Abstract
The human patatin-like phospholipase domain-containing-3 (PNPLA3) gene rs738409 C>G polymorphism is associated with several types of liver disease. The aim of this meta-analysis was to assess the risk of cirrhosis on the basis of rs738409 allele frequency and genotype. Medline, the Cochrane Library, EMBASE, and Google Scholar were searched for prospective and retrospective studies assessing the effect of the rs738409 polymorphism on liver cirrhosis. Seven studies, involving 2,023 patients with cirrhosis, were included. The G allele was associated with a significantly increased risk of cirrhosis versus the C allele [pooled odds ratio (OR) = 1.86, 95% confidence interval (CI) = 1.64–2.12, Z = 9.55, P < 0.001]. Both the GC and GG genotypes were associated with a significantly increased risk of cirrhosis versus the CC genotype (GC vs. CC: pooled OR = 1.73, 95% CI = 1.51–1.98, Z = 7.86, P < 0.001; GG vs. CC: pooled OR = 3.41, 95% CI = 2.77–4.18, Z = 11.65, P < 0.001). There was no evidence of publication bias. Our findings suggest that patients at risk for liver cirrhosis may benefit from PNPLA3 genotyping and thus more intensive monitoring if the rs738409 C>G polymorphism is identified.
Keywords: patatin-like phospholipase domain-containing-3 gene, polymorphism, cirrhosis
Liver cirrhosis is the 12th leading cause of death in the United States, with about half of these deaths being associated with alcohol (1). The precise worldwide incidence of liver cirrhosis is unknown; however, epidemiological data from Europe suggest that there are 15 to 133 cases per 100,000 population per year (2). Although alcohol use increases the risk of cirrhosis, there appear to be other factors that influence disease progression as only 10–20% of heavy drinkers develop cirrhosis (3). Other factors purported to affect the development of cirrhosis include gender, insulin resistance, steatosis, environmental factors, body mass index, chronic viral infection, and genetics (4–12). One such genetic factor is the patatin-like phospholipase domain-containing-3 (PNPLA3) gene, also known as adiponutrin.
Although the precise physiological role of PNPLA3 in the liver is currently unclear (13), the PNPLA3 gene encodes a 481-amino-acid protein that contains a highly conserved patatin-like domain with lipolytic activity toward triglycerides (13, 14). Biochemical analysis indicates that the PNPLA3 protein exerts lipase activity and may play a role in the hydrolysis of glycerolipids, with maximum enzymatic activity against triglycerides, diacylglyerol, and monacylglycerol (15). A genome-wide association study of a multiethnic population found that a single rs738409 C>G polymorphism of PNPLA3, which encodes I148M, is strongly associated with hepatic fat content and conferred susceptibility to nonalcoholic fatty liver disease (NAFLD) (11). Other studies have substantiated and extended these findings by demonstrating an association of rs738409 with fatty liver and alcoholic liver diseases (16–22). Further to this, several studies have investigated the influence of rs738409 on the clinical, biochemical, and histological parameters of liver diseases across different patient populations (20–28). The rs738409 variant has been reported to be associated with fibrosis (20, 29–31), histological disease severity (19, 29–32), steatosis (29), and elevated levels of liver enzymes in healthy adults (12, 21). Of note, a number of studies have reported that rs738409 is also a risk factor for cirrhosis (20–25, 27, 33). Interestingly, the magnitude and strength of the effect of this variant on cirrhosis varied among studies, with odds ratios (ORs) ranging from 1.47 (27) to 3.26 (33). The differences across the studies likely reflect differences in patient characteristics.
No meta-analysis to date has examined the association between the aforementioned rs738409 variant and cirrhosis. Therefore, to gain a more comprehensive understanding of the strength of this association, we reviewed the available published literature and carried out meta-analysis to assess the risk of cirrhosis on the basis of rs738409 allele frequency (C vs. G) and genotype (CC vs. GG vs. GC).
METHODS
The literature search, data extraction, review, and data analysis were performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (34).
