Abstract
Recently, several publications have surfaced describing methods to manipulate mitochondrial genomes in tissues and embryos. With them, a somewhat sensationalistic uproar about the generation of children with ‘three parents’ has dominated the discussion in the lay media. It is important that society understands the singularities of mitochondrial genetics to judge these procedures in a rational light, so that this ongoing discussion does not preclude the helping of patients and families harboring mutated mitochondrial genomes.
Mitochondrial genome disorders
Defects in mitochondrial function can be devastating and can cause disease in almost every tissue of the body. Because there are more than 1000 proteins in mitochondria, the genetic cause of these diseases is variable. Nuclear DNA encodes the vast majority of mitochondrial proteins. By contrast, the much smaller mitochondrial DNA (mtDNA) contributes to only 13 of these proteins, all critical subunits of the oxidative phosphorylation system (OXPHOS) complexes. The mitochondrial genome is present in multiple copies per cell, ranging from approximately 1000 in somatic cells to almost 100 000 in oocytes. Sperm mtDNA is degraded before fertilization and, consequently, mtDNA is inherited exclusively through the maternal lineage [1]. Due to the high number of mtDNA copies in cells, mtDNA mutations can frequently occur in a subset of the total mtDNA pool, a condition known as mtDNA heteroplasmy. Because these pathogenic mutations in mtDNA can be present in the germline or arise in oocytes, the resulting offspring can inherit them. These mutations can cause severe decreases in OXPHOS function, leading to many diseases that currently lack effective treatments, such as mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS syndrome), myoclonus epilepsy with ragged red fibers (MERRF syndrome), and Kearns–Sayre syndrome (KSS) [2].
In the past two decades, it has become clear that the ratio of wild type to mutated mtDNA in the cell is a critical factor in disease severity [2]. Following this realization, numerous approaches have been attempted to change the balance of mtDNA heteroplasmy toward the healthy, wild type population (reviewed in [2]).
Approaches to correct mtDNA mutations in patients
Because of its location inside the double membrane of the mitochondrion, no technique is available to alter the sequence of mtDNA in vertebrate cells; however, because of this physical separation from nuclear DNA, cell-biology techniques have been devised to replace the whole mtDNA molecule in mammalian cells. Pioneered by Michael King and the late Guiseppe Attardi in 1989, exogenous mitochondria (with mtDNA) can be introduced into cell lines devoid of mtDNA (ρ° cells) by fusing the latter with enucleated cells (cytoplasts) from patients harboring mtDNA mutations [3]. This technique allowed the production of cell clones with different ratios of mutated and wild type mtDNA from patient cells. The possibility of replacing mutated genes or gene products has been also explored, including the import of tRNA [4] and mRNA molecules into mitochondria [5] for their translation into mitochondrial ribosomes. Although promising, the general applicability of these approaches remains to be determined.
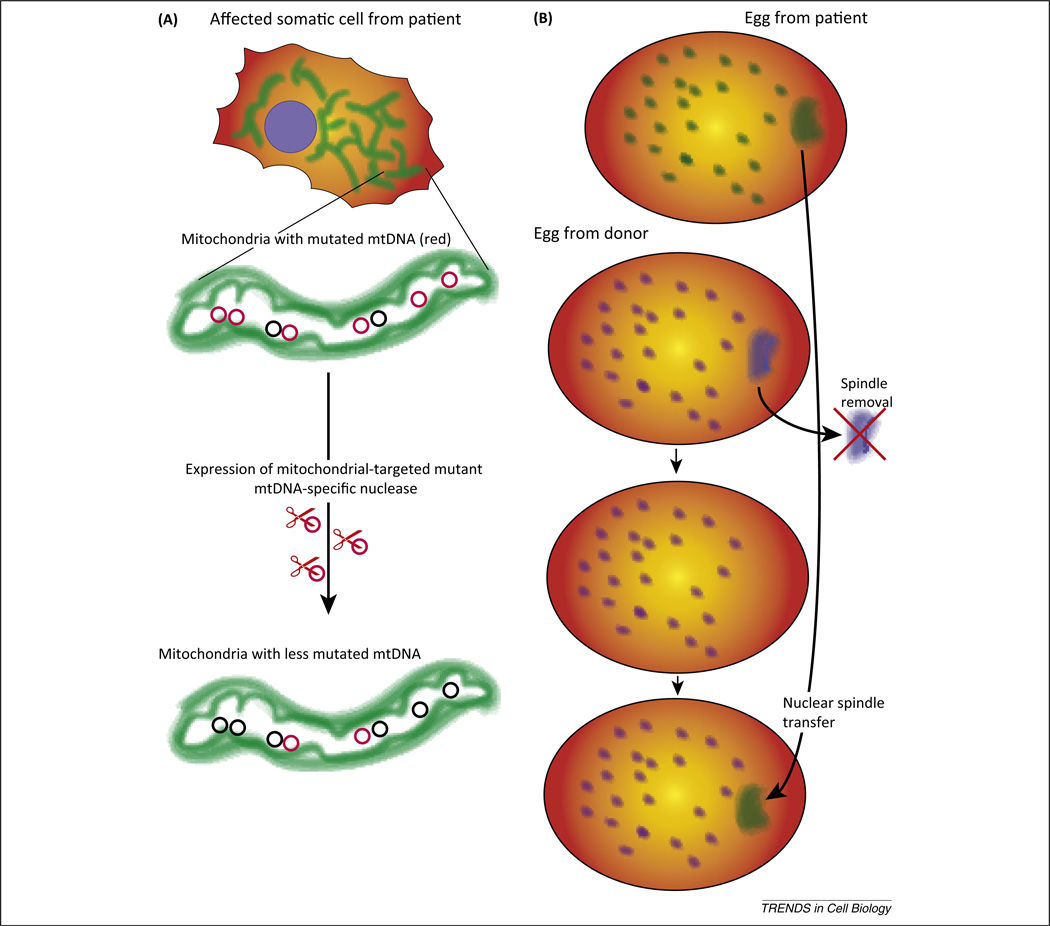
