated endocytosis and defense gene expression in plant cells. FEBS Lett. 586, 3293–3298. 10.1016/j.febslet.2012.06.053 [DOI] [PubMed] [Google Scholar]
- Afzal A. J., Kim J. H., Mackey D. (2013). The role of NOI-domain containing proteins in plant immune signaling. BMC Genomics 14:327. 10.1186/1471-2164-14-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander T., Toth R., Meier R., Weber H. (1989). Dynamics of arbuscule development and degeneration in onion, bean, and tomato with reference to vesicular–arbuscular mycorrhizae in grasses. Can. J. Bot. 67, 2505–2513 10.1139/b89-320 [DOI] [Google Scholar]
- An Q., Hückelhoven R., Kogel K. H., van Bel A. J. (2006). Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell. Microbiol. 8, 1009–1019. 10.1111/j.1462-5822.2006.00683.x [DOI] [PubMed] [Google Scholar]
- Anders N., Jürgens G. (2008). Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65, 3433–3445. 10.1007/s00018-008-8227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthikala M. K., Sánchez-López R., Nava N., Santana O., Cárdenas L., Quinto C. (2014). RbohB, a Phaseolus vulgaris NADPH oxidase gene, enhances symbiosome number, bacteroid size, and nitrogen fixation in nodules and impairs mycorrhizal colonization. New Phytol. 202, 886–900. 10.1111/nph.12714 [DOI] [PubMed] [Google Scholar]
- Assaad F. F., Qiu J. L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., et al. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15, 5118–5129. 10.1091/mbc.E04-02-0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo H., Conde C., Gerós H., Tavares R. M. (2006). The non-host pathogen Botrytis cinerea enhances glucose transport in Pinus pinaster suspension-cultured cells. Plant Cell Physiol. 47, 290–298. 10.1093/pcp/pci248 [DOI] [PubMed] [Google Scholar]
- Bar M., Avni A. (2009). EHD2 inhibits ligand-induced endocytosis and signaling of the leucine-rich repeat receptor-like protein LeEix2. Plant J. 59, 600–611 10.1111/j.1365-313X.2009.03897.x [DOI] [PubMed] [Google Scholar]
- Bassham D. C., Blatt M. R. (2008). SNAREs: cogs and coordinators in signaling and development. Plant Physiol. 147, 1504–1515. 10.1104/pp.108.121129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Zhou J., Faulkner C., MacLean D., Robatzek S. (2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24, 4205–4219. 10.1105/tpc.112.100263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R. A., Miklis M., Schmelzer E., Schulze-Lefert P., Panstruga R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. U.S.A. 102, 3135–3140. 10.1073/pnas.0500012102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhor V. M., Sivakami S. (2003). Regional variations in intestinal brush border membrane fluidity and function during diabetes and the role of oxidative stress and non-enzymatic glycation. Mol. Cell. Biochem. 252, 125–132. 10.1023/A:1025599126840 [DOI] [PubMed] [Google Scholar]
- Bitterlich M., Krügel U., Boldt-Burisch K., Franken P., Kühn C. (2014). The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 78, 877–889. 10.1111/tpj.12515 [DOI] [PubMed] [Google Scholar]
- Böhlenius H., Mørch S. M., Godfrey D., Nielsen M. E., Thordal-Christensen H. (2010). The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 22, 3831–3844. 10.1105/tpc.110.078063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Genre A. (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1, 48. 10.1038/ncomms1046 [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153–166 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Bourque S., Lemoine R., Sequeira-Legrand A., Fayolle L., Delrot S., Pugin A. (2002). The elicitor cryptogein blocks glucose transport in tobacco cells. Plant Physiol. 130, 2177–2187. 10.1104/pp.009449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt T. O., Richardson A., Dagdas Y. F., Mongrand S., Kamoun S., Raffaele S. (2014). The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol. 165, 1005–1018. 10.1104/pp.114.235804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas J. L., Furt F., Le Guedard M., Schmitter J. M., Bure C., Gerbeau-Pissot P., et al. (2012). Lipids of plant membrane rafts. Prog. Lipid Res. 51, 272–299. 10.1016/j.plipres.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Caillaud M. C., Piquerez S. J., Fabro G., Steinbrenner J., Ishaque N., Beynon J., et al. (2012). Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 69, 252–265. 10.1111/j.1365-313X.2011.04787.x [DOI] [PubMed] [Google Scholar]