Search strategy
Medline, the Cochrane Library, EMBASE, and Google Scholar were searched for articles published up to August 2013 that assessed the effect of the rs738409 (also called I148) variant in the PNPLA3 gene on cirrhosis using a differing combination of the following terms: PNPLA3, I148, rs738409, adiponutrin, and cirrhosis. Prospective and retrospective studies were included if subjects were 18 years of age and older and had liver cirrhosis, regardless of cause. All studies had to be in English. Excluded studies included letters, comments, editorials, or case reports. Two independent reviewers screened potentially relevant studies, and both had to agree for inclusion of an article. If there was disagreement between reviewers, it was resolved by a third reviewer.
Data extraction
The following information was extracted from the included studies: the name of the first author, year of publication, study design, number of participants in each genotype group, participants’ age and gender, diagnoses, and number or association with cirrhosis. The data were extracted by two reviewers. A third reviewer collated and checked the pooled data prior to the data being assessed by a statistician.
Quality assessment
The quality of included studies was evaluated using the Newcastle-Ottawa Scale, which is a validated technique to assess the quality of nonrandomized studies (35). The assessed outcomes for the seven included studies are listed in Table 1.
TABLE 1.
Summary of basic characteristics of studies included in the meta-analysis
| First Author (Year) | Race | Study Groupsa | Na | Sexa (M/F) | Agea | PNPLA3 Genotypea | MAF | OR (95% CI) | Newcastle-Ottawa Scale Score | ||||
| CC | GC | GG | G versus C | GC versus CC | GG versus CC | ||||||||
| Dutta (2013) | North Indian | Alcoholic cirrhosis Versus healthy control | 60 versus 100 | NA | 49.5 (10.3) versus 32.9 (9.5) | 24 versus 62 | 21 versus 69 | 15 versus 9 | G: 42.5%g | 2.41 (1.48, 3.92) | 1.87 (0.90, 3.89) | 4.31 (1.66, 11.15) | 8 |
| Nischalke (2011) | Caucasian | Alcoholic or HCV cirrhosis versus healthy control | 322 versus 190 | 230/92 versus 106/84 | 56 (37–82) versus 40.5 (20–86)c | 134 versus 112 | 142 versus 69 | 46 versus 9 | G: 36.3%g | 1.92 (1.44, 2.56) | 1.72 (1.17, 2.52) | 4.27 (2.00, 9.11) | 7 |
| Falleti (2011) | Caucasian | Cirrhosis versus healthy control | 483 versus 428 | 344/139 versus 314/114 | 57 (22–86) versus 49 (18–77)c | 168 versus 218 | 220 versus 175 | 95 versus 35 | G: 42.4%g | 1.84 (1.51, 2.24) | 1.63 (1.23, 2.16) | 3.52 (2.28, 5.44) | 7 |
| Trépo (2011) | Caucasian | ALD with cirrhosis versus healthy control | 265 versus 328 | NA | NA | 100 versus 181 | 130 versus 118 | 35 versus 29 | G: 37.7%g | 1.65 (1.29, 2.12) | 1.99 (1.41, 2.83) | 2.18 (1.26, 3.78) | 7 |
| Stickel (2011) | Caucasian | Alcoholic cirrhosis versus alcoholic control | 210 versus 439 | 167/43 versus 439/0 | 50 (43–55) versus 39 (34–45)d | 90 versus 264 | 93 versus 153 | 27 versus 22 | G: 35.0%g | 1.86 (1.44, 2.40) | 1.78 (1.25, 2.53) | 3.60 (1.95, 6.64) | 8 |
| Krawczyk (2011) | NA | CLD cirrhosisfversus CLD control | 201 versus 698 | 547/352 | 50 (17–83)c | 91 versus 394 | 90 versus 261 | 20 versus 43 | G: 32.3%g | 1.45 (1.14, 1.84) | 1.49 (1.07, 2.08) | 2.01 (1.13, 3.59) | 8 |
| Tian (2010) | Mestizo Mexicansb | Alcoholic cirrhosis versus alcoholic control | 482 versus 305 | 411/71 versus 257/48 | 52 (11.6) versus 39 (12.7)e | 59 versus 83 | 264 versus 198 | 371 versus 111 | C: 27.5%g | 2.28 (1.90, 2.74) | 1.88 (1.28, 2.75) | 4.70 (3.17, 6.98) | 8 |
ALD, alcoholic liver disease; CI, confidence interval; CLD, chronic liver disease; F, female; HCV, hepatitis C virus; M, male; MAF, minor allele frequency; NA, not available.
