Abstract
Klebsiella pneumoniae liver abscess with metastatic complications is an emerging infectious disease in Taiwan. To identify genes associated with liver infection, we used a DNA microarray to compare the transcriptional profiles of three strains causing liver abscess and three strains not associated with liver infection. There were 13 clones that showed higher RNA expression levels in the three liver infection strains, and 3 of these 13 clones contained a region that was absent in MGH 78578. Sequencing of the clones revealed the replacement of 149 bp of MGH 78578 with a 21,745-bp fragment in a liver infection strain, NTUH-K2044. This 21,745-bp fragment contained 19 open reading frames, 14 of which were proven to be associated with allantoin metabolism. The K2044 (ΔallS) mutant showed a significant decrease of virulence in intragastric inoculation of BALB/c mice, and the prevalence of this chromosomal region was significantly higher in strains associated with liver abscess than in those that were not (19 or 32 versus 2 of 94; P = 0.0001 [χ2 test]). Therefore, the 22-kb region may play a role in K. pneumoniae liver infection and serve as a marker for rapid identification.
Klebsiella pneumoniae is a common hospital-acquired pathogen which is often involved in nosocomial pneumonia, urinary tract infections, and bacteremia in immunocompromised humans. However, liver abscesses caused by this single pathogen have exhibited an upward trend over the last two decades in Taiwan (5, 7, 9, 15, 23, 26, 40). K. pneumoniae liver abscess typically presents as community-acquired primary liver abscess without biliary tract disease (23, 40), and about 10 to 12% of patients develop serious extrahepatic complications such as metastatic meningitis or endophthalmitis (7, 9, 15, 26). Diabetes mellitus is thought to be an important risk factor in patients with K. pneumoniae liver abscess, with a prevalence of approximately 50% (5, 7, 9, 15, 23, 26, 40), and a similar situation has been reported in other countries, such as the United States, Japan, and Singapore (11, 38, 39, 41, 43). Even when standardized treatment, including pigtail catheter drainage plus antimicrobial therapy, is performed, there is still significant morbidity and an at least 10% mortality, especially in patients with metastatic complications (7, 9, 15, 23, 26, 40). The timing of appropriate antimicrobial therapy is an important factor related to the survival rate (8, 13, 15). Therefore, rapid diagnosis is helpful to prevent exacerbation of disease. However, the pathogenesis of the newly emerging infectious disease remains unclear. The aim of this study was to identify genetic loci specifically associated with liver infection strains of K. pneumoniae by comparing strains causing primary liver abscess with strains from patients without liver abscess or other tissue invasions. We constructed a DNA microarray from a genomic library of a clinical K. pneumoniae isolate to investigate the genes with increased RNA expression levels in liver infection strains of K. pneumoniae.
MATERIALS AND METHODS
Bacterial strains.
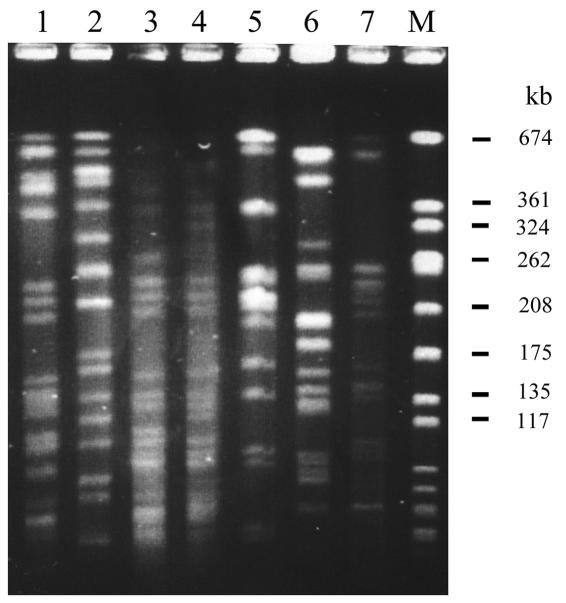
K. pneumoniae isolates from patients with liver abscess, meningitis, or endophthalmitis were defined as liver infection strains, and bacterial isolates from patients without any tissue invasions other than bacteremia were defined as non-liver infection strains. A total of 126 K. pneumoniae strains were used in this study (Table 1). Sixty strains were collected from National Taiwan University Hospital (NTUH) during 1996 to 2001. Of these strains, 31 liver infection strains were isolated from patients with primary liver abscess, including three isolates (NTUH-K2044, A5011, and A3021) from patients who had metastatic meningitis or endophthalmitis in addition to liver abscess. These three isolates from patients with metastatic complication were chosen for microarray analysis. The other 29, non-liver infection strains were isolated from patients without clinical symptoms of liver abscess, meningitis, or endophthalmitis. Three of the 29 strains (N3423, N3529, and N5322) were randomly chosen for microarray experiments. All of the patients with primary liver abscess were confirmed by sonography-guided aspiration or surgical drainage, and patients without liver abscess were confirmed by abdominal sonography or computed tomography. Most of the isolates, including the three liver infection strains and three non-liver infection strains selected for microarray experiments, were not genetically related, which was confirmed by pulsed-field gel electrophoresis (5, 42) (Fig. 1).
TABLE 1.
K. pneumoniae strains used in this study and prevalence of the 22-kb region
| Strain or isolate (no. of isolates) | Origin | Patient symptom(s) | PCR (slot blot) resultsa
|
|
|---|---|---|---|---|
| No. positive for 22-kb region | No. negative for 22-kb region | |||
| Liver infection strains (32) | Taiwan | |||
| NTUH strains (28) | Liver abscess | 16 (16) | 12 (12) | |
| K2044 and A5011 | Liver abscess and meningitis | 2 (2) | 0 (0) | |
| A3021 | Liver abscess and endophthalmitis | 1 (1) | 0 (0) | |
| ECK strain (1) | Meningitis | 0 (0) | 1 (1) | |
| Non-liver infection strains (94) | Other than liver abscess | |||
| NTUH strains (29) | Taiwan | 1 (1) | 28 (28) | |
| FEM strains (32) | Taiwan | 0 (0) | 30 (32) | |
| ECK strains (11) | Taiwan | 1 (1) | 10 (10) | |
| ATCC strains (22) | United States | 0 (0) | 20 (22) | |
A total of 122 strains were consistent by PCR. Positive, positive with inside primers and negative with outside primers, negative, negative with inside primers and positive with outside region primers (see text). Four strains (two FEM strains and two ATCC strains) were PCR negative with both inside and outside primer pairs and were proven to be negative by slot blot hybridization.
FIG. 1.
