Abstract
In vertebrates, the neural plate border (NPB) is established by a group of transcription factors including Dlx3, Msx1 and Zic1. The crosstalk between these NPB specifiers governs the separation of the NPB region into placode and neural crest (NC) territories and also their further differentiation. Understanding the mechanisms of NPB formation and NC development is critical for our knowledge of related human diseases. Here we identified Nkx6.3, a transcription factor of the Nkx family, as a new NPB specifier required for neural crest development in Xenopus embryos. XNkx6.3 is expressed in the ectoderm of the neural plate border region at neurula stages, covering the epidermis, placode and neural crest territories, but not the neural plate. Inhibition of Nkx6.3 by dominant negative construct or specific morpholino leads to neural crest defects, while overexpression of Nkx6.3 induces ectopic neural crest in the anterior neural fold. In animal caps, Nkx6.3 alone is able to initiate the whole neural crest regulatory network and induces neural crest fate robustly. We showed that overexpression of Nkx6.3 affects multiple signaling pathways, creating a high-Wnt, low-BMP environment required for neural crest development. Gain- and loss-of-function of Nkx6.3 have compound effects on the expression of known NPB genes, which is largely opposite to that of Dlx3. Overexpression of Dlx3 blocks the NC inducing activity of Nkx6.3. The crosstalk between Nkx6.3, Dlx3 and Msx1 is likely crucial for proper NPB formation and neural crest development in Xenopus.
Introduction
Neural crest (NC) cells are a multipotent, migratory cell population arising at the neural plate border (NPB) in vertebrates, which give rise to various cell lineages including craniofacial bones and cartilages, melanocytes and peripheral neurons [1]. Understanding the mechanisms of neural crest development is critical for our knowledge of related human diseases, including defects in pigmentation, craniofacial and heart development.
The development of neural crest is regulated by a multi-step gene regulatory network (GRN), involving complicated interactions of multiple signaling molecules and tissues [2]–[4]. The determination of NPB by a group of signaling molecules is the first step in neural crest development. During gastrulation, a mediolateral gradient of BMP activity is established in the ectoderm through the action of BMP antagonists diffusing from the underlying notochord, such that the medial ectoderm with low BMP activity develops into neural plate, and the lateral ectoderm with high BMP signal becomes epidermis [5]–[7]. The region in between with intermediate BMP activity will become the NPB region which is crucial for neural crest development [8]. Wnt and FGF signals play key roles to position the neural crest territory along the anterior-posterior axis [9]. Signals from neural and non-neural territory inhibit each other to sharpen and refine the NPB region [2], [10]–[15]. The signaling events that establish the neural plate border control the broad expression of a set of transcription factors, including members of the Zic, Pax, Dlx and Msx families [4], [13], [15]–[17]. These factors, known as neural plate border specifiers, further control the expression of a group neural crest specific genes, including Snail1, Slug, FoxD3, Sox10, Sox9, AP-2 and c-Myc [4]. The neural crest specifier genes collectively control the expression of several downstream effector genes, which confer certain properties such as migration and multipotency before their terminal differentiation [18].
Another group of cells, the placodes, also originate from NPB, which are crucial for the development of the cranial sensory systems in vertebrates [19]. At early neurula stages, the neural crest territory occupies the medial side of NPB, within the trunk and head regions except the anteriormost neural folds [2]. The pre-placodal ectoderm, which expresses the panplacodal markers Six1 and Eya1, occupies the lateral part of the NPB, forming a “U” shape pattern at the anteriormost of head [20]. It has been proposed that neural crest and placodes develop from a common group of ancestor cells [19], which were then specified by different signals. However, recent studies support a binary competence model, according to which neural crest and placode originate differently from the neural and non-neural ectoderm respectively [21]. Dlx3, which is expressed in the placodal part of the NPB, is critical in the regulation of non-neural competence [13], [17], [21], [22]. Dlx and Msx can inhibit each other to determine the NPB cell identity, to become neural crest (Msx high and Dlx low) or placode (Msx low and Dlx high) [23]. However, the regulatory network to discriminate placode and neural crest fates and determine the sharp border between them remains elusive.
The Nkx family transcription factors are involved in a variety of developmental processes. Of the Nkx6 subfamily genes, Nkx6.1 and Nkx6.2 have been implicated in the control of cell differentiation in the central nervous system and pancreas [24]–[28]. Nkx6.3, a third member of this subfamily, is expressed in the anterior neural plate border region at neurula stages in Xenopus embryos [25]. Here, we analyzed the role of Nkx6.3 in Xenopus neural crest development and NPB formation by gain and loss of function studies. We showed evidence that XNkx6.3 is required for neural crest development and is able to induce neural crest fates dependent on Wnt signaling. Nkx6.3 is also involved in neural plate border formation and antagonizes the function of Dlx3.
Results
XNkx6.3 is expressed in the neural plate border ectoderm
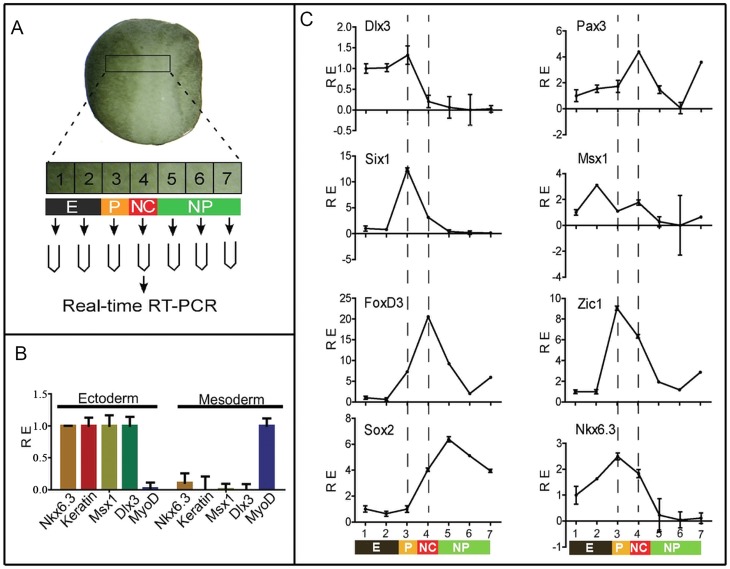
By in situ hybridization, we showed previously that XNkx6.3 is expressed in the non-neural ectoderm at cleavage to gastrula stages in Xenopus, and at neurula stages, its expression is gradually restricted to neural plate border regions [25]. However, due to its relative weak expression, we failed to verify its detailed expression in comparison to other neural plate border/neural crest markers using double in situ hybridization. We thus compared the expression patterns of XNkx6.3 with other neural plate border/neural crest markers in serial dissected pieces of ectodermal tissues along the medial-lateral axis of the neural plate border regions by real-time RT-PCR. A transverse slice of tissue of the neural plate border region of single stage 17 embryos was dissected out and separated sequentially into 7 continuous pieces, which were then proceeded to real-time RT-PCR analysis. The 7 pieces of tissues were expected to represent epidermis, placode, neural crest and neural plate identities respectively (Fig. 1A). We first checked whether XNkx6.3 is expressed in the ectodermal or mesodermal tissues. A piece of tissue corresponding to the placode/neural crest region was dissected out and separated into surface ectodermal and deep mesodermal parts. The identities of the tissues were confirmed by the relative expression levels of known epidermis marker keratin and mesodermal marker myoD (Fig. 1B). As expected, Dlx3 and Msx1, two genes involved in neural plate border formation, were predominantly expressed in the ectodermal regions. In such experiments, the expression of Nkx6.3 was always found to be expressed predominantly in the ectodermal part and only very weakly if any in the mesoderm.