Data presented as cirrhosis group versus control group.
Mixed European and Native American ancestry.
Median (range).
Median [25th percentile and 75th percentile (Q1–Q3)].
Mean (standard deviation).
CLD patients with cirrhosis were defined by liver stiffness values ≥13.0 kPa.
Indicates that the allele frequencies were in Hardy-Weinberg equilibrium (Fisher’s exact test).
Statistical analysis
The primary outcome measure was the association of rs738409 with cirrhosis. The OR with 95% CI was calculated for the primary outcome in the cirrhosis group compared with the control group. Heterogeneity among the eligible studies was assessed by determining the Cochran Q and the I2 statistic. For the Q statistic, P < 0.10 was considered to indicate statistically significant heterogeneity. For the I2 statistic, which indicates the percentage of the observed between-study variability due to heterogeneity rather than chance, no heterogeneity was indicated by 0% to 25%, moderate heterogeneity was indicated by 25% to 50%, large heterogeneity was indicated by 50% to 75%, and extreme heterogeneity was indicated by 75% to 100%. Because the random effects model assumes that different studies may contain different underlying effects and takes into consideration both within- and between-study variation, the random effects model was adopted for the current study. Hence, pooled OR of the outcome was calculated by the DerSimonian-Laird method (36). Allele frequencies of the PNPLA3 I148M were tested for consistency with Hardy-Weinberg equilibrium (HWE) using the Fisher’s exact test. A two-sided P value < 0.05 indicated statistical significance. All statistical analyses were performed using the statistical software Comprehensive Meta-Analysis, version 2.0 (Biostat, Englewood, NJ).
RESULTS
Study selection
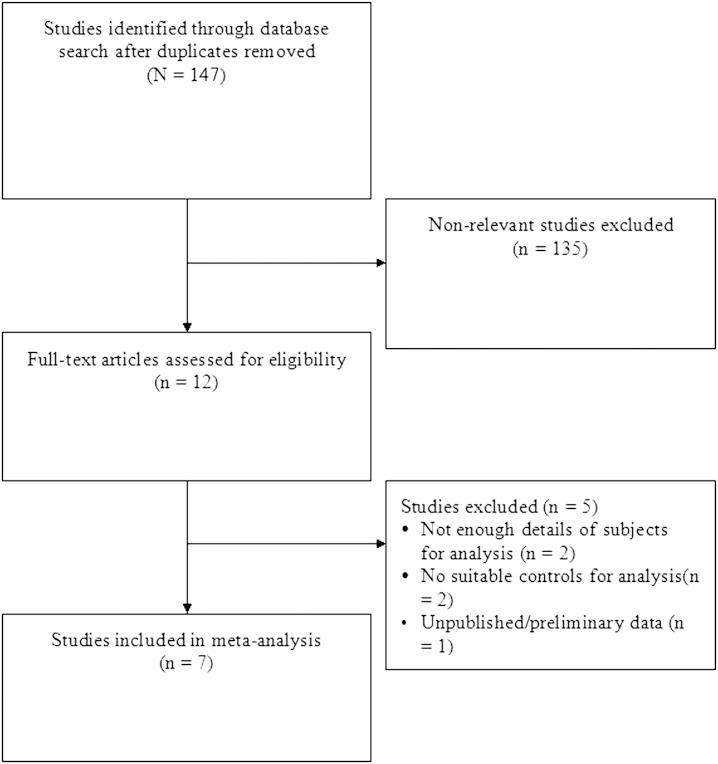
We identified 147 potential studies and excluded 135 for not being relevant or not meeting the inclusion criteria (Fig. 1). Of the 12 remaining studies, 2 were excluded due to lack of details of subjects needed for the analysis, another 2 did not have a suitable control arm, and 1 contained preliminary unpublished data. A total of seven studies were included; all were prospective in design (20–25, 37).
