Summary
TGF-β signaling is essential in many processes, including immune surveillance, and its dysregulation controls various diseases, including cancer, fibrosis, and inflammation. Studying the innate host defense, which functions in most cell types, we found that RLR signaling represses TGF-β responses. This regulation is mediated by activated IRF3, using a dual mechanism of IRF3-directed suppression. Activated IRF3 interacts with Smad3, thus inhibiting TGF-β-induced Smad3 activation, and, in the nucleus, disrupts functional Smad3 transcription complexes by competing with co-regulators. Consequently, IRF3 activation by innate antiviral signaling represses TGF-β-induced growth inhibition, gene regulation and epithelial-mesenchymal transition, and the generation of Treg effector lymphocytes from naïve CD4+ lymphocytes. Conversely, silencing IRF3 expression enhances epithelial-mesenchymal transition, TGF-β-induced Treg cell differentiation upon virus infection, and Treg cell generation in vivo. We present a novel mode of regulation of TGF-β signaling by the antiviral defense, with evidence for its role in immune tolerance and cancer cell behavior.
Keywords: TGF-β, innate antiviral host defense, IRF3, Smad, RIG-I-like receptor, epithelial-mesenchymal transition, Treg differentiation
Introduction
The immune system is a central context in which TGF-β controls cell differentiation and function (Li and Flavell, 2008; Yang et al., 2010). For example, TGF-β regulates the activation of naïve T cells following antigen recognition, and their differentiation into effector T cells to combat pathogens. Specifically, TGF-β controls differentiation of regulatory T (Treg) lymphocytes and T helper-17 (Th17) effector T cell subsets, while restricting the generation of Th1 and Th2 cells (Li and Flavell, 2008). In the presence of interleukin (IL)-2, TGF-β induces expression of the transcription factor Foxp3, which drives Treg cell differentiation from naïve T cells (Chen et al., 2003), and exposure to TGF-β with IL-6 induces Th17 cells differentiation (Bettelli et al., 2006).
TGF-β signaling also controls cancer progression by inducing an epithelial plasticity response that often leads to epithelial-mesenchymal transition (EMT) (Ikushima and Miyazono, 2010; Heldin et al., 2012). EMT dissolves epithelial junctions, downregulates epithelial and activates mesenchymal gene expression, and increases motility and invasion. Increased TGF-β signaling and EMT, or at a minimum an epithelial plasticity response, are increasingly also seen as prerequisites in the development of fibrosis (Chapman, 2011).
The different roles of TGF-β derive from the versatility of TGF-β signaling and its regulation by other signaling pathways (Feng and Derynck, 2005; Massagué, 2012; Xu et al., 2012). TGF-β initiates signaling through cell surface complexes of two pairs of transmembrane kinases. Upon ligand binding, the TβRII kinases phosphorylate and induce conformation changes in TβRI, enabling recruitment of Smads and phosphorylation of two C-terminal serines by TβRI. The receptor-activated (R-) Smads then dissociate from the receptors, and form trimers with one Smad4 that translocate into the nucleus, where they activate or repress transcription of target genes through association with high affinity DNA binding transcription factors at regulatory gene sequences, and recruitment of co-activators or co-repressors (Feng and Derynck, 2005; Massagué, 2012). Activation of transcription requires direct R-Smad interactions with the histone acetyltransferases CBP or p300 that are stabilized by Smad4, which consequently also serves as coactivator (Feng et al., 1998; Janknecht et al., 1998; Feng and Derynck, 2005). GRIP1, discovered as a coactivator of the glucocorticoid receptor, also acts as Smad3 coactivator in activating gene responses (Li et al., 2006). The cooperation of Smads with other transcription factors sets the stage for extensive versatility in transcription, and crosstalk with other signaling pathways (Feng and Derynck, 2005), and explains the context-dependent responses of hundreds of target genes (Koinuma et al., 2009). TGF-β also induces non-Smad signaling pathways, such as MAP kinase pathways or the PI3K-Akt-TOR pathway, that target Smad signaling for further regulation and activate non-transcription responses (Derynck and Zhang, 2003; Zhang, 2009).
Metazoans developed innate defense mechanisms to recognize pathogens and defend against infection. Viral double-stranded RNA can be sensed by Toll-like receptors (TLRs) in endosomes or cytoplasmic RIG-I-Like receptors (RLRs) (Akira et al., 2006). Binding of viral dsRNA to these receptors leads to activation of the kinases TBK1 and/or IKKε that C-terminally phosphorylate, and thus activate, the signaling mediator IRF3 (Fitzgerald et al., 2003; Sharma et al., 2003). Following dimerization and nuclear translocation, activated IRF3 acts as DNA-binding transcription factor (Belgnaoui et al., 2011; Kawasaki et al., 2011). TLR and RLR activation by dsRNA also induces the NF-κB pathway. IRF3 and NF-κB then cooperate to activate interferon-β expression, which initiates an antiviral response through IRF7 expression, IRF7 activation by TBK1 or IKKε, and coordinate regulation of IRF7-and IRF3-responsive genes(Belgnaoui et al., 2011).
The transactivation domain of IRF3 has substantial structural similarity with the transactivation domain, i.e. the MH2 domain, of Smads, in organization of α-helices and β-sheets, and three-dimensional structure, albeit much less in sequence; however, IRF3 forms dimers, while Smads form trimers. IRF3 and the related IRF7 also show similarities in activation mechanism with R-Smads (Qin et al., 2003; Takahasi et al., 2003). IRF3 and IRF7 are activated by phosphorylation of multiple C-terminal serines, resulting in reorganization of auto-inhibitory elements and functional unmasking of the transactivation domain (Qin et al., 2003), while R-Smads are activated by phosphorylation of two C-terminal serines (Chacko et al., 2004). These similarities raise the question whether Smads can associate with IRF3 or IRF7, thus enabling functional crosstalk between innate immune signaling through IRF3 and TGF-β signaling through Smad activation.
Here we show that IRF3 activation in response to RLR signaling regulates the activation of Smad signaling in response to TGF-β. We propose a dual mechanism for IRF3-mediated inhibition of Smads, i.e. by preventing association of Smad3 with the TβRI receptor, thus decreasing TGF-β-induced Smad3 activation, and by interfering with functional Smad transcription complexes in the nucleus. The repression of TGF-β signaling by antiviral immune signaling controls TGF-β physiology, apparent by the decreased target gene activation by TGF-β, impaired EMT, and decreased Treg leukocyte differentiation.
Results
RLR signaling suppresses TGF-β-induced Smad responses
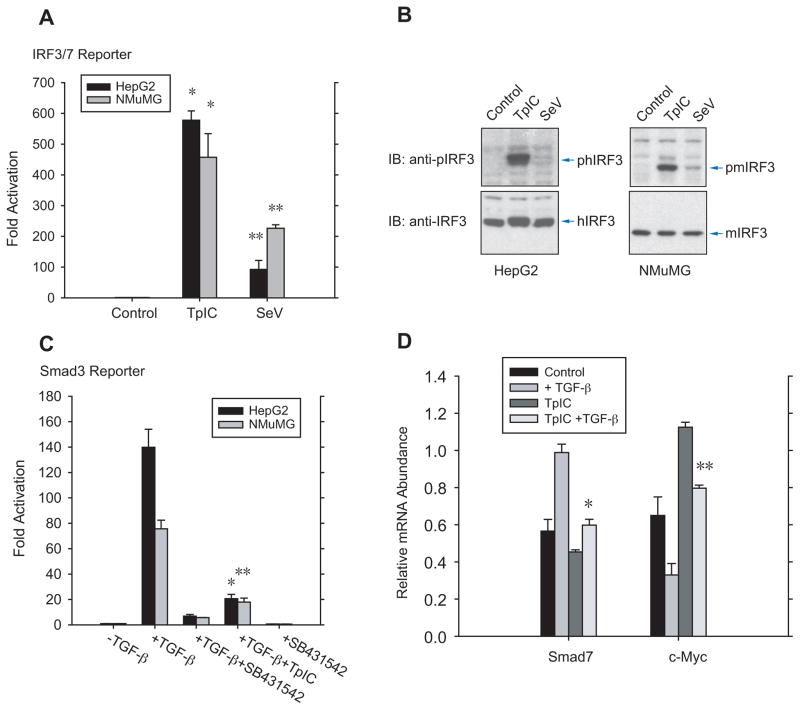
Transfection of double-stranded poly (I:C) RNA, designated TpIC, and infection with Sendai virus (SeV) are commonly used to activate RLR signaling (Belgnaoui et al., 2011; Kato et al., 2011; Kawasaki et al., 2011). To study the effect of RLR signaling on TGF-β signaling, we used human HepG2 hepatoma cells and mouse NMuMG mammary epithelial cells, which are generally used in studies of TGF-β signaling mechanisms. TpIC and SeV induced luciferase expression from an IRF3/7-responsive reporter (Qin et al., 2003), with TpIC acting more strongly than SeV infection (Fig. 1A). Accordingly, TpIC activated IRF3 much more efficiently than SeV, as shown by immunoblotting for C-terminally phosphorylated IRF3 (Fig. 1B). These results indicate that RLR signaling activates IRF3 in epithelial cells. Conversely, SeV infection, but not TpIC, induced IRF3 activation efficiently in human CD4+ cells (Fig. S1A). The lack of IRF3 activation by TpIC in these cells was due to a failure of liposomal poly (I:C) delivery (data not shown). TGF-β induced transcription from a Smad3-responsive promoter (Zhang et al., 1998) in HepG2 and NMuMG cells, which was inhibited by the TβRI kinase inhibitor SB431542 (Laping et al., 2002) (Fig. 1C).
Figure 1. RLR signaling suppresses TGF-β responsiveness.
(A), Transfection of poly (I:C) (TpIC) or Sendai virus (SeV) infection increased transcription from an IRF3/7-responsive promoter in HepG2 and NMuMG cells. N=3 experiments. * and **, P < 0.001, compared with control, by Student’s t-test. (B), Immunoblotting of C-terminally phosphorylated IRF3 and total IRF3 revealed IRF3 activation in response to TpIC or SeV infection in HepG2 or NMuMG cells. Note the slightly decreased electorophoretic mobility of human IRF3, but not mouse IRF3, after C-terminal phosphorylation. (C), TGF-β-induced transcription from a Smad3-responsive promoter in HepG2 or NMuMG cells, and inhibition of this induction by TpIC. SB431542 blocked TGF-β-induced transcription. N=4 experiments. * and **, P< 0.001, compared with control, by Student’s t -test. (D), TpIC blocked TGF-β-induced Smad7 mRNA expression and TGF-β-induced repression of c-Myc mRNA expression in HepG2 cells. TpIC also repressed the basal Smad7 mRNA and enhanced the basal c-Myc mRNA expression under autocrine TGF-β signaling control. mRNA levels were quantified by qRT-PCR. N=3 experiments. *, P < 0.001, and **, P < 0.01, compared with control with TGF-β treatment, by Student’s t-tests.