- Catalano C. M., Czymmek K. J., Gann J. G., Sherrier D. J. (2007). Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta 225, 541–550. 10.1007/s00425-006-0369-y [DOI] [PubMed] [Google Scholar]
- Catalano C. M., Lane W. S., Sherrier D. J. (2004). Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25, 519–531. 10.1002/elps.200305711 [DOI] [PubMed] [Google Scholar]
- Chen L., Shiotani K., Togashi T., Miki D., Aoyama M., Wong H. L., et al. (2010a). Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 51, 585–595. 10.1093/pcp/pcq024 [DOI] [PubMed] [Google Scholar]
- Chen L. Q., Hou B. H., Lalonde S., Takanaga H., Hartung M. L., Qu X. Q., et al. (2010b). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. 10.1038/nature09606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Irani N. G., Friml J. (2011). Clathrin-mediated endocytosis: the gateway into plant cells. Curr. Opin. Plant Biol. 14, 674–682. 10.1016/j.pbi.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Chen X., Naramoto S., Robert S., Tejos R., Löfke C., Lin D., et al. (2012). ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr. Biol. 22, 1326–1332. 10.1016/j.cub.2012.06.032 [DOI] [PubMed] [Google Scholar]
- Cherfils J., Zeghouf M. (2013). Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309. 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- Choi S. W., Tamaki T., Ebine K., Uemura T., Ueda T., Nakano A. (2013). RABA members act in distinct steps of subcellular trafficking of the FLAGELLIN SENSING2 receptor. Plant Cell 25, 1174–1187. 10.1105/tpc.112.108803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N. C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J. L., et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. 10.1038/nature02076 [DOI] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Dörmann P., Kim H., Ott T., Schulze-Lefert P., Trujillo M., Wewer V., et al. (2014). Cell-autonomous defense, re-organization and trafficking of membranes in plant–microbe interactions. New Phytol. 204, 815–822. 10.1111/nph.12978 [DOI] [PubMed] [Google Scholar]
- Du Y., Tejos R., Beck M., Himschoot E., Li H., Robatzek S., et al. (2013). Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc. Natl. Acad. Sci. U.S.A. 110, 7946–7951. 10.1073/pnas.1220205110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi F., Krause C., Hiller U., Stierhof Y. D., Mayer U., Conner L., et al. (2013). SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell 24, 1593–1601. 10.1091/mbc.E13-02-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschen-Lippold L., Landgraf R., Smolka U., Schulze S., Heilmann M., Heilmann I., et al. (2012). Activation of defense against Phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol. 193, 985–996. 10.1111/j.1469-8137.2011.04024.x [DOI] [PubMed] [Google Scholar]
- Feddermann N., Muni R. R., Zeier T., Stuurman J., Ercolin F., Schorderet M., et al. (2010). The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J. 64, 470–481. 10.1111/j.1365-313X.2010.04341.x [DOI] [PubMed] [Google Scholar]
- Feechan A., Jermakow A. M., Ivancevic A., Godfrey D., Pak H., Panstruga R., et al. (2013). Host cell entry of powdery mildew is correlated with endosomal transport of antagonistically acting VvPEN1 and VvMLO to the papilla. Mol. Plant Microbe Interact. 26, 1138–1150. 10.1094/MPMI-04-13-0091-R [DOI] [PubMed] [Google Scholar]
- Fendrych M., Synek L., Pecenková T., Drdová E. J., Sekeres J., de Rycke R., et al. (2013). Visualization of the exocyst complex dynamics at the plasma membrane of Arabidopsis thaliana. Mol. Biol. Cell 24, 510–520. 10.1091/mbc.E12-06-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin L. K., Krishnamurthy N., Tör M., Sjölander K. V., Jones J. D. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138, 611–623. 10.1104/pp.104.054452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Hamada S., Hiratsuka M., Fukao Y., Kawasaki T., Shimamoto K. (2009). Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol. 50, 1191–1200. 10.1093/pcp/pcp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Uemura T., Ebine K., Nishimori Y., Ueda T., Nakano A., et al. (2014). Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 55, 781–789. 10.1093/pcp/pcu038 [DOI] [PubMed] [Google Scholar]