Fig. 1.
Flow chart of study selection.
Study characteristics
The key characteristics of the studies included in the meta-analysis are summarized in Table 1. Four of the studies included subjects of Caucasian race (21, 23–25), while one each included subjects who were North Indian (37) and Mestizo Mexican (22), respectively. The study reported by Krawczyk et al. (20) did not include information on race, although all subjects were noted to be “European individuals.” Across all seven studies, most subjects were male, and ages ranged from 33 to 57 years. The number of subjects in the studies ranged from 60 to 483 (total = 2,023) in the cirrhosis groups and 100 to 698 (total = 2,488) in the control groups. Except for the study of Tian et al. (22), the CC and GC genotypes of rs738409 were more common than the GG genotype in both the cirrhosis and control groups; in these studies, the ranges of the number of patients with CC, GC, or GG genotypes for the cirrhosis group were 24–168, 21–220, and 15–95, respectively, and for the control groups were 62–394, 29–261, and 9–43. For Tian et al. (22), the numbers of patients with the CC, GC, and GG genotypes were 59, 264, and 371, respectively, in the cirrhosis group and 83, 198, and 11 in the control group. G was the minor allele in six of the seven studies (20, 23–25, 37), with frequencies ranging from 32.3% to 42.5%. In the study reported by Tian et al. (22), C was the minor allele at a frequency of 27.5%. All seven studies included in the meta-analysis were in HWE. Call rates were reported in three studies and were 99%, 100%, and 98.7%, respectively (20–22).
Evaluation of the relationship of rs738409 alleles and genotypes with cirrhosis
The OR (95% CI) for the comparison of the frequency of the G allele compared with the C allele in the cirrhosis group ranged from 1.45 (1.14, 1.84) to 2.41 (1.48, 3.92) across the studies. For the GC and GG genotypes compared with the CC genotype, the OR (95% CI) ranged from 1.49 (1.07, 2.08) to 1.99 (1.41, 2.83) and from 2.01 (1.13, 3.59) to 4.70 (3.17, 6.98), respectively.
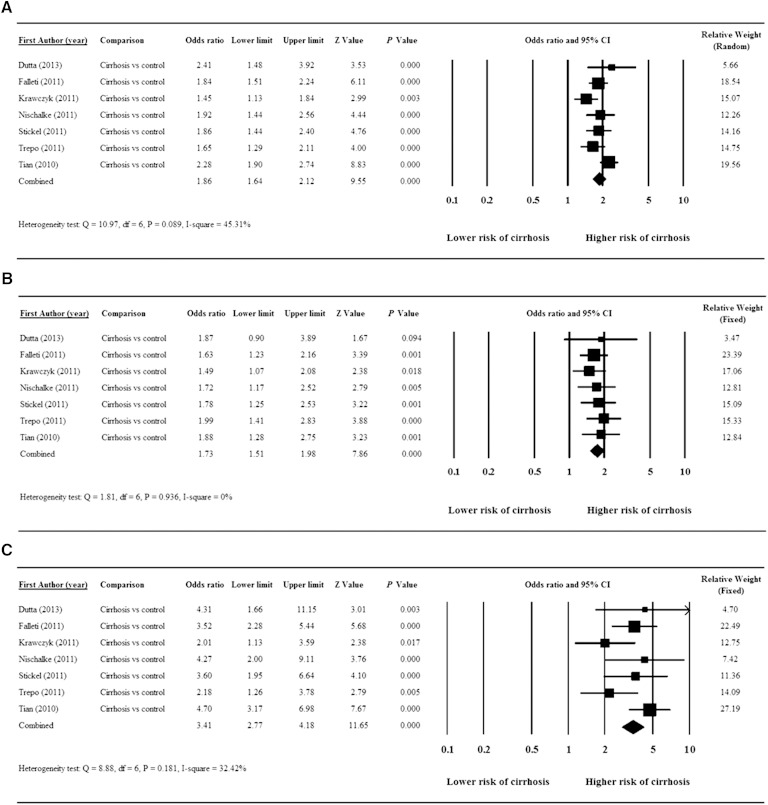
All seven studies were included in the meta-analyses. Significant heterogeneity was found in the pooled data for evaluation of the G compared with the C allele pooled [Q = 10.97, degrees of freedom (df) = 6, P = 0.089, I2 = 45.31%]. This analysis found that the G allele was associated with a significantly increased risk of cirrhosis versus the C allele (pooled OR = 1.86, 95% CI = 1.64–2.12, Z = 9.55, P < 0.001) (Fig. 2A).