Combining RLR activation with TGF-β signaling, TpIC inhibited TGF-β/Smad3-induced transcription from a Smad3-dependent reporter (Fig. 1C). Smad signaling directly controls Smad7 and c-Myc expression, with Smad7 mRNA expression increased and c-Myc mRNA expression decreased in response to TGF-β (Nakao et al., 1997; Seoane et al., 2001). TpIC inhibited the induction of endogenous Smad7 mRNA expression, and repression of c-Myc mRNA expression, in response to TGF-β (Fig. 1D). The decreased basal Smad7 mRNA and increased basal c-Myc mRNA expression in response to TpIC (Fig. 1D) are consistent with the autocrine TGF-β signaling control of gene expression in cultured cells.
IRF3 activation represses TGF-β-induced transcription
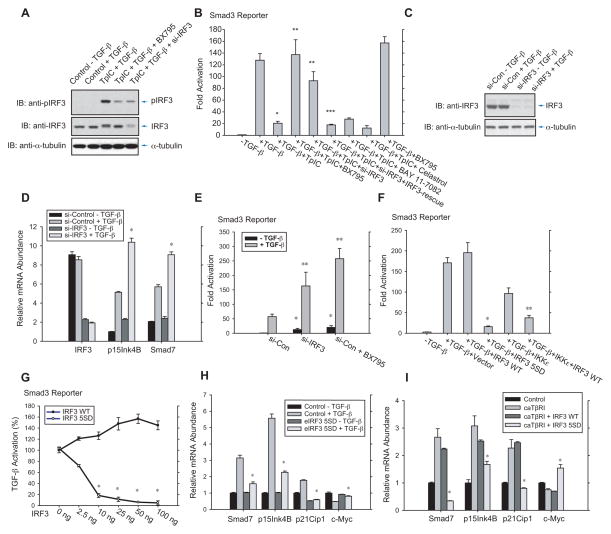
RLR-induced IRF3 activation results from the activities of TBK1 or IKKε that phosphorylate and thus activate IRF3 (Fitzgerald et al., 2003; Sharma et al., 2003). BX795, an inhibitor of TBK1 and IKKε (Clark et al., 2009), inhibited the activation of IRF3 (Fig. 2A, 4th lane), and restored the inhibition of TGF-β-induced transcription by TpIC (Fig. 2B, lanes 3, 4). In contrast, BAY 11-7082 and Celastrol, which inhibit the NF-κB pathway, failed to reverse the TpIC-induced repression of TGF-β/Smad3 signaling (Fig. 2B, lanes 7, 8). These results suggested that RLR signaling inhibits TGF-β/Smad signaling through IRF3 activation. Accordingly, silencing IRF3 expression using siRNA (Fig. 2A, 5th lane) largely rescued the repression of the TGF-β/Smad pathway by RLR activation (Fig. 2B, lanes 3, 5, 6, Fig. S2A).
Figure 2. IRF3 activation controls TGF-β signaling.
(A), Immunoblotting for C-terminally phosphorylated IRF3 revealed IRF3 activation, and inhibition of IRF3 activation by BX795, or siRNA-mediated depletion of IRF3, in HepG2 cells. (B), BX795 and silencing IRF3 expression (si-IRF3), but not NF-κB inhibition by BAY11-7082 or Celastrol, inhibited TGF-β-induced transcription from a Smad3-responsive promoter. N=3 experiments. *, P < 0.001, compared with control with TGF-β treatment; ** and ***, P < 0.001, compared with samples with TGF-β and TpIC treatments, by Student’s t-test. (C)–(E), HaCaT cells, transfected with control or IRF3 siRNA, were treated or not with TGF-β, and subjected to immunoblotting for IRF3 (C), qRT-PCR quantification of IRF3, p15Ink4B or Smad7 mRNA (D), or reporter assay of Smad3-responsive transcription (E). In (D), siRNA to IRF3 mRNA enhanced basal and TGF-β-induced gene expression. N=3 experiments. *, P < 0.01, compared with control siRNA with TGF-β treatment, by Student’s t-test. In (E), TGF-β-induced, Smad3-mediated transcription was enhanced by silencing IRF3 expression, or preventing IRF3 activation by BX795. N=3 experiments. *, P < 0.001, compared with control siRNA without TGF-β treatment; **, P < 0.01, compared with control siRNA with TGF-β treatment, by Student’s t-test. (F), TGF-β-induced transcription is inhibited in HepG2 cells by activated IRF3 5SD, but not wild-type IRF3, or IRF3 activation by IKKε. N=3 experiments. * and **, P < 0.01, compared with control with TGF-β treatment with vector or wild-type IRF3, by Student’s t-test. (G), Dose-dependent inhibition of Smad3-mediated transcription by IRF3 5SD, but not wild-type IRF3. N=3 experiments. *, P < 0.001, compared with wild-type IRF3 with TGF-β treatment by Student’s t-test. (H) and (I), IRF3 5SD, but not wild-type IRF3, suppressed TGF-β-induced activation of Smad7, p15Ink4B and p21Cip1 mRNA expression or repression of c-Myc mRNA expression. In (H), HaCaT cells, stably expressing GFP-tagged IRF3 5SD, were used. In (I), TGF-β signaling was activated in 293T cells by coexpressing activated TβRI (caTβRI). N=3 experiments. *, P < 0.01, compared with control with TGF-β treatment by Student’s t-test.
Evaluating several cell lines, human epithelial HaCaT cells showed constitutive IRF3 activation (Fig. S1B), allowing us to study its control of TGF-β signaling. Transduction of cells with siRNA for IRF3, thus decreasing IRF3 protein (Fig. 2C) and mRNA expression (Fig. 2D), enhanced the TGF-β responsiveness, apparent by increased p15Ink4B and Smad7 mRNA expression from direct TGF-β/Smad3 responsive genes (Nakao et al., 1997; Seoane et al., 2001) (Fig. 2D) and increased transcription from a Smad3-dependent reporter (Fig. 2E). Silencing IRF3 expression also enhanced their basal levels without adding TGF-β (Fig. 2D, 2E), consistent with autocrine TGF-β responsiveness. Blocking IRF3 activation using BX795 similarly increased the basal and TGF-β-induced transcription from the Smad3-responsive luciferase promoter (Fig. 2E).
Our data using BX795 implicated IRF3 activation in the suppression of the TGF-β pathway by RLR signaling. We therefore evaluated, in the absence of RLR activation, the effects of an activated form of IRF3, known as IRF3 5SD, in which the activating phosphorylation of five serines is mimicked by replacing these with aspartic acids (Lin et al., 1998). Wild-type IRF3 did not repress TGF-β-induced, Smad3-dependent activity, but IRF3 5SD expression resulted in >90% inhibition of the TGF-β/Smad response (Fig. 2F; Fig. S1C). Expression of IKKε, which activates IRF3, also inhibited TGF-β-induced transcription, and this inhibition was enhanced when wild-type IRF3 was co-expressed (Fig. 2F). The differential effects of IRF3 5SD versus wild-type IRF3 were striking in a dose-dependent comparison using TGF-β/Smad3-activated transcription as read-out (Fig. 2G). IRF3 5SD repressed transcription at low levels (>80% using 10 ng plasmid DNA), progressing to >97% repression at high levels, and wild-type IRF3 did not repress TGF-β/Smad3 responsiveness even at 100 ng DNA (Fig. 2G).
Finally, we compared wild-type IRF3 and IRF3 5SD for their effects on TGF-β target genes, and measured basal, i.e. autocrine TGF-β-dependent, and TGF-β-induced mRNA expression of Smad7, p15Ink4B and p21Cip1, three direct Smad3 targets that are induced by TGF-β (Feng et al., 2000; Moustakas and Kardassis, 1998; Nakao et al., 1997), and c-Myc, which is directly repressed by Smad3 in response to autocrine or added TGF-β (Seoane et al., 2001). In HaCaT and 293 cells, IRF3 5SD abolished or decreased the TGF-β-induced increase of Smad7, p15Ink4B and p21Cip1, and decrease of c-Myc mRNA expression (Fig. 2H, 2I). In contrast, wild-type IRF3 expression had only a minimal effect (Fig. 2I). These results indicate that IRF3 phosphorylation is critical for repression of Smad-dependent TGF-β activation by innate antiviral signaling.
IRF3 activation represses TGF-β-induced Smad3 activation
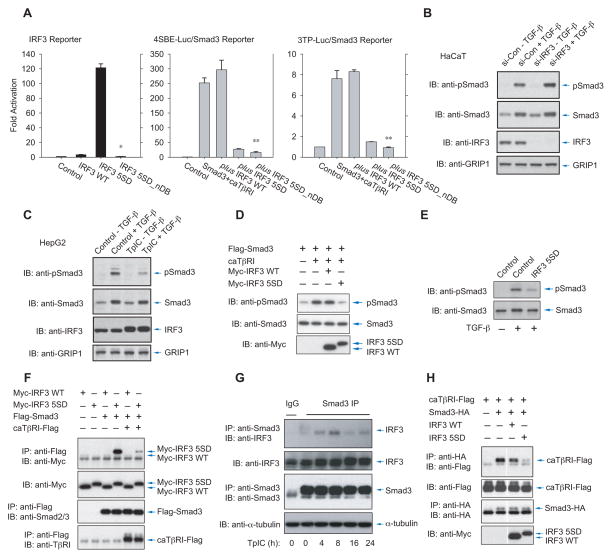
The repression of Smad3-mediated transcription in response to TGF-β by RLR-activated IRF3 might result from gene regulation by IRF3, e.g. from the expression of IRF7 (Marie et al., 1998). However, TpIC did not induce IRF7 expression in HepG2 cells (Fig. S2B), and siRNA to IRF7 did not affect the repression of Smad3-mediated transcription by TpIC (Fig. S2C). Furthermore, TpIC did not affect much the interferon-β mRNA expression, which can be directed by IRF3 (Wathelet et al., 1998) (Fig. S2B). Increased expression of Smad7, an inhibitory Smad that binds TβRI, that can be induced by Jak/STAT signaling (Ulloa et al., 1999) might also inhibit TGF-β-induced Smad3 activation; however, TpIC decreased Smad7 mRNA expression (Fig. 1D). Moreover, a derivative of IRF3 5SD defective in DNA binding, IRF3 5SD_nDB, retained its ability to inhibit TGF-β/Smad3-induced transcription (Fig. 3A, middle, right panels), even though it was transcriptionally inactive (Fig. 3A, left). This result suggested that activated IRF3 itself, and not one or several IRF3 target genes, confers the inhibition.