- Gage D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300. 10.1128/MMBR.68.2.280-300.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Robatzek S. (2008). Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol. 147, 1565–1574. 10.1104/pp.108.120287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Bonfante P. (1999). Cytoskeleton-related proteins in tobacco mycorrhizal cells: gamma-tubulin and clathrin localisation. Eur. J. Histochem. 43, 105–111. [PubMed] [Google Scholar]
- Genre A., Ivanov S., Fendrych M., Faccio A., Zársky V., Bisseling T., et al. (2012). Multiple exocytotic markers accumulate at the sites of perifungal membrane biogenesis in arbuscular mycorrhizas. Plant Cell Physiol. 53, 244–255. 10.1093/pcp/pcr170 [DOI] [PubMed] [Google Scholar]
- Guether M., Balestrini R., Hannah M., He J., Udvardi M. K., Bonfante P. (2009). Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytologist. 182, 200–212. 10.1111/j.1469-8137.2008.02725.x [DOI] [PubMed] [Google Scholar]
- Hakoyama T., Oi R., Hazuma K., Suga E., Adachi Y., Kobayashi M., et al. (2012). The SNARE protein SYP71 expressed in vascular tissues is involved in symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Physiol. 160, 897–905. 10.1104/pp.112.200782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C. H., Long S. R. (2010). Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 478–483. 10.1073/pnas.0910081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C. H., Riely B. K., Tricoli D. M., Cook D. R., Ehrhardt D. W., Long S. R. (2011). Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23, 2774–2787. 10.1105/tpc.111.086389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Fan L., Chen T., Li R., Li X., He Q., et al. (2014). Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26, 1729–1745. 10.1105/tpc.113.122358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. J. (1999). Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 361–389. 10.1146/annurev.arplant.50.1.361 [DOI] [PubMed] [Google Scholar]
- Harrison M. J. (2012). Cellular programs for arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 15, 691–698. 10.1016/j.pbi.2012.08.010 [DOI] [PubMed] [Google Scholar]
- Harrison M. J., Dewbre G. R., Liu J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. 10.1105/tpc.004861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R., Fodor J., Preis C., Kogel K. H. (1999). Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119, 1251–1260. 10.1104/pp.119.4.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N., Ueda T. (2014). Membrane trafficking pathways and their roles in plant–microbe interactions. Plant Cell Physiol. 55, 672–686. 10.1093/pcp/pcu046 [DOI] [PubMed] [Google Scholar]
- Ivanov S., Fedorova E., Bisseling T. (2010). Intracellular plant microbe associations: secretory pathways and the formation of perimicrobial compartments. Curr. Opin. Plant Biol. 13, 372–377. 10.1016/j.pbi.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Ivanov S., Fedorova E. E., Limpens E., De Mita S., Genre A., Bonfante P., et al. (2012). Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. U.S.A. 109, 8316–8321. 10.1073/pnas.1200407109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Zhou F., Katirai F., Li P. L. (2011). Lipid raft redox signaling: molecular mechanisms in health and disease. Antioxid. Redox Signal. 15, 1043–1083. 10.1089/ars.2010.3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson O. N., Fantozzi E., Fahlberg P., Nilsson A. K., Buhot N., Tör M., et al. (2014). Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J. 79, 466–476. 10.1111/tpj.12571 [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Kalde M., Nühse T. S., Findlay K., Peck S. C. (2007). The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. U.S.A. 104, 11850–11855. 10.1073/pnas.0701083104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Jelenska J., Cecchini N. M., Li Y., Lee M. W., Kovar D. R., et al. (2014). HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLoS Pathog. 