Pulsed-field gel electrophoresis of K. pneumoniae isolates digested with XbaI. Lanes 1 to 4, four liver infection strains (NTUH-K2044, A1208, A3021, and A5011, respectively). Three of these four liver infection strains (excluding A1208) were chosen for microarray hybridization. Lanes 5 to 7, three non-liver infection strains (N3423, N3529, and N5322, respectively), which were chosen for microarray hybridization. Lane M, molecular size marker.
For purposes of comparison, 66 strains were obtained from other areas. Thirty-two non-liver infection strains (FEM strains) were provided by Far Eastern Memorial Hospital, Banciao, Taiwan. Eleven non-liver infection strain isolates and one liver infection strain (ECK strains) were provided by En Chu Kong Hospital, Sansia, Taiwan. All clinical isolates were identified as K. pneumoniae according to standard clinical microbiologic methods (1). Twenty-two American strains, including MGH 78578, which was chosen for full genome sequencing by the Genomic Sequencing Center at Washington University (St. Louis, Mo.), were purchased from the American Type Culture Collection (ATCC).
Growth conditions.
Bacteria were grown in Luria-Bertani medium at 37°C, except those for the allantoin utilization assay. For allantoin analysis, bacteria were grown in allantoin minimal medium (4), consisting of 34 mM NaH2PO4, 64 mM K2HPO4, 1 μM FeSO4, 0.1 mM MgSO4, and 10 μM CaCl2 and supplemented with 60 mM allantoin. Aerobic growth was carried out in a 37°C incubator with shaking. Anaerobic growth was carried out at 37°C in a microaerophilic chamber (Don Whitley, West Yorkshire, England) containing 10% CO2, 5% O2, and 85% N2.
DNA extraction and library construction.
Genomic DNA of a liver infection K. pneumoniae strain, NTUH-K2044, was extracted from 3 ml of overnight culture by using a DNA isolation kit (Puregene, Minneapolis, Minn.) according to the manufacturer's instructions. Aliquots of 20 μg of genomic DNA were partially digested with Sau3AI, and DNA fragments of 3 to 5 kb that had been harvested from an agarose gel were ligated to the BamHI site of a λ-ZAP-II vector (Stratagene, La Jolla, Calif.). The phagemid library was obtained by using a Gigapack III Gold packaging kit (Stratagene) as previously described (12). A total of 5,760 clones were amplified by PCR with primers in vector (Table 2), and 3,146 of these clones were randomly selected for the microarray. To test the redundancy of library, 798 of the 3,146 clones were randomly selected for sequencing. The 798 clones revealed 678 distinct sequences (i.e., 15% redundancy).
TABLE 2.
DNA primers used in this study
| Primer | Sequence | nt
|
Localization region | Use | |
|---|---|---|---|---|---|
| 5′ | 3′ | ||||
| 336F | TCTGATTTATCCCACATT | 1131 | 1114 | 22-kb region | Prevalence of 22-kb region |
| 1416R | CCGTTAGGCAATCCAGAC | 42 | 59 | 22-kb region | Prevalence of 22-kb region |
| 2801769F | GTACACCAGCGGCCTGTTCTGGGCGCAATC | 2362637 | 2362666 | MGHa contig 3591 | Prevalence of 22-kb region |
| 2803831R | CAGAACAGCATTTACTATGATGTGGTG | 2364699 | 2364673 | MGH contig 3591 | Prevalence of 22-kb region |
| 252R | GACGTTGGGAAGGGGAAC | 1208 | 1225 | 22-kb region | Inverse PCR |
| 1729F | TCGCATATCGTCCAGCAG | −271 | −288 | Upstream of 22-kb region | Inverse PCR |
| 92F | GTGAAAACGCTGTACTTTGC | 1368 | 1349 | 22-kb region | Pooled PCR |
| 422R | GAGCATCGCCATATTCCTCG | 1038 | 1057 | 22-kb region | Pooled PCR |
| R1-14F | CCTCTCTCCAGCGCCTGTGCGCC | 1905 | 1883 | 22-kb region | Pooled PCR, allS and cap deletion construct |
| R1-466R | GTCACCGAACTGGAGCGCCATCC | 1453 | 1475 | 22-kb region | Pooled PCR |
| R2-5F | AGTCGGCCTGGGGTTTAAGG | 2642 | 2623 | 22-kb region | Pooled PCR and slot blot hybridization for prevalence of 22-kb region |
| R2-475R | CAGTCAACGTGGCGATTCGC | 2072 | 2191 | 22-kb region | Pooled PCR and slot blot hybridization for prevalence of 22-kb region |
| A1 | GACCTCCGGTCCGGCGGGTA | 2953 | 2972 | 22-kb region | Pooled PCR |
| A2 | CAAACGCCAGCTGAACGCAG | 4220 | 4201 | 22-kb region | Pooled PCR |
| B1F | CATGGTTTAATCGCTACCGC | 5281 | 5300 | 22-kb region | Pooled PCR |
| B1R | CACGTTGGCCGAGGAGAGCG | 6751 | 6732 | 22-kb region | Pooled PCR |
| B2F | CTGAGTATCGCTAATGCCGG | 6638 | 6657 | 22-kb region | Pooled PCR |
| B2R | TCAGAAATGCCAGCGATCCG | 8086 | 8067 | 22-kb region | Pooled PCR |
| C1 | GTTTGGCTTGCAATGCTACG | 8046 | 8065 | 22-kb region | Pooled PCR |
| C2 | CGCCAGGGTGTTACGTTTAT | 10861 | 10842 | 22-kb region | Pooled PCR |
| D1 | CATGGTGGTATTGCCGCTAA | 10822 | 10841 | 22-kb region | Pooled PCR |
| D2 | CTTTATGGGCGCTTATTCTC | 13612 | 13593 | 22-kb region | Pooled PCR |
| E1F | GCCTGTAGGGAATAAGCGCC | 13586 | 13605 | 22-kb region | Pooled PCR |
| E1R | GGGGCAGCAAAAATATCTTC | 15135 | 15116 | 22-kb region | Pooled PCR |
| E2F | GCTGCCCCAGAAGACGTAAG | 15128 | 15147 | 22-kb region | Pooled PCR and z0673-z0674 deletion construct |
| E2R | CCTGCGCCAGGGCGGATAAG | 17209 | 17190 | 22-kb region | Pooled PCR |
| F1 | GAAGCGGACAAAGTCAGAGT | 17210 | 17229 | 22-kb region | Pooled PCR |
| F2 | TGTAAACATAGGGACCTCTTATTG | 18612 | 18589 | 22-kb region | Pooled PCR |
| G1 | GGTTGAAAACTGGCATTACAC | 19713 | 19732 | 22-kb region | Pooled PCR |
| G2 | TCTACCGTCAGCGCTTCTCC | 20751 | 20732 | 22-kb region | Pooled PCR and z0673-z0674 deletion construct |
| 616R | ATACTCGCCGTATGCCTG | −798 | −781 | Upstream of 22-kb region | aIIS deletion construct |
| 2803749R | GTATTAATGGCGATTACG | 2364617 | 2364600 | MGH contig 3591 | cap deletion construct |
| T3 | ATTAACCCTCACTAAAG | Plasmid pBKCMV | Microarray PCR | ||
| T7 | TAATACGACTCACTATAGGG | Plasmid pBKCMV | Microarray PCR | ||
MGH, MGH 78578.
