Abstract
Bacterial recognition of host sialic acid-containing receptors plays an important role in microbial colonization of the human oral cavity. The sialic acid-binding adhesin of Streptococcus gordonii DL1 was previously associated with the hsa gene encoding a 203-kDa protein. The predicted protein sequence consists of an N-terminal nonrepetitive region (NR1), including a signal sequence, a relatively short serine-rich region (SR1), a second nonrepetitive region (NR2), a long serine-rich region (SR2) containing 113 dodecapeptide repeats, and a C-terminal cell wall anchoring domain. In the present study, the contributions of SR1, NR2, and SR2 to Hsa-mediated adhesion were assessed by genetic complementation. Adhesion of an hsa chromosomal deletion mutant to sialic acid-containing receptors was restored by plasmids containing hsa constructs encoding Hsa that lacked either the N- or C-terminal portion of SR2. In contrast, hsa constructs that lacked the coding sequences for SR1, NR2, or the entire SR2 region failed to restore adhesion. Surface expression of recombinant Hsa was not affected by removal of SR1, NR2, or a portion of SR2 but was greatly reduced by complete removal of SR2. Wheat germ agglutinin, a probe for Hsa-specific glycosylation, reacted with recombinant Hsa lacking SR1, NR2, or SR2 but not with recombinant Hsa lacking both SR1 and SR2. Significantly, the aggregation of human platelets by S. gordonii DL1, an interaction implicated in the pathogenesis of infective endocarditis, required the expression of hsa. Moreover, neuraminidase treatment of the platelets eliminated this interaction, further supporting the hypothesis that Hsa plays an essential role in the bacterium-platelet interaction.
Strains of Streptococcus gordonii, Streptococcus sanguinis, and Streptococcus oralis adhere to saliva-coated hydroxyapatite, an experimental model of the tooth surface, and also attach to host cells, including erythrocytes (RBC) and polymorphonuclear leukocytes (5, 6, 11, 16, 19). A common mechanism involved in these interactions is recognition of surface-associated host sialoglycoconjugates. Binding of S. gordonii DL1 to such receptors depends on the Hs antigen, a high-molecular-weight streptococcal surface component that appears to be glycosylated based on the reaction of this component with wheat germ agglutinin (WGA) and its labeling following periodate oxidation (25). The gene encoding this antigen, designated hsa, was identified by its expression in Escherichia coli by using anti-Hs antibody and was subsequently deleted from the S. gordonii DL1 chromosome (23). This deletion eliminated Hs antigen production and bacterial adhesion to immobilized sialic acid-containing receptors. Moreover, expression of hsa from a streptococcal plasmid restored these properties, firmly associating this gene with the sialic acid-binding adhesin of strain DL1 (23).
The hsa gene encodes a large (203-kDa) serine-rich repeat protein composed of 2,178 amino acid residues. The predicted protein sequence includes an N-terminal nonrepetitive region (NR1), a serine-rich repeat region (SR1), another nonrepetitive region (NR2), another serine-rich repeat region (SR2), and a C-terminal cell wall anchoring domain. Most of NR1 consists of a putative signal sequence, and this region along with SR1 and NR2 comprises the first 450 amino acid residues of the Hsa sequence. The remainder of this sequence includes SR2, which contains more than 100 dodecapeptide repeats with the consensus sequence SASTSASVSASE, and the cell wall anchoring domain (23). Serine-rich repeat proteins also occur on other viridans group streptococci; these proteins include Fap1 of Streptococcus parasanguinis, which mediates bacterial adhesion to saliva-coated hydroxyapatite (29), and GspB of S. gordonii M99, which mediates bacterial binding of human platelets (1). The occurrence of the latter interaction raises the possibility that serine-rich repeat proteins play a role in the pathogenesis of streptococcus-induced infective endocarditis, which is thought to involve platelet-streptococcus interactions (8). The structural specificity of these interactions is not yet known.
It has previously been suggested that the putative sialic acid-binding domain of Hsa is associated with the N-terminal region of the protein, through NR2, and that this domain is presented on the bacterial surface at the end of a long molecular stalk formed by SR2 (23). To assess this model, in this study we examined hsa constructs that specifically lack the coding regions for SR1, NR2, or SR2 in order to determine their abilities to restore Hsa-mediated adhesion of an hsa deletion mutant of strain DL1. In addition, by using genetic complementation we found that the biological role of Hsa-mediated adhesion extends to the aggregation of human platelets, thereby raising the possibility that sialic acid-binding adhesins of oral viridans group streptococci contribute not only to oral colonization but also to the pathogenesis of infective endocarditis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All streptococci and plasmids used in this study are listed in Table 1. Streptococci were cultured overnight at 37°C in complex medium containing 0.5% tryptone, 0.5% yeast extract, 0.5% K2HPO4, 0.05% Tween 80, and 0.2% glucose (15) or on plates of brain heart infusion agar (Difco Laboratories, Detroit, Mich.). These media were supplemented, as needed, with 200 μg of spectinomycin dihydrochloride per ml or 8 μg of chloramphenicol per ml, both of which were obtained from Sigma-Aldrich, St. Louis, Mo.