10:e1004232. 10.1371/journal.ppat.1004232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk L. M., Hachey J., Lynch D. R., Kulcsar F., van Rooijen G., Waterer D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 6511–6515. 10.1073/pnas.091114198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath N. F., Kierszniowska S., Lorek J., Bourdais G., Kessler S. A., Asano H., et al. (2010). PAMP-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285, 39140–39149. 10.1074/jbc.M110.160531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiirika L. M., Bergmann H. F., Schikowsky C., Wimmer D., Korte J., Schmitz U., et al. (2012). Silencing of the Rac1 GTPase MtROP9 in Medicago truncatula stimulates early mycorrhizal and oomycete root colonizations but negatively affects rhizobial infection. Plant Physiol. 159, 501–516. 10.1104/pp.112.193706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., O’Connell R., Maekawa-Yoshikawa M., Uemura T., Neumann U., Schulze-Lefert P. (2014). The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J. 79, 835–847. 10.1111/tpj.12591 [DOI] [PubMed] [Google Scholar]
- Kobae Y., Hata S. (2010). Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 51, 341–353. 10.1093/pcp/pcq013 [DOI] [PubMed] [Google Scholar]
- Koh S., André A., Edwards H., Ehrhardt D., Somerville S. (2005). Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 44, 516–529. 10.1111/j.1365-313X.2005.02545.x [DOI] [PubMed] [Google Scholar]
- Krügel U., He H. X., Gier K., Reins J., Chincinska I., Grimm B., et al. (2012). The potato sucrose transporter StSUT1 interacts with a DRM-associated protein disulfide isomerase. Mol. Plant 5, 43–62. 10.1093/mp/ssr048 [DOI] [PubMed] [Google Scholar]
- Kwon C., Neu C., Pajonk S., Yun H. S., Lipka U., Humphry M., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. 10.1038/nature06545 [DOI] [PubMed] [Google Scholar]
- Kwon S. I., Cho H. J., Kim S. R., Park O. K. (2013). The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161, 1722–1736. 10.1104/pp.112.208108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C., Dixon R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. 10.1146/annurev.arplant.48.1.251 [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N., Adam T., Bouhidel K. (2010). Endocytosis in plant–microbe interactions. Protoplasma 247, 177–193. 10.1007/s00709-010-0195-8 [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N., Lherminier J., Der C., Fromentin J., Houot V., Simon-Plas F. (2008). The plant defense elicitor cryptogein stimulates clathrin-mediated endocytosis correlated with reactive oxygen species production in bright yellow-2 tobacco cells. Plant Physiol. 146, 1255–1266. 10.1104/pp.107.111716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leborgne-Castel N., Luu D.-T. (2009). Regulation of endocytosis by external stimuli in plant cells. Plant Biosyst. 143, 630–635 10.1080/11263500903233375 [DOI] [Google Scholar]
- Leckie C. P., Callow J. A., Green J. R. (1995). Reorganization of the endoplasmic reticulum in pea leaf epidermal cells infected by the powdery mildew fungus Erysiphe pisi. New Phytol. 131, 211–221 10.1111/j.1469-8137.1995.tb05722.x [DOI] [Google Scholar]
- Lee H. Y., Bowen C. H., Popescu G. V., Kang H. G., Kato N., Ma S., et al. (2011). Arabidopsis RTNLB1 and RTNLB2 reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 23, 3374–3391. 10.1105/tpc.111.089656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rubio M. C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412. 10.1016/j.cell.2013.02.045 [DOI] [PubMed] [Google Scholar]
- Lefebvre B., Klaus-Heisen D., Pietraszewska-Bogiel A., Hervé C., Camut S., Auriac M. C., et al. (2012). Role of N-glycosylation sites and CXC motifs in trafficking of Medicago truncatula Nod factor perception protein to plasma membrane. J. Biol. Chem. 287, 10812–10823. 10.1074/jbc.M111.281634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B., Timmers T., Mbengue M., Moreau S., Hervé C., Tóth K., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348. 10.1073/pnas.0913320107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonnier P., Gaillard C., Veillet F., Verbeke J., Lemoine R., Coutos-Thevenot P., et al. (2014). Expression of Arabidopsis sugar transport protein STP13 differentially affects glucose transport activity and basal resistance to Botrytis cinerea. Plant Mol. Biol. 85, 473–484. 10.1007/s11103-014-0198-5 [DOI] [PubMed] [Google Scholar]