Fig. 2.
Forest plots showing results for the meta-analysis of OR: G versus C allele (A); GC versus CC genotype (B); and GG versus CC genotype (C).
There was no significant heterogeneity of the pooled data for the evaluation of the association of the GC or the GG genotypes compared with the CC genotype with cirrhosis (Q = 1.81, df = 6, P = 0.936, I2 = 0% for GC genotype and Q = 8.88, df = 6, P = 0.181, I2 = 32.42% for the GG genotype). The meta-analysis showed that the GC genotype was associated with a significantly increased risk of cirrhosis versus the CC genotype (pooled OR = 1.73, 95% CI = 1.51–1.98, Z = 7.86, P < 0.001) (Fig. 2B). Similarly, the GG genotype was associated with a significantly increased risk of cirrhosis versus the CC genotype (pooled OR = 3.41, 95% CI = 2.77–4.18, Z = 11.65, P < 0.001) (Fig. 2C).
Sensitivity analysis
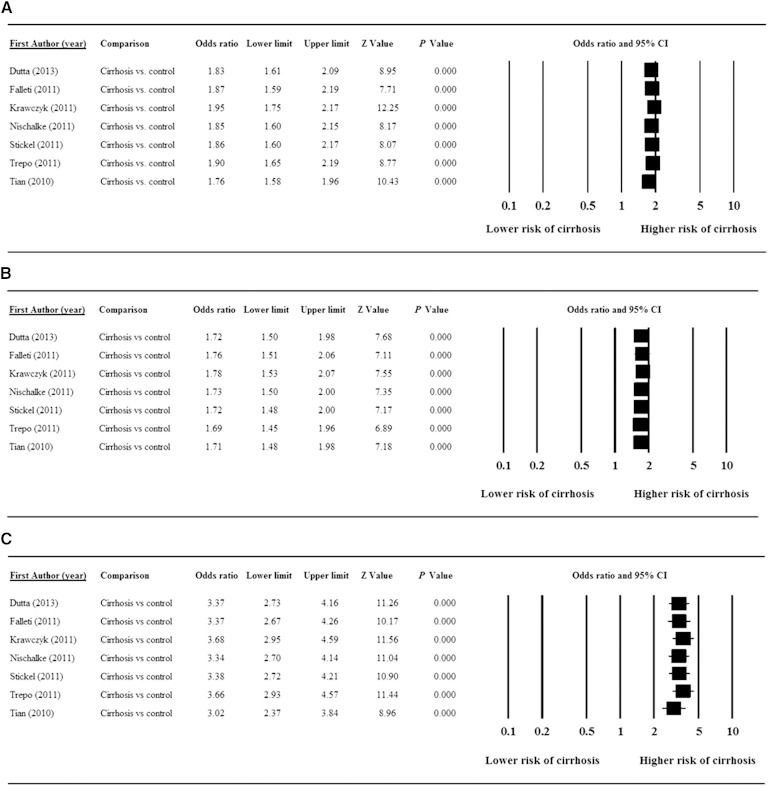