RNA isolation and probe preparation.
Total RNA was purified from log-phase cultures of K. pneumoniae by CsCl gradient centrifugation (33). Aliquots of 40 μg of total RNAs were labeled as previously described (2). Briefly, total RNA was mixed with 12 μM random primers; 1 mM (each) dCTP, dATP, and dGTP; 80 μM dTTP; 80 μM biotin-16-dUTP; RNase inhibitor (0.5 U/μl) (Roche, Mannheim, Germany); 10 mM dithiothreitol; and 600 U of Superscript II reverse transcriptase (Invitrogen/Gibco BRL, Carlsbad, Calif.) in 62.5 μl of solution at 42°C for 4 h. The reaction was stopped by heating to 95°C for 5 min, and RNA was degraded in the presence of NaOH and neutralized by addition of acetic acid. The labeled cDNA was precipitated with isopropanol and washed with 75% ethanol. Immediately before use, the probes were resuspended in hybridization buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% N-lauroylsarcosine, 0.1% sodium dodecyl sulfate (SDS), 1% blocking reagent (Roche), and 40 μg of herring sperm DNA per μl.
Microarray construction.
Colonies picked from the phagemid library were inoculated into 96-well microtiter plates. After overnight culture at 37°C, 1 μl of bacteria containing DNA fragments of K. pneumoniae was amplified by PCR with primers in vector. The PCR conditions were, for the first step, 96°C for 5 min, followed by 30 cycles of 96°C for 30 s, 56°C for 30 s, and 74°C for 2 min, carried out in a volume of 50 μl for each well in a V-bottom 96-well polycarbonate microtiter plate. The PCR products were concentrated by evaporation at 95°C to 2 μg/μl. A total of 3,146 clones were amplified by PCR and spotted onto a positive-charge nylon membrane (Roche). An arraying machine with a personal computer-controlled XYZ translation system (PM500; Newport Inc., Fountain Valley, Calif.) was outfitted with Teflon-coated tool steel pins (Teflon-AF; DuPont, Wilmington, Del.) for sample delivery (6). Samples were held at the tips of the six pins for delivery by the action of surface tension.
Hybridization of microarray.
The microarray-carrying membrane was prehybridized in 2 ml of hybridization buffer at 65°C for 2 h and hybridized at 63°C for 16 h. The membrane was washed twice with 2× SSC containing 0.1% SDS at room temperature for 5 min and then washed three times with 0.1× SSC containing 0.1% SDS at 65°C for 15 min each time.
Colorimetry detection and image analysis.
After hybridization and washing, the membrane was blocked with 2 ml of blocking buffer consisting of 7% casein (Sigma, St. Louis, Mo.) in 1× PBST buffer (1× phosphate-buffered saline, 0.05% Tween 20) at room temperature for 1 h and then incubated with 2 ml of a mixture containing 2,500× diluted alkaline phosphatase-conjugated streptavidin (Gibco BRL), 4% polyethylene glycol 8000 (Sigma), and 1% bovine serum albumin in PBST buffer for 1 h. The membrane was then washed with 1× PBST buffer four times for 5 min each and incubated with 2.5 ml nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate substrate buffer (Pierce, Rockford, Ill.) at room temperature for 40 min. Color development was stopped with 1× phosphate-buffered saline containing 20 mM EDTA. Image analysis was as described previously (2). Briefly, the signals in the membrane were scanned and converted to gray levels. Densitometry was performed, and 23S rRNA signals were used as an internal standard.
Construction of K. pneumoniae mutants.
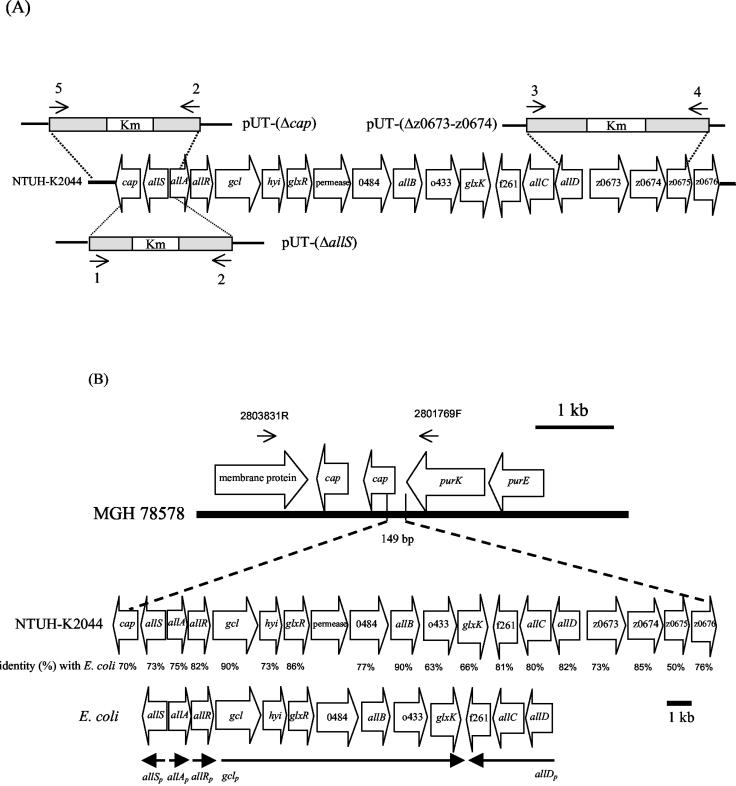
For gene replacement of allS, a PCR fragment generated by primers R1-14F and 616R (Table 2 and Fig. 2A) was cloned into a PCRII vector (Invitrogen, San Diego, Calif.), and the 677-bp region from the HincII to the EcoRV site located at allS was replaced with a kanamycin cassette. The deletion construct was then cloned into the suicide vector pUT (21). A second marker, for spectinomycin, was cloned into a pUT-(ΔallS) vector as a negative selective marker to distinguish between single and double crossovers for chromosomal integration. pUT-(ΔallS) was transferred to NTUH-K2044 by conjugation. The same procedures were used for the cap and z0673-z0674 deletion constructs. To construct the pUT-(Δz0673-z0674) mutant (Fig. 2A), a PCR fragment containing the allD-z0675 region was first generated with primers E2F and G2 (Table 2 and Fig. 2A), and then an 1,874-bp fragment located from open reading frame (ORF) z0673 nucleotide (nt) 90 to ORF z0674 nt 286 was replaced with a kanamycin cassette. To construct the pUT-(Δcap) mutant (Fig. 2A), a PCR fragment was generated with primers 2803749R and R1-14F (Table 2), and then a 634-bp fragment located at putative cap nt 146 to nt 779 was replaced with a kanamycin cassette. Chromosomal deletion of mutants was confirmed by PCR with multiple primer pairs.