- Levine A., Tenhaken R., Dixon R., Lamb C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593 10.1016/0092-8674(94)90544-4 [DOI] [PubMed] [Google Scholar]
- Levitan I., Fang Y., Rosenhouse-Dantsker A., Romanenko V. (2010). Cholesterol and ion channels. Subcell. Biochem. 51, 509–549. 10.1007/978-90-481-8622-8_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lherminier J., Elmayan T., Fromentin J., Elaraqui K. T., Vesa S., Morel J., et al. (2009). NADPH oxidase-mediated reactive oxygen species production: subcellular localization and reassessment of its role in plant defense. Mol. Plant Microbe Interact. 22, 868–881. 10.1094/MPMI-22-7-0868 [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Li R., Liu P., Wan Y., Chen T., Wang Q., Mettbach U., et al. (2012). A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 24, 2105–2122. 10.1105/tpc.112.095695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesche J., He H. X., Grimm B., Schulz A., Kühn C. (2010). Recycling of Solanum sucrose transporters expressed in yeast, tobacco, and in mature phloem sieve elements. Mol. Plant 3, 1064–1074. 10.1093/mp/ssq059 [DOI] [PubMed] [Google Scholar]
- Liu P., Li R. L., Zhang L., Wang Q. L., Niehaus K., Baluška F., et al. (2009). Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J. 60, 303–313. 10.1111/j.1365-313X.2009.03955.x [DOI] [PubMed] [Google Scholar]
- Liu Q., Yuan M., Zhou Y., Li X., Xiao J., Wang S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969. 10.1111/j.1365-3040.2011.02391.x [DOI] [PubMed] [Google Scholar]
- Lota F., Wegmüller S., Buer B., Sato S., Bräutigam A., Hanf B., et al. (2013). The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12, a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicus. Plant J. 74, 280–293. 10.1111/tpj.12120 [DOI] [PubMed] [Google Scholar]
- Lu Y. J., Schornack S., Spallek T., Geldner N., Chory J., Schellmann S., et al. (2012). Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell. Microbiol. 14, 682–697. 10.1111/j.1462-5822.2012.01751.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D., Andrio E., Danchin E. G., Oger E., Gucciardo S., Lambert A., et al. (2011). A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189, 580–592. 10.1111/j.1469-8137.2010.03509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendgen K., Bachem U., Starkurnau M., Xu H. (1995). Secretion and endocytosis at the interface of plants and fungi. Can. J. Bot. 73, S640–S648 10.1139/b95-306 [DOI] [Google Scholar]
- Meyer D., Pajonk S., Micali C., O’Connell R., Schulze-Lefert P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57, 986–999. 10.1111/j.1365-313X.2008.03743.x [DOI] [PubMed] [Google Scholar]
- Micali C. O., Neumann U., Grunewald D., Panstruga R., O’Connell R. (2011). Biogenesis of a specialized plant–fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell. Microbiol. 13, 210–226. 10.1111/j.1462-5822.2010.01530.x [DOI] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. 10.1016/j.pbi.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Mongrand S., Morel J., Laroche J., Claverol S., Carde J. P., Hartmann M. A., et al. (2004). Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279, 36277–36286. 10.1074/jbc.M403440200 [DOI] [PubMed] [Google Scholar]
- Montiel J., Nava N., Cárdenas L., Sánchez-López R., Arthikala M. K., Santana O., et al. (2012). A Phaseolus vulgaris NADPH oxidase gene is required for root infection by Rhizobia. Plant Cell Physiol. 53, 1751–1767. 10.1093/pcp/pcs120 [DOI] [PubMed] [Google Scholar]
- Morel J., Claverol S., Mongrand S., Furt F., Fromentin J., Bessoule J.-J., et al. (2006). Proteomics of plant detergent-resistant membrane. Mol. Cell. Proteomics 5, 1396–1411. 10.1074/mcp.M600044-MCP200 [DOI] [PubMed] [Google Scholar]
- Murray J. D., Muni R. R., Torres-Jerez I., Tang Y., Allen S., Andriankaja M., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65, 244–252. 10.1111/j.1365-313X.2010.04415.x [DOI] [PubMed] [Google Scholar]