Figure 1. XNkx6.3 is expressed in the neural plate border ectoderm.
(A) Experimental strategy to verify the expression domain of Nkx6.3 by qPCR. A transverse slice of tissue of the potential neural plate border region of single stage 17 embryos was dissected out and separated sequentially into 7 pieces, which were expected to represent epidermis, placode, neural crest and neural plate identities respectively. The explants were then processed to real-time RT-PCR analysis. E, epidermis; P, placode; NC, neural crest; NP, neural plate. (B) Nkx6.3 is expressed predominantly in the ectoderm. A piece of tissue corresponding to region 3 in (A) was dissected out and separated into surface ectodermal and deep mesodermal parts. The expression of Nkx6.3 and known ectodermal (Keratin, Dlx3 and Msx1) and mesodermal (MyoD) genes were monitored by qPCR. (C) The expression of Nkx6.3 and known neural plate border markers in a representative series of dissected epidermis, placode, neural crest, and neural plate explants from a single embryo at stage 17. The series of explants from each embryo were checked first for the expression of Dlx3, Six1, FoxD3 and Sox2, and only those with relative clean separation of the epidermis, placode, neural crest and neural plate were further analyzed for the expression of Nkx6.3 and additional markers. RE, relative expression.
The expression of XNkx6.3 was then examined in the dissected neural plate border tissues using real-time PCR (Fig. 1C). To verify the success of the separation of the tissues, the expression known markers of different tissues, Dlx3 for epidermis and placode, Six1 for placode, FoxD3 for neural crest and Sox2 for neural plate, were first examined in such tissue serials. In successful series, the expression of these markers peaked in different pieces of tissues as expected, such that Dlx3 was expressed in the epidermis (pieces 1–2) and placode (piece 3), Six1 and FoxD3 peaked in the placode (piece 3) and neural crest (piece 4) respectively, and Sox2 was detected in the neural plate (pieces 5–7) and neural crest region (piece 4), but not in the placodal region (piece 3) (Fig. 1C). Of the neural plate border specifiers, Pax3 and Zic1 are both highly expressed in the placode and neural crest regions, peaked in the neural crest and placode regions respectively. Msx1 was found to be expressed in the epidermis and neural crest regions, and at a weaker level in the placode. In such a serial of tissues, XNkx6.3 was found to be mainly expressed in the placode and neural crest regions, and to a weaker extent, also in the epidermal regions (Fig. 1C).
Nkx6.3 is required for neural crest development
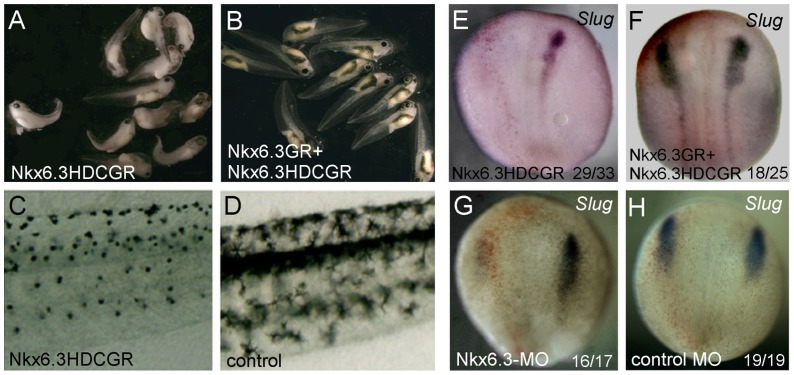
Nkx6 proteins are believed to act as transcriptional repressors with its eh1 domain as the repressor domain [29]. We constructed an eh1 deletion construct (the HDC construct) which should lack repressor activity and antagonize the function of endogenous Nkx6.3. As injection of wild type Nkx6.3 or the HDC-Nkx6.3 generally leads to gastrula defects, we constructed glucocorticoid receptor (GR) fusion constructs of Nkx6.3 and HDC, the nuclear translocation and activity of which could be induced by addition of dexamethasone (DEX). When induced after gastrulation stage (stage 12), embryos injected with the HDC construct showed severe defects in pigmentation at tadpole stages (Fig. 2A, 2C), a hallmark of neural crest development. This effect was nicely rescued by co-expression of the Nkx6.3-GR construct (Fig. 2B). At early neurula stages, the expression of neural crest marker Slug was abolished in HDC-Nkx6.3 injected embryos (Fig. 2E), but not when Nkx6.3-GR mRNA was co-expressed (Fig. 2F).
Figure 2. Nkx6.3 is required for neural crest development in Xenopus.
