Abstract
Many of the target molecules that reside in blood are also present in oral fluids, albeit at lower concentrations. Oral fluids are, however, relatively easy and safe to collect without the need for specialized equipment and training. Thus, oral fluids provide convenient samples for medical diagnostics. Recent advances in lab-on-a-chip technologies have made minute, fully integrated diagnostic systems practical for an assortment of point-of-care tests. Such systems can perform either immunoassays or molecular diagnostics outside centralized laboratories within time periods ranging from minutes to an hour. The article briefly reviews recent advances in devices for point-of-care testing with a focus on work that has been carried out by the authors as part of a NIH program.
Keywords: lab-on-a-chip, point-of-care testing, oral-based diagnostics, microfluidics
Introduction
In recent years, there has been a growing interest in point-of-care testing (PoCT) because of its advantages over standard laboratory procedures, of which we list just a few examples. PoCT provides timely information to medical teams, facilitating rational, time-critical decisions, and has been demonstrated to improve patient outcomes in critical care settings (Birkhahn et al, 2010). PoCT facilitates personalized medicine, allowing the caregiver to customize the therapy according to the patient’s needs rather than using standardized one-size-fits-all protocols. PoCT reduces turn-around time for results, which avoids the problem of patients not following up with their caregiver after a test (Pothier et al, 2010). This is particularly problematic in developing countries and underserved populations in developed nations. PoCT eliminates some of the costs associated with sample handling, packaging, tracking, and shipping to centralized laboratories and reduces the likelihood of samples being contaminated, mixed up, lost, and/or degraded (Lee-Lewandrowski and Lewandrowski, 2009). PoCT also facilitates real-time monitoring of the spread of infectious diseases, and food and water contaminants; enables rapid identification of the contamination sources; and provides critical information to public health workers and policy and decision makers. In short, providing relatively sophisticated tests at locations lacking laboratory facilities, such as rural areas, homes, border points, and first-responders, PoCT has the potential to save lives, improve the quality of life, and increase public safety. Given the above, it is not surprising that recent years have seen rapidly growing interest in PoCT.
Most PoCT devices, such as the Abbott iSTAT® (Abbott Point of Care, Princeton, NJ, USA) blood analyzer, the Johnson & Johnson OneTouch® (Life-Scan, a Johnson & Johnson Company, Milpitas, CA, USA) glucose meter, and the Roche CoaguCheck® (Roche, Basel, Switzerland) blood coagulation tester, utilize blood samples. In some cases, blood is the only option, but for quite a few analytes, oral fluids provide a suitable alternative. Oral fluid–based tests have been proposed to monitor growth factors, steroids, drugs of abuse, infectious diseases (Corstjens and Malamud, 2008; Malamud, 2011), and oral cancer (Ziober et al, 2008). A number of commercial salivary tests are available to detect antibodies to HIV (OraSure Technologies, Inc. steroid hormones (Salivary Assay – Salivary Cortisol – Salimetrics), and alcohol and drugs of abuse (Dräger USA – Home), as well as for forensic and genetic analyses (IFI Independent Forensics, Hillside, IL, USA). Although most analytes are present in saliva at lower concentrations than in blood (Malamud and Niedbala, 2007), in many cases oral fluids offer offsetting advantages. The collection of oral samples is noninvasive, does not require a trained phlebotomist and special facilities, and is ideal for vulnerable populations including pediatric, geriatric, and hemophiliacs. In contrast to urine samples, it is easier to maintain a chain of custody for oral samples, and oral fluid collection is less hazardous to both the patients and the health personnel than blood collection. Thus, oral fluids are ideal for PoCT.
Point-of-care testing assays must be highly automated to minimize errors, sample contamination, and sample degradation because it is often desirable that minimally trained personnel be able to operate these devices. Early PoCT was mostly based on lateral flow (LF) assays (also known as ‘dip sticks’). LF assays have the great advantage of simplicity, but lack the capabilities for carrying out complicated operations such as may be needed, for example, for nucleic acid diagnostics. In order to be able to perform sophisticated laboratory procedures at the point of testing, significant engineering challenges must be overcome such as developing means to autonomously transport liquids and control fluid flow in small devices, facilitate mixing and thermal control, dry store reagents for prolonged periods of time (greater than a year), and provide means for result detection.
Recent advances in microfluidics make it possible to miniaturize, integrate, and automate various bench-top procedures into credit card-sized cassettes or chips. Microfluidics offers greater functionality and more sophisticated flow control than LF devices. The field is also often referred to as micro-total analysis (µTAS) or lab-on-a-chip (LoC). LoC has two subdisciplines: one is concerned with automating laboratory procedures with the aim of enhancing discoveries through the high throughput parallel processing in the laboratory that may be needed for combinatorial biology and chemistry, drug screening, and other biological studies (Paegel and Joyce, 2010); and the other is aimed at creating autonomous, fully integrated PoCT devices, which are often disposable and aimed at assaying one or a few analytes within a relatively short period of time.
PoCT microfluidic devices can be further classified as instrumented or un-instrumented devices (Weigl et al, 2008). Instrumented devices typically consist of a disposable cassette accompanied by a portable, durable analyzer. The disposable cassette hosts a microfluidic circuit with reaction chambers and interconnecting channels as well as required reagents. The analyzer provides means for pumping, thermal control for incubation and enzymatic amplification, and detection via optical signals, electrochemical signals, or added mass. Un-instrumented devices are a reincarnation of the LF strips with added capabilities and sophistication. The actuators, such as finger-actuated pouches (Qiu et al, 2009) and exothermic reaction chambers, are integrated into the disposable cassette, and no external processor is needed.
Yet, another classification of PoCT devices is based on the target analytes, e.g. proteins (detected via an immunoassay) or nucleic acids. PoCT for nucleic acids is referred to as molecular diagnostics. Molecular diagnostics devices often include enzymatic amplification such as polymerase chain reaction (PCR), and they are generally more complicated than immunoassays. Below, we will first discuss devices for immunoassays and then devices for molecular diagnostics. However, because of space limitations, it is not possible to provide an extensive survey of the field, and the sections below are biased toward research carried out in the authors’ laboratories.