- Nielsen E., Cheung A. Y., Ueda T. (2008). The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 147, 1516–1526. 10.1104/pp.108.121798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot E., Der C., Lherminier J., Robert F., Moricova P., Kiêu K., et al. (2014). Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J. Exp. Bot. 65, 5011–5022. 10.1093/jxb/eru265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y. N., Imboden L. A., Chang J. H., He S. Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 10774–10779. 10.1073/pnas.1103338108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nziengui H., Schoefs B. (2009). Functions of reticulons in plants: what we can learn from animals and yeasts. Cell. Mol. Life Sci. 66, 584–595. 10.1007/s00018-008-8373-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R. J., Panstruga R. (2006). Tete a tete inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol. 171, 699–718. 10.1111/j.1469-8137.2006.01829.x [DOI] [PubMed] [Google Scholar]
- Opalski K. S., Schultheiss H., Kogel K. H., Hückelhoven R. (2005). The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J. 41, 291–303. 10.1111/j.1365-313X.2004.02292.x [DOI] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K. M., Wang W., Tada Y., Oka N., Tucker C. L., Fonseca J. P., et al. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112. 10.1016/j.cub.2011.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajonk S., Kwon C., Clemens N., Panstruga R., Schulze-Lefert P. (2008). Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J. Biol. Chem. 283, 26974–26984. 10.1074/jbc.M805236200 [DOI] [PubMed] [Google Scholar]
- Pathuri I. P., Zellerhoff N., Schaffrath U., Hensel G., Kumlehn J., Kogel K. H., et al. (2008). Constitutively activated barley ROPs modulate epidermal cell size, defense reactions and interactions with fungal leaf pathogens. Plant Cell Rep. 27, 1877–1887. 10.1007/s00299-008-0607-9 [DOI] [PubMed] [Google Scholar]
- Pečenková T., Hála M., Kulich I., Kocourková D., Drdová E., Fendrych M., et al. (2011). The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 62, 2107–2116. 10.1093/jxb/erq402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W. A. (2011). “Plant cell monographs,” in The Plant Plasma Membrane, eds Murphy A. S., Peer W. A., Schulz B. (Heidelberg: Springer-Verlag; ), 31–56. [Google Scholar]
- Peleg-Grossman S., Volpin H., Levine A. (2007). Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J. Exp. Bot. 58, 1637–1649. 10.1093/jxb/erm013 [DOI] [PubMed] [Google Scholar]
- Popescu S. C. (2012). A model for the biosynthesis and transport of plasma membrane-associated signaling receptors to the cell surface. Front. Plant Sci. 3:71. 10.3389/fpls.2012.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poraty-Gavra L., Zimmermann P., Haigis S., Bednarek P., Hazak O., Stelmakh O. R., et al. (2013). The Arabidopsis Rho of plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiol. 161, 1172–1188. 10.1104/pp.112.213165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Mondo S. J., Topp S., Starker C. G., Gantt J. S., Harrison M. J. (2010). Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61, 482–494. 10.1111/j.1365-313X.2009.04072.x [DOI] [PubMed] [Google Scholar]
- Pumplin N., Zhang X., Noar R. D., Harrison M. J. (2012). Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc. Natl. Acad. Sci. U.S.A. 109, E665–E672. 10.1073/pnas.1110215109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo A., Pauly N., Boscari A., Mandon K., Brouquisse R. (2013). Hydrogen peroxide and nitric oxide: key regulators of the Legume-Rhizobium and mycorrhizal symbioses. Antioxid. Redox Signal. 18, 2202–2219. 10.1089/ars.2012.5136 [DOI] [PubMed] [Google Scholar]
- Puri N., Roche P. A. (2006). Ternary SNARE complexes are enriched in lipid rafts during mast cell exocytosis. Traffic 7, 1482–1494. 10.1111/j.1600-0854.2006.00490.x [DOI] [PubMed] [Google Scholar]