Injection of a inducible dominant negative form of Nkx6.3, Nkx6.3-HDC, induced severe defects in pigmentation at tadpole stages (A) when dexamethasone was added, which was rescued by co-expression of the wild type Nkx6.3-GR construct (B). The pigmentation patterns of the trunk region of an Nkx6.3-HDC injected embryo and a control embryo were highlighted in (C) and (D) respectively. Injection of Nkx6.3HDC-GR reduced the expression of Slug (E), which can be restored by co-injected Nkx6.3GR mRNA (F). (G) and (H) Injection of specific morpholino against Nkx6.3 but not a control morpholino impaired the expression of the neural crest marker Slug. The injected sides in (E)–H) are on the left, labeled by the red staining of the co-injected tracing lacZ. In (E)–H), the numbers of embryos showing similar changes of gene expression and total injected embryos in each group are indicated.
We also tried to block the function of endogenous Nkx6.3 using one specific morpholino (MO) against its ATG start cordon region. We confirmed that the MO efficiently blocked the expression of a reporter GFP construct harboring the targeted sequence at its 5′ ATG region (data not shown). Embryos injected with the Nkx6.3 MO but not the control MO showed impaired expression of the neural crest marker Slug (Fig. 2G, 2H), supporting a role of Nkx6.3 in neural crest development. However, the effect of the Nkx6.3 MO on neural crest induction was poorly rescued by co-injection of either wild type Nkx6.3 or the Nkx6.3-GR (induced at stage 11–12). This could be due to the fact that Nkx6.3 actually inhibits NC development when injected into the NC territory itself (see below) and when induced by DEX, the transient nuclear Nkx6.3-GR level would be much higher than just compensating the loss of endogenous Nkx6.3.
Nkx6.3 is able to induce ectopic neural crest dependent on Wnt signaling
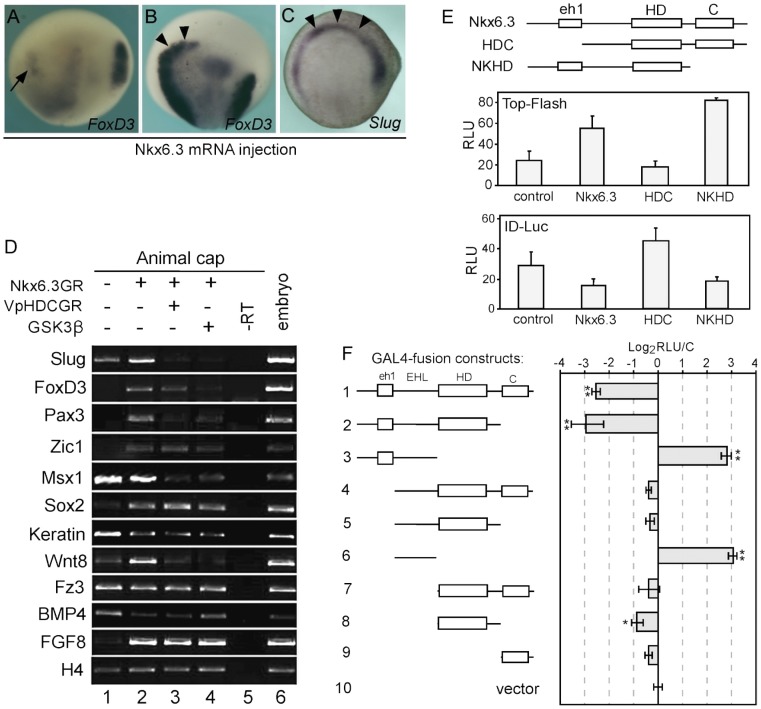
As Nkx6.3 is required for neural crest development, we next tested whether overexpression of Nkx6.3 is sufficient to induce ectopic neural crest. However, when Nkx6.3-GR was injected at 2-cell stage and DEX was added at stage 11, the neural crest marker expression was rather inhibited than induced (Fig. 3A). We then tried injections at 32-cell stage, targeting the dorsal neural plate region. Interestingly, when the anterior neural fold regions were targeted, ectopic neural crest was frequently observed (Fig. 3B, 3C). We further tested the neural crest induction activity of Nkx6.3 in animal cap explants. Interestingly, in animal caps, XNkx6.3 alone is sufficient to induce the expression of the whole panel of neural crest genes, including Zic1, Pax3, Slug and FoxD3 (Fig. 3D).
Figure 3. Nkx6.3 is able to induce neural crest dependent on Wnt signaling.
(A) Nkx6.3GR mRNA was injected into one blastomere at 2-cell stage and DEX was added at stage 11, the expression of neural crest marker FoxD3 was inhibited on the injected side (arrow). (B, C) Local injection of Nkx6.3 mRNA at 32-cell stage induced neural crest (arrowheads) in the anterior neural folds. (D) In animal caps, overexpression of Nkx6.3 induced the expression of neural crest markers and that of Wnt8 and FGF8 (lanes 1, 2). The VpHDC construct, in which the eh1 repressor domain was replaced by the Vp16 activation domain, repressed the neural crest inducing effect of Nkx6.3 (compare lanes 2, 3). Inhibiting Wnt signaling by GSK3β blocked most of the effects of Nkx6.3 on marker gene expression (lane 4). (E) The effect of different Nkx6.3 constructs on the expression of luciferase reporter genes of Wnt (Top-Flash, middle panel) and BMP (ID-Luc, lower panel) signaling in Xenopus embryos. The domain structures of Nkx6.3 and the HDC, NKHD constructs were shown in the upper panel. eh1, eh1 repressor domain; HD, homeodomain; C, C-terminal domain. (F) Effects of the fusion constructs of GAL4 and various Nkx6.3 domains on the expression of a GAL4 luciferase reporter gene. Wild type Nkx6.3 and that lacking the C terminal domain worked as repressors (bars 1, 2) while the EHL region with or without the eh1 domain both activated the transcription of the reporter (bars 3, 6). EHL, the linker region between the eh1 repressor domain and the HD domain. *, p<0.05; **, p<0.01. The expression of the different constructs were confirmed by Western blot (data not shown).