Although mitochondrial transfer techniques are useful for understanding mtDNA disorders, they cannot be used to replace or reduce the levels of mutated mtDNA in patients’ organs, which would ultimately cure them of these diseases. A more feasible approach would be the active elimination of mutated mitochondrial genomes. Accordingly, several groups have made a considerable effort to develop site-specific nucleases targeted to mitochondria for the elimination of mutated mtDNA. Although mitochondrion-targeted restriction endonucleases have been used successfully to change mtDNA heteroplasmy in mice [6], a recent breakthrough in this area came with the development of designer nucleases, namely the TAL effector nucleases (TALEN) and zinc-finger nucleases (ZFN). These modular site-specific nucleases allow the development of specific nucleases against essentially any DNA sequence, overcoming the main limitation of restriction endonucleases, which have strict and limited DNA sequence-recognition capacity. Once targeted to mitochondria, designer nucleases can effectively eliminate pathogenic mtDNA mutations, as demonstrated for large mtDNA deletions [7,8] and point mutations [7] (Figure 1A). Although the use of mitoTALENs has been restricted to cultured cells, the expression of these proteins in patients’ tissues has a strong therapeutic potential.
Figure 1.
Novel approaches for the manipulation of mitochondrial genomes in tissues and embryos. In the past few years, promising approaches have been developed for the genetic therapy and prevention of mitochondrial DNA (mtDNA) disorders. (A) Use of mitochondrion-targeted designer nucleases to eliminate or reduce levels of mutated mitochondrial genomes. Because mitochondria have a copy number-control mechanism, the elimination of a subpopulation of mtDNA is quickly compensated by an increase in replication of the residual mtDNA, in this case mostly wild type molecules. (B) Replacement of the nuclear spindle in an oocyte that functions as a mtDNA donor.
The delivery of mitoTALENs to tissues would be likely to involve gene therapy, an approach considered risky. Indeed, traditional gene therapy of nuclear DNA defects has only started to make a comeback after having been plagued with bad news in its infancy [9]. Considering these problems, is there any compelling reason to use gene therapy for mtDNA diseases? In fact, there are some good arguments to advocate its use, because the mitochondrial genome plays by different rules. In contrast to nuclear-gene defects, the mutation-specific mitochondrion-targeted nuclease does not need to be expressed throughout the life of the patient. A single dose, which may be active for only a few weeks, could permanently alter mtDNA heteroplasmy toward wild type. The development of transient gene-delivery methods that do not include integration of the transgene into chromosomes would reduce the primary concern related to gene therapy: insertional activation of oncogenes [10]. Furthermore, due to the small size of the mitochondrial genome, there is less concern about off-target cleavage by specific nucleases. Of course, transient delivery of these mitochondrial nucleases to affected organs remains a considerable challenge, particularly for the bulky mitoTALENs and mitoZFNs.
Approaches to correct mtDNA mutations in the oocyte or embryo
The past few years also saw the development of embryo-manipulation techniques that can essentially eliminate mutated mtDNA. Preimplantation genetic diagnosis and selection of embryos with low mtDNA mutation load, which would have a lower probability of developing clinical symptoms later in life, has been described (reviewed in [11]).