- Robatzek S. (2007). Vesicle trafficking in plant immune responses. Cell. Microbiol. 9, 1–8. 10.1111/j.1462-5822.2006.00829.x [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542. 10.1101/gad.366506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615. 10.1105/tpc.022475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. 10.1146/annurev-arplant-050213-040251 [DOI] [PubMed] [Google Scholar]
- Rutherford S., Moore I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5, 518–528 10.1016/S1369-5266(02)00307-2 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A. A., Assaad F. F., Raikhel N. V. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124, 1558–1569. 10.1104/pp.124.4.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S. M., Kuhn H., Micali C., Liller C., Kwaaitaal M., Panstruga R. (2014). Interaction of a Blumeria graminis f. sp. hordei effector candidate with a barley ARF-GAP suggests that host vesicle trafficking is a fungal pathogenicity target. Mol. Plant Pathol. 15, 535–549. 10.1111/mpp.12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler I., Milon A., Nakatani Y., Ourisson G., Albrecht A. M., Benveniste P., et al. (1991). Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proc. Natl. Acad. Sci. U.S.A. 88, 6926–6930. 10.1073/pnas.88.16.6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharfman M., Bar M., Ehrlich M., Schuster S., Melech-Bonfil S., Ezer R., et al. (2011). Endosomal signaling of the tomato leucine-rich repeat receptor-like protein LeEix2. Plant J. 68, 413–423. 10.1111/j.1365-313X.2011.04696.x [DOI] [PubMed] [Google Scholar]
- Spallek T., Beck M., Ben Khaled S., Salomon S., Bourdais G., Schellmann S., et al. (2013). ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 9:e1004035. 10.1371/journal.pgen.1004035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth E. B., Imboden L., Hauck P., He S. Y. (2009). Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol. 149, 1824–1837. 10.1104/pp.108.132092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislas T., Bouyssie D., Rossignol M., Vesa S., Fromentin J., Morel J., et al. (2009). Quantitative proteomics reveals a dynamic association of proteins to detergent-resistant membranes upon elicitor signaling in tobacco. Mol. Cell. Proteomics 8, 2186–2198. 10.1074/mcp.M900090-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R. G., Ichimura K., Pecenková T., Reuter P., Zársky V., et al. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24, 4703–4716. 10.1105/tpc.112.104463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sup Yun H., Yi C., Kwon H., Kwon C. (2013). Model for regulation of VAMP721/722-mediated secretion: growth vs. stress responses. Plant Signal. Behav. 8, e27116. 10.4161/psb.27116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L., Sekereš J., Zárský V. (2014). The exocyst at the interface between cytoskeleton and membranes in eukaryotic cells. Front. Plant Sci. 4, 543. 10.3389/fpls.2013.00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Gapper C., Kaya H., Bell E., Kuchitsu K., Dolan L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319, 1241–1244. 10.1126/science.1152505 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19, 391–397. 10.1016/j.cub.2009.01.057 [DOI] [PubMed] [Google Scholar]
- Tarkka M. T., Herrmann S., Wubet T., Feldhahn L., Recht S., Kurth F., et al. (2013). OakContigDF159.1, a reference library for studying differential gene expression in Quercus robur during controlled biotic interactions: use for quantitative transcriptomic profiling of oak roots in ectomycorrhizal symbiosis. New Phytol. 199, 529–540. 10.1111/nph.12317 [DOI] [PubMed] [Google Scholar]
- Teh O. K., Hofius D. (2014). Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J. Exp. Bot. 65, 1297–1312. 10.1093/jxb/ert441 [DOI] [PubMed] [Google Scholar]
- Tintor N., Saijo Y. (2014). ER-mediated control for abundance, quality, and signaling of transmembrane immune receptors in plants. Front. Plant Sci. 5:65. 10.3389/fpls.2014.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. A. (2010). ROS in biotic interactions. Physiol. Plant. 138, 414–429. 10.1111/j.1399-3054.2009.01326.x [DOI] [PubMed] [Google Scholar]
- Torres M. A., Jones J. D., Dangl J. L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37, 1130–1134. 10.1038/ng1639 [DOI] [PubMed] [Google Scholar]