During development, the presumptive neural crest territory is induced at the neural plate border through the interplay of different signaling pathways including BMPs, Wnts and FGFs. We tested whether components of these signaling pathways were attenuated in Nkx6.3 injected animal cap explants. The results showed that overexpression of Nkx6.3 strongly induced the expression of Wnt8 and FGF8 while inhibited that of BMP4 in animal caps (Fig. 3D). The expression of the Wnt receptor, Frizzled 3, which has also been shown to regulate neural crest development [30], did not change. We then tested the effect of different Nkx6.3 constructs on Wnt and BMP signaling using reporter assays in Xenopus embryos (Fig. 3E). Consistent with above results, wild type Nkx6.3 activates the Wnt reporter expression while inhibiting the BMP reporter expression. The HDC construct, which lacks the eh1repressor domain, shows the opposite effects, inhibiting Wnt while activating BMP signaling. The NKHD construct, which lacks the C-terminal domain, works similarly to wild type Nkx6.3 (Fig. 3E). These data suggest that overexpression of Nkx6.3 in animal caps attenuated the signaling environment to promote neural crest development.
In animal caps, the induction of neural crest genes is dependent on Wnt signaling, since co-expression of a Wnt signaling inhibitor, Gsk3β, completely abolished its activity on neural crest induction (Fig. 3D). Interestingly, the expression of FGF8 was also strongly induced by Nkx6.3, which can not be blocked by GSK3β. Also, the VpHDCGR construct, in which the eh1 repressor domain of Nkx6.3 was replaced by the VP16 activation domain, effectively blocked most of the activities of Nkx6.3 on neural crest genes expression in animal caps, yet it failed to repress the induction of FGF8 (Fig. 3D). These results suggested that Nkx6.3 works mainly as a transcriptional repressor to induce neural crest genes expression, including that of Wnt8. However, it likely works as an activator to stimulate the expression of FGF8. To check the possibility that Nkx6.3 is able to work both as a transcriptional repressor and an activator, we tested the activities of a series of fusion constructs of Nkx6.3 deletions with GAL4 DNA binding domain on the expression of a luciferase reporter harboring GAL4 binding sites (Fig. 3F). In the reporter assay, full length Nkx6.3 and the NKHD construct effectively inhibited the reporter expression, while the construct containing only the linker region between the eh1 repressor domain and the HD domain (EHL, #6 in Fig. 3F) works as a transcriptional activator. The construct containing the eh1 domain and the EHL region (#3 in Fig. 3F) also activates the reporter expression while the other constructs most showed very weak repressor activities. Thus Nkx6.3 is potentially able to work as either a repressor or an activator in different contexts.
In order to find the immediate-early response genes to Nkx6.3, we performed RT-qPCR to test the expression of these genes in animal caps treated with cycloheximide to block protein synthesis. In the presence of cycloheximide, after addition of dexamethasone, the induction of most of the neural crest genes was clearly blocked. However, the expression of Msx1 remained stimulated to more than 2 folds (Fig. 4A). However, in our previous semi-quantitative analysis, the induction of Msx1 by Nkx6.3 was less clear (Fig. 3D, lane 2). We then tested the time course of Msx1 induction in Nkx6.3 injected animal caps (Fig. 4B). Our results showed that the induction of Msx1 was transient, which went up in 1 hour but then declined to control level in about 3 hours. As the two genes are co-expressed in the neural plate border region (Fig. 1), we suggest that Msx1 is potentially one of the direct targets of Nkx6.3.
Figure 4. Msx1 is an immediate target gene in response to Nkx6.3.
(A) 1 ng Nkx6.3GR mRNA was injected into each cell at 2-cell stage. Animal caps were dissected at Stage 9, treated with cycloheximide for 30 minutes to inhibit protein synthesis, and then cultured in media containing dexamethasone for 2 hours before processed for RT-qPCR to monitor the expression of the marker genes indicated. Among the tested genes, Msx1 was the only one up-regulated clearly under such condition. (B) Dynamic induction of Msx1 by Nkx6.3. The induction of Msx1 was monitored at different time points after induction as described in (A). The expression level of Msx1 was up-regulated in one hour, but was then down-regulated at later stages.
Nkx6.3 is a neural plate border specifier
The above data showed that at least in animal cap assays, Nkx6.3 is able to modulate Wnt, BMP and FGF signaling, which are required for proper neural border formation in vivo. We then tested systematically the effects of Nkx6.3 overexpression and inhibition on the expression of neural plate border genes as well as the placode and neural crest marker genes.
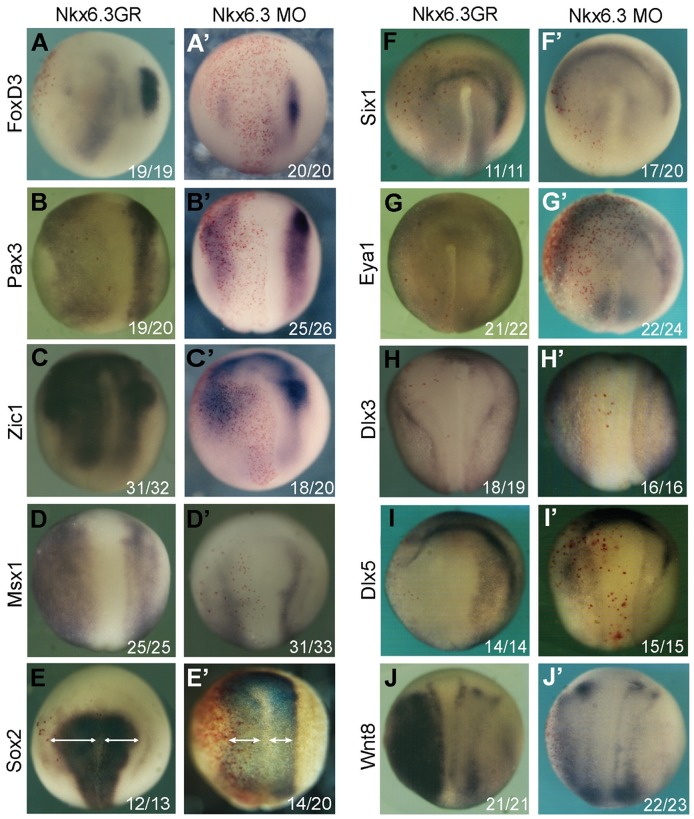
In the overexpression experiments, the embryos were injected with Nkx6.3GR mRNA at 4-cell stage, induced at the end of stage 11 and processed for in situ hybridization at stage 15. As mentioned above, overexpession Nkx6.3 strongly inhibited the expression of the neural crest marker FoxD3 (Fig. 5A). Consistent with the results in animal cap assays, Nkx6.3 also stimulated the expression of neural plate border specifiers Zic1 and Msx1(Fig. 5C, 5D). The expression level of Pax3, however, was clearly reduced in Nkx6.3 injected sides (Fig. 5B), although its expression domain was expanded. The expression of the pan neural marker Sox2 expanded slightly (Fig. 5E). The expression of the panplacodal markers Six1 and Eya1, general nonneural markers Dlx3 and Dlx5, was all reduced in Nkx6.3 injected areas (Fig. 5F–I), suggesting a general failure of neural plate border development. Interestingly, as in animal caps, overexpression of Nkx6.3 induced strong Wnt8 expression in injected sides (Fig. 5J), which might be partially responsible for the patterning defects of the neural plate border.