Devices for immunoassays
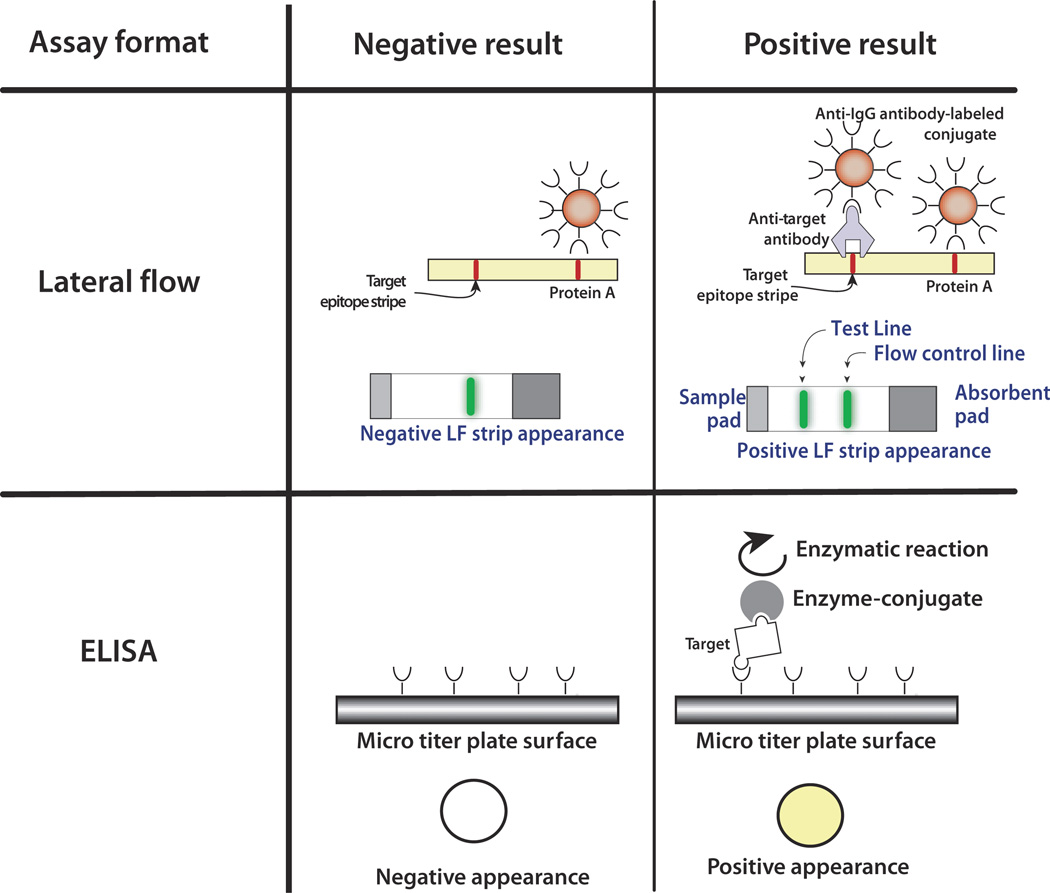
Immunoassays test for the presence of small molecules and proteins and take advantage of the high specificity of antibody binding to their target antigen ligand. A wide variety of assay formats, platforms, and detection methods exist. In laboratory settings, the enzyme-linked immunoassay (ELISA) is the gold standard (Ashihara et al, 2007). This method is robust, relatively easy to use, and has excellent sensitivity and specificity. However, the ELISA requires several lengthy steps and a spectrophotometer or a fluorometer for detection; therefore, it is not commonly used in the point-of-care setting. A schematic of the ELISA method is shown in Figure 1.
Figure 1.
A schematic comparing enzyme-linked immunoassay (ELISA) and lateral flow (LF) assays. The ELISA uses an enzyme such as horseradish peroxidase or alkaline phosphatase linked to an antibody to generate either a color change or an electrochemical signal. The ELISA requires multiple washes. The LF assay often uses visual labels such as gold nanoparticles and includes a specific capture line and a control line. The control line binds the labeled conjugate directly without a target and ensures that the sample has flowed up the LF strip
For example, an ELISA to detect the presence of an antigen is carried out by transmitting a sample containing target molecules along a surface functionalized with appropriate antibodies. The targets bind to the immobilized antibodies. The rest of the sample is washed away. Then, the bound antigens are labeled with an antibody linked to an enzyme to form the sandwich antibody–antigen–antibody. Subsequent to a second wash, a material known as substrate is introduced. The enzyme reacts with the substrate in solution to produce a measurable signal such as color change or an electrochemical signal (Liu et al, 2009b). The enzyme acts as a signal amplifier. When one wishes to detect antibodies, a similar procedure is employed except that surface is functionalized with the antigen. There are many possible variants of the ELISA test. The ligands can be immobilized within porous beads such as agarose beads to increase the ligand density and test sensitivity or attached to magnetic beads that are initially suspended in solution to improve reaction kinetics and that eventually can be immobilized with the aid of an external magnetic field to facilitate washing. The main challenge for adapting the ELISA test for point-of-care applications is the need to facilitate multiple flow steps.
The most ubiquitous point-of-care immunoassay is the LF strip, also known as the immunochromato-graphic strip (Wong and Tse, 2008). The LF test is inexpensive and sufficiently easy to use to be accessible to the public. LF–based tests are used for, among other things, detecting pregnancy at home, infectious diseases, cardiac and emergency conditions, drugs of abuse, and food- and water-borne pathogens. Samples may include body fluids, such as urine, saliva, blood, and stool, or water and food products. The popularity of LF assays partially stems from the fact that they do not require any active means to pump fluids and control fluid flow. In its simplest form, the test employs gold or carbon particles as reporters, and the test results can be detected by eye without any instrumentation. For better sensitivity and to enable quasi-quantification of test results, readers (or developers) can be employed. In contrast to an ELISA test, the typical LF test lacks signal amplification, and thus, its sensitivity falls short of that of the ELISA test.