FIG. 2.
Organization of genes in the 22-kb chromosomal region. (A) Schematic diagram of deletion constructs. The primers (also shown in Table 2) are indicated by arrows: 1, 616R; 2, R1-14F; 3, E2F; 4, G2; 5, 2803749R. The deletion region was replaced with a kanamycin cassette. (B) The 149-bp chromosome in MGH 78578 was replaced by a 21,745-bp fragment in NTUH-K2044. The boldface line at the top represents a genomic fragment of MGH 78578. Alignment of the gene cluster between the 22-kb region of NTUH-K2044 and E. coli is shown, and the amino acid identity is shown below the gene. The extents and directions of the genes are indicated by open arrows, and designations are according to those proposed for E. coli (10, 35). Arrows on the top represent primer locations outside the 22-kb region for PCR confirmation of strains without a 22-kb region. The lines at the bottom show the transcriptional units in E. coli.
Slot blot hybridization.
For cap and allD region detection, the PCR products of each locus were transferred onto a nylon membrane by passive vacuum pressure, and a PCR product of 23S rRNA was used as internal control for each membrane. Twenty micrograms of total RNA was reverse transcribed and labeled with biotin. For determination of the 22-kb prevalence, 5 μg of K. pneumoniae genomic DNA was transferred onto a nylon membrane; the probe was a biotin-labeled PCR fragment (Table 2) located in the 22-kb region. The membrane was prehybridized with hybridization buffer at 65°C for 2 h and hybridized with biotin-labeled cDNA probe at 65°C for 16 h. Detection was performed with the Southern-Light chemiluminescent detection system (Tropix, Bedford, Mass.) according to the manufacturer's instructions.
DNA sequencing of flanking chromosomal regions.
The primers used in this study are listed in Table 2. Inverse PCR and pooled PCR were used to extend the flanking region of clone 1. For inverse PCR, the genomic DNA was digested with KpnI and then self-ligated. Inverse PCR was carried out with primers 1729F and 252R (Table 2). The PCR fragments were cloned into a pGEM-Teasy vector (Promega, Madison, Wis.), and the sequence was determined by BigDye termination methods. For pooled PCR, 20 bacterial colonies were randomly selected from the genomic library of NTUH-K2044 and pooled in each well of a 96-well plate. After an overnight culture, 1 μl of bacterial culture from each well was transferred to a V-bottom 96-well plate and amplified by PCR with primers derived from sequences of the 5′ end (Table 2). Positive wells were broken down, and clones indicated to be positive by PCR were picked up for sequencing. The procedures were repeated until the 5′ end sequences matched sequences in MGH 78578.
Animal inoculation.
Six-week-old female BALB/c mice were used in this experiment. Groups of four mice were each infected intraperitoneally and intragastrically (18) with the indicated doses of K. pneumoniae in 0.5 and 0.2 ml of saline solution, respectively. Mice were given 102, 103, and 104 CFU of wild-type NTUH-K2044 or mutants or were given 106 and 107 CFU of MGH 78578 intraperitoneally. Mice were given 104 to 106 CFU of wild-type NTUH-K2044 or mutants intragastrically. The 50% lethal dose (LD50) was calculated as described by Reed and Muench (34). The exact inoculation dose was confirmed by examining CFU on Luria-Bertani agar plates.
RESULTS
Comparison of expression levels by microarray analysis.
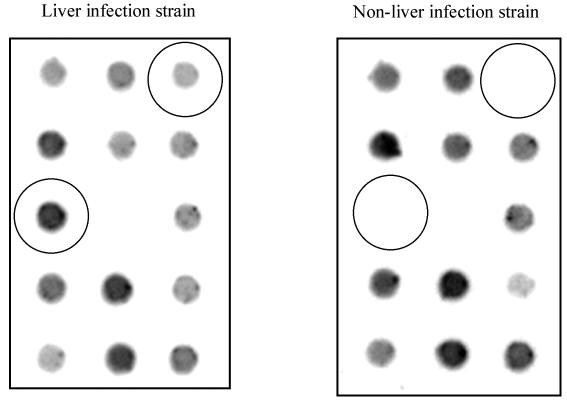
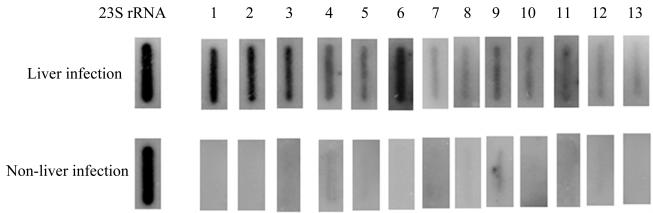
Microarray analysis was performed to compare the RNA profiles of three liver infection (A3021, A5011, and NTUH-K2044) and three non-liver infection (N3423, N3529, and N5322) K. pneumoniae strains from NTUH. Parts of the microarray with different expression levels were magnified and are shown in Fig. 3. There were 27 clones that had at least twofold-decreased RNA expression levels in liver infection strains. In contrast, there were 13 clones (Table 3) that revealed an increase of RNA expression levels of greater than or equal to twofold in liver infection strains. Slot blot analysis confirmed the results of the microarray experiments (Fig. 4). Because sequences of all of the 27 clones were present in MGH 78578, we chose to study genes exhibiting increased RNA expression levels in liver infection strains. The nucleotide sequences of these 13 clones were analyzed. There were three clones (clone 1, 2, and 6) that showed no sequence similarity in the 5′ end with the 10× shotgun sequence of K. pneumoniae MGH 78578. The three clones contained the same loci, only with different lengths in the 3′ end (Table 3). Although the other 10 clones had sequences corresponding to those of MGH 78578, several ORFs were similar to some virulence genes in other pathogens. Clone 5 includes two ORFs homologous to etfB and etfC in Edwardsiella tarda (Table 3). Both etfB and etfC are responsible for the synthesis of fimbrial protein (37). Clone 7 encodes a protein that is homologous to glutathione peroxidase, an enzyme which has been proven to be important in the cellular metabolism or defense processes in Neisseria meningitidis (29). One of the ORFs in clone 9 showed amino acid similarity to ferredoxin-NADP-reductase (encoded by fpr). Disruption of fpr has been shown to render Salmonella enterica serovar Typhimurium hypersensitive to paraquat (32). The two ORFs included in clone 11 showed amino acid sequence identity to a component of Clp protease and an ABC-type efflux carrier, respectively. The Clp proteolytic complexes have been demonstrated to be important for survival during stress and play a role in virulence in many pathogens (14, 16, 25, 31), whereas the ABC-type efflux carrier is an antibiotic transporter that confers erythromycin resistance in Escherichia coli (24). The actual roles of these ORFs in K. pneumoniae-related liver infections await further investigation; however, our data provide an important basis for future studies.