Figure 3. IRF3 activation enables Smad3 association and inhibits TGF-β-induced Smad3 activation.
(A), Activated IRF-3 (IRF3 5SD), but not its derivative that lacks DNA binding (IRF3 5SD_nDB), activates an IRF3-responsive transcription reporter (left), yet both IRF-3 mutants block Smad3-mediated transcription from the 4SBE (middle) or 3TP (right) reporter. N=3 experiments. *, P < 0.001, compared with IRF3 5SD; **, P < 0.001, compared with IRF3 WT coexpression, by Student’s t-test. (B), Depletion of IRF3 expression using IRF3 siRNA enhanced TGF-β-induced Smad3 activation in HaCaT cells. Smad3 activation was shown by immunoblotting of phospho-Smad3 (top panel) versus total Smad3 in the nuclear fraction (second panel). IRF3 in the nuclear fraction was detected by anti-IRF3 immunoblotting (third panel), and GRIP1 levels (lowest panel) served as loading control of nuclear fraction proteins. (C), TpIC induced a decrease in Smad3 activation in HepG2 cells, assessed by immunoblotting for phospho-Smad3 (top panel) versus total Smad3 (second panel) in the nuclear fraction, and increased IRF3 accumulation in the nuclear fraction. GRIP1 levels (lowest panel) served as loading control of nuclear proteins. (D) and (E), Activated IRF3 5SD, but not wild-type IRF3, reduced Smad3 activation, assessed by immunoblotting for phospho-Smad3, in 293T cells expressing activated TβRI (caTβRI) (D), or HaCaT cells treated with TGF-β (E). (F), Flag-tagged Smad3 associated with Myc-tagged activated IRF3 (IRF3 5SD), but not Myc-tagged wild-type IRF3 in transfected 293T cells. This association was reduced when Smad3 was activated by caTβRI. (G), Time-dependent association of endogenous Smad3 with endogenous IRF3 in HepG2 cells that were treated with TpIC. (H), Activated, but not wild-type IRF3 interfered with Smad3 association with caTβRI, shown by coimmunoprecipitation of HA-tagged Smad3 with Flag-tagged caTβRI.
The inhibition of TGF-β-induced transcription by activated IRF3 might be due to either direct repression of Smad3 activation, or inhibition of Smad3-mediated transcription. We first explored whether IRF3 activation decreased the C-terminal phosphorylation of Smad3 by TβRI. Silencing IRF3 expression in HaCaT cells with their constitutive IRF3 activation enhanced TGF-β-induced Smad3 activation, determined by immunoblotting of nuclear phospho-Smad3 (Fig. 3B). Conversely, treatment of HepG2 cells with TpIC decreased TGF-β-induced Smad3 activation, concomitantly with activation of IRF3, apparent from its slower migration on gel (Fig. 3C) or immunoblotting for phospho-IRF3 (Fig. 1B, 2A). Since IRF3 activation correlated with decreased Smad3 activation and Smad3-mediated transcription (Fig. 1C, 2B, 2F, and 3C), we compared the effects of wild-type IRF3 and IRF3 5SD on Smad3 activation. Transfected wild-type IRF3 did not affect Smad3 activation in 293T cells, but similar levels of activated IRF3 5SD strongly inhibited Smad3 activation (Fig. 3D, S3A), not requiring IRF3’s DNA binding domain (Fig. S3A). Additionally, activated IRF3 5SD repressed TGF-β-induced Smad3 activation in HaCaT cells (Fig. 3E), and TpIC induced wild-type IRF3, but not a mutant IRF3 SA that cannot be activated, to repress Smad3 activation (Fig. S3B). These results correlate IRF3 activation with repression of Smad3 activation.
The transactivation domain of IRF3, which in its non-activated form is masked by an auto-inhibitory conformation (Qin et al., 2003), and mediates IRF3 dimerization (Takahasi et al., 2003), structurally resembles the MH2 transactivation domain, which mediates Smad trimerization (Chacko et al., 2004). Additionally, IRF7 can associate with Smad3 through its MH2 domain (Qing et al., 2004). Given the striking similarities between IRF3 and Smad3 in structure and activation through C-terminal phosphorylation (Qin et al., 2003; Takahasi et al., 2003), we hypothesized that IRF3 activation may inhibit Smad3 activation by TβRI through association with Smad3. Indeed, when coexpressed with Smad3, activated IRF3 5SD, but not wild-type IRF3, associated with Smad3 (Fig. 3F, lanes 3 vs. 4). This occurred predominantly in the cytoplasm, but was also seen in the nucleus (Fig. S3C). Activated IRF3 5SD did not interact with Smad4, while wild-type IRF3 did (Fig. S3D). Furthermore, TpIC promoted cytoplasmic IRF3 association with Smad3 (Fig. S3E) and dissociation from Smad4 (Fig. S3D) in transfected cells, and association of endogenous IRF3 and Smad3 in HepG2 cells (Fig. 3G). However, TβRI activation, which confers Smad3 activation, reduced the IRF3 5SD association with Smad3 (Fig. 3F, lanes 4 vs 6, Fig. S3F). These results strongly indicate that, upon activation, IRF3 interacts with non -activated Smad3.
Association of activated IRF3 with non-activated Smad3 (Fig. 3F) explains the decreased Smad3 phosphorylation upon IRF3 activation (Fig. 3C–E). Since Smad3 activation results from transient interaction with TβRI, which cannot be visualized at endogenous levels, we compared the effects of wild-type or activated IRF3 on the interaction of tagged Smad3 with activated TβRI. Activated IRF3 5SD, but not wild-type IRF3 or IRF3 SA, which cannot be activated, strongly decreased the association of Smad3 with the receptor (Fig. 3H, S3G, S3H). Enhanced Smad3-TβRI association in the presence of IRF3 SA (Fig. S3H) may reflect interference with autocrine inhibition by endogenous IRF3. Collectively, these data show that IRF3 activation controls Smad3 phosphorylation, through association of activated IRF3 with non-activated Smad3, thus interfering with the Smad3 interaction with TβRI that is required for TGF-β-induced Smad activation.
Activation of IRF3 disrupts the Smad3 transcription complex
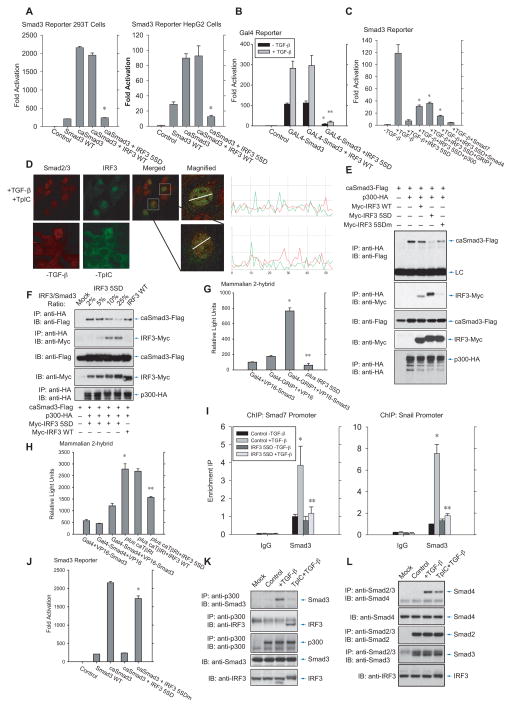
Since activated IRF3 was also seen to associate with Smad3 in the nucleus (Fig. S3C), we evaluated whether IRF3 disrupts the Smad3 transcription complex. For this purpose, we used an activated Smad3 with the C-terminal serine phosphorylation mimicked through substitution for glutamic acid (Chipuk et al., 2002). In reporter assays, IRF3 5SD, but not wild-type IRF3, repressed transcription directed by activated Smad3 (Fig. 4A), or Smad3 fused to a Gal4 DNA binding domain at a Gal4 sequence-controlled promoter (Fig. 4B). Enhanced expression of the Smad coactivators Smad4, p300 or GRIP1 partially rescued the repression of Smad3-mediated transcription by activated IRF3 (Fig. 4C), supporting the notion that in these assays the repression by activated IRF3 results from inhibition of the transcription complex.
Figure 4. IRF3 activation decreases Smad3 transcription complex formation and promoter binding.