Briefly, the LF strip is fabricated from a porous, paperlike material, such as nitrocellulose. Antigens to the target antibody (or antibodies) are immobilized as a dried stripe (test line) across the strip. The strip can accommodate a few test lines to allow the concurrent detection of multiple targets (multiplexing). Alternatively, when it is not necessary to identify a particular protein, the test line can be comprised of a mixture of antigens. In addition to the test(s) line(s), the strip is equipped with a flow control line, composed of a generic capture agent, located downstream of and parallel to the test line. The sample (typically mixed with a buffer) is applied at one end of the strip and wicks by capillary action toward the test and control lines. Along the way, the sample hydrates and suspends dried, functionalized labels, which bind to the target molecules. As the sample flows past the test line, the target molecule (e.g. a specific antibody) label complex binds to the immobilized ligands to form a sandwich (Qian and Bau, 2003). When a sufficient number of reporter particles have been captured, they form a visible line. A positive test consists of a visible test line and a visible control line. A negative test shows labels bound only to the control line. Absence of signal at the control line indicates that the sample did not wick up the strip, the label-bound proteins degraded, or the labels did not hydrate, and the test must be repeated. For a visual test, the labels are usually gold or carbon particles that can easily be seen with the naked eye. Because there is no need for pumps to transmit the fluids and equipment to read the signal, the LF strip format is amazingly simple. This simplicity, however, comes at the cost of lower sensitivity and rigidity of assay format.
The sandwich assay is not the only possible assay, and sometimes, one may elect to operate in a competitive mode where the absence of the signal indicates a positive test (Qian and Bau, 2004). Also, occasionally, LF assays are utilized with a reader (sometimes referred to as a detector or developer). The reader can extend the repertoire of reporter particles to include fluorophores, quantum dots (QDots), and phosphor nanocrystals that require excitation at specific wavelengths. The reader provides excitation and detects the light emitted from the reporter particles. In the sandwich assay, the detected light intensity correlates with the number of captured target molecules. The use of such labels can improve test sensitivity, provide objective documentation of test results, and facilitate quantification. Usually, the test results are reported as the ratio of the signal intensity detected from the test and control lines.
In its simplest manifestation, the LF test requires the user to bring the LF strip into contact with the sample and the process carries out on its own without any further user intervention. On occasion, it may be desirable to alter the test sequence. For example, it may be desirable to introduce the reporter particles only after the target analytes have had the opportunity to bind to the test line and the other proteins in the sample were washed away. This is the consecutive flow format (Corstjens et al, 2007; Liu et al, 2009a). The consecutive flow format requires the user to bring the LF strip into contact with a sample mixed with a buffer, wait awhile (i.e. a couple of minutes), then bring the strip in contact with a wash solution and wait again, and finally bring the strip in contact with a suspension of reporter particles. Although the steps are simple, there is a greater risk of operator’s error. To minimize user mistakes, it is desirable to automate the various processing steps. We will use the consecutive flow assay as a simple example to illustrate how microfluidics can be used to automate the process.
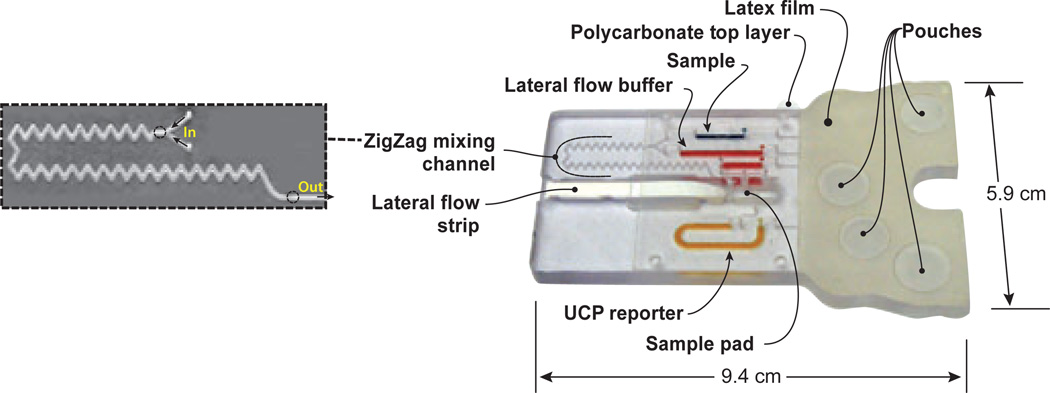
Figure 2 shows a photograph of a plastic cassette designed to carry out LF–based, consecutive flow assays (Liu et al, 2009a). The plastic cassette houses four air-filled pouches, storage chambers, conduits, an injection port, a zig-zag mixing chamber, and a LF strip. The storage chambers are located downstream of the pouches, and the LF strip is located downstream of the storage chambers. During storage, the inlets and outlets of the storage chambers are sealed. Immediately prior to use, the cassette is loaded with a sample and the connections upstream and downstream of the storage chambers are opened to bring the storage chambers into connection with the upstream pouches and the downstream LF strip. At the start of the process, two of the pouches are simultaneously compressed to concurrently transmit the sample and diluent through the zig-zag path, which serves to mix the sample with diluent. The mixture is then ejected onto the LF strip sample pad and migrates up the strip by capillary action. As the sample-diluent mixture passes through the test line, target molecules bind specifically to the immobilized ligands. After a predetermined time interval, a third pouch is compressed to discharge wash solution onto the strip. The wash solution removes any unbound proteins. The last pouch is then compressed to discharge reporter particles functionalized with protein A, which binds to all IgG molecules. The particles wick up the strip and bind to the immobilized ligand–target complex as well as to the control line. The actuation of the pouches can be carried out manually (Qiu et al, 2009), with a mechanical actuator consisting of a modified spring-driven timer (Liu et al, 2009a) or with computer-controlled motors (Qiu et al, 2011).
Figure 2.