The transfer of pronuclei between zygotes [12] or nuclear spindles between oocytes has shown promising results [13] (Figure 1B), suggesting it could soon be used in humans. Specifically, chromosomes from the enucleated donor egg are removed and replaced with the would-be mother’s chromosomes, thereby allowing the donor egg to contribute only its mtDNA. Zygotes created by the transfer of nuclear material from a donor zygote to a second enucleated donor zygote was found to contain less than 2% mtDNA carry-over from the nuclear donor [12]. Oocytes that had nuclear spindles transferred to enucleated donor oocytes showed less than 1% (often undetectable) carry-over of mtDNA [13]. At the same time, manipulated human oocytes could be fertilized and developed into normal embryonic stem cells [13]. A similar approach in macaques gave rise to normal offspring harboring the mtDNA from the enucleated donor oocytes [14].
Although research remains lacking, it is tempting to speculate that the engineered nucleases described above may also be able to correct the mtDNA mutation load in the patient’s own oocytes, which would bypass the use of a mtDNA donor and any ethical concern related to the use of DNA from a third person.
Are we ready to use these approaches to treat or prevent mitochondrial diseases?
Most people would not have problems with replacing a kidney, a heart, or multiple organs. However, they may raise an eyebrow when considering modifying or replacing genetic material due to perceived ethical concerns about borrowing genes from a third person or modifying the basic material that makes us who we are. However, like a heart transplant, alterations to the mtDNA background can improve a patient’s overall health while being unlikely to change the physical and personality traits that define him or her. To many, a change toward improved health is usually welcome and reaching this goal for patients suffering from mtDNA disorders should be no exception. Still, fears related to the uncertainties of genetic therapy remain very real today. ‘Insertional oncogenesis’ is at the top of the unintended consequences of genetic therapy, particularly that involving integrating viral vectors [10]. However, the possibility of treating a mitochondrial disease with transient exposure to a therapeutic gene product that would not activate oncogenes by not inserting into chromosomes should challenge us to think outside the box on how to deliver mitochondrial nucleases safely.
Embryo manipulation of any kind is a more controversial issue, particularly because the genetic alteration will be transmitted through the germline. However, assisted reproduction is an established approach and improvements in outcome are constantly being implemented. Replacement of mtDNA can be seen as one more technique to help ensure the health of the developing embryo and child. As with any procedure, the techniques have to be tested to ensure safety and reliability, but other than that there seem to be no major biological barriers to moving it forward. A detailed UK-based discussion and report on the ethical issues of mtDNA replacement (http://www.nuffield-bioethics.org/mitochondrial-dna-disorders) found that, because of their limited genetic contribution, mtDNA donors should not be considered ‘third parents’, a conclusion fully backed by our biological knowledge. The report also emphasized that, with informed consent and further experimentation to assure safety, these procedures should be available to families attempting to have children unaffected by mtDNA mutations. Moreover, the Human Fertilisation and Embryology Authority (HFEA) gathered the public’s views on the social and ethical impact of making these techniques available to patients and found overwhelming support, as long as the procedures are safe (http://www.hfea.gov.uk/6896.html).
Mitochondrial genetics play by different rules, in many cases making it harder but in others facilitating intervention. We should take advantage of the latter to focus on strategies to help patients and families in need.
References
- 1.Luo SM, et al. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13038–13043. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schon EA, et al. Therapeutic prospects for mitochondrial disease. Trends Mol. Med. 2010;16:268–276. doi: 10.1016/j.molmed.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 4.Gowher A, et al. Induced tRNA import into human mitochondria: implication of a host aminoacyl-tRNA-synthetase. PLoS ONE. 2013;8:e66228. doi: 10.1371/journal.pone.0066228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G, et al. Correcting human mitochondrial mutations with targeted RNA import. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4840–4845. doi: 10.1073/pnas.1116792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacman SR, et al. Manipulation of mtDNA heteroplasmy in all striated muscles of newborn mice by AAV9-mediated delivery of a mitochondria-targeted restriction endonuclease. Gene Ther. 2012;19:1101–1106. doi: 10.1038/gt.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacman SR, et al. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minczuk M, et al. Construction and testing of engineered zinc-finger proteins for sequence-specific modification of mtDNA. Nat. Protoc. 2010;5:342–356. doi: 10.1038/nprot.2009.245. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann KB, et al. Gene therapy on the move. EMBO Mol. Med. 2013;5:1642–1661. doi: 10.1002/emmm.201202287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deichmann A, Schmidt M. Biosafety considerations using gamma-retroviral vectors in gene therapy. Curr. Gene Ther. 2013;13:469–477. doi: 10.2174/15665232113136660004. [DOI] [PubMed] [Google Scholar]
- 11.Smeets HJ. Preventing the transmission of mitochondrial DNA disorders: selecting the good guys or kicking out the bad guys. Reprod. Biomed. Online. 2013;27:599–610. doi: 10.1016/j.rbmo.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Paull D, et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature. 2013;493:632–637. doi: 10.1038/nature11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana M, et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature. 2013;493:627–631. doi: 10.1038/nature11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tachibana M, et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]