FIG. 3.
Colorimetric detection of total K. pneumoniae RNA expression on a microarray. The dots showing increased RNA expression levels in the liver infection strain are circled.
TABLE 3.
Predicted ORFs of the 13 clones
| Clone no. | Predicted protein (accession no.) | Length of comparable amino acid sequence (% identity) | RNA level increase (fold) |
|---|---|---|---|
| 1 | LysR-type transcriptional regulator (P77702) | 269 (73) | 15.5 |
| Capsule-anchoring protein (NP_459508) | 349 (73) | ||
| 2 | Same as clone 1 except 600 bp less at 3′ end | 11.1 | |
| 3 | Amino acid ABC transporter (NP_470183) | 194 (64) | 12.3 |
| Hypothetical protein (NP_458927) | 200 (60) | ||
| 4 | Putative 2-ketogluconate transporter (NP_745517) | 246 (69) | 8.8 |
| 2-Ketoaldonate reductase (P37666) | 322 (81) | ||
| 5 | Fimbrial chaperone protein (BAC55513) | 222 (45) | 4.9 |
| Fimbrial usher protein (BAD00163) | 762 (45) | ||
| 6 | Same as clone 1 | 13.4 | |
| 7 | Hypothetical protein (NP_456227) | 316 (90) | 3.7 |
| Glutathione peroxidase (ZP_00066077) | 158 (58) | ||
| 8 | Hypothetical protein (NP_804847) | 217 (89) | 2.9 |
| Putative nucleolar protein (NP_416349) | 462 (79) | ||
| 9 | Ferredoxin-NADP reductase (NP_290553) | 248 (87) | 4.7 |
| Hypothetical protein (CAD98874) | 200 (83) | ||
| Triosephosphate isomerase (CAD98875) | 237 (99) | ||
| 10 | Regulator of gluconate operon (NP_312314) | 331 (92) | 3.8 |
| 11 | ATP-binding component of serine protease (NP_752948) | 277 (97) | 4.0 |
| Macrolide-specific ABC-type efflux carrier (P75831) | 468 (76) | ||
| 12 | Same as clone 9 | 3.8 | |
| 13 | Possible drug efflux protein (NP_457613) | 318 (83) | 2.8 |
FIG. 4.
RNA expression levels of the 13 clones (Table 3) by slot blot hybridization. Blots 1 to 13, hybridization with PCR products from clones 1 to 13, respectively. Biotin-labeled cDNA probes were from either a liver infection strain or a non-liver infection strain. The 23S rRNA was used as an internal control.
Clone 1 and flanking regions.
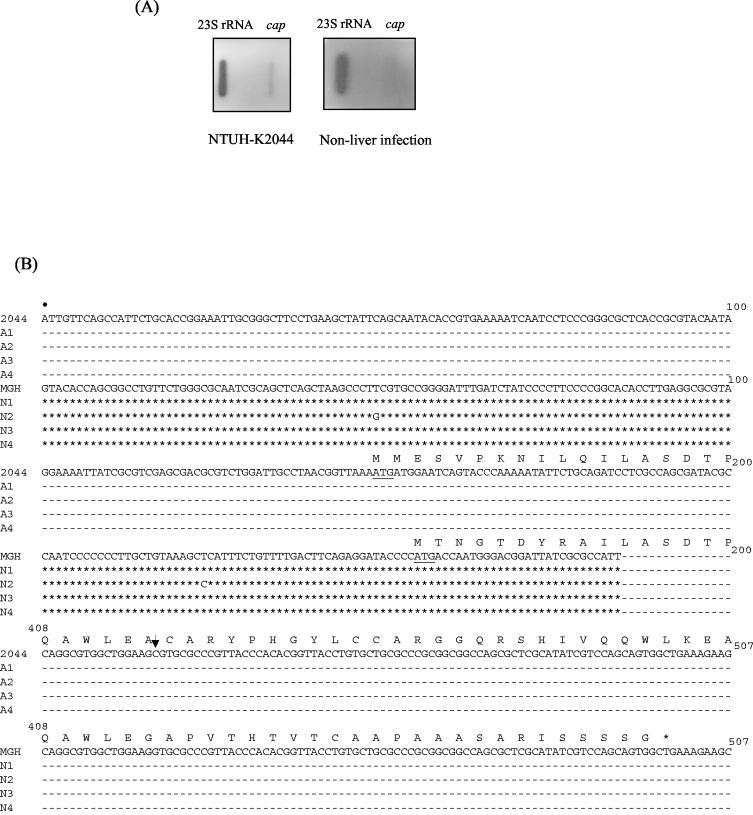
Clone 1 was selected for further study because of the absence of a 5′ sequence in MGH 78578. It also showed the highest difference in expression level and contained a 2,752-bp fragment and two ORFs, one of which (putative allS) showed 73% amino acid sequence identity to the AllS activator for allantoin regulon and the other of which (putative cap) showed 73% amino acid sequence identity to a capsule-anchoring protein (Table 3). Slot blot hybridization showed that the putative cap could be detected in only NTUH-K2044 and not in a non-liver infection strain (Fig. 5A). Therefore, four non-liver infection strains were randomly selected, and the cap sequence compared with four liver infection strains. The results demonstrated that the four liver infection strains shared the same putative promoter region and had identical deduced amino acid sequences; however, the putative promoter region and part of the N-terminal residues in these liver infection strains were totally different from those of the non-liver infection strains (Fig. 5B). In addition, a frameshift was observed in MGH 78578 (http://genome.wustl.edu/gsc), resulting in a early stop codon (Fig. 5B). The sequence of the clone 1 flanking regions and the comparison with MGH 78578 revealed that 149 bp in MGH 78578 (contig 3591, nt 2362672 to 2362820) was replaced by a 21,745-bp fragment in NTUH-K2044 (Fig. 2B) (GenBank accession no. AB115590). The 21,745-bp region contained 19 ORFs, including a partial fragment of the putative cap gene. Analysis done by using the National Center for Biotechnology Information BLAST server revealed that the alignment of the 19 ORFs was similar to the chromosomal region z0657 to z0676 in E. coli. Thirteen genes (corresponding to allS to allD) located in the region z0658 to z0672 have been proven to be responsible for the anaerobic assimilation of nitrogen from allantoin and the glyoxylate pathway in E. coli (10, 35) (Fig. 2B). In NTUH-K2044, the region containing 14 ORFs was similar to the region responding to the use of allantoin in E. coli (Fig. 2B), and a protein encoded between glxR and 0484 showed 73% amino acids identity to a permease of S. enterica serovar Typhimurium that was absent in E. coli. Of the remaining five ORFs, one was the putative cap and the other four were similar to chromosome region z0673 to z0676 in E. coli (Fig. 2B).