A pouch-based immunoassay cassette. The zigzag conduit (enlarged) facilitates the mixing of the sample with lateral flow (LF) buffer prior to application to the LF strip’s sample pad. The various storage chambers are filled with food coloring for clarity. The assay employs up converting phosphor particles (UCP) as reporters
Another common assay that is employed at the point of care is based on microbeads. The microbead-based assays require somewhat more complicated flow control than the LF assay. Microbeads with various function-alizations are ubiquitous solid supports for capturing target molecules in both bench-top and microfluidic systems (Derveaux et al, 2008; Verpoorte, 2003; Christodoulides et al, 2005; Blicharz et al, 2009; Thompson et al, 2010). Typically, the beads are polymeric (e.g. polystyrene or agarose), range in diameter from a few micrometers to a few hundred micrometers, and can be readily purchased from many vendors with prefunctionalized surfaces to bind various targets such as antibodies of various types and oligonucleotides. Functionalized beads can be placed in an array to concurrently detect multiple targets. These arrays may include a few beads to detect a handful of analytes (Ali et al, 2003) or thousands of beads capable of detecting hundreds of target molecules simultaneously and constructing a sample’s pattern or ‘finger print’, where the emphasis is on the relative concentration of various molecules in the sample (Steemers and Gunderson, 2007).
A number of recent studies have demonstrated the value of microbead-based detection in complex biological samples. For instance, bead-based microfluidic systems have been used to detect DNA and inflammatory cytokines in saliva (Blicharz et al, 2009; Bowden et al, 2005) and blood (Hashmi et al, 2005), C-reactive protein in saliva (Christodoulides et al, 2005), human secretory immunoglobulin A (Sato et al, 2000), carcinoembryonic antigen (Sato et al, 2001), and interferon-gamma (Sato et al, 2002).
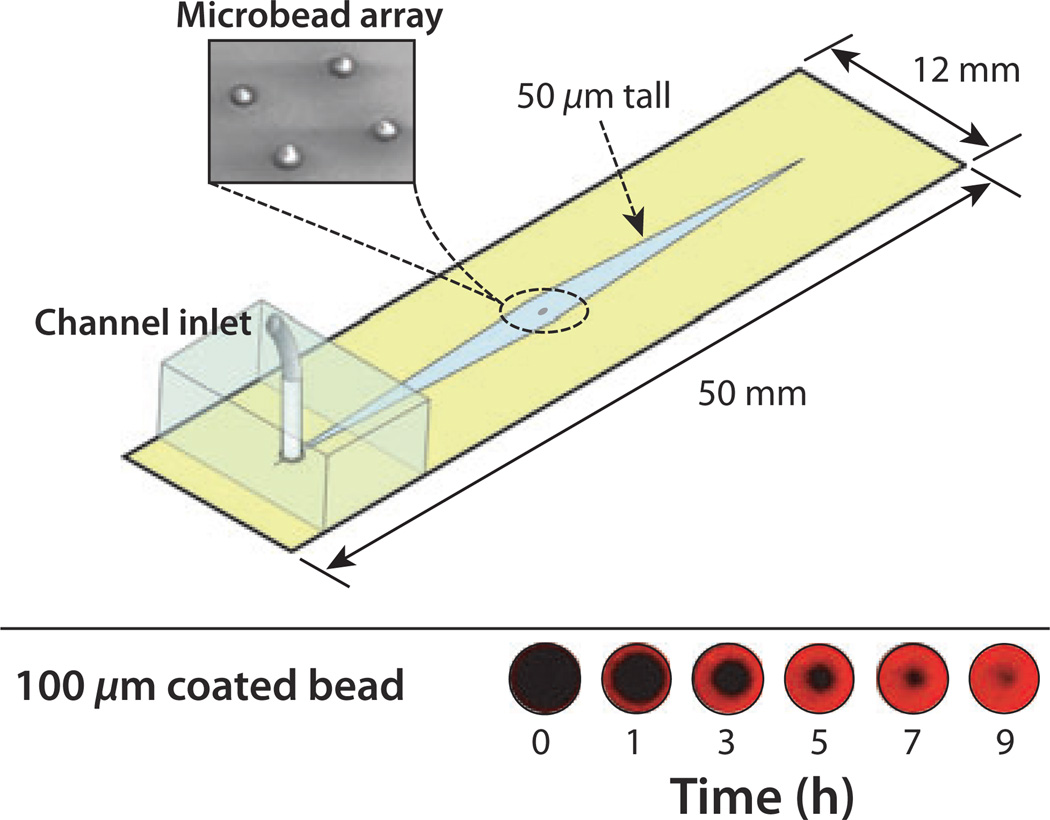
In the schematic shown in Figure 3, four porous agarose microbeads are arrayed in the chip and capture QDot labels pumped through the channel (Thompson and Bau, 2010a,b). One of the four beads is shown (Figure 3, bottom) imaged over time using a confocal microscope (at the bead’s mid-height plane). The high surface-are-to-volume ratio facilitates high binding density, which improves signal intensity compared to non-porous beads and planar surfaces. This advantage allows the agarose beads to accumulate labels faster and over longer periods of time.
Figure 3.
A schematic depiction of a microbead-based immunoassay chip. The chip contains four porous, agarose beads within a 50-µm tall channel. The beads are sandwiched in place between the floor and ceiling of the channel and remain stationary during continuous flow. The lower image shows the binding of biotin-coated quantum dot labels to the mid-height cross-section of a streptavidin-coated bead as a function of time. The porous structure of the bead and its accessibility to the labels increases the number of binding events that take place in a small volume and improves signal intensity over that obtained with solid beads, which rely solely on peripheral interactions at their surface
Molecular diagnostics
Molecular diagnostics comprises numerous tests based on the sequence-specific detection of target nucleic acids. These tests can be used to determine the presence of a virus, bacterium, or parasite in a clinical specimen such as serum, saliva, urine, feces, and nasal swabs. Nucleic acid tests are also used for genetic identification of forensic samples; determination of the gene profile and identification of alterations in expressed genes in cancer cells; and genotyping, e.g. detecting mutations and other genetic anomalies. In many cases, when the patient has been pre-exposed to a pathogen, antibody tests cannot provide the necessary information and, often, nucleic acid tests are used. For example, the response of HIV patients to therapy is often monitored by detecting viral load with a nucleic acid test.