(A), IRF3 5SD, but not wild-type IRF3, inhibits transcription induced by constitutively activated Smad3 (caSmad3) in 293T cells (left) or HepG2 cells (right). N=3 experiments. *, P < 0.001, compared with control with caSmad3 expression, by Student’s t-test. (B), IRF3 5SD, but not wild-type IRF3, inhibits Gal4-responsive transcription from a nuclear Gal4 DNA binding domain-fused Smad3 (Gal4-Smad3) in HepG2 cells. N=3 experiments. *, P < 0.001, compared with control Gal4-Smad3 expression, and **, P < 0.001, compared with Gal4-Smad3 expression and TGF-β treatment, by Student’s t-test. (C), Inhibition of Smad3-mediated transcription by activated IRF3 5SD in HepG2 cells is partially rescued by expression of the coregulators p300, GRIP1 or Smad4. N=3 experiments. *, P < 0.01, compared with TGF-β treatment and IRF3 5SD coexpression. (D), Subnuclear distribution, assessed by confocal immunofluorescence, of endogenous IRF3 and Smad3 in HepG2 cells, treated with TGF-β and TpIC to activate both IRF3 and Smad3. The punctate IRF3 and Smad3 patterns showed minimal overlap, as illustrated with Imageplus analysis. (E), Activated, Flag-tagged Smad3 (caSmad3) associated with HA-tagged p300 in 293T cells, and this interaction was disrupted by activated IRF3 5SD, but not by wild-type IRF3 or the basic patch mutant of IRF3 5SD (IRF3 5SDm) (top panel). Under these conditions, activated IRF3 5SD, but not wild-type IRF3 or IRF3 5SDm, interacted strongly with p300 (2nd panel). (F), Increasing levels of IRF3 5SD disrupt the interaction of Flag-Smad3 with HA-p300 in 293T cells, with parallel formation of an IRF3 5SD complex with p300. (G), IRF3 5SD disrupts the interaction of VP16-fused Smad3 (VP-Smad3) with Gal4 DNA binding domain-fused GRIP1 (Gal4-GRIP1) in HepG2 cells. (H), Activated TβRI promotes the interaction of VP-Smad3 with Gal4 DNA binding domain-fused Smad4 (Gal4-Smad4) in HepG2 cells, and this increase is prevented by activated IRF3 5SD, but not wild-type IRF3. For (H) and (I), N=3 experiments;*, P < 0.01, compared with both controls, **, P < 0.01, compared with coexpression of fusion proteins, by Student’s t-test. (I), CHIP assay revealed that IRF3 5SD largely abolishes the TGF-β-induced Smad3 association with Smad7 (left) or Snail (right) promoter sequences in NMuMG cells. N=3 experiments. *, P < 0.01, compared with control without TGF-β treatment, and **, P < 0.01, compared with control with TGF-β treatment but without IRF3 5SD coexpression, by Student’s t-test. (J), Activated IRF3 5SD, but not its basic patch mutant IRF3 5SDm, inhibited Smad3-mediated transcription by activated Smad3 (caSmad3). (K) and (L), TGF-β-induced association of endogenous Smad3 with p300 (K) or Smad4 (L) in HepG2 cells is disrupted (K) or decreased (L) upon IRF3 activation in response to TpIC, concomitant with endogenous p300-IRF3 complex formation (K).
Visualizing the subcellular localization of endogenous Smad3 and IRF3, combined treatment with TGF-β and TpIC did not induce colocalization of activated Smad3 with activated IRF3 in a punctate pattern (Fig. 4D). This is consistent with our observations that Smad3 activation by TβRI strongly decreased the association of activated IRF3 with non-activated Smad3 (Fig. 3F, lanes 4 and 6), and suggests that no complexes of activated IRF3 with activated Smad3 are formed.
Since Smad4, p300 and GRIP1 partially rescued the inhibition of Smad3-mediated transcription (Fig. 4C), we explored whether IRF3 activation disrupts the Smad3 transcription complex by displacing these coactivators. Shown by coimmunoprecipitation, IRF3 5SD interfered with the Smad3-p300 association (Fig. 4E, F, top panels), whereas wild-type IRF3 or IRF SA had much less effect (Fig. 4E; S4A). While decreasing the Smad3-p300 interaction, activated IRF3 interacted with p300 (Fig. 4E, F, second panels, S4B, S4C), consistent with the roles of CBP and p300 as IRF3 coactivators (Wathelet et al., 1998; Weaver et al., 1998), and the structure of this interface (Qin et al., 2005). Similarly to the Smad3-p300 interaction, activated IRF3 5SD, but not wild-type IRF3, interfered with the association of Smad3 with GRIP1 (Fig. 4G) or Smad4 (Fig. 4H), shown in two-hybrid assays. Finally, IRF3 5SD decreased the TGF-β-induced interaction of Smad3 at endogenous Smad7 and Snail promoter regulatory sequences (Fig. 4I), further showing that IRF3 activation impacts the functional integrity of the Smad3 transcription complex.
The interference of activated IRF3 5SD with Smad3-coactivator interactions likely involves a basic amino acid patch in IRF3’s transactivation domain that provides a structural interface with several coregulators (Qin et al., 2003; Takahasi et al., 2003). Indeed, replacement of four basic amino acids in this patch with alanines, thus generating IRF3 5SDm, abolished the inhibition of activated Smad3-mediated transcription by IRF3 5SD (Fig. 4J, last two lanes), without affecting its ability to inhibit Smad3 recruitment to TβRI (Fig. S4D) or Smad3 activation (Fig. S4E), and decreased the interaction of IRF3 5SD with p300 (Fig. 4E, compare last two lanes).
Finally, we evaluated the effect of TpIC on the integrity of the Smad3 transcription complex. As reported (Feng et al., 1998; Janknecht et al., 1998), TGF-β induced the interaction of Smad3 with Smad4 and with p300. TpIC treatment decreased the TGF-β-induced Smad3 association with p300 (Fig. 4K, top panel), concomitantly with association of IRF3 with p300 (Fig. 4K, second panel), and of Smad3 with Smad4 (Fig. 4L).
Together, these results illustrate that activation of IRF3 directly impacts the integrity of the functional Smad3 nucleoprotein complex, required for TGF-β-induced transcription activation, through interference with Smad3-coactivator interactions.
IRF3 activation represses TGF-β-induced EMT
Since IRF3 controls TGF-β-induced gene expression, we evaluated whether IRF3 activation regulates TGF-β-induced EMT. HaCaT cells transition into a mesenchymal phenotype in response to TGF-β, with increased expression of Slug, a Snail-related transcription factor that drives EMT, dispersion of E-cadherin from junctions, decreased epithelial and increased mesenchymal gene expression, and changes in actin organization and cell shape (Lamouille and Derynck, 2007; Thuault et al., 2006). TGF-β-activated Smad3 directly controls Slug and several other EMT-regulated genes (Brandl et al., 2010; Xu et al., 2009). In contrast to many cell lines, HaCaT cells do not repress E-cadherin expression during EMT (data not shown).
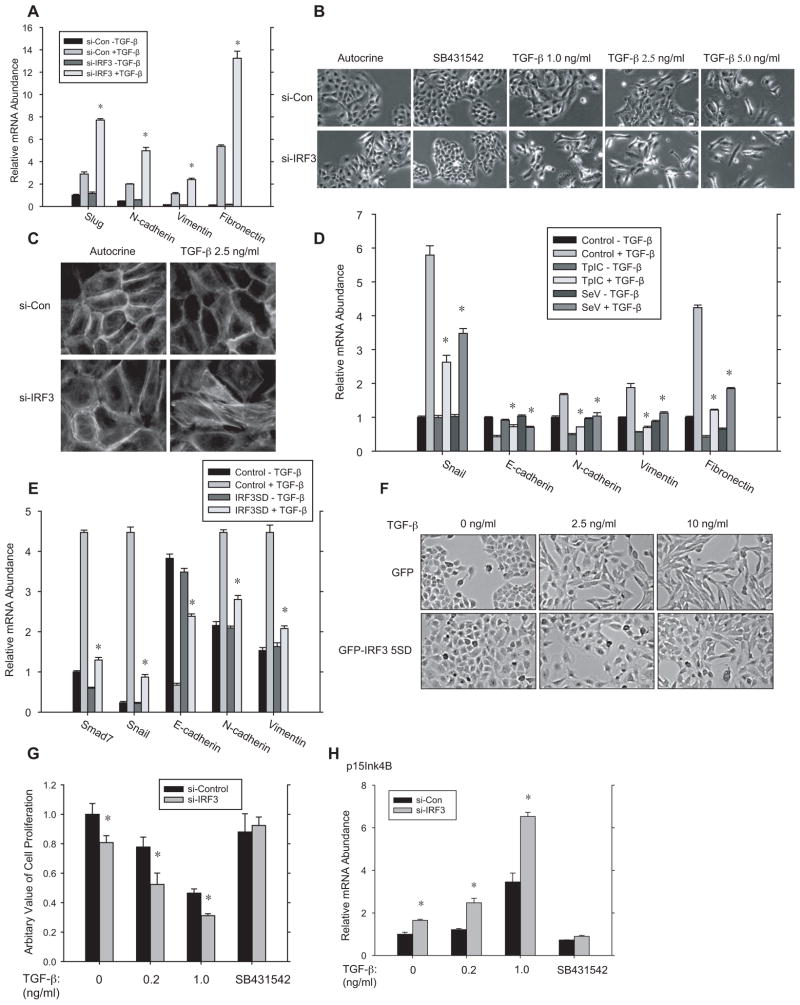
Compared to control HaCaT cells, downregulation of IRF3 expression enhanced the TGF-β-induced mesenchymal expression of Slug, fibronectin, N-cadherin and vimentin (Fig. 5A), and the responsiveness to TGF-β-induced morphology changes Without adding TGF-β, control HaCaT cells had an epithelial cobblestone-like morphology, and increasing TGF-β levels resulted in loss of the epithelial phenotype and change toward an elongated cell shape (Fig. 5B). Cells with silenced IRF3 expression acquired a fibroblast phenotype at lower TGF-β concentrations than control HaCaT cells (Fig. 5B), and showed less cortical actin and more stress fiber formation (Fig. 5C), indicating a more robust EMT response to TGF-β when IRF3 signaling is silenced.
Figure 5. IRF3 controls TGF-β-induced epithelial-mesenchymal transition and growth inhibition.
(A)–(C), HaCaT cells, transfected with control or IRF3 siRNA, were treated or not with TGF-β to induce EMT. (A) qRT-PCR quantification of Slug, N-cadherin, vimentin and fibronectin mRNA at 2 h after adding TGF-β. N=3 experiments. *, P < 0.01, compared with control siRNA, by Student’s t-test. (B), Changes in cell morphology show transition from a cuboidal epithelial to an elongated mesenchymal appearance in response to increasing TGF-β levels for 48 h. Suppressed IRF3 expression using siRNA allows for EMT at lower TGF-β levels. (C), F-actin staining in HaCaT cells treated with 2.5 ng/ml TGF-β for 48 h. At this concentration, control HaCaT cells showed cortical actin organization, characteristic of epithelial cells, whereas suppression of IRF3 expression allowed actin reorganization into stress fibers, as seen in mesenchymal cells. (D), NMuMG cells, exposed or not to TpIC or SeV, were treated or not with TGF-β to induce EMT. qRT-PCR quantified Snail, E-cadherin, N-cadherin, and vimentin mRNA at 2 h after adding TGF-β. N=3 experiments. *, P < 0.01, compared with samples treated with TGF-β only, by Student’s t-test. (E) and (F), NMuMG cells expressing activated IRF3 5SD were analyzed for EMT marker mRNA expression at 2 h after adding TGF-β (E), or for cell morphology at 24 h after adding TGF-β (F). IRF3 5SD expression renders the cells less sensitive to EMT. (G)–(H), Silencing IRF3 expression enhances TGF-β-induced growth inhibition (G) and p15Ink4B mRNA expression (H) in HaCaT cells. In (G), cell proliferation was assessed by BrdU incorporation after 48 h of TGF-β treatment, normalized to untreated, control cells. N=3 experiments. *, P < 0.05, compared with control cells treated with the same TGF-β concentration, by Student’s t-test. In (H), p15Ink4B mRNA was quantified by qRT-PCR after 72 h of TGF-β treatment, and mormalized to untreated control cells. P < 0.05, compared with control cells, by Student’s t-test.