FIG. 5.
(A) RNA expression levels of cap determined by slot blot hybridization. The 23S rRNA served as an internal control. Probes were derived from NTUH-K2044 and from a non-liver infection strain. (B) Comparison of the putative promoter regions of cap in liver infection and non-liver infection strains. Dashes represent 100% sequence identity with NTUH-K2044, and asterisks represent 100% sequence identity with MGH 78578. The first nucleotide (marked by a dot) corresponds to contig 3591 nt 2362637 in MGH 78578 and nt 184 in the 22-kb fragment of NTUH-K2044. The putative start codon is underlined. Translated amino acids are shown above the DNA sequence. nt 201 to 407 are omitted because the sequences were same in all investigated strains. The arrow represents a deoxycytidine deletion in MGH 78578, and thus a stop codon (asterisk) occurred in MGH 78578. MGH, MGH 78578; N1 to N4, four non-liver infection strains; 2044, NTUH-K2044; A1 to A4, four liver infection strains.
Characterization of the 22-kb chromosomal region.
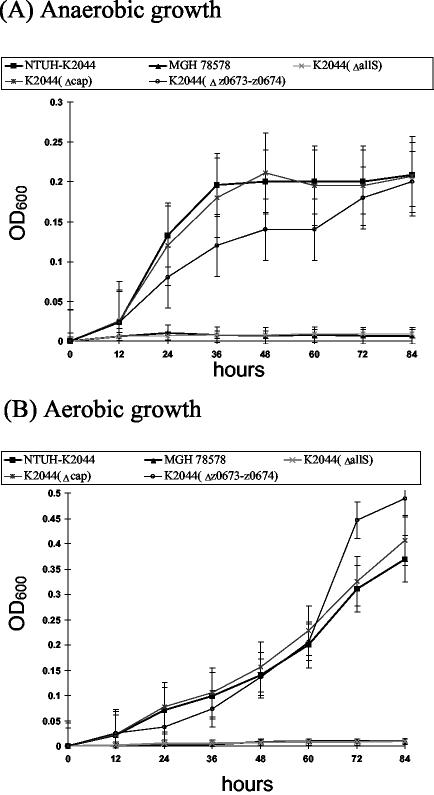
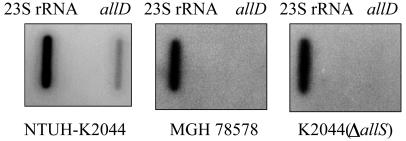
The growth of K2044(ΔallS) was compared with that of the wild-type NTUH-K2044 under the condition that allantoin was used as the sole carbon and nitrogen source. The results showed that K2044(ΔallS) could not grow in allantoin minimal medium. The same result occurred with MGH 78578 and a non-liver infection strain, which do not contain the allantoin regulon region, whereas the wild-type strain NTUH-K2044 grew in such a medium (Fig. 6). Interestingly, K. pneumoniae NTUH-K2044 was able to utilize allantoin as the sole carbon and nitrogen source even under aerobic conditions (Fig. 6B). Because AllS is essential for the expression of the allD operon in E. coli (35), slot blotting was performed to detect RNA expression of the putative allD operon. RNA expression of the allD operon could be detected in NTUH-K2044 but not in MGH 78578 or K2044(ΔallS) (Fig. 7). To examine whether the remaining five ORFs are related to allantoin utilization, two deletion clones, K2044(Δcap) and K2044(Δz0673-z0674), were grown in allantoin minimal medium, and the growth curves of the mutants were investigated. The K2044(Δcap) mutant exhibited the same growth rate as the wild-type strain under either aerobic or anaerobic conditions (Fig. 6). However, K2044(Δz0673-z0674) grew faster than the parent strain, NTUH-K2044, after 60 h under aerobic conditions (Fig. 6B). Although K2044(Δz0673-z0674) grew more slowly than the NTUH-K2044 strain during 12 to 72 h under anaerobic conditions, the difference was not statistically significant (Fig. 6A).
FIG. 6.
Growth study of wild-type K. pneumoniae and mutant strains in allantoin minimal medium. (A) Bacteria were grown under anaerobic conditions. (B) Bacteria were grown under aerobic conditions. The experiments were repeated three times and error bars indicate standard deviations.
FIG. 7.
RNA expression levels of allD determined by slot blot hybridization. The 23S rRNA served as an internal control for each membrane. Probes were derived from NTUH-K2044, MHG 78578, and K2044(ΔallS).
Animal study.
Inoculation of the NTUH-K2044, K2044(ΔallS), K2044(Δcap), and K2044(Δz0673-z0674) strains in mice showed the same LD50 (102 CFU) as for the wild-type strain, whereas the mice remained healthy 6 weeks after inoculation of 107 CFU of MGH 78578 (LD50 of >107). However, intragastric inoculations of mice showed a significant difference in LD50 between the wild-type strain and K2044(ΔallS) mutant (104 for the wild-type strain and 105 to 106 for the mutant).
Prevalence of 22-kb region.
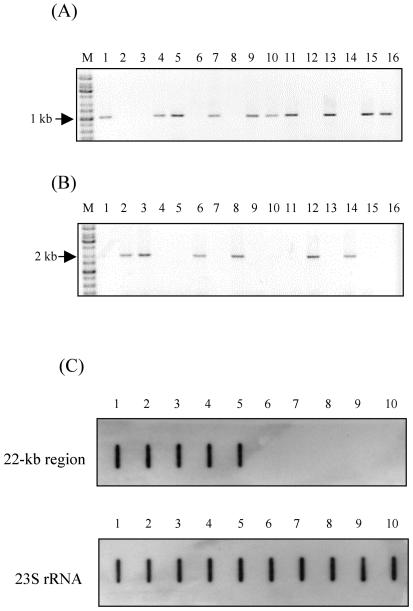
To detect the prevalences of the 22-kb region in different K. pneumoniae strains, PCR with a primer pair (336F and 1416R [Table 2]) inside the 22-kb region (inside primers) was performed with 126 K. pneumoniae isolates. A second primer pair (2801769F and 2803831R [Table 2 and Fig. 2B]) outside the 22-kb region (outside primers) was then used to perform PCR. The PCR with the second primer pair could generate a 2-kb fragment only in the absence of the 22-kb region, because the primer pair was too far apart in the genome (Fig. 8A and B). Consistent results with the two primer pairs were found for 122 strains, while for four non-liver infection strains PCR products could not be obtained with both sets of primers (Table 1). The PCR results for the 126 strains were reconfirmed by slot blot hybridization (Table 1 and Fig. 8C). The four PCR-negative strains were proven to be negative for the 22-kb region by slot blot hybridization. The prevalence of the 22-kb region was significantly higher in liver infection strains than in non-liver infection strains (19 of 32 versus 2 of 94; P = 0.0001 by χ2 test) (Table 1).