Figure 5. Nkx6.3 is a neural plate border specifier.
The effects of Nkx6.3 overexpression (A–J) or knockdown (A′–J′) on the expression of indicated neural and non-neural ectodermal markers and that of Wnt8. The injected sides were all on the left marked by red β-galactosidase staining. In (E) and (E′), the arrowed lines indicate the width of the neural plates. The numbers of embryos showing similar changes of gene expression and total injected embryos in each group are indicated.
When Nkx6.3 was blocked by specific morpholino, the expression levels of the neural crest markers FoxD3, neural plate border specifier Pax3, Zic1 and Msx1 were all reduced (Fig. 5A′–D′), with the expression domain of Pax3 and Zic1 expanded. The expression of Sox2 also expanded slightly (Fig. 5E′). In the Nkx6.3 morphants, the placode markers Six1 and Eya1 became slightly stronger (Fig. 5F′, 5G′). Interestingly, the expression border of Dlx3 and Dlx5 became blurred when Nkx6.3 was knocked-down (Fig. 5H′, 5 I′). The expression of Wnt8 had no clear change in Nkx6.3 morphants (Fig. 5J′).
The above gain- and loss-of-function phenotypes of Nkx6.3 are largely opposite to that of Dlx3 [21], suggesting opposite roles of the two genes in neural plate border development. We then tested whether Dlx3 could inhibit the neural crest induction activity of Nkx6.3. Indeed, co-expression of Dlx3 largely inhibited the activity of Nkx6.3 to induce neural crest markers in animal cap assay (Fig. 6). Interestingly, overexpression of Nkx6.3 also reduced the expression of endogenous Dlx3 and Dlx5 (Fig. 6). These data suggest that Nkx6.3 and Dlx3 have opposite roles as regards to neural crest development. Dlx3 is expressed in the placodal region where Nkx6.3 is co-expressed. Dlx3 has been suggested to regulate the non-neural competence [13], [21] and we suggest that the function of Dlx3 is dominant in vivo in the placodal region. In the neural crest territory, Nkx6.3 but not Dlx3 is expressed (Fig. 1), where it functions to regulate the signaling environment to promote neural crest development.
Figure 6. Dlx3 blocks the neural crest induction activity of Nkx6.3.
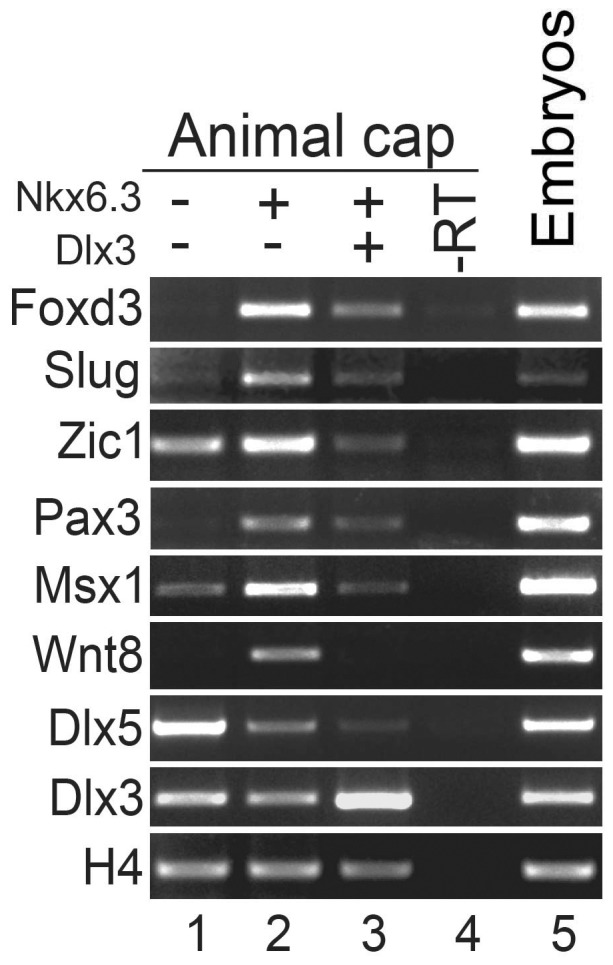
RT-PCR analysis in animal cap assay showing the effect of Dlx3 on the expression of the indicated neural crest genes induced by Nkx6.3. Note that the expression of endogenous Dlx3 and Dlx5 were all reduced upon Nkx6.3 over-expression. -RT, negative control with reverse-transcriptase omitted in the RT reaction; embryo, RNA template from whole embryos was used as a positive control.