Consistent with the effect of silencing IRF3 expression in HaCaT cells, RLR signaling in response to TpIC or SeV infection (Fig. 5D), or activated IRF3 5SD expression (Fig. 5E) attenuated the EMT responses in NMuMG cells. Thus, TpIC and SeV attenuated the decrease in E-cadherin, and increases in Snail, N-cadherin and vimentin expression that accompany EMT in NMuMG cells (Fig. 5D), and the change in cell morphology toward an elongated spindle phenotype (Fig. 5F). For example, at 10 ng/ml TGF-β NMuMG cells had a spindle cell phenotype, whereas IRF3 activation enabled the cells to largely maintain their cuboidal epithelial phenotype. These data demonstrate that IRF3 activation controls the TGF-β-induced EMT response in HaCaT and NMuMG cells.
IRF3 signaling controls the growth inhibitory effect of TGF-β
Since TGF-β also inhibits epithelial cell proliferation, we addressed the effect of IRF3 activation on the growth inhibitory response of TGF-β. Silencing IRF3 expression enhanced the inhibition of HaCaT cell proliferation by TGF-β (Fig. 5G), and, the expression of the cdk inhibitor p15Ink4B (Fig. 5H). These effects of IRF3 siRNA were already seen without adding TGF-β, suggesting either regulation of autocrine TGF-β signaling by IRF3, or a direct effect of IRF3 on growth control, as proposed (Kim et al., 2007). Since IRF3 siRNA did not affect basal cell proliferation (Fig. 5G) or basal p15Ink4B mRNA expression (Fig. 5H) in the presence of the TβRI kinase inhibitor SB431542, we conclude that IRF3 controls growth inhibition through its control of TGF-β signaling.
RLR-IRF3 activation regulates Treg lymphocyte differentiation
TGF-β controls the differentiation of multiple immune cell lineages. Notably, TGF-β induces Treg cell differentiation from naïve CD4+ T cells, marked by the expression of Foxp3, which drives Treg cell differentiation (Li and Flavell, 2008; Yang et al., 2010). Since antiviral signaling mobilizes the immune response through interferon induction and inflammation, we evaluated whether RLR signaling through IRF3 affects induced (i)Treg lymphocyte development.
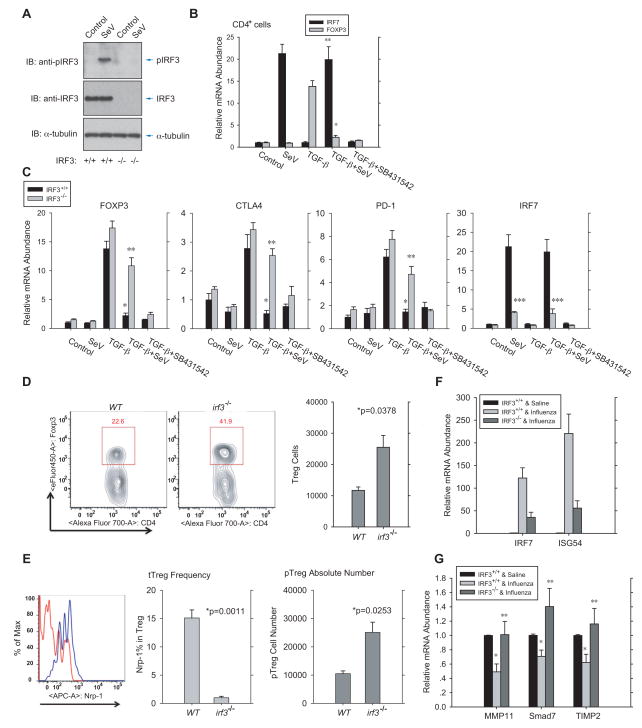
As in primary human CD4+ cells (Fig. S5A), SeV infection elicited robust IRF3 activation in primary mouse CD4+ T cells, assessed by immunoblotting for C-terminally phosphorylated IRF3 (Fig. 6A). We activated TCR signaling in naïve CD4+ T cells using anti-CD3 and anti-CD28 antibodies, and treated the cells with TGF-β and IL-2, which resulted in activation of Foxp3, CTLA4 and PD-1 expression, as a measure of CD4+T cell differentiation into iTreg cells. iTreg cell induction was blocked by SB431542 (Fig. 6B, 6C). SeV infection prior to this treatment prevented induction of Foxp3, CTLA4 and PD-1 mRNA expression, and induced IRF7 mRNA expression, known to result from IRF3 activation (Fig. 6B, 6C), indicating that SeV-induced RLR signaling represses the generation of iTreg cells in culture.
Figure 6. Virus-induced IRF3 activation represses Treg lymphocyte differentiation and TGF-β/Smad3 target genes in mice.
(A), SeV induced IRF3 activation, assessed by immunoblotting for C-terminally phosphorylated IRF3, in primary Irf3+/+ or Irf3−/− mouse CD4+ cells at 12 h after infection. (B), In vitro Treg cell differentiation of naïve CD4+ T cells from wild-type mice with or without SeV infection for 1 h. TGF-β-induced Foxp3 mRNA expression, detected by qRT-PCR after 48 h, was largely abolished by the short exposure to SeV that resulted in IRF7 mRNA expression. * and **, P < 0.001, compared with TGF-β-treated control cells, by Student’s t-test. (C), Effect of SeV-induced RLR signaling on Treg differentiation of naïve CD4+ T cells isolated from wild-type or Irf3−/− mice. Expression and activation of endogenous IRF3 in these cells were shown in (A). Foxp3, CTLA4, PD-1 and IRF7 mRNA levels were determined by qRT-PCR after 48 h of TGF-β treatment. *, P < 0.01, compared with wild-type cells without SeV infection, ** and ***, p<0.01, compared with wild-type cells after SeV infection, by Student’s t-test. (D), Increased CD4+Foxp3+ Treg cells in the colonic lamina propria of Irf3−/− mice, compared to wild-type control B6 mice. Treg lymphocytes were quantified by flow cytometer following labeling with tagged, fluorescence-conjugated antibodies. N=3 mice/group. *, P < 0.05, by Student’s t-test. (E), Flow cytometry using fluorescence-conjugated antibodies distinguished thymic-derived (tTreg, Nrp-1-positive) and periphery-derived (pTreg, Nrp-1-negative) Treg lymphocytes from the lamina propria. Irf3−/− mice showed decreased tTreg cell frequency (left) and increased pTreg cell number (right). N=3 mice/group. *, P < 0.05, **, p<0.001, by Student’s t-test. (F), Influenza virus infection of wild-type and Irf3−/− mice induced IRF7 and ISG56 mRNA expression in immune cells of broncho-alveolar lavage, detected by qRT-PCR48 h after infection. (G), Influenza viral infection of wild-type or Irf3−/− mice changed lung mRNA expression of TGF-β/Smad3 target genes encoding MMP11, Smad7, and TIMP2, detected by qRT-PCR at 48 h after infection. N=4 mice/group, *, P < 0.001, compared with wild-type mice without viral infection, and **, p<0.05, compared with wild-type mice after viral infection, by Student’s t-test.
To validate the role of RLR-IRF3 signaling in Treg differentiation, we also isolated primary CD4+ naïve T cells from age- and sex-matched Irf3−/− mice. As T cells from this strain lacked IRF3 expression and SeV was therefore unable to induce IRF3 activation (Fig. 6A), SeV induced only a minimal level of IRF7 mRNA expression, compared to wild-type CD4+ cells (Fig. 6C, right). TGF-β and IL-2 induced Foxp3 mRNA expression in Irf3−/− cells to a higher level than in wild-type CD4+ T cells (Fig. 6C, left), and, unlike wild-type cells in which SeV infection repressed iTreg differentiation, the TGF-β-induced Foxp3, CTLA4, and PD-1 mRNA levels were only minimally reduced by SeV infection (Fig. 6C). These results argue that RLR signaling represses TGF-β-induced differentiation of Treg cells through IRF3.
We also assessed the Treg generation in the colonic lamina propria, which is inherently prone to RLR activation and TGF-β signaling (Curotto de Lafaille and Lafaille, 2009; Sheridan and Lefrancois, 2011). The number of CD4+ Foxp3 expressing Treg cells was increased in Irf3−/− mice, compared with wild-type mice (Fig. 6D), with an increase in peripheral Treg cells and proportional decrease in thymic-derived Treg cells (Fig. 6E). These results are consistent with increased TGF-β/Smad signaling in Irf3−/− mice, and illustrate attenuation of TGF-β-induced Treg cell generation by activated IRF3 in vivo, with effects on peripheral and thymic T cells.
Viral infection represses TGF-β-induced gene expression in mice
To evaluate whether viral infection represses the TGF-β response in vivo, we took advantage of the high basal TGF-β activity that is normally seen in the lung and is required for normal lung physiology and remodeling (Sheppard, 2006). RLR-IRF3 antiviral signaling was induced in lung cells by intranasal delivery of influenza A virus (strain A/Puerto Rico/8/1934; H1N1) (Kumar et al., 2006). As shown in Fig. 6F, intranasal delivery of the virus induced a dramatic increase in IRF7 and ISG54 mRNA expression in pulmonary immune cells obtained by bronchoalveolar lavage, indicating strong activation of IRF3. Much weaker activation of IRF7 and ISG54 expression was observed in Irf3−/− mice (Fig. 6F), consistent with our data in Irf3−/− CD4 + T cells (Fig. 6C, right).
Among the genes known to be targeted by TGF-β-activated Smads, we found that the genes encoding Smad7 (Nakao et al., 1997), MMP11 (Barrasa et al., 2012) and TIMP2 (unpublished data) are expressed in lung epithelium. Their mRNA expression was significantly attenuated following influenza virus infection in the lungs of wild-type mice, but not in lungs of Irf3−/− mice (Fig. 6G), consistent with the decreased Smad3 activation in response to influenza infection in wild-type, but not Irf3−/− lungs (Fig. S5B). These data support the inhibitory cross-talk of virus-induced IRF3 signaling on TGF-β-induced gene responses in vivo.