FIG. 8.
PCR and slot blot hybridization for determination of 22-kb prevalence. (A and B) The PCR was performed for 16 liver infection strains with primers included in the 22-kb region (A) or outside the 22-kb region (B). All of the PCR results that are negative in panel A are positive in panel B. Lanes M, molecular size markers. (C) Slot blot hybridization with a probe located in the 22-kb region. The 23S rRNA gene served as a control. Lanes 1 to 5, five liver infection strains; lanes 6 to 10, five non-liver infection strains.
DISCUSSION
Clone 1 includes allS and cap. Although cap can be found in the non-liver infection strains, the RNA expression levels of cap in liver infection strains and non-liver infection strains are different. Sequence extension of clone 1 revealed that allS is included in a 22-kb fragment, and PCR showed that this region is absent in most of the non-liver infection strains. The same condition was reported for S. enterica (17). Garaizar et al. (17) compared S. enterica serovar [4,5,12:i:−] strains with serovar Typhimurium LT2 by microarray analysis and reported that a region including 13 genes responsible for the anaerobic assimilation of allantoin as a nitrogen source was absent in all S. enterica serovar [4,5,12:i:−] strains but was present in serovar Typhimurium LT2 strains. However, the physiological implication of the deletion in S. enterica remains unclear. The region was large enough to be a pathogenicity island (10 to 200 kb) (20) and was present in the genomes of liver infection strains but was absent from the most genomes of non-liver infection strains, and the G+C content of this region (54.2%) was different from that of the rest of the genome (57.5%). Direct repeats (GTTGCCGATAGCGC) were found at nt 4457 and 20087 of the 22-kb region. However, this 22-kb region seems not to be a pathogenicity island, since no integrase or other mobility locus was observed in this region and most of the genes have the same function associated with allantoin metabolism in this region.
A chromosomal region containing 13 genes associated with allantoin metabolism has been identified in E. coli (10, 35). The allantoin regulon was formed by three structural operons expressed from promoters allAp, gclp, and allDp (Fig. 2B) and two regulators encoded by allR and allS. AllR is a repressor for the regulon, whereas AllS is an activator which interacts only with allDp. However, E. coli cannot use allantoin as a sole carbon source but is capable of using allantoin as a sole nitrogen source anaerobically. In this study, it was shown that K. pneumoniae use allantoin as the sole source of carbon, nitrogen, and energy under either aerobic or anaerobic condition. Nitrogen is a major component of nearly all of the macromolecules in microorganisms; therefore, a delicate system was developed to provide a constant supply of nitrogen. Certain nitrogenous compounds are preferred as nitrogen sources by microorganisms; however, when these primary nitrogen sources are not available or are in low concentrations, different nitrogen sources such as purines, proteins, or allantoin can be used (27, 36). Therefore, the allantoin-utilizing capability in K. pneumoniae may help its competition for nitrogen sources. In addition, the allantoin level is increased in certain patients with diseases such as rheumatoid arthritis (19), chronic lung disease (28, 30), bacterial meningitis (22), and non-insulin-dependent diabetes mellitus with early peripheral vascular disease (3). Because primary liver abscess caused by K. pneumoniae frequently occurs in diabetes mellitus patients, an increased allantoin concentration might benefit bacteria that can use allantoin.
A decrease of virulence in the deletion mutant was found only in the mouse model in comparison with that of the wild-type strain with intragastric infection, and not with intraperitoneal injection. This supported the importance of the infection route, since natural liver abscess is probably due to the resident K. pneumoniae in the gastrointestinal tract. In addition, the strong association of the 22-kb region with liver infection strains provides indirect evidence for the association of the 22-kb region with virulence.
In conclusion, we have identified 13 loci which expressed higher mRNA levels in liver infection strains, and one of these loci is associated with allantoin metabolism. The virulence was attenuated in a mouse model with intragastric infection. The prevalence of the 22-kb region is significantly higher in K. pneumoniae strains that cause liver infections than in those that do not. The 22-kb region may play a role in infections caused by liver infection K. pneumoniae strains and may serve as a marker for rapid identification.
Acknowledgments
This study was supported by grants from the National Science Council (NSC-91-2314-B-002-178 and NSC-92-3112-B-002-002), the National Taiwan University Hospital (NTUH-90-A09), and the Liver Disease Prevention and Treatment Research Foundation, Taiwan.
We thank Shih-Si Wang and Jann-Tay Wang for their generous gifts of the K. pneumoniae strains, Shih-Feng Tsai for sequencing of 798 microarray clones, and Chia-Rong Wu and colleagues for their help in constructing the library.