Nucleic acid testing (NAT) currently provides the highest levels of sensitivity and specificity of all clinical methods. Single-molecule limits of detection have been demonstrated, both in bench-top and microfluidic assays (Yung et al, 2009; Nakamura et al, 1993). NAT can provide three to four orders of magnitude greater sensitivity than immunoassays. This high sensitivity is because of the fact that nucleic acids are amenable to enzymatic amplification. In other words, it is possible to amplify the number of target molecules and increase their concentration to detectable levels. Unfortunately, these performance advantages come at a price as molecular diagnostics requires elaborate sample processing, often longer test times, and more expensive reagents. Moreover, the tests are susceptible to contamination and inhibitors, which may lead to false negatives, and to false-positive results because of tests’ complexity.
The typical NAT involves the sequential steps of lysis to release and solubilize nucleic acids from viruses and cells; isolation to extract, purify, and concentrate the nucleic acids; reverse transcription (for RNA targets only); enzymatic amplification with primers that target a specific nucleic acid sequence; and detection of the amplicons. A cell-sorting or cell-fractionating step may need to precede the lysis process when it is necessary to analyze a specific subpopulation of cells in the sample. The latter is essential when one desires to detect the gene profile of rare cells such as cancer cells (Ziober et al, 2008).
Most NA amplification methods are based on PCR, which requires precisely controlled cycling of the reaction temperatures. Lately, isothermal (constant temperature) amplification methods have been gaining popularity for many applications. Isothermal amplification allows a considerable simplification of the process and is often more robust than PCR.
Although it would be desirable to cast NAT in a format similar to the LF test, and important progress in this direction has been made, current NAT procedures require a substantial amount of flow and thermal control. To demonstrate how a NAT can be implemented in a microfluidic format, we describe below one device that was developed in our group (Mauk et al, 2010). As in the case of our immunoassay cassette, we facilitate fluid pumping with pouches and store all the reagents on the chip. The pouch-based cassette evolved from an earlier design (Wang et al, 2006), in which the reagents were supplied externally with syringe pumps.
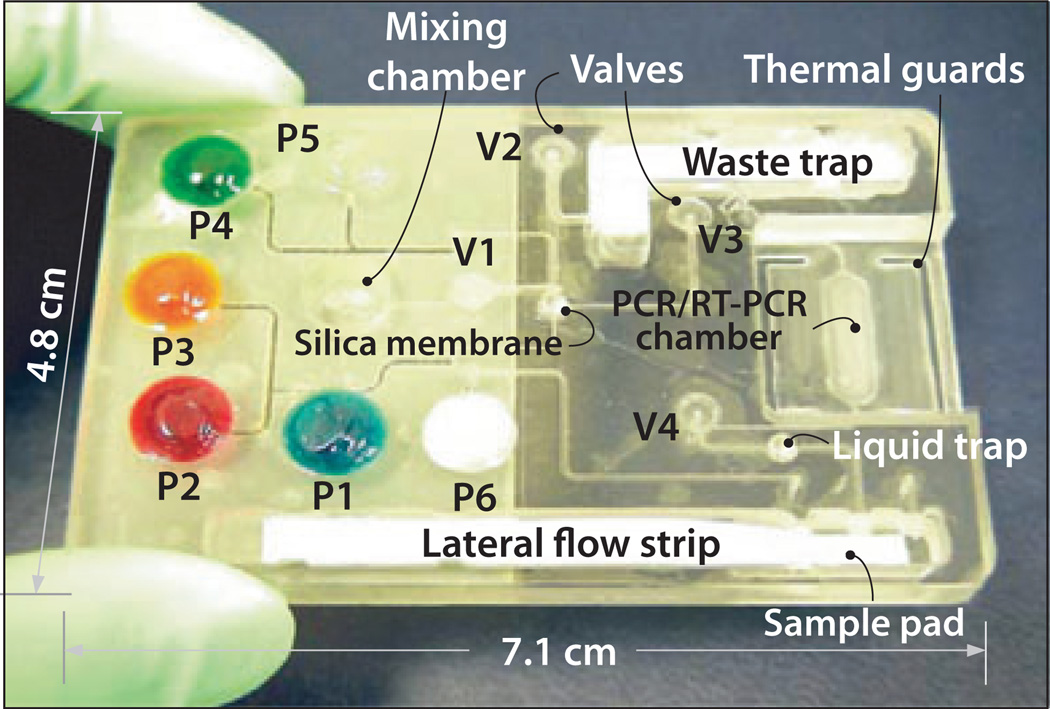
Figure 4 shows a single-use, disposable NAT cassette. The various buffers are stored in liquid form in flexible pouches on the chip. Flexible membranes serve as diaphragm valves for flow control (Chen et al, 2010). The chip is preloaded with dried PCR reagents that are reconstituted, just in time, during the test (Kim et al, 2009). Briefly, the sample is mixed and incubated with lysis agents. The mixture is then filtered through a porous silica membrane that binds the nucleic acids in the sample. The lysate filters through the silica membrane and is discharged into a waste chamber. The silica membrane is then thoroughly washed to remove residual lysis agents. Subsequent to the wash, the membrane is dried at an elevated temperature to evaporate volatile components of the wash solution such as ethanol. The nucleic acids are then eluted with water and transferred to the amplification chamber. The reagents needed for PCR and detection are encapsulated in wax and prestored in the reaction chamber while being protected from the liquids that flow through the chamber (Kim et al, 2009). The reaction chamber temperature is controlled with a thermoelectric unit. During the initial heating, the wax encapsulation melts and the reagents are released and hydrated. When the target analyte is RNA (as in the case of HIV), a reverse transcription step precedes the amplification. The amplification chamber’s temperature is then cycled among 95, 55, and 70°C. Typically, 25–35 temperature cycles are used. The amplification process can be monitored, in real time, with appropriate dyes such as SYBR green which emits light only when intercalated within a double-stranded DNA. Thus, the emitted light intensity increases as the concentration of the amplified DNA increases. In certain instances, primers can form double-stranded dimers, which will interact with the intercalating dye to generate a signal, which is indistinguishable from the one emitted by the amplicons and which may cause a false-positive reading.