- Torres M. A., Onouchi H., Hamada S., Machida C., Hammond-Kosack K. E., Jones J. D. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370. 10.1046/j.1365-313X.1998.00136.x [DOI] [PubMed] [Google Scholar]
- Truernit E., Schmid J., Epple P., Illig J., Sauer N. (1996). The sink-specific and stress-regulated Arabidopsis STP4 gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8, 2169–2182. 10.1105/tpc.8.12.2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P. S., De Vries K. J., De Boer-Brouwer M., Garcia-Gil N., Van Gestel R. A., Colenbrander B., et al. (2007). Syntaxin and VAMP association with lipid rafts depends on cholesterol depletion in capacitating sperm cells. Mol. Membr. Biol. 24, 313–324. 10.1080/09687860701228692 [DOI] [PubMed] [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. 10.1016/j.pbi.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., Sato M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49–65. 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- Vitale A., Boston R. S. (2008). Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9, 1581–1588. 10.1111/j.1600-0854.2008.00780.x [DOI] [PubMed] [Google Scholar]
- Wang K., Senthil-Kumar M., Ryu C. M., Kang L., Mysore K. S. (2012). Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol. 158, 1789–1802. 10.1104/pp.111.189217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang Y., Wen Y., Berkey R., Ma X., Pan Z., et al. (2013). A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. Plant Cell 25, 4242–4261. 10.1105/tpc.113.117226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Deng L., Chang H., Dubcovsky J., Feng H., Han Q., et al. (2014). Wheat TaNPSN SNARE homologues are involved in vesicle-mediated resistance to stripe rust (Puccinia striiformis f. sp. tritici). J. Exp. Bot. 65, 4807–4820. 10.1093/jxb/eru241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win J., Chaparro-Garcia A., Belhaj K., Saunders D. G., Yoshida K., Dong S., et al. (2012). Effector biology of plant-associated organisms: concepts and perspectives. Cold. Spring Harb. Symp. Quant. Biol. 77, 235–247. 10.1101/sqb.2012.77.015933 [DOI] [PubMed] [Google Scholar]
- Yalovsky S., Bloch D., Sorek N., Kost B. (2008). Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 147, 1527–1543. 10.1104/pp.108.122150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I. M., Hughson F. M. (2010). Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26, 137–156. 10.1146/annurev.cellbio.042308.113327 [DOI] [PubMed] [Google Scholar]
- Yun H. S., Kwaaitaal M., Kato N., Yi C., Park S., Sato M. H., et al. (2013). Requirement of vesicle-associated membrane protein 721 and 722 for sustained growth during immune responses in Arabidopsis. Mol. Cells 35, 481–488. 10.1007/s10059-013-2130-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zárský V., Kulich I., Fendrych M., Pečenková T. (2013). Exocyst complexes multiple functions in plant cells secretory pathways. Curr. Opin. Plant Biol. 16, 726–733. 10.1016/j.pbi.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Zeng M. H., Liu S. H., Yang M. X., Zhang Y. J., Liang J. Y., Wan X. R., et al. (2013). Characterization of a gene encoding clathrin heavy chain in maize up-regulated by salicylic acid, abscisic acid and high boron supply. Int. J. Mol. Sci. 14, 15179–15198. 10.3390/ijms140715179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Feechan A., Pedersen C., Newman M. A., Qiu J. L., Olesen K. L., et al. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49, 302–312. 10.1111/j.1365-313X.2006.02961.x [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shrestha J., Tateda C., Greenberg J. T. (2014). Salicylic acid signaling controls the maturation and localization of the Arabidopsis defense protein ACCELERATED CELL DEATH6. Mol. Plant 7, 1365–1383. 10.1093/mp/ssu072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Bednarek S. Y., Sanderfoot A. A., Alonso J., Ecker J. R., Raikhel N. (2002). NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE1. Plant Physiol. 129, 530–539. 10.1104/pp.003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D., Boller T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium mediated transformation. Cell 125, 749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]