Discussion
TGF-β signaling plays pervasive roles in the regulation of cell proliferation, differentiation and functions, with the outcome dependent on other signaling pathways, and cell and tissue type. The functional availability of TGF-β receptors and the Smad activities are regulated through post-translational modifications, and cooperation of Smads with DNA-binding transcription factors and co-regulators sets the stage for signaling crosstalk of Smads at nucleoprotein complexes. We now present a novel mode of regulation of TGF-β/Smad responsiveness, i.e. through activation of IRF3 in response to RLR signaling. This crosstalk emanates from the innate antiviral host response, and represses TGF-β-induced Smad signaling. Considering the expression of IRF3 and RLRs in many cell types, we surmise that IRF3-mediated repression affects many TGF-β responses in many cell types, dependent on the level of IRF3 and its activation in response to extracellular cues.
Mechanism for IRF3-mediated inhibition of Smad signaling
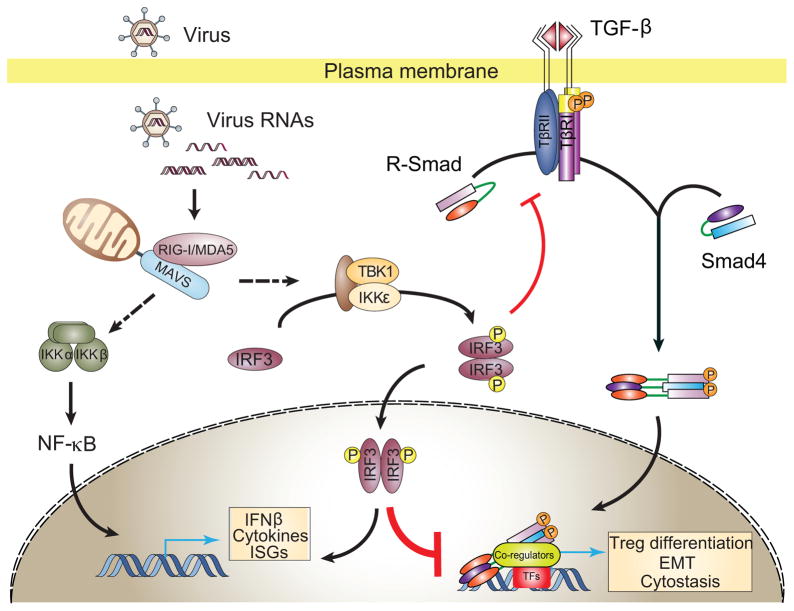
Inhibition of TGF-β/Smad signaling by IRF3 involves a dual mechanism, i.e. inhibition of Smad3 activation in response to TGF-β, and functional interference with Smad transcription complexes (Fig. 7). Interference with Smad3 activation by TβRI results from association of activated IRF3, but not inactive IRF3, with non-activated Smad3, thus preventing Smad3 recruitment to TβRI, and decreasing Smad3 activation. The structural basis for this association is suggested by the remarkably similar three-dimensional structures of the IRF3 and Smad3 transactivation domains (Qin et al., 2003; Takahasi et al., 2003). Both have a central β-sandwich and a loop-helix region that binds phosphoserines, and are followed by C-terminal serines. Phosphorylation of two C-terminal serines by TβRI results in Smad activation and trimer formation (Chacko et al., 2004), and phosphorylation of multiple serines by TBK1 or IKKε confers IRF3 activation, through reorganization of auto-inhibitory elements, and dimerization(Qin et al., 2003; Takahasi et al., 2003). Smad trimer and IRF3 dimer formation involve in either case a conserved basic cleft with negatively charged amino acids (Chacko et al., 2004; Qin et al., 2003; Takahasi et al., 2003). Association of activated IRF3 with Smad3 is consistent with the interaction of the Smad3 MH2 domain with the transactivation domain of the IRF3-related IRF7 (Qing et al., 2004).
Figure 7. Model for the roles of RLR-induced IRF3 activation in suppression of TGF-β signaling.
Without activation of innate antiviral signaling, IRF3 resides primarily in the cytosol and is not associated with Smad3. Upon RLR activation by virus RNA, IRF3 is C-terminally phosphorylated by TBK1 or IKKε. Activated IRF3 then mostly forms a dimer, translocates into the nucleus to activate transcription from IRE-containing promoters. Some activated IRF3 attenuates TGF-β signaling by dual mechanisms, based on the remarkable structural similarity of its transactivation domain with the Smad3 MH2 domain, thus preventing its association with TβRI and attenuating TGF-β-induced Smad3 activation. In the nucleus, activated IRF3 competes with Smad3/4 coregulators, thus disrupting the formation of functional Smad complexes and their binding to Smad -responsive promoters.
The interference of activated IRF3 with Smad3 transcription complexes may result from shared use of CBP or p300 (Feng et al., 1998; Janknecht et al., 1998; Qing et al., 2004; Wathelet et al., 1998; Weaver et al., 1998), and GRIP1 (Li et al., 2006; Reily et al., 2006) as transcription coactivators. Activated IRF3 repressed Smad3-mediated transcription by interfering with the interactions of Smad3 with p300, GRIP1, and Smad4, which stabilizes R-Smad-CBP/p300 interactions. Conversely, TGF-β-induced Smad3 activation did not inhibit IRF3 activation, and activated Smad3 did not interfere with the IRF3-p300 association, nor repress IRF3-mediated transcription. Our findings complement previous results showing TGF-β/Smad3-mediated increase of transcription by IRF7 at IRF3/7-binding gene sequences, as found in the interferon-β gene (Qing et al. 2004). Collectively, the structural similarities between the transactivation domains of Smads and IRF3/7 enable multiple levels of crosstalk that may extend to other IRFs.
The inhibition of TGF-β/Smad signaling by IRF3 activation resembles the dual repression by inhibitory Smads, i.e. Smad6 and Smad7. Inhibitory Smads interact with R-Smads and activated type I receptors, thus antagonizing R-Smad recruitment to the receptor, and inhibiting R-Smad activation (Hayashi et al., 1997; Imamura et al., 1997). Similarly, activated IRF3 interacted with Smad3, interfering with Smad3 recruitment to TβRI, although we did not detect IRF3 association with TβRI. Inhibitory Smads can also directly repress transcription at gene regulatory sequences (Miyake et al., 2010; Zhang et al., 2007), providing a second mode of repression, although the underlying mechanism at endogenous genes requires further study. IRF3-mediated repression of TGF-β/Smad signaling is as effective as repression by inhibitory Smads, with the latter not known to be inducible and the former fully dependent on activation of RLR or TLR signaling.
Roles of IRF3 activation in the control of EMT and T cell differentiation
The repression of TGF-β responsiveness by IRF3 may substantially affect many processes that are controlled by TGF-β. RLRs, TBK1 and/or IKKε, and IRF3 are widely expressed, and IRF3 can be activated by DNA damage, membrane fusion, and ER stress, in addition to virus infection. Additionally, pathogens and extracellular stimuli other than viruses also activate IRF3 via STING, TLR and RLR signaling (Goubau et al., 2013; Collins and Mossman, 2014).
Among the TGF-β-regulated processes, the repression of EMT-associated gene reprogramming may be particularly relevant in epithelial healing, fibrosis and cancer progression. In wound healing, epithelial cells undergo a transient epithelial plasticity response that enables wound closure (Heldin et al., 2012; Xu et al., 2009). Epithelial plasticity also contributes to fibrosis, with EMT-associated reprogramming activated by injury and inflammation (Chapman, 2011). In carcinomas, EMT, or at a minimum an epithelial plasticity response, is seen as prerequisite for tumor cell invasion (Heldin et al., 2012), whereas EMT is also integral to the generation of cancer stem cells (Katsuno et al., 2013; Scheel and Weinberg, 2012). In all three contexts, increased TGF-β signaling is thought to drive EMT. Reprogramming of gene expression during EMT involves Smad3-mediated activation of Snail or other transcription factors that drive EMT, and Smad3-mediated repression of epithelial genes and activation of mesenchymal genes (Heldin et al., 2012; Xu et al., 2009). Accordingly, RLR signaling leading to IRF3 activation represses gene reprogramming during EMT. We hypothesize that RLR or TLR signaling affects wound healing, fibrosis and carcinoma progression, through repression of Smad signaling by activated IRF3.
TGF-β also controls suppression of immune surveillance, and regulates the differentiation and functions of T cell lineages. TGF-β-induced Smad signaling drives Foxp3 induction, and suppression of IL-2, IL-4, and interferon-γ expression. CD4+CD25+Foxp3+ Treg lymphocytes differentiate from naïve T cells in response to TGF-β and IL-2, and prevent pathological self-reactivity, i.e. autoimmune disease (Li and Flavell, 2008; Yang et al., 2010). They also enforce tumor immune surveillance and are critical for preventing cancer metastasis (Curiel, 2007). We show that IRF3 activation represses Smad3-mediated control of T cell differentiation, and postulate that the antiviral defense attenuates immunosuppressive functions of TGF-β, e.g. in immune tolerance or tumor surveillance, by inhibiting Smad-dependent Treg cell generation and function.
In conclusion, we unveiled a role of innate antiviral host defense in Treg cell differentiation and EMT, and a novel mode of functional control of TGF-β-induced Smad signaling, i.e. through RLR or TLR signaling resulting in IRF3 activation, enabling innate antiviral signaling to suppress the TGF-β pathway. This repression may potently affect many processes that are regulated by TGF-β, perhaps with most relevance to epithelial plasticity responses, as seen in wound healing and cancer progression, and to the regulation of immune responses.
Materials and Methods
(more detailed information is in Supplemental Information)
Luciferase reporter and mammalian two -hybrid assays
HepG2, NMuMG or HaCaT cells were transfected and used for luciferase assays. In brief, cells were cultured for 12 h post transfection, stimulated by transfection with poly(I:C), then after 8 h treated overnight with TGF-β at the indicated concentration, and/or pharmacological inhibitors. Luciferase assays were performed using a dual luciferase system, quantified with SpectraMax M5 luminometer, and normalized to the internal Renilla luciferase control. Mammalian two-hybrid assays were performed as described (Feng et al., 1998), by coexpressing Gal4-DBD-fused GRIP1 or Smad4 with VP16-fused Smad3, and the luciferase reporter, using the Mammalian Matchmaker two-hybrid kit.