Figure 4.
A disposable nucleic acid–processing cassette. The plastic cassette hosts a microfluidic network for lysis, nucleic acid isolation by solid-phase extraction, polymerase chain reaction (PCR), and detection of phosphor-labeled PCR products on a lateral flow strip. On-board, fluid-filled pouches are compressed judiciously to propel fluids in the cassette. The actuators of the pouches and valves, the heating, and (optional) real-time detection are housed in a durable analyzer/developer (not shown). The cassette does not exchange any fluids with the analyzer, eliminating the possibility of cross-contamination
To minimize false positives, the intercalating dye can be replaced with hairpin-shaped molecular beacons. Each molecular beacon consists of a fluorophore and a quencher. Upon specific binding to a target sequence, the beacon’s hairpin opens up, the fluorophore and quencher separate, and the fluorophore emits light. In contrast to the intercalating dye, the molecular beacons can emit light only when they bind specifically to the target sequence and, therefore, the test is free from false positives.
Alternatively, with the use of labeled primers, the PCR products can be detected with a LF strip at the conclusion of the amplification process. The LF–based detection precludes quantification but has the advantage of reduced cost as the test results can be read by eye. The cassette shown in Figure 4 provides for both real-time and LF–based detections. When real-time detection is used, one can also obtain a melting curve at the conclusion of the PCR process to assure that the amplification products have the appropriate number of base pairs. When the melting temperature is inconsistent with the target sequence, the test is interpreted as false positive.
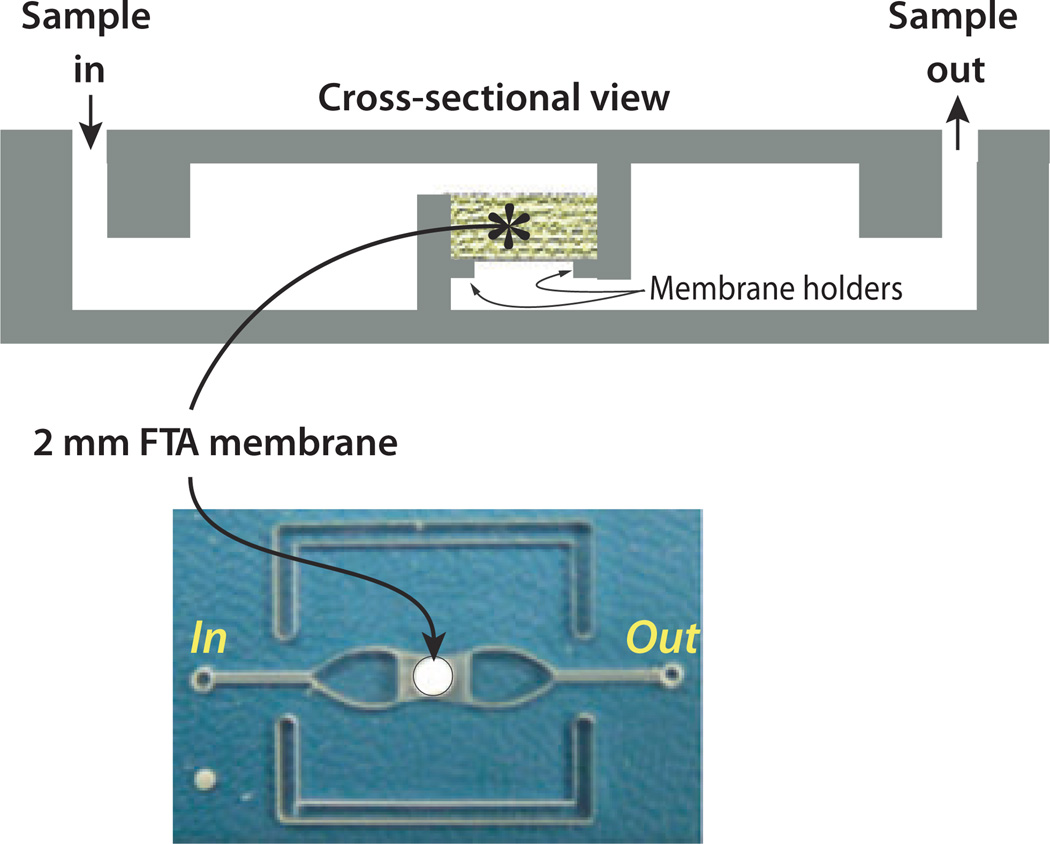
In recent years, the trend has been to reduce the complexity of the nucleic acid assay without sacrificing its performance by integrating multiple functions into a single-reaction chamber and avoiding the necessity for shuttling samples and reagents from one chamber to another. To this end, we have integrated a solid-state immobilization membrane into the reaction chamber (Kim et al, 2010; Liu et al, 2011). Figure 5 shows a chip that incorporates a Whatman FTA® (GE Healthcare, Piscataway, NJ, USA), nitrocellulose membrane for nucleic acid binding into the reaction chamber. In this chip, lysis, nucleic acid isolation, amplification, and detection are all carried out in a single chamber. The sample is mixed with lysing reagents and filtered through the FTA membrane to extract the nucleic acid targets. The sample is then discharged into a waste chamber. The membrane-bound nucleic acid is then washed. Instead of eluting the isolated nucleic acids from the membrane, the bound nucleic acids are used directly as the templates for the amplification reaction, thus eliminating the need for a separate elution step. The chip is further simplified by using fluorescence-based, real-time detection of the amplification products. The chip shown in Figure 5 was used for isothermal amplification (LAMP). Isothermal amplification requires much simpler thermal control than PCR and is more tolerant of temperature variations. As in the case of PCR, with appropriately functionalized primers, one can use a LF strip to detect the reaction products instead of a real-time detector, thus reducing the cost of the device. The temperature needed for the isothermal amplification (approximately 65°C) can be provided with an exothermic reaction such as between magnesium and water, allowing the amplification process to be carried out without any electrical power.