TABLE 1.
S. gordonii strains and plasmids
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| DL1 (= Challis) | Wild type | 11 |
| EM230 | DL1 hsa::ermAM | 23 |
| CM100 | DL1 Δhsa cat | 23 |
| Plasmids | ||
| pAS40S | P15Aori pVA380-1rep, Spr, resident streptococcal plasmid | 21 |
| pAS8741 | pAS40S carrying hsa within a 7.4-kb HindIII-SphI cloned fragment of S. gordonii DL1 genomic DNAb | 23 |
| pAS8748 | pAS8741 with the 1.6-kb HindIII-BglII fragment replaced by DNA fragment F8748; encodes HsaΔ105-446b | This study |
| pAS8744 | pAS8741 with the 1.6-kb HindIII-BglII fragment replaced by DNA fragment F8744; encodes HsaΔ105-245b | This study |
| pAS8746 | pAS8741 with the 1.6-kb HindIII-BglII fragment replaced by DNA fragment F8746; encodes HsaΔ246-446b | This study |
| pAS8749 | pAS8741 with the 2.9-kb BglII-SalI fragment replaced by the A8749 adapter sequence; encodes HsaΔ448-1459b | This study |
| pAS8375 | pAS8741 with the 3.7-kb SalI-SphI fragment replaced by DNA fragment F05; encodes HsaΔ1092-2143b | This study |
| pAS8165 | pAS8741 with the 5.8-kb BglII-SphI fragment replaced by DNA fragment F05; encodes HsaΔ448-2143b | This study |
| pAS8164 | pAS8741 the 7.4-kb HindIII-SphI fragment replaced by DNA fragment F8164; encodes HsaΔ105-245, Δ448-2143b | This study |
DNA manipulations.
Restriction enzymes, T4 DNA ligase, and calf intestinal alkaline phosphatase were purchased from Takara Bio Inc. (Kusatsu, Japan) and were used according to the supplier's instructions. PCR was performed with the primer pairs and templates listed in Table 2 by using Vent DNA polymerase (New England Biolabs Inc., Beverly, Mass.). Other procedures were performed by using standard methods (20).
TABLE 2.
PCR-amplified DNA fragments and adapter used for preparation of hsa deletion constructs
| DNA fragment (coding region)a | PCR primer pair or DNA adapter sequenceb | PCR template |
|---|---|---|
| F8748 (P-NR1) | GCCAAGCTTGGCTACAGTTATAGCTG, CGCGGATCCCGTGTATCAATTACTTTTTC | pAS8741 |
| F8744UP (P-NR1) | GCCAAGCTTGGCTACAGTTATAGCTG, GAGGAGCTTCTGTGTCCCGTGTATCAATTACTTTTTC | pAS8741 |
| F8744DN (NR2) | GATACACGGGACACAGAAGCTCCTCAAGTGAAATC, AATCTGAAGATCTTTAACTACTCTG | pAS8741 |
| F8744 (P-NR1-NR2) | GCCAAGCTTGGCTACAGTTATAGCTG, AATCTGAAGATCTTTAACTACTCTG | F8744UP/F8744DN mixture |
| F8746 (P-NR1-SR1) | GCCAAGCTTGGCTACAGTTATAGCTG, GCCTGATCATTCGCGGCTACCGCACGCC | pAS8741 |
| F05 (CWAD) | GCGAGATCTGTCGACGCTTCCTCGAACAGGTGAATCTG, GGCGCATGCTAACAAAACACTCTGTTGG | pAS8741 |
| F8164DN (NR2-CWAD) | GATACACGGGACACAGAAGCTCCTCAAGTGAAATC, GGCGCATGCTAACAAAACACTCTGTTGG | pAS8165 |
| F8164 (P-NR1-NR2-CWAD) | GCCAAGCTTGGCTACAGTTATAGCTG, GGCGCATGCTAACAAAACACTCTGTTGG | F8744UP/F8164DN mixture |
| A8749 (adapter) | GATCTTTCCGCG, TCGACGCGGAAA | None |
P, putative promoter region of hsa; NR1 and NR2, nonrepetitive regions; SR1 and SR2, serine-rich repetitive regions; CWAD, cell wall anchoring domain.
Incorporated restriction sites are underlined.
Preparation of hsa deletion constructs.