Quantitative RT-PCR assay
Total RNA was extracted using an RNAeasy extraction kit. cDNA was generated using the one-step iScript cDNA synthesis kit, and quantitative real-time PCR was performed using the iQ SYBR green supermix and CFX96 real-time PCR system. Relative quantification was expressed as 2-ΔCt, where ΔCt is the difference between the main Ct value of triplicates of the sample and that of an endogenous L19 or GAPDH mRNA control.
Coimmunoprecipitations, nuclear extract preparation, and immunoblotting
HepG2 or 293T cells, transfected with plasmids encoding Myc-, Flag-, or HA-tagged Smad3, caSmad3, IRF3, caTβRI or p300, were treated with TGF-β and/or TpIC, lysed, and subjected to immunoprecipitation using anti-Flag or anti-HA antibodies for transfected proteins, or anti-Smad2/3, anti-IRF3, or anti-p300 antibodies for endogenous proteins. After extensive washing, adsorbed proteins were analyzed by gradient SDS-PAGE and immunoblotting. Nuclear and cytoplasmic extracts were prepared using NE -PER Nuclear and Cytoplasmic Extraction kit.
RNA interference
HaCaT or HepG2 cells were transfected with double stranded siRNA targeting the human IRF3 mRNA using Lipofectamine RNAiMAX for 48 h before assay, or 24 h before adding TGF-β in the EMT assay. Reverse transfection was used to reach optimal efficiency.
Immunofluorescence and microscopy
C2C12 cells were treated as indicated, fixed, permeabilized, blocked, and incubated sequentially with primary antibodies (anti-Smad2/3 or anti-IRF3) and Alexa-labeled secondary antibodies with extensive washing. Slides were then mounted with Vectorshield and stained with DAPI. Images were obtained and analyzed using a Leica SP5 AOBS Upright 2 laser scanning confocal microscope. For F-actin staining, HaCaT cells were fixed, washed, and then incubated with Alexa-546 phalloidin at a 1:500 dilution. Images were obtained and analyzed by a Leica DMI 4000 B inverted microscope.
Chromatin immunoprecipitation assays
NMuMG cells were treated with 2 ng/ml TGF-β for 1 h and fixed, then resuspended in SDS lysis buffer. After sonication, samples were incubated overnight at 4°C with anti-Smad3 antibody or control IgG. After adding Protein A Dynabeads, immunoprecipitates were sequentially washed once with low-salt buffer, then high-salt buffer, and LiCl buffer, and twice with TE buffer. DNA-protein complexes were eluted and crosslinked by heating, digested with proteinase K and RNase A. DNA was recovered and subjected to RT-PCR analysis using primers for the Smad7 or Snail promoter regions.
In vitro Treg cell differentiation
Naïve CD4+CD25− cells were isolated from wild-type or Irf3−/− C57BL/6 mice as described (Fantini et al., 2007), and seeded in 24-well plates coated with 10 μg/ml anti-CD3 antibody in the presence of 2 μg/ml anti-CD28 antibody. After 48 h, cells were infected with SeV for 1 h, with or without inhibitors and/or TGF-β1 (10 ng/ml), as indicated. Induction of Foxp3, CTLA4, PD-1 and IRF7 mRNA expression after 48 h was quantified by RT-PCR.
Leukocyte isolation from colonic lamina propria and FACS
Colons were harvested from 8–10 week old wild-type or Irf3−/− C57BL/6 mice, sliced into pieces, and digested as described (Yadav et al., 2012). Cells were then resuspended in 40% Percoll and carefully underlaid with 80% Percoll. After centrifuging, the interface containing the leukocytes was collected for surface staining for CD4 and Nrp-1, and intracellular staining for Foxp3. Stained cells were FACS analyzed on LSR II and by FlowJo software.
Cell proliferation assays
HaCaT cells were transfected with siRNA to IRF3 mRNA or control siRNA, and were seeded 24 h later in a 48-well plate without or with TGF-β or SB431542. After 72 h, cells were incubated with BrdU for 8 h, and incorporated BrdU was measured using aBrdU Cell Proliferation Assay kit.
Statistics
Quantified data from at least three independent experiments are presented as mean ± standard error of mean (SEM). Data shown as fold change or percentage were log-transformed before statistical analysis. When appropriate, statistical differences between groups were analyzed using an unpaired Student’s t-test by Sigmaplot 10.0. Differences were considered significant at *p < 0.05.
Supplementary Material
Supplemental Figure S1 (related to Fig. 1 and 2): (A), IRF3 activation, assessed by immunoblotting for C-terminally phosphorylated IRF3, in response to TpIC or SeV infection in primary human CD4+ cells. The lower panel shows immunoblotting for total IRF3 in the cell lysates. Note the slightly decreased mobility of human IRF3 after C-terminal phosphorylation. (B), Luciferase reporter activities of an IRF3/7-responsive promoter show high basal activation in HaCaT and HeLa cells, but not in HepG2 and NMuMG cells. The reporter was activated by IRF3 5SD in all cell lines. N=3 experiments. (C), TGF-β-induced transcription from the Smad7 promoter, measured by luciferase expression, was repressed by IRF3 5SD, but not wild type IRF3 in the absence of RLR activation. N=3 experiments. *, p<0.001, compared with control samples with vector and TGF-β treatment, by Student’s t-test.
Supplemental Figure S2 (preceding Fig. 3): (A), Silencing IRF3 expression by either of two siRNAs (si-IRF3-144 and si-IRF3-400), or the TBK1/IKKε inhibitor BX795, rescued the TpIC-induced repression of TGF-β/Smad3-activated transcription in HepG2 cells. N=3 experiments. * and **, P < 0.01, compared with TGF-β + TpIC treatment, by Student’s t-test. (B), Endogenous IRF3, IRF7, ISG54, and interferon-β1 mRNA in HepG2 cells after 12 h of control or TpIC treatment, determined by qRT-PCR. (C), Silencing IRF7 expression in HepG2 by siRNA did not restore the inhibition of Smad signaling by TpIC. N=3 experiments. *, P < 0.001, compared with samples with si-IRF3 + TpIC treatment, by Student’s t-test.
Supplemental Figure S3 (related to Fig. 3): (A), Activated IRF3 5SD and its DNA-binding defective mutant, IRF3 5SD_nDB, inhibited TGF-β-induced Smad3 activation, shown by anti-phospho-Smad3 immunoblotting, in HepG2 cells. (B), Wild-type IRF3, but not IRF3 that cannot be activated (IRF3 SA, with S396 and S398 replaced by alanines), enabled the TpIC-induced inhibition of Smad3 activation in HepG2 cells. N=3 experiments; *, P < 0.01, compared with wild-type IRF3 + TpIC, by Student’s t-test. (C), Association of Smad3 with activated IRF3 (IRF3 5SD) in the cytoplasmic and nuclear fractions of transfected HepG2 cells. α-tubulin and lamin A/C immunoblotting mark the cytoplasmic and nuclear fractions of the HepG2 cells used. (D) Smad4 association with wild-type IRF3 in 293T cells decreased in response to TpIC or when IRF3 is mutationally activated (IRF3 5SD). (E), TpIC induces association of Smad3 with wild-type IRF3 in the cytoplasm. (F), Reciprocal co-immunoprecipitation, compared to Fig. 3F, showing association of IRF3 5SD with Smad3. Smad3 activation by caTβRI prevents association of HA-tagged Smad3 with Myc-tagged activated IRF3 (IRF3 5SD) in 293T cells. (G) and (H), Activated IRF3 (IRF3 5SD), but not wild-type IRF3 or inactive IRF3 SA, inhibited the association of Smad3 with caTβRI, shown by co-immunoprecipitation, in 293T cells.
Supplemental Figure S4 (related to Fig. 4): (A), Association of Flag-tagged activated Smad3 (caSmad3) with HA-tagged p300 in 293T cells was disrupted by activated IRF3 5SD, but not IRF3 SA, even after TpIC treatment, which induced wild-type IRF3 to interfere with the caSmad3:p300 complex. (B), Association of activated IRF3 (IRF3 5SD) or Smad3 (caSmad3) with HA-p300 in transfected 293T cells. (C), The Myc-tagged p300 IBiD domain, located within the reported Smad3 interacting p300 segment (Lin et al. 2001), associated with activated IRF3 (IRF3 5SD), but not with activated Smad3 (caSmad3) in transfected 293T cells. (D), Activated IRF3 5SD, or its basic patch mutant (IRF3 5SDm), but not wild-type IRF3, interfered with the association of HA-tagged Smad3 with Flag-tagged caTβRI. (E), Activated IRF3 5SD or its basic patch mutant (IRF 5SDm), but not wild-type IRF3, reduced Smad3 activation, assessed by phospho-Smad3 immunoblotting, in 293T cells expressing activated TβRI (caTβRI).
Supplemental Figure S5 (related to Fig. 6): (A), SeV-induced IRF3 activation, assessed by immunoblotting for C-terminally phosphorylated IRF3 in primary human CD4+ cells at 12 h after infection. IRF3 activation was blocked by the T K1/IKKε inhibitor BX795. (B), Activated Smad3, and total Smad3 and IRF3 levels in mouse lung tissue, visualized by immunoblot.
Supplemental Table: DNA sequences of primers used in qRT-PCR.