Figure 5.
Single-chamber chip with an integrated flow through FTA membrane for lysis, nucleic acid isolation, and real-time amplification. Top: Chip’s cross-section. Bottom: chip’s photograph
Conclusion and outlook
The convergence of microfluidics and oral-based diagnostics has made it possible to detect diseases and monitor other conditions at the point of testing. Point-of-testing diagnostics is likely to reduce the spread of diseases, facilitate intelligent, individualized treatments, and improve peoples’ standard of living and safety. Similar devices are useful for assuring the safety of water and food products as well as monitoring the environment. The devices can be used in remote areas, where laboratory facilities are lacking or overburdened, to carry out sophisticated laboratory procedures, which previously could be carried out only in centralized laboratories. The devices can also be used at home and in the clinic to tailor treatments to patients’ individual needs. In all these cases, oral fluids (saliva, gingival crevicular fluid, or mucosal transudates) are ideal samples as the collection of the fluid does not require any specialized equipment or training and presents minimal risk to the healthcare worker.
Although rapid progress has been made in recent years, the field is still in its infancy and much more can and should be done. Advances are needed in technology, in particular in the development of means for large-scale production of integrated devices and the merging of diagnostic devices with cell telephone technology to facilitate two way flow of information (the transmission of test results to the patient’s files and the accessibility of the patient’s records to the health provider at the point of testing). Advances are also needed in biochemistry. Developments in biochemistry are likely to simplify the sample preparation steps and lead to simpler, more affordable, and more reliable devices.
Point-of-care testing devices may be of special interest to the dental community as patients typically visit their dentists more often than their primary care physician. Initial screening tests could be performed and completed while the patient is still in the clinic, and, when necessary, the patient can be referred to the appropriate medical professional for confirmatory testing and subsequent treatment.
Acknowledgements
The work was supported, in part, by NIH/NIDCR Grant U01DE017855.
Footnotes
Author contribution
R. Hart and M. Mauk collected the necessary information and prepared earlier drafts of the paper. M. Mauk, R. Hart, C. Liu and X. Qiu prepared the material on the immunoassays. J. Thompson wrote the part on microbead arrays. D. Chen contributed to the part on molecular diagnostics. D. Malamud and W. Abrams contributed material on saliva-based diagnostics. H. Bau directed the engineering aspects of the project and edited the final version of the paper.
References
- Ali MF, Kirby R, Goodey AP, et al. DNA hybridization and discrimination of single-nucleotide mismatches using chip-based microbead arrays. Anal Chem. 2003;75:4732–4739. doi: 10.1021/ac034106z. [DOI] [PubMed] [Google Scholar]
- Ashihara Y, Kasahara Y, Nakamura R. Immunoassay and immunochemistry. Chapter 43. In: McPherson RA, Pincus MR, editors. Henry’s clinical diagnosis and management by laboratory methods. Philadelphia, PA, USA: Saunders Elsevier; 2007. pp. 793–818. [Google Scholar]
- Birkhahn RH, Haines E, Wen W, Reddy L, Briggs WM, Datillo PA. Estimating the clinical impact of bringing a multimarker cardiac panel to the bedside in the ED. [accessed on 5 January 2011];Am J Emerg Med. 2010 doi: 10.1016/j.ajem.2011.01.003. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20825823. [DOI] [PubMed]
- Blicharz TM, Siqueira WL, Helmerhorst EJ, et al. Fiber-optic microsphere-based antibody array for the analysis of inflammatory cytokines in saliva. Anal Chem. 2009;81:2106–2114. doi: 10.1021/ac802181j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden M, Song L, Walt DR. Development of a microfluidic platform with an optical imaging microarray capable of attomolar target DNA detection. Anal Chem. 2005;77:5583–5588. doi: 10.1021/ac050503t. [DOI] [PubMed] [Google Scholar]
- Chen D, Mauk M, Qiu X, et al. An integrated, self-contained microfluidic cassette for isolation, amplification, and detection of nucleic acids. Biomed Microdevices. 2010;12:705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulides N, Mohanty S, Miller CS, et al. Application of microchip assay system for the measurement of C-reactive protein in human saliva. Lab Chip. 2005;5:261–269. doi: 10.1039/b414194f. [DOI] [PubMed] [Google Scholar]
- Corstjens PLAM, Malamud D. Point-of-care diagnostics for infectious diseases. In: Wong DT, editor. Saliva Diagnostics. Ames: Wiley-Blackwell; 2008. pp. 136–149. [Google Scholar]
- Corstjens PLAM, Chen Z, Zuiderwijk M, et al. Rapid assay format for multiplex detection of humoral immune responses to infectious disease pathogens (HIV, HCV, and TB) Ann N Y Acad Sci. 2007;1098:437–445. doi: 10.1196/annals.1384.016. [DOI] [PubMed] [Google Scholar]
- Derveaux S, Stubbe BG, Braeckmans K, et al. Syner-gism between particle-based multiplexing and microfluidics technologies may bring diagnostics closer to the patient. Anal Bioanal Chem. 2008;391:2453–2467. doi: 10.1007/s00216-008-2062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [accessed on 5 January 2011];Dräger USA – Home. Available at: http://www.draeger.com/US/en_US/
- Hashmi G, Shariff T, Seul M, et al. A flexible array format for large-scale, rapid blood group DNA typing. Transfusion. 2005;45:680–688. doi: 10.1111/j.1537-2995.2005.04362.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Byun D, Mauk MG, Bau HH. A disposable, self-contained PCR chip. Lab Chip. 2009;9:606–612. doi: 10.1039/b807915c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Mauk M, Chen D, et al. A PCR reactor with an integrated alumina membrane for nucleic acid isolation. Analyst. 2010;135:2408–2414. doi: 10.1039/c0an00288g. [DOI] [PubMed] [Google Scholar]
- Lee-Lewandrowski E, Lewandrowski K. Perspectives on cost and outcomes for point-of-care testing. Clin Lab Med. 2009;29:479–489. doi: 10.1016/j.cll.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Liu C, Qiu X, Ongagna S, et al. A timer-actuated immunoassay cassette for detecting molecular markers in oral fluids. Lab Chip. 2009a;9:768–776. doi: 10.1039/b814322f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Schrlau MG, Bau HH. Single bead-based electrochemical biosensor. Biosens Bioelectron. 2009b;25:809–814. doi: 10.1016/j.bios.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Geva E, Mauk M, et al. An isothermal amplification reactor with an integrated isolation membrane for point-of-care detection of infectious diseases. Analyst. 2011 doi: 10.1039/c1an00007a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud D. Saliva as a diagnostic fluid. Dent Clin North Am. 2011;55:159–178. doi: 10.1016/j.cden.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud D, Niedbala RS. Oral-based diagnostics. Ann N Y Acad Sci. 2007;1098:xiv–xxv. doi: 10.1196/annals.1384.024. [DOI] [PubMed] [Google Scholar]
- Mauk M, Qiu X, Liu C, et al. An integrated, self-contained microfluidic cassette for isolation, amplification, and detection of nucleic acids. Biomed Microdevices. 2010;12:705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Katamine S, Yamamoto T, et al. Amplification and detection of a single molecule of human immunodeficiency virus RNA. Virus Genes. 1993;7:325–338. doi: 10.1007/BF01703389. [DOI] [PubMed] [Google Scholar]
- [accessed on 5 January 2011];OraSure Technologies, Inc. Available at: http://www.orasure.com/
- Paegel BM, Joyce GF. Microfluidic compartmentalized directed evolution. Chem Biol. 2010;17:717–724. doi: 10.1016/j.chembiol.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothier K, Kirtland M, Gupta S. Has point-of-care come of age? Point Care. 2010;9:147–150. [Google Scholar]
- Qian S, Bau HH. A mathematical model of lateral flow bioreactions applied to sandwich assays. Anal Biochem. 2003;322:89–98. doi: 10.1016/j.ab.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Qian S, Bau HH. Analysis of lateral flow biodetectors: competitive format. Anal Biochem. 2004;326:211–224. doi: 10.1016/j.ab.2003.12.019. [DOI] [PubMed] [Google Scholar]
- Qiu X, Thompson JA, Chen Z, et al. Finger-actuated, self-contained immunoassay cassettes. Biomed Microdevices. 2009;11:1175–1186. doi: 10.1007/s10544-009-9334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Liu C, Mauk MG, et al. A portable analyzer for pouch-actuated, immunoassay cassettes. Sens Actuators B Chem. 2011 doi: 10.1016/j.snb.2011.08.012. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [accessed on 5 January 2011];Salivary Assay – Salivary Cortisol – Salimetrics. Available at: http://www.salimetrics.com/
- Sato K, Tokeshi M, Odake T, et al. Integration of an immunosorbent assay system: analysis of secretory human immunoglobulin A on polystyrene beads in a microchip. Anal Chem. 2000;72:1144–1147. doi: 10.1021/ac991151r. [DOI] [PubMed] [Google Scholar]
- Sato K, Tokeshi M, Kimura H, Kitamori T. Determination of carcinoembryonic antigen in human sera by integrated bead-bed immunoassay in a microchip for cancer diagnosis. Anal Chem. 2001;73:1213–1218. doi: 10.1021/ac000991z. [DOI] [PubMed] [Google Scholar]
- Sato K, Yamanaka M, Takahashi H, Tokeshi M, Kimura H, Kitamori T. Microchip-based immunoassay system with branching multichannels for simultaneous determination of interferon-gamma. Electrophoresis. 2002;23:734–739. doi: 10.1002/1522-2683(200203)23:5<734::AID-ELPS734>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Steemers FJ, Gunderson KL. Whole genome genotyping technologies on the BeadArray platform. Biotechnol J. 2007;2:41–49. doi: 10.1002/biot.200600213. [DOI] [PubMed] [Google Scholar]
- Thompson JA, Bau HH. Microfluidic, bead-based assay: theory and experiments. J Chromatogr B Analyt Technol Biomed Life Sci. 2010a;878:228–236. doi: 10.1016/j.jchromb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Bau HH. Microfluidic, porous bead-based assay: theory and experiments. Proceedings of USNCTAM2010. 2010b doi: 10.1016/j.jchromb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Du X, Grogan JM, Schrlau MG, Bau HH. Polymeric microbead arrays for microfluidic applications. J Micromech Microeng. 2010;20:115017. [Google Scholar]
- Verpoorte E. Beads and chips: new recipes for analysis. Lab Chip. 2003;3:60N–68N. doi: 10.1039/b313217j. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen Z, Corstjens PLAM, Mauk MG, Bau HH. A disposable microfluidic cassette for DNA amplification and detection. Lab Chip. 2006;6:46–53. doi: 10.1039/b511494b. [DOI] [PubMed] [Google Scholar]
- Weigl B, Domingo G, LaBarre P, Gerlach J. Towards non- and minimally instrumented, microfluidics-based diagnostic devices. Lab Chip. 2008;8:1999–2014. doi: 10.1039/b811314a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R, Tse H. Lateral flow immunoassay. 1st edn. New York: Humana Press; 2008. [Google Scholar]
- Yung TKF, Chan KCA, Mok TSK, Tong J, To K, Lo YMD. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- Ziober BL, Mauk MG, Falls EM, Chen Z, Ziober AF, Bau HH. Lab-on-a-chip for oral cancer screening and diagnosis. Head Neck. 2008;30:111–121. doi: 10.1002/hed.20680. [DOI] [PubMed] [Google Scholar]