Editor: F. C. Fang
REFERENCES
- 1.Abbott, S. 1999. Klebsiella, Enterobacter, Citrobacter, and Serratia. p. 475-482. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 2.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benzie, I. F. F., Chung, W. Y., and B. Tomlinson. 1999. Simultaneous measurement of allantoin and urate in plasma: analytical evaluation and potential clinical application in oxidant:antioxidant balance studies. Clin. Chem. 45:901-904. [PubMed] [Google Scholar]
- 4.Boronat, A., and J. Aguilar. 1979. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, S. C., C. T. Fang, P. R. Hsueh, Y. C. Chen, and K. T. Luh. 2000. Klebsiella pneumoniae isolates causing liver abscess in Taiwan. Diagn. Microbiol. Infect. Dis. 37:279-284. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. W., R. Wu, P. C. Yang, J. Y. Huang, Y. P. Sher, M. H. Han, W. C. Kao, P. J. Lee, T. F. Chiu, F. Chang, et al. 1998. Profiling expression patterns and isolating differentially expressed genes by cDNA microarray system with colorimetry detection. Genomics 51:313-324. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, D. L., Y. C. Liu, M. Y. Yen, C. Y. Liu, and R. S. Wang. 1991. Septic metastatic lesions of pyogenic liver abscess: association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch. Intern. Med. 151:1557-1559. [PubMed] [Google Scholar]
- 8.Cheng, H. P., L. K. Siu, and F. Y. Chang. 2003. Extended-spectrum cephalosporin compared to cefazolin for treatment of Klebsiella pneumoniae-caused liver abscess. Antimicrob. Agents Chemother. 47:2088-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, C. T., D. Y. Lin, and Y. F. Liaw. 1988. Metastatic septic endophthalmitis in pyogenic liver abscess. J. Clin. Gastroenterol. 10:524-527. [DOI] [PubMed] [Google Scholar]
- 10.Cusa, E., N. Obradors, L. Baldoma, J. Badia, and J. Aguilar. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 181:7479-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Figlia, S. E., and C. Cramer. 1951. Friedlander's bacillus meningitis in a case with liver abscess and recurrent bacteremia and analysis of cases receiving specific therapy. N.Y. State J. Med. 15:761-765. [PubMed] [Google Scholar]
- 12.Fang, C. T., H. C. Chen, Y. P. Chuang, S. C. Chang, and J. T. Wang. 2002. Cloning of a cation efflux pump gene associated with chlorhexidine resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 46:2024-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, C. T., Y. C. Chen, S. C. Chang, W. Y. Sau, and K. T. Luh. 2000. Klebsiella pneumoniae meningitis: timing of antimicrobial therapy and prognosis. QJM 93:45-53. [DOI] [PubMed] [Google Scholar]
- 14.Frees, D., S. N. A. Qazi, P. J. Hill, and H. Ingmer. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565-1578. [DOI] [PubMed] [Google Scholar]
- 15.Fung, C. P., F. Y. Chang, S. C. Lee, B. S. Hu, B. I. Kuo, C. Y. Liu, M. Ho, and L. K. Siu. 2002. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50:420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 17.Garaizar, J., S. Porwollik, A. Echeita, A. Rementeria, S. Herrera, R. M. Wong, J. Frye, M. A. Usera, and M. McClelland. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gort, A. S., and V. L. Miller. 2000. Identification and characterization of Yersinia enterocolitica genes induced during systemic infection. Infect. Immun. 68:6633-6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grootveld, M., and B. Hallwell. 1987. Measurement of allantoin and uric acid in human body fluids: a potential index of free-radical reactions in vivo? Biochem. J. 243:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 21.Herrero, M., V. Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastenbauer, S., Y. Koedel, B. F. Becker, and H. W. Pfister. 2002. Oxidative stress in bacterial meningitis in humans. Neurology 58:186-191. [DOI] [PubMed] [Google Scholar]
- 23.Ko, W. C., D. L. Paterson, A. J. Sagnimeni, D. S. Hansen, A. Von Gottberg, S. Mohapatra, J. M. Casellas, H. Goossens, L. Mulazimoglu, G. Trenholme, et al. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, N., K. Nishino, and A. Yamaguchi. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruger, E., E. Witt, S. Ohlmeier, R. Hanschke, and M. Hecker. 2000. The Clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 182:3259-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Y. C., D. L. Cheng, and C. L. Lin. 1986. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146:1913-1916. [PubMed] [Google Scholar]
- 27.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moison, R., A. de Beaufort, A. Hassnoot, T. Dubbelman, D. van-Zoeren-Grubben, and H. Berger. 1997. Uric acid and ascorbic acid redox ratios in plasma and tracheal aspirate of preterm babies with acute and chronic lung disease. Free Radic. Biol. Med. 23:226-234. [DOI] [PubMed] [Google Scholar]
- 29.Moore, T. D. E., and P. F. Sparling. 1995. Isolation and identification of a glutathione peroxidase homolog gene, gpxA, present in Neisseria meningitidis but absent in Neisseria gonorrhoeae. Infect. Immun. 63:1603-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogihara, T., H. Kim, K. Hirano, M. Imanishi, H. Ogihara, H. Tamai, R. Okamoto, and M. Mino. 1998. Oxidation products of uric acid and ascorbic acid in preterm infants with chronic lung disease. Biol. Neonate 73:24-33. [DOI] [PubMed] [Google Scholar]
- 31.Pederson, K. J., S. Carlson, and D. E. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 32.Pomposiello, P. J., and B. Demple. 2000. Identification of SoxS-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy, K. J., and M. Gilman. 1987. Preparation and analysis of RNA, p. 4.4.1-4.4.7. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y. [Google Scholar]
- 34.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 35.Rintoul, M. R., E. Cusa, L. Baldoma, J. Badia, L. Reitzer, and J. Aguilar. 2002. Regulation of the Escherichia coli allantoin regulon: coordinated function of the repressor AllR and the activator AllS. J. Mol. Biol. 324:599-610. [DOI] [PubMed] [Google Scholar]
- 36.Rouf, M. A., and R. f. Lomprey. 1968. Degradation of uric acid by certain aerobic bacteria. J. Bacteriol. 96:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai, T., K. Kanai, K. Osatomi, and K. Yoshikoshi. 2003. Identification of a 19.3-kDa protein in MRHA-positive Edwardsiella tarda: putative fimbrial major subunit. FEMS Microbiol. Lett. 226:127-133. [DOI] [PubMed] [Google Scholar]
- 38.Salky, B. A., A. Kaynon, J. J. Bauer, I. M. Gelernt, and I. Kreel. 1982. Ruptured hepatic abscess: a rare cause of spontaneous pneumoperitoneum. Am. J. Gastroenterol. 77:880-881. [PubMed] [Google Scholar]
- 39.Seeto, R. K., and D. C. Rocky. 1996. Pyogenic liver abscess: change in etiology, management, and outcome. Medicine 75:99-113. [DOI] [PubMed] [Google Scholar]
- 40.Wang, J. H., Y. C. Liu, S. S. Lee, M. Y. Yen, Y. S. Chen, J. H. Wang, S. R. Wang, and H. H. Lin. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26:1434-1438. [DOI] [PubMed] [Google Scholar]
- 41.Yanagawa, T., H. Nakamura, I. Takei, H. Maruyama, K. Kataoka, T. Saruta, and Y. Kobayashi. 1989. Klebsiella pneumoniae meningitis associated with liver abscess: a case report. Jpn. J. Antibiot. 42:2135-2140. [PubMed] [Google Scholar]
- 42.Yeh, Y. C., K. C. Chang, C. T. Fang, J. C. Yang, and J. T. Wang. 2002. Association of metronidazole resistance and natural competence in Helicobacter pylori. Antimicrob. Agents Chemother. 46:1564-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeoh, K. G., I. Yap, S. T. Wong, A. Wee, R. Guan, and J. Y. Kang. 1997. Tropical liver abscess. Postgrad. Med. J. 73:89-92. [DOI] [PMC free article] [PubMed] [Google Scholar]