Discussion
Nkx6.3 is required for neural crest development
In this study, we provided several lines of evidence that Nkx6.3 is required for neural crest development in Xenopus. First, Nkx6.3 is expressed in the ectoderm of the neural plate border region at neurula stages, covering the epidermis, placode and neural crest territories, but not the neural plate (Fig. 1). Second, inhibition of Nkx6.3 either by dominant negative construct or specific morpholino leads to neural crest defects, including pigmentation defects and loss of neural crest marker expression (Fig. 2). Third, overexpression of Nkx6.3 promotes ectopic neural crest development in the anterior neural fold, although it inhibits endogenous neural crest when injected at the NC territory itself (Fig. 3). Furthermore, in animal caps, we showed that Nkx6.3 alone is able to initiate the whole neural crest regulatory network and induce neural crest fate robustly. In animal caps, Nkx6.3 strongly induced the expression of Wnt8 and FGF8 while inhibited that of BMP4, thus created a high-Wnt, low-BMP environment required for neural crest development. We confirmed that Nkx6.3 overexpression also induces robust Wnt8 expression in whole embryos (Fig. 5J), which has been shown to be a NC inducer [9]. We showed that Nkx6.3 mainly works as a transcriptional repressor in neural crest induction, as an activator version of Nkx6.3 (VpHDC) works as a dominant negative form in this process (Fig. 3D). Thus its induction of Wnt8 is most likely indirect. As Nkx6.3 has a compound effect on various signaling pathways, its overexpression in the neural crest territory itself likely interferes with the signaling environment required for NC development and thus inhibits NC induction. Active Wnt signaling is required for Nkx6.3 in neural crest induction, as co-expression of Gsk3β, a Wnt signaling inhibitor, abolished its activity on neural crest induction. Although in animal caps, Nkx6.3 is able to stimulate FGF8 expression likely as a transcriptional activator, it can not in the presence of protein synthesis inhibitor, suggesting its effect is also indirect. The only potential direct gene of Nkx6.3 implicated from our study is Msx1, which was still activated in the presence of cycloheximide. Interestingly, the induction of Msx1 by Nkx6.3 was transient, which went down to control level in about 3 hours. As Msx1 is an established target of BMP signaling, we assume that this effect is likely a feedback of the inhibition of BMP signaling by Nkx6.3 at later stages.
Nkx6.3 as a neural plate border specifier
Its expression pattern and gain- and loss-of function effects on NPB formation support Nkx6.3 as a new neural plate border specifier. Nkx6.3 is expressed in the non-neural ectoderm and neural crest territory at the neural plate border of neurula stage embryos (Fig. 1). Overexpression of Nkx6.3 upregulates while loss of function of Nkx6.3 reduces the expression of the NPB genes Zic1 and Mxs1 (Fig. 5C, 5D). However, either overexpression or inhibition of Nkx6.3 leads to reduced expression of Pax3, although its expression domain became expanded (Fig. 5B). This could be the reason that Nkx6.3 overexpression inhibits rather than induces NC development when injected in the NC territory (Fig. 2A).
Interestingly, the gain- and loss-of-function phenotypes of Nkx6.3 on NPB markers are largely opposite to that of Dlx3 [21], which is expressed in the placodal part of the NPB and is critical for placode development. Nkx6.3 and Dlx3 seem to promote placode and neural crest development respectively and antagonize the function of each other when overexpressed. Indeed, co-expression of Dlx3 largely inhibited the NC induction activity of Nkx6.3 assay (Fig. 6). When injected into the NC domain, Nkx6.3 also inhibits the expression of neural crest as well as neural plate border marker genes [21]. On the other hand, overexpression of Nkx6.3 reduced the expression of Dlx3 and Dlx5 and also the neural placodal markers Six1 and Eya1 (Fig. 5F–I). Dlx3 has been shown to work as a transcription activator to repress neural fates [13] and is able to repress Wnt-β-catenin signaling when overexpressed [31]. Thus it is possible that Nkx6.3 and Dlx3 might share common target genes or regulatory units. Indeed, we also observed activation of Wnt8 expression in Dlx3 morphants (data not shown), as in the case of Nkx6.3 overexpression. Nkx6.1, which contains a homeodomain highly similar to that of Nkx6.3, has been shown to bind elements containing the homeodomain core-binding site (5′-TAAT-3′or 5′-ATTA-3′) [32], [33], similar to that of Dlx3 [34]. Thus it is possible that the Nkx6.3 and Dlx3 might directly compete for common target gene regulation.
Another possibility is that Nkx6.3 inhibits the function of Dlx3 indirectly through induction of Msx1, which has been shown to antagonize Dlx3 through forming heterodimers with it [35]. In zebrafish, the mutual antagonism between Msx and Dlx proteins has also been shown to be required for normal placode development [23]. In mouse, Msx1 is able to repress the pre-placodal marker Six1 through direct binding to its enhancer while the binding of Dlx5 activates it [36]. Thus the stimulation of Msx1 by Nkx6.3 might account for its activity to inhibit placodal development. Nkx6.3 and Msx1 are both co-expressed with Dlx3 in the placodal region and might contribute to the fine tuning of the signaling environment for placode development in vivo. However, other factors must be involved in its induction of NC fates, since Nkx6.3 works mainly as a repressor to induce NC and that overexpression of Msx1 expands NC development in whole embryos [16], unlike Nkx6.3.
In summary, our study established Nkx6.3 as a neural plate border specifier required for neural crest development. Together with Dlx3 and Msx1, it is likely involved in the regulation of local signaling environment for proper NPB formation and also downstream events of neural crest development.
Materials and Methods
Ethics Statement
The care of Xenopus laevis (Nasco), in vitro fertilization procedure and embryos study were performed according to protocols approved by the Ethics Committee of Kunming Institute of Zoology, Chinese Academy of Sciences (permit number: SYDW20070301001).
Microinjection and in situ hybridization
In vitro fertilization, embryo culture, whole mount in situ hybridization,preparation of mRNA, and microinjection were carried out as described [37]. The sequence of the antisense morpholino oligo (MO) for XNkx6.3 used was: 5′- TAGGCCTTCTGCTCTCTCAACATGG -3′, which was obtained from Gene Tools (OR). A standard control oligo was used as a negative control morpholino which targets a human beta-globin intron mutation (Gene Tools). For in situ hybridization, the probes for Slug, FoxD3, Pax3, Zic1, Sox2, Msx1, Six1, Eya1, Dlx3, Dlx5 and Wnt8 were used as described [11], [16], [38]–[42].
RNA isolation and reverse transcriptase PCR assay
Total RNAs were extracted using the Trizol total RNA extract kit (Tiangen) and reverse transcribed using the Fermentas RevertAid First Strand cDNA Synthesis Kit to prepare templates for semi-quantitative or real-time quantitative PCR (qPCR).