Acknowledgments
We are grateful to Dr. John Hiscott for IRF3 plasmids, Dr. Tadatsugu Taniguchi for Irf3−/− mice and Dr. Jeffrey Gotts for technical support. This research was sponsored by NIH grants RO1-CA63101 & -CA136690 to R.D, MOST 973 Plan 2015CB553800, NSFC Project 81472665, and the Fundamental Research Funds for the Central Universities 2014QN81002 to P.X., NIH grants RO1-AI50834 and P30-DK63720 to J.A.B, RO1-HL44712 to H.A.C., and RO1-HL64353, -HL53949, -HL083950 and-AI024674 to D.S. S.B.-B. was supported by an American Diabetes Association Mentor Based Award, J. L. was supported by a Muscular Dystrophy Association Scientist Development Award, D.D. was supported by a Human Frontiers Science Program postdoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrasa JI, Olmo N, Santiago-Gomez A, Lecona E, Anglard P, Turnay J, Lizarbe MA. Histone deacetylase inhibitors upregulate MMP11 gene expression through Sp1/Smad complexes in human colon adenocarcinoma cells. Biochim Biophys Acta. 2012;1823:570–581. doi: 10.1016/j.bbamcr.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Brandl M, Seidler B, Haller F, Adamski J, Schmid RM, Saur D, Schneider G. IKKα controls canonical TGFβ-SMAD signaling to regulate genes expressing SNAIL and SLUG during EMT in panc1 cells. J Cell Sci. 2010;123:4231–4239. doi: 10.1242/jcs.071100. [DOI] [PubMed] [Google Scholar]
- Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, De Caestecker M, Lin K. Structural basis of heteromeric smad protein assembly in TGF-β signaling. Mol Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Chapman HA. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol. 2011;73:413–435. doi: 10.1146/annurev-physiol-012110-142225. [DOI] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Cornelius SC, Pultz NJ, Jorgensen JS, Bonham MJ, Kim SJ, Danielpour D. The androgen receptor represses transforming growth factor-β signaling through interaction with Smad3. J Biol Chem. 2002;277:1240–1248. doi: 10.1074/jbc.M108855200. [DOI] [PubMed] [Google Scholar]
- Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IκB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284:14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SE, Mossman KL. Danger, diversity and priming in inate antiviral immunity. Cytokine Growth Factor Rev. 2014 doi: 10.1016/j.cytogfr.2014.07.002. in press. [DOI] [PubMed] [Google Scholar]
- Curiel TJ. Tregs and rethinking cancer immunotherapy. J Clin Invest. 2007;117:1167–1174. doi: 10.1172/JCI31202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Fantini MC, Dominitzki S, Rizzo A, Neurath MF, Becker C. In vitro generation of CD4+ CD25+ regulatory cells from murine naive T cells. Nat Protoc. 2007;2:1789–1794. doi: 10.1038/nprot.2007.258. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15Ink4B transcription in response to TGF -β. Embo J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Goubau D, Deddouche S, Reise Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFβ in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-β superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Janknecht R, Wells NJ, Hunter T. TGF-β-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Katsuno YL, Lamouille S, Derynck R. TGF-β signaling and epithelial–mesenchymal transition in cancer progression. Current Opinion in Oncology. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Lee JS, Oh SY, Jin X, Choi YJ, Lee TH, Lee E, Choi YK, You S, Chung YG, et al. Direct transcriptional activation of promyelocytic leukemia protein by IFN regulatory factor 3 induces the p53-dependent growth inhibition of cancer cells. Cancer Res. 2007;67:11133–11140. doi: 10.1158/0008-5472.CAN-07-1342. [DOI] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol Cell Biol. 2009;29:172–186. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Derynck R. Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, Martin W, Fornwald J, Lehr R, Harling J, et al. Inhibition of transforming growth factor (TGF)-β1-induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity: SB-431542. Mol Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- Li G, Heaton JH, Gelehrter TD. Role of steroid receptor coactivators in glucocorticoid and transforming growth factor β regulation of plasminogen activator inhibitor gene expression. Mol Endocrinol. 2006;20:1025–1034. doi: 10.1210/me.2005-0145. [DOI] [PubMed] [Google Scholar]
- Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Hare BJ, Wagner G, Harrison SC, Maniatis T, Fraenkel E. A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies. Mol Cell. 2001;8:581–590. doi: 10.1016/s1097-2765(01)00333-1. [DOI] [PubMed] [Google Scholar]
- Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T, Alli NS, McDermott JC. Nuclear function of Smad7 promotes myogenesis. Mol Cell Biol. 2010;30:722–735. doi: 10.1128/MCB.01005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci U S A. 1998;95:6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Qin BY, Liu C, Lam SS, Srinath H, Delston R, Correia JJ, Derynck R, Lin K. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol. 2003;10:913–921. doi: 10.1038/nsb1002. [DOI] [PubMed] [Google Scholar]
- Qin BY, Liu C, Srinath H, Lam SS, Correia JJ, Derynck R, Lin K. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13:1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Qing J, Liu C, Choy L, Wu RY, Pagano JS, Derynck R. Transforming growth factor β/Smad3 signaling regulates IRF-7 function and transcriptional activation of the beta interferon promoter. Mol Cell Biol. 2004;24:1411–1425. doi: 10.1128/MCB.24.3.1411-1425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. EMBO J. 2006;25:108–117. doi: 10.1038/sj.emboj.7600919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massagué J. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Sheppard D. Transforming growth factor β: a central modulator of pulmonary and airway inflammation and fibrosis. Proc Am Thorac Soc. 2006;3:413–417. doi: 10.1513/pats.200601-008AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan BS, Lefrancois L. Regional and mucosal memory T cells. Nat Immunol. 2011;12:485–491. doi: 10.1038/ni.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Suzuki NN, Horiuchi M, Mori M, Suhara W, Okabe Y, Fukuhara Y, Terasawa H, Akira S, Fujita T, Inagaki F. X-ray crystal structure of IRF-3 and its functional implications. Nat Struct Biol. 2003;10:922–927. doi: 10.1038/nsb1001. [DOI] [PubMed] [Google Scholar]
- Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-β employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa L, Doody J, Massagué J. Inhibition of transforming growth factor-β/SMAD signalling by the interferon-γ/STAT pathway. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Weaver BK, Kumar KP, Reich NC. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lamouille S, Derynck R. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Liu J, Derynck R. Post-translational regulation of TGF-β receptor and Smad signaling. FEBS Lett. 2012;586:1871–1884. doi: 10.1016/j.febslet.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–1722. S1711–1719. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Pang Y, Moses HL. TGF-β and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-β signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1 (related to Fig. 1 and 2): (A), IRF3 activation, assessed by immunoblotting for C-terminally phosphorylated IRF3, in response to TpIC or SeV infection in primary human CD4+ cells. The lower panel shows immunoblotting for total IRF3 in the cell lysates. Note the slightly decreased mobility of human IRF3 after C-terminal phosphorylation. (B), Luciferase reporter activities of an IRF3/7-responsive promoter show high basal activation in HaCaT and HeLa cells, but not in HepG2 and NMuMG cells. The reporter was activated by IRF3 5SD in all cell lines. N=3 experiments. (C), TGF-β-induced transcription from the Smad7 promoter, measured by luciferase expression, was repressed by IRF3 5SD, but not wild type IRF3 in the absence of RLR activation. N=3 experiments. *, p<0.001, compared with control samples with vector and TGF-β treatment, by Student’s t-test.
Supplemental Figure S2 (preceding Fig. 3): (A), Silencing IRF3 expression by either of two siRNAs (si-IRF3-144 and si-IRF3-400), or the TBK1/IKKε inhibitor BX795, rescued the TpIC-induced repression of TGF-β/Smad3-activated transcription in HepG2 cells. N=3 experiments. * and **, P < 0.01, compared with TGF-β + TpIC treatment, by Student’s t-test. (B), Endogenous IRF3, IRF7, ISG54, and interferon-β1 mRNA in HepG2 cells after 12 h of control or TpIC treatment, determined by qRT-PCR. (C), Silencing IRF7 expression in HepG2 by siRNA did not restore the inhibition of Smad signaling by TpIC. N=3 experiments. *, P < 0.001, compared with samples with si-IRF3 + TpIC treatment, by Student’s t-test.
Supplemental Figure S3 (related to Fig. 3): (A), Activated IRF3 5SD and its DNA-binding defective mutant, IRF3 5SD_nDB, inhibited TGF-β-induced Smad3 activation, shown by anti-phospho-Smad3 immunoblotting, in HepG2 cells. (B), Wild-type IRF3, but not IRF3 that cannot be activated (IRF3 SA, with S396 and S398 replaced by alanines), enabled the TpIC-induced inhibition of Smad3 activation in HepG2 cells. N=3 experiments; *, P < 0.01, compared with wild-type IRF3 + TpIC, by Student’s t-test. (C), Association of Smad3 with activated IRF3 (IRF3 5SD) in the cytoplasmic and nuclear fractions of transfected HepG2 cells. α-tubulin and lamin A/C immunoblotting mark the cytoplasmic and nuclear fractions of the HepG2 cells used. (D) Smad4 association with wild-type IRF3 in 293T cells decreased in response to TpIC or when IRF3 is mutationally activated (IRF3 5SD). (E), TpIC induces association of Smad3 with wild-type IRF3 in the cytoplasm. (F), Reciprocal co-immunoprecipitation, compared to Fig. 3F, showing association of IRF3 5SD with Smad3. Smad3 activation by caTβRI prevents association of HA-tagged Smad3 with Myc-tagged activated IRF3 (IRF3 5SD) in 293T cells. (G) and (H), Activated IRF3 (IRF3 5SD), but not wild-type IRF3 or inactive IRF3 SA, inhibited the association of Smad3 with caTβRI, shown by co-immunoprecipitation, in 293T cells.
Supplemental Figure S4 (related to Fig. 4): (A), Association of Flag-tagged activated Smad3 (caSmad3) with HA-tagged p300 in 293T cells was disrupted by activated IRF3 5SD, but not IRF3 SA, even after TpIC treatment, which induced wild-type IRF3 to interfere with the caSmad3:p300 complex. (B), Association of activated IRF3 (IRF3 5SD) or Smad3 (caSmad3) with HA-p300 in transfected 293T cells. (C), The Myc-tagged p300 IBiD domain, located within the reported Smad3 interacting p300 segment (Lin et al. 2001), associated with activated IRF3 (IRF3 5SD), but not with activated Smad3 (caSmad3) in transfected 293T cells. (D), Activated IRF3 5SD, or its basic patch mutant (IRF3 5SDm), but not wild-type IRF3, interfered with the association of HA-tagged Smad3 with Flag-tagged caTβRI. (E), Activated IRF3 5SD or its basic patch mutant (IRF 5SDm), but not wild-type IRF3, reduced Smad3 activation, assessed by phospho-Smad3 immunoblotting, in 293T cells expressing activated TβRI (caTβRI).
Supplemental Figure S5 (related to Fig. 6): (A), SeV-induced IRF3 activation, assessed by immunoblotting for C-terminally phosphorylated IRF3 in primary human CD4+ cells at 12 h after infection. IRF3 activation was blocked by the T K1/IKKε inhibitor BX795. (B), Activated Smad3, and total Smad3 and IRF3 levels in mouse lung tissue, visualized by immunoblot.
Supplemental Table: DNA sequences of primers used in qRT-PCR.