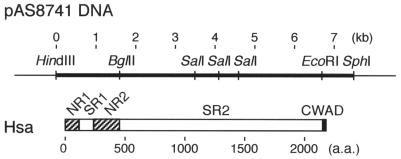
Each hsa deletion plasmid listed in Table 1 was prepared from pAS8741, which carries hsa in a 7.4-kb fragment of cloned S. gordonii DL1 genomic DNA (Fig. 1). Deletions in hsa were created by removal of a specific DNA fragment by restriction endonuclease digestion, followed by in-frame ligation of a PCR product or DNA adapter (Table 2), thereby creating a new plasmid construct (Table 1 and Fig. 2). The coding sequences of NR1 and NR2 in DNA fragments F8744 and F8164 were linked by overlap extension PCR (10) by using template fragments prepared with appropriately designed primers. Specifically, the 5′ end of the reverse primer used to synthesize upstream DNA fragment F8744UP was designed so that it was complementary to the 5′ ends of the forward primers used to synthesize downstream DNA fragments F8744DN and F8164DN (Table 2). Plasmid constructs were transformed as previously described (18) into S. gordonii CM100 and were purified by the method of Takamatsu et al. (26) from transformants grown with spectinomycin and chloramphenicol. Each deletion in hsa was confirmed by digestion of the corresponding plasmid with appropriate restriction endonucleases. In addition, the reading frame and fidelity of each PCR-amplified region introduced into a plasmid construct was verified by DNA sequencing performed with an AutoRead sequencing kit (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) and an A.L.F. DNA sequencer (Amersham Pharmacia Biotech).
FIG. 1.
Alignment of the physical map of the 7.4-kb HindIII-SphI fragment of S. gordonii DL1 chromosomal DNA in pAS8741 with the nonrepetitive regions (NR1 and NR2), serine-rich repetitive regions (SR1 and SR2), and cell wall anchoring domain (CWAD) of the encoded Hsa protein. a.a, amino acid.
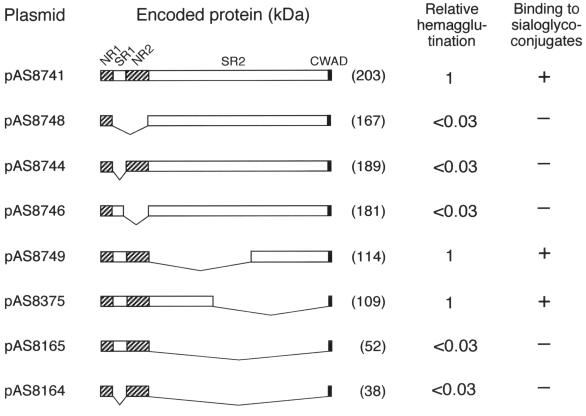
FIG. 2.
Restoration of Hsa-mediated adhesion by genetic complementation of S. gordonii CM100 with plasmids expressing hsa (i.e., pAS8741) or hsa deletion constructs. The positions of the nonrepetitive regions (NR1 and NR2), serine-rich repetitive regions (SR1 and SR2), and cell wall anchoring domain (CWAD) of Hsa are indicated above the diagram of the complete protein encoded by pAS8741. The V-shaped lines indicate the regions removed from recombinant proteins by in-frame deletions in plasmid-borne hsa. The hemagglutinating activity of each plasmid-bearing transformant is expressed relative to that of CM100(pAS8741). Bacterial binding to immobilized sialoglycoconjugates is indicated by a plus or minus sign.
Immunological procedures and lectin overlay.
Rabbit antisera against the Hs antigen (anti-Hs) and S. gordonii DL1 (anti-DL1) have been described previously (25). Biotinylated succinyl WGA (WGA-biotin) was purchased from EY Laboratories, Inc., San Mateo, Calif.
Dot immunoblotting was performed to detect Hs antigen on bacterial cells. Nitrocellulose membranes were spotted with 1 μl of each streptococcal cell suspension (1 × 109 cells per ml), blocked, incubated with primary antibody or biotinylated WGA, and developed with peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Bio-Rad Laboratories, Richmond, Calif.) or avidin D-horseradish peroxidase (Vector Laboratories, Inc., Burlingame, Calif.), respectively, as previously described (25).
Bacteria harvested from cultures in the logarithmic phase of growth were digested with mutanolysin (Sigma-Aldrich) in the presence of raffinose (26%, wt/vol) as previously described (4). Solubilized cell wall proteins and protoplasts were collected in separate fractions following centrifugation of each digest. The spent culture medium was also collected, filtered to remove residual bacteria, and concentrated by ultrafiltration by using a Vivapore 10/20 concentrator (Vivascience Limited, Gloucestershire, United Kingdom). Bacterial antigens were also prepared by sonication of streptococci as previously described (23, 25). The protein concentrations in all samples were determined by using the bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.). Samples were boiled for 10 min in reducing sample buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using 5 or 10% polyacrylamide gels (12), and electrophoretically transferred to nitrocellulose membranes (27) for Western blotting with anti-Hs (1:100 dilution) or for lectin overlay analysis with WGA-biotin (50 μg/ml) as previously described (25).
Bacterial adhesion.
Bacterium-mediated hemagglutination was performed as previously described (25) by mixing serial dilutions of each transformant, starting with 1 × 109 bacteria per ml, with a constant amount of human type O RBC. The relative hemagglutinating activity of each plasmid-bearing transformant was calculated with respect to the hemagglutinating activity of strain CM100 harboring pAS8741, which exhibited an endpoint titer of 1:32. Adhesion of NHS-LC-biotin (Pierce)-labeled bacteria to sialoglycoconjugates immobilized on nitrocellulose has also been described previously (25). The glycoconjugates tested as adhesion receptors included fetuin (Sigma-Aldrich) and NeuAcα2-3Galβ1-4(Glc)-HSA (Accurate Chemical and Scientific Corp., Westbury, N.Y.), a neoglycoprotein prepared by conjugation of the reduced oligosaccharide to human serum albumin through an acetylphenylenediamine spacer.
The platelet bacterial adhesion assay (2, 9) was performed by mixing 50 μl of washed platelets (5 × 108 platelets/ml) with 50 μl of bacterial suspension (2 × 109 cells/ml) in individual wells of microtiter plates. Supernatants containing unaggregated bacteria and platelets were harvested following low-speed centrifugation (35 × g) and were diluted 1:2 prior to measurement of A620. The platelets utilized in these assays were harvested by centrifugation of platelet-rich plasma from citrated type O blood and were washed with Tris-buffered saline (TBS) containing 10 mM EDTA or 0.1 mM CaCl2. Adhesion assays were also performed with platelets that were washed following treatment with 1 U of type X neuraminidase from Clostridium perfringens (Sigma-Aldrich) per ml for 30 min at 37°C to remove sialic acid.
RESULTS
Characterization of hsa deletion constructs by genetic complementation and Western blotting of recombinant Hsa proteins.
The expression of hsa by pAS8741 was evident from the ability of this plasmid to complement the hsa deletion in S. gordonii CM100, which restored bacterial adhesion of the resulting transformant to RBC and immobilized sialoglycoconjugates (23) (Fig. 2). Plasmid derivatives of pAS8741 with in-frame deletions in the coding regions for SR1, NR2, and SR2 of Hsa were also examined for genetic complementation in strain CM100. The coding region for NR1, which includes a predicted (17) 90-amino-acid signal sequence, was maintained in each hsa construct to facilitate surface expression of the encoded recombinant protein in S. gordonii. Likewise, the coding region for the cell wall anchoring domain was maintained in each construct as previous deletion of this region from hsa in pAS8741 eliminated the ability of this plasmid to restore Hsa-mediated adhesion in strain CM100 (23).
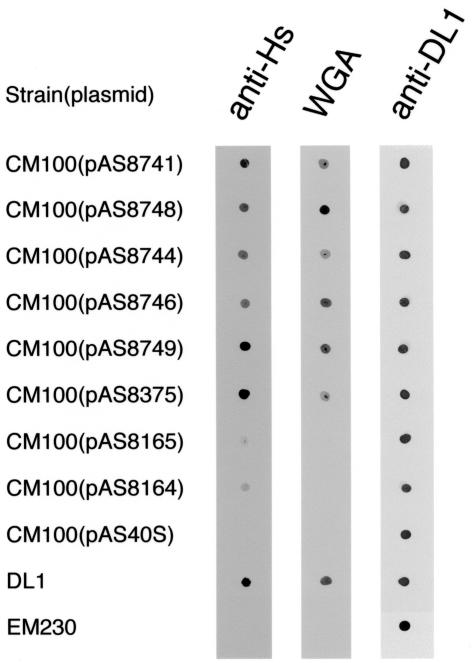
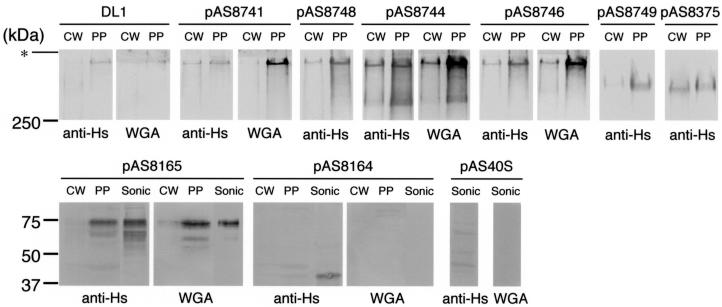
Expression of hsa constructs that lacked the coding regions for SR1 and/or NR2 failed to restore adhesion. Transformants harboring these plasmids (pAS8748, pAS8744, and pAS8746) lacked hemagglutinating activity and failed to bind immobilized sialoglycoconjugates (Fig. 2). Each transformant did, however, react with anti-Hs (Fig. 3), suggesting that there was surface expression of an immunoreactive protein. Such proteins also were readily detected by Western blotting of concentrated culture supernatants (results not shown), as well as cell wall and protoplast proteins prepared by mutanolysin digestion of the transformants (Fig. 4). However, the electrophoretic mobilities of these proteins were similar and indistinguishable from the mobility of full-length Hsa produced by wild-type strain DL1 or strain CM100 harboring pAS8741. In all cases, the major band of anti-Hs immunoreactive protein migrated above the 250-kDa marker, near the top of the 5% gel (Fig. 4), rather than in the 180- to 200-kDa region expected based on the gene sequences of the various constructs examined (Fig. 2).
FIG. 3.
Binding of anti-Hs (1:100), WGA-biotin (50 μg/ml), and anti-DL1 (1:1,000) to S. gordonii CM100 harboring different plasmids. Wild-type strain DL1 and the hsa insertional mutant strain EM230 were also included. Washed bacteria were spotted on three separate nitrocellulose membranes. Each membrane was incubated with primary antibody followed by peroxidase-conjugated goat anti-rabbit IgG or with WGA-biotin followed by avidin. The membrane labeled with anti-DL1 was included as a positive control for each strain.
FIG. 4.
Western and lectin blotting of streptococcal cell wall (CW) and protoplast (PP) proteins of S. gordonii DL1 and strain CM100 harboring different plasmids. The material in each sample was obtained from 1 ml of bacterial culture. Sonic extracts (Sonic) (50 μg of total protein/lane) of CM100(pAS8165), CM100(pAS8164), and CM100(pAS40S) were also examined. SDS-PAGE was performed with 5% polyacrylamide gels (upper panels) or 10% polyacrylamide gels (lower panels). Nitrocellulose transfers were incubated with anti-Hs (1:100) followed by peroxidase-conjugated goat anti-rabbit IgG or with WGA-biotin (50 μg/ml) followed by avidin. The boundary between the stacking and separating gels (asterisk) and the positions of molecular mass markers are indicated on the left.
Plasmids pAS8749 and pAS8375, with overlapping deletions in the coding region for SR2, restored Hsa-mediated adhesion in strain CM100 (Fig. 2). Accordingly, anti-Hs immunoreactive proteins were detected on transformants harboring these plasmids (Fig. 3) and in the cell wall and protoplast proteins prepared from these bacteria (Fig. 4). During SDS-PAGE, the recombinant proteins encoded by pAS8749 and pAS8375 migrated between full-length Hsa and the 250-kDa marker, far above the region expected based on the calculated molecular masses of the proteins (114 and 109 kDa, respectively) (Fig. 2).
Plasmid pAS8165, obtained by removal of the entire coding region for SR2, did not restore Hsa-mediated adhesion in strain CM100 (Fig. 2). Anti-Hs reacted weakly or not at all with CM100(pAS8165) (Fig. 3) and with the cell wall proteins solubilized by mutanolysin digestion of this transformant (Fig. 4), indicating that surface-expressed recombinant Has was absent. The protoplast fraction and sonic extract prepared from this transformant did, however, contain recombinant Hsa (Fig. 4). The major immunoreactive protein migrated with the 75-kDa marker, significantly above the calculated molecular mass (52 kDa).
Plasmid pAS8164, which lacked the coding regions for both SR1 and SR2, also failed to restore Hsa-mediated adhesion (Fig. 2). An anti-Hs immunoreactive protein, although not detected on the surface of CM100(pAS8164) (Fig. 3 and 4), was detected as a faint band in the protoplast fraction and more strongly in a sonic extract of this transformant (Fig. 4). A similar band was not observed in the corresponding control sonic extract prepared from strain CM100 harboring pAS40S, which lacked hsa. The recombinant Hsa protein encoded by pAS8164 migrated slightly above the 37-kDa marker, near the position expected based on the predicted sequence of Hsa lacking both serine-rich regions.
Hsa-specific glycosylation detected by binding of WGA.
The previously observed reaction of WGA with the Hs antigen of S. gordonii DL1 prompted further consideration of this GlcNAc-binding lectin as a possible probe for Hsa-specific glycosylation. WGA reacted specifically with each strain that surface expressed Hsa (Fig. 3) and with solubilized wild-type and recombinant Hsa that lacked SR1, NR2, or SR2 (Fig. 2 and 4) (the recombinant proteins produced from pAS8744, pAS8746, and pAS8765, respectively). This lectin also reacted with recombinant Hsa produced from pAS8748, which lacked SR1 and NR2, and with the proteins encoded by pAS8749 and pAS8375, which lacked portions of SR2 (results not shown). However, recombinant Hsa that lacked both SR1 and SR2 (i.e., the protein produced from pAS8164) failed to react with WGA. This protein was detected by Western blotting with anti-Hs in the sonic extract of CM100(pAS8164) and was not detected following incubation of an identical transfer with WGA (Fig. 4). Thus, binding of WGA appeared to involve the serine-rich regions of Hsa.
Role of Hsa in bacterial adhesion to human platelets.
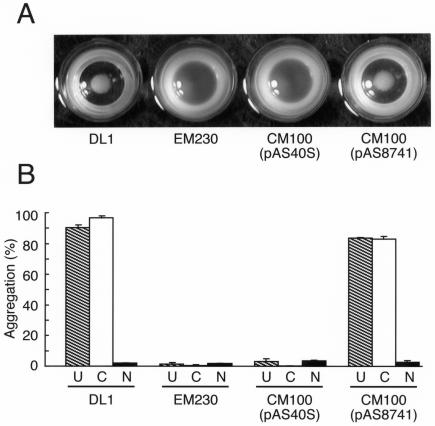
The recent finding that GspB, the serine-rich repeat protein of S. gordonii M99, mediates the adhesion of this strain to human platelets (1) prompted us to examine the possible role of Hsa in platelet binding. Strong aggregation occurred when S. gordonii DL1 was mixed with human platelets (Fig. 5A). This interaction was noted in both EDTA- and Ca2+-containing buffers but not following pretreatment of the platelets with neuraminidase (Fig. 5B). In identical assays, strain EM230, an hsa insertional mutant, and strain CM100 harboring control plasmid pAS40S failed to interact with platelets (Fig. 5). Significantly, platelet-aggregating activity was fully restored in strain CM100 by expression of hsa from plasmid pAS8741 (Fig. 5), firmly establishing the role of this gene and its encoded protein in the bacterium-platelet interaction.
FIG. 5.
Essential role of Hsa in adhesion of S. gordonii DL1 to human platelets. (A) Microtiter wells containing washed platelets and bacteria, showing visible aggregation in wells containing bacteria that surface express Hsa (wild-type strain DL1 and strain CM100 harboring pAS8741) and no aggregation in wells containing bacteria that do not produce Hsa (strain EM230, an hsa insertional mutant, and strain CM100 harboring control plasmid pAS40S). (B) Aggregation of untreated platelets in TBS-EDTA (U) (cross-hatched bars) or in TBS-Ca2+ (C) (open bars) and aggregation of neuraminidase-treated platelets (N) (solid bars).
DISCUSSION
The present findings provide insight into the distinct roles of the SR1, NR2, and SR2 regions in Hsa-mediated bacterial adhesion. As shown in Fig. 2, adhesion of the hsa chromosomal deletion mutant S. gordonii CM100 to sialic acid-containing receptors was restored by expression of hsa on pAS8741. Plasmids expressing copies of this gene with in-frame deletions that removed the coding regions for SR1, NR2, or SR2 did not restore adhesion. The recombinant proteins lacking SR1 or NR2 were surface expressed, suggesting that their failure to mediate adhesion most likely reflected their failure to bind receptor. In contrast, recombinant Hsa lacking SR2 (i.e., the protein encoded by pAS8165) was not surface expressed (Fig. 3 and 4), precluding any assessment of receptor binding. Adhesion was, however, restored by recombinant proteins that contained either the N- or C-terminal portion of SR2, in addition to SR1 and NR2. The finding that Hsa-mediated adhesion does not require a specific segment of SR2 suggests that this region is not directly involved in receptor binding. Instead, our findings support the hypothesis that SR2 functions in presentation of the adhesin binding site, which requires both SR1 and NR2 for activity. However, we cannot exclude the possibility that the loss of adhesion associated with the removal of either SR1 or NR2 depends on altered folding of the resulting recombinant protein. Clearly, further studies are required to define the roles of SR1 and NR2 in receptor binding, as well as the minimum length of SR2 that is required for Hsa-mediated adhesion.
The finding that WGA reacted with recombinant proteins that lacked SR1, NR2, or SR2 but not with the recombinant protein that lacked both SR1 and SR2 strongly suggests that the serine-rich regions of Hsa are glycosylated. This possibility is also consistent the anomalous migration of Hsa in SDS-PAGE, which was observed with recombinant proteins that contained either SR1 or SR2 but not with the recombinant protein encoded by pAS8164, which lacked both serine-rich regions. Glycosylation of the serine-rich regions in another serine-rich repeat protein, Fap1 of S. parasanguinis, has also been suggested by the results of a comparative carbohydrate analysis of this protein and a Fap1 glycopeptide (22). Likewise, the previously observed binding of WGA to a fimbrial glycoprotein of Streptococcus salivarius (13) is of interest since this reaction may involve glycosylated peptide repeats similar to those found in Hsa. Extensive O glycosylation, like that noted for mucin-type glycoproteins (3, 7), may confer an extended rod-shaped conformation on the serine-rich regions of Hsa, especially SR2, which would enable this region to function as a molecular stalk for cell surface presentation of the putative receptor binding domain.
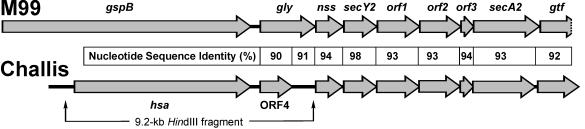
The synthesis and export of GspB, the serine-rich repeat protein of S. gordonii M99, depend on an accessory sec locus immediately downstream of gspB (1) (Fig. 6). Interestingly, a similar locus occurs downstream of hsa (Fig. 6), as shown by analysis of this region obtained from the genomic database for S. gordonii Challis (i.e., DL1) available at The Institute of Genomic Research website (http://www.tigr.org/tdb/mdb/mdbinprogress.html). The only significant difference between these regions in the two strains that has been found involves the gene identified as gly in strain M99, which encodes a putative glycosyltransferase. In strain M99, this gene appears to be complete, while in strain Challis it is interrupted by deletion of a single adenine at position 1083 of the complete 2,049-bp sequence, which gives rise to a terminator codon 133 nucleotides downstream of the deletion. Thus, while gly may have a role in GspB synthesis, the truncated gene in strain DL1, which was previously designated ORF4 (23), is most likely inactive. Indeed, the deletion of hsa and virtually all the gly-related sequence from strain CM100, which lacks a 9.2-kb HindIII fragment of chromosomal DNA (Fig. 6), is complemented by pAS8741 (Fig. 1), which contains hsa and only 0.2 kb of the gly-related sequence. In strain M99, two genes further downstream, secY2 and secA2, encode proteins that function in the specific export of GspB. Disruption of either gene eliminated surface expression of GspB, resulting in accumulation of this protein in the cytoplasm (1). Interestingly, the recombinant protein encoded by pAS8165, which lacked SR2 (Fig. 2), also accumulated in the cytoplasm, raising the possibility that this region contains the signal for secY2-secA2-dependent export of Hsa from strain DL1.
FIG. 6.
Comparison of the accessory sec locus downstream of gspB in S. gordonii M99 (1) with a similar locus downstream of hsa in S. gordonii Challis (i.e., strain DL1). Nucleotide sequence identities between corresponding genes were calculated with the program BESTFIT (GCG Wisconsin package). The deletion mutant strain CM100, which was used in genetic complementation studies, lacked the 9.2-kb HindIII fragment identified in the open reading frame diagram of strain Challis.
The corresponding NR1 sequences and serine-rich repeats of GspB and Hsa are nearly identical, while the NR2 sequences of these proteins are less than 50% similar (1). Despite the difference in NR2, both GspB and Hsa mediate bacterial adhesion to human platelets (1) (Fig. 5). While the receptor binding specificity of GspB is unknown, Hsa binds α2-3-linked sialic acid termini of O-glycosylated mucin-type glycoproteins, including salivary mucin MG2 (23, 24, 25) and leukosialin, the major surface glycoprotein of human polymorphonuclear leukocytes (19). Significantly, the major surface glycoprotein of human platelets, GP1b, also contains sialylated mucin-like domains (28), making this component a potential receptor for sialic acid-binding bacterial adhesins. In previous studies (11), bacterium-mediated hemagglutinating activity was observed with 41 of 48 S. sanguinis, S. gordonii, and S. oralis strains, and in virtually all cases pretreatment of the RBC with neuraminidase eliminated these interactions. Interestingly, the interactions between these bacteria and RBC from different mammalian species were not identical but instead varied from strain to strain in a manner similar to that noted in early studies of viral hemagglutinins that bind different sialic acid-containing structures (14). Moreover, representative streptococcal strains that possessed hemagglutinating activity varied widely in their reactions with specific antibody against the Hs antigen of strain DL1 (25). These observations suggest that there is heterogeneity among the sialic acid-binding adhesins of these bacteria, both in terms of their receptor binding specificities and in terms of their antigenic properties. Further studies are needed to determine whether these adhesins are associated with different but structurally related serine-rich repeat proteins and, if so, whether there are differences in the interactions of individual proteins with sialoglycoconjugate receptors on oral surfaces, neutrophils (19), and human platelets. The results of such studies may provide important insights into the pathogenesis of infective endocarditis induced by oral viridans group streptococci.
Acknowledgments
We thank Paul Kolenbrander and Robert Palmer for their helpful comments during preparation of the manuscript.
This work was supported by grant-in-aid for scientific research 12771098 from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Editor: J. N. Weiser
REFERENCES
- 1.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081-1094. [DOI] [PubMed] [Google Scholar]
- 2.Burnette-Curley, D., V. Wells, H. Viscount, C. L. Munro, J. C. Fenno, P. Fives-Taylor, and F. L. Macrina. 1995. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect. Immun. 63:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlstedt, I., H. Lindgren, and J. K. Sheehan. 1983. The macromolecular structure of human cervical-mucus glycoproteins. Studies on fragments obtained after reduction of disulphide bridges and after subsequent trypsin digestion. Biochem. J. 213:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demuth, D. R., Y. Duan, W. Brooks, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403-413. [DOI] [PubMed] [Google Scholar]
- 5.Gibbons, R. J., I. Etherden, and E. C. Moreno. 1983. Association of neuraminidase-sensitive receptors and putative hydrophobic interactions with high-affinity binding sites for Streptococcus sanguis C5 in salivary pellicles. Infect. Immun. 42:1006-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbons, R. J., I. Etherden, and E. C. Moreno. 1985. Contribution of stereochemical interactions in the adhesion of Streptococcus sanguis C5 to experimental pellicles. J. Dent. Res. 64:96-101. [DOI] [PubMed] [Google Scholar]
- 7.Gururaja, T. L., N. Ramasubbu, P. Venugopalan, M. S. Reddy, K. Ramalingam, and M. J. Levine. 1998. Structural features of the human salivary mucin, MUC7. Glycoconj. J. 15:457-467. [DOI] [PubMed] [Google Scholar]
- 8.Herzberg, M. C. 1996. Platelet-streptococcal interactions in endocarditis. Crit. Rev. Oral Biol. Med. 7:222-236. [DOI] [PubMed] [Google Scholar]
- 9.Herzberg, M. C., K. L. Brintzenhofe, and C. C. Clawson. 1983. Aggregation of human platelets and adhesion of Streptococcus sanguis. Infect. Immun. 39:1457-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 11.Hsu, S. D., J. O. Cisar, A. L. Sandberg, and M. Kilian. 1994. Adhesive properties of viridans streptococcal species. Microb. Ecol. Health Dis. 7:125-137. [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Levesque, C., C. Vadeboncoeur, F. Chandad, and M. Frenette. 2001. Streptococcus salivarius fimbriae are composed of a glycoprotein containing a repeated motif assembled into a filamentous nondissociable structure. J. Bacteriol. 183:2724-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markwell, M. A. K. 1986. Viruses as hemagglutinins and lectins, p. 21-54. In D. Mirelman (ed.), Microbial lectins and agglutinins: properties and biological activity. John Wiley & Sons, Inc., New York, N.Y.
- 15.Maryanski, J. H., and C. L. Wittenberger. 1975. Mannitol transport in Streptococcus mutans. J. Bacteriol. 124:1475-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, P. A., M. J. Levine, L. A. Tabak, and M. S. Reddy. 1982. Specificity of salivary-bacterial interactions. II. Evidence for a lectin on Streptococcus sanguis with specificity for a NeuAcα2,3Galβ1,3GalNAc sequence. Biochem. Biophys. Res. Commun. 106:390-396. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 18.Perry, D., L. M. Wondrack, and H. K. Kuramitsu. 1983. Genetic transformation of putative cariogenic properties in Streptococcus mutans. Infect. Immun. 41:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruhl, S., J. O. Cisar, and A. L. Sandberg. 2000. Identification of polymorphonuclear leukocyte and HL-60 cell receptors for adhesins of Streptococcus gordonii and Actinomyces naeslundii. Infect. Immun. 68:6346-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Shiroza, T., N. Shinozaki, M. Hayakawa, T. Fujii, T. Oguma, M. Kobayashi, K. Fukushima, and Y. Abiko. 1998. Application of the resident plasmid integration technique to construct a strain of Streptococcus gordonii able to express the Bacillus circulans cycloisomaltooligosaccharide glucanotransferase gene, and secrete its active gene product. Gene 207:119-126. [DOI] [PubMed] [Google Scholar]
- 22.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43:147-157. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi, Y., K. Konishi, J. O. Cisar, and M. Yoshikawa. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi, Y., S. Ruhl, J. W. Yoon, A. L. Sandberg, and J. O. Cisar. 2002. Adhesion of viridans group streptococci to sialic acid-, galactose- and N-acetylgalactosamine-containing receptors. Oral Microbiol. Immunol. 17:257-262. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi, Y., A. L. Sandberg, S. Ruhl, J. Muller, and J. O. Cisar. 1997. A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to α2-3-linked sialic acid-containing receptors. Infect. Immun. 65:5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takamatsu, D., M. Osaki, and T. Sekizaki. 2000. Sequence analysis of a small cryptic plasmid isolated from Streptococcus suis serotype 2. Curr. Microbiol. 40:61-66. [DOI] [PubMed] [Google Scholar]
- 27.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuji, T., S. Tsunehisa, Y. Watanabe, K. Yamamoto, H. Tohyama, and T. Osawa. 1983. The carbohydrate moiety of human platelet glycocalicin. J. Biol. Chem. 258:6335-6339. [PubMed] [Google Scholar]
- 29.Wu, H., and P. M. Fives-Taylor. 1999. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol. Microbiol. 34:1070-1081. [DOI] [PubMed] [Google Scholar]