For traditional RT-PCR analysis, the primers for Pax3, Slug, Msx1, Zic1, FoxD3 and FGF8 and Fz3 were used as described [8], [43]–[45]. Primers for XWnt8 were: forward primer: 5′GACAAGATGCCAGAGCCCTAA; reverse primer: 5′TAAGTTCAGACCCGGCCACA. H4 was used as a loading control. The Qiagene QuantiNova probe PCR kit was used to monitor the expression of Nkx6.3 and the reference GAPDH by probe method. The primers and probes used were: Nkx6.3: forward primer: 5′CCCATCATCCTGGAGCATTT; reverse primer: 5′TGGCATCCAGAAGATTTCATTTC; probe: 5′TGCTCCCATCCTACTC, labeled with FAM and MGB; GAPDH: forward primer: 5′ GTCTGGCTCCTCTCGCAAAG, reverse primer: 5′GTCATGAGTCCCTCAACAATGC; Probe: 5′TCATCAACGACAACTTT, labeled with VIC and MGB. The primers and probes were synthesized by Invitrogen. The expression of other genes was examined by SYBR green qPCR using the following primers:
Dlx3F: 5′ TCGGCCGTTTGTCCATTACA 3′, R: 5′GGTTTCGGGCTCTTCCTTCA 3′; Wnt8F: 5′GTCGGGTAACAGTGCTGACA3′, R: 5′ATAAGTTCAGACCCGGCCAC3′;
Six1F:5′ CTTACTCCCTGAGCGCACTT 3′, R: 5′ GGTCGCTCTTACGATCCCAG 3′.
The primers for GAPDH, Keratin, MyoD, Six1, Sox2, FoxD3, Pax3, Zic1, Msx1 and Bmp4 were used as described [15], [46]–[48]
Plasmids construction
The full open reading frame of XNkx6.3 and that without the eh1 domain coding region were cloned into pCS2-GR vector to create the Nkx6.3GR and Nkx6.3HDCGR constructs. The VpHDCGR construct was prepared by cloning the HDCGR fragment into a VP16 expression vector [49]. Different fragments XNkx6.3 were cloned into the pBIND vector (Promega), which contains the yeast GAL4 DNA-binding domain, for the expression of GAL4 fusion proteins with different XNkx6.3 domains.
Luciferase reporter assays
For luciferase reporter assay in Xenopus embryos to monitor the effect of Nkx6.3 on Wnt signaling, the reporter plasmids (25pg of TOP-flash and 5pg of pTK-renilla) and 0.5ng mRNAs of different XNkx6.3 constructs (Nkx6.3GR, VpHDCGR and NKHDGR) were injected into animal poles of all blastomeres at 4-cell stage. dexamethasone was added immediately after injection to activate of the Nkx6.3 constructs. The embryos were harvested at Stage 10, divided into 3 groups (>10 embryos each group), lysed and analyzed using the Dual-Luciferase Reporter Assay System (Promega). The effect of Nkx6.3 on BMP signaling was examined in animal caps using the ID-reporter (gift from Prof. Jing). The reporter plasmids (25pg of ID-reporter and 5pg of pTK-renilla) and 0.5 ng Nkx6.3GR mRNA were injected into animal poles of all blastomeres at 4-cell stage and cultured in media containing dexamethasone. Animal caps were cut at Stage 9, cultured till Stage 12 and lysed for reporter activity measurement. The control embryos were cultured without dexamethasone.
To test the transcriptional activity of various XNkx6.3 constructs, HEK293T cells in 96-well plates were transfected with the pG5-Luc (100ng) which contains GAL4 binding sites in its promoter region and pRL-TK (10ng) reporters (Promega) and the GAL4-fusion Nkx6.3 constructs in pBIND. The luciferase activities were analyzed 24 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). Statistical significance test was done using Student's t-test.
Acknowledgments
We thank Profs. Naihe Jing, Daniel Weinstein, Thomas D. Sargent and Aaron M. Zorn for plasmids; and Guimei Li and Shuangjuan Yang at Kunming Biological Diversity Regional Center of Large Apparatus and Equipments for help with real-time PCR analysis.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by grants from the CAS Key Project (KSCX2-EW-R-05) to BM, National Natural Science Foundation of China (81102519) and the Chongqing Science and Technology Committee (cstc2012jjA0147) to YS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sauka-Spengler T, Bronner-Fraser M (2008) A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol 9:557–568. [DOI] [PubMed] [Google Scholar]
- 2. Milet C, Monsoro-Burq AH (2012) Neural crest induction at the neural plate border in vertebrates. Dev Biol 366:22–33. [DOI] [PubMed] [Google Scholar]
- 3. Stuhlmiller TJ, Garcia-Castro MI (2012) Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci 69:3715–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meulemans D, Bronner-Fraser M (2004) Gene-regulatory interactions in neural crest evolution and development. Dev Cell 7:291–299. [DOI] [PubMed] [Google Scholar]
- 5. Marchant L, Linker C, Ruiz P, Guerrero N, Mayor R (1998) The inductive properties of mesoderm suggest that the neural crest cells are specified by a BMP gradient. Dev Biol 198:319–329. [PubMed] [Google Scholar]
- 6. Patthey C, Edlund T, Gunhaga L (2009) Wnt-regulated temporal control of BMP exposure directs the choice between neural plate border and epidermal fate. Development 136:73–83. [DOI] [PubMed] [Google Scholar]
- 7. Tucker JA, Mintzer KA, Mullins MC (2008) The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev Cell 14:108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R (2003) Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development 130:6441–6452. [DOI] [PubMed] [Google Scholar]
- 9. Hong CS, Park BY, Saint-Jeannet JP (2008) Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development 135:3903–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Groves AK, LaBonne C (2014) Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev Biol 389:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo T, Matsuo-Takasaki M, Sargent TD (2001) Distinct roles for Distal-less genes Dlx3 and Dlx5 in regulating ectodermal development in Xenopus. Mol Reprod Dev 60:331–337. [DOI] [PubMed] [Google Scholar]
- 12. McLarren KW, Litsiou A, Streit A (2003) DLX5 positions the neural crest and preplacode region at the border of the neural plate. Dev Biol 259:34–47. [DOI] [PubMed] [Google Scholar]
- 13. Woda JM, Pastagia J, Mercola M, Artinger KB (2003) Dlx proteins position the neural plate border and determine adjacent cell fates. Development 130:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA (2004) Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development 131:5871–5881. [DOI] [PubMed] [Google Scholar]
- 15. Hong CS, Saint-Jeannet JP (2007) The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell 18:2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monsoro-Burq AH, Wang E, Harland R (2005) Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell 8:167–178. [DOI] [PubMed] [Google Scholar]
- 17. Feledy JA, Beanan MJ, Sandoval JJ, Goodrich JS, Lim JH, et al. (1999) Inhibitory patterning of the anterior neural plate in Xenopus by homeodomain factors Dlx3 and Msx1. Dev Biol 212:455–464. [DOI] [PubMed] [Google Scholar]
- 18. Theveneau E, Mayor R (2012) Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev Biol 366:34–54. [DOI] [PubMed] [Google Scholar]
- 19. Baker CV, Bronner-Fraser M (2001) Vertebrate cranial placodes I. Embryonic induction. Dev Biol 232:1–61. [DOI] [PubMed] [Google Scholar]
- 20. Saint-Jeannet JP, Moody SA (2014) Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol 389:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pieper M, Ahrens K, Rink E, Peter A, Schlosser G (2012) Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development 139:1175–1187. [DOI] [PubMed] [Google Scholar]
- 22. Ahrens K, Schlosser G (2005) Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev Biol 288:40–59. [DOI] [PubMed] [Google Scholar]
- 23. Phillips BT, Kwon HJ, Melton C, Houghtaling P, Fritz A, et al. (2006) Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev Biol 294:376–390. [DOI] [PubMed] [Google Scholar]
- 24. Dichmann DS, Harland RM (2011) Nkx6 genes pattern the frog neural plate and Nkx6.1 is necessary for motoneuron axon projection. Dev Biol 349:378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao S, Jiang H, Wang W, Mao B (2007) Cloning and developmental expression of the Xenopus Nkx6 genes. Dev Genes Evol 217:477–483. [DOI] [PubMed] [Google Scholar]
- 26. Ma P, Xia Y, Ma L, Zhao S, Mao B (2013) Xenopus Nkx6.1 and Nkx6.2 are required for mid-hindbrain boundary development. Dev Genes Evol 223:253–259. [DOI] [PubMed] [Google Scholar]
- 27. Binot AC, Manfroid I, Flasse L, Winandy M, Motte P, et al. (2010) Nkx6.1 and nkx6.2 regulate alpha- and beta-cell formation in zebrafish by acting on pancreatic endocrine progenitor cells. Dev Biol 340:397–407. [DOI] [PubMed] [Google Scholar]
- 28. Taylor BL, Liu FF, Sander M (2013) Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep 4:1262–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith ST, Jaynes JB (1996) A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development 122:3141–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Deardorff MA, Tan C, Saint-Jeannet JP, Klein PS (2001) A role for frizzled 3 in neural crest development. Development 128:3655–3663. [DOI] [PubMed] [Google Scholar]
- 31. Beanan MJ, Feledy JA, Sargent TD (2000) Regulation of early expression of Dlx3, a Xenopus anti-neural factor, by beta-catenin signaling. Mech Dev 91:227–235. [DOI] [PubMed] [Google Scholar]
- 32. Jorgensen MC, Vestergard Petersen H, Ericson J, Madsen OD, Serup P (1999) Cloning and DNA-binding properties of the rat pancreatic beta-cell-specific factor Nkx6.1. FEBS Lett 461:287–294. [DOI] [PubMed] [Google Scholar]
- 33. Mirmira RG, Watada H, German MS (2000) Beta-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J Biol Chem 275:14743–14751. [DOI] [PubMed] [Google Scholar]
- 34. Feledy JA, Morasso MI, Jang SI, Sargent TD (1999) Transcriptional activation by the homeodomain protein distal-less 3. Nucleic Acids Res 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang H, Hu G, Wang H, Sciavolino P, Iler N, et al. (1997) Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol 17:2920–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato S, Ikeda K, Shioi G, Ochi H, Ogino H, et al. (2010) Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev Biol 344:158–171. [DOI] [PubMed] [Google Scholar]
- 37. Zeng W, Kong Q, Li C, Mao B (2010) Xenopus RCOR2 (REST corepressor 2) interacts with ZMYND8, which is involved in neural differentiation. Biochem Biophys Res Commun 394:1024–1029. [DOI] [PubMed] [Google Scholar]
- 38. Aybar MJ, Nieto MA, Mayor R (2003) Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development 130:483–494. [DOI] [PubMed] [Google Scholar]
- 39. Sasai N, Mizuseki K, Sasai Y (2001) Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development 128:2525–2536. [DOI] [PubMed] [Google Scholar]
- 40. Sato T, Sasai N, Sasai Y (2005) Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development 132:2355–2363. [DOI] [PubMed] [Google Scholar]
- 41. David R, Ahrens K, Wedlich D, Schlosser G (2001) Xenopus Eya1 demarcates all neurogenic placodes as well as migrating hypaxial muscle precursors. Mech Dev 103:189–192. [DOI] [PubMed] [Google Scholar]
- 42. Pandur PD, Moody SA (2000) Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech Dev 96:253–257. [DOI] [PubMed] [Google Scholar]
- 43. Monsoro-Burq AH, Fletcher RB, Harland RM (2003) Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development 130:3111–3124. [DOI] [PubMed] [Google Scholar]
- 44. Nakata K, Nagai T, Aruga J, Mikoshiba K (1998) Xenopus Zic family and its role in neural and neural crest development. Mech Dev 75:43–51. [DOI] [PubMed] [Google Scholar]
- 45. Li J, Shi Y, Sun J, Zhang Y, Mao B (2011) Xenopus reduced folate carrier regulates neural crest development epigenetically. PLoS One 6:e27198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nichane M, de Croze N, Ren X, Souopgui J, Monsoro-Burq AH, et al. (2008) Hairy2-Id3 interactions play an essential role in Xenopus neural crest progenitor specification. Dev Biol 322:355–367. [DOI] [PubMed] [Google Scholar]
- 47. Nakagawa T, Iwabuchi J (2012) Brain-specific promoter/exon I.f of the cyp19a1 (aromatase) gene in Xenopus laevis. J Steroid Biochem Mol Biol 132:247–255. [DOI] [PubMed] [Google Scholar]
- 48. Schohl A, Fagotto F (2003) A role for maternal beta-catenin in early mesoderm induction in Xenopus. EMBO J 22:3303–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suri C, Haremaki T, Weinstein DC (2004) Inhibition of mesodermal fate by Xenopus HNF3beta/FoxA2. Dev Biol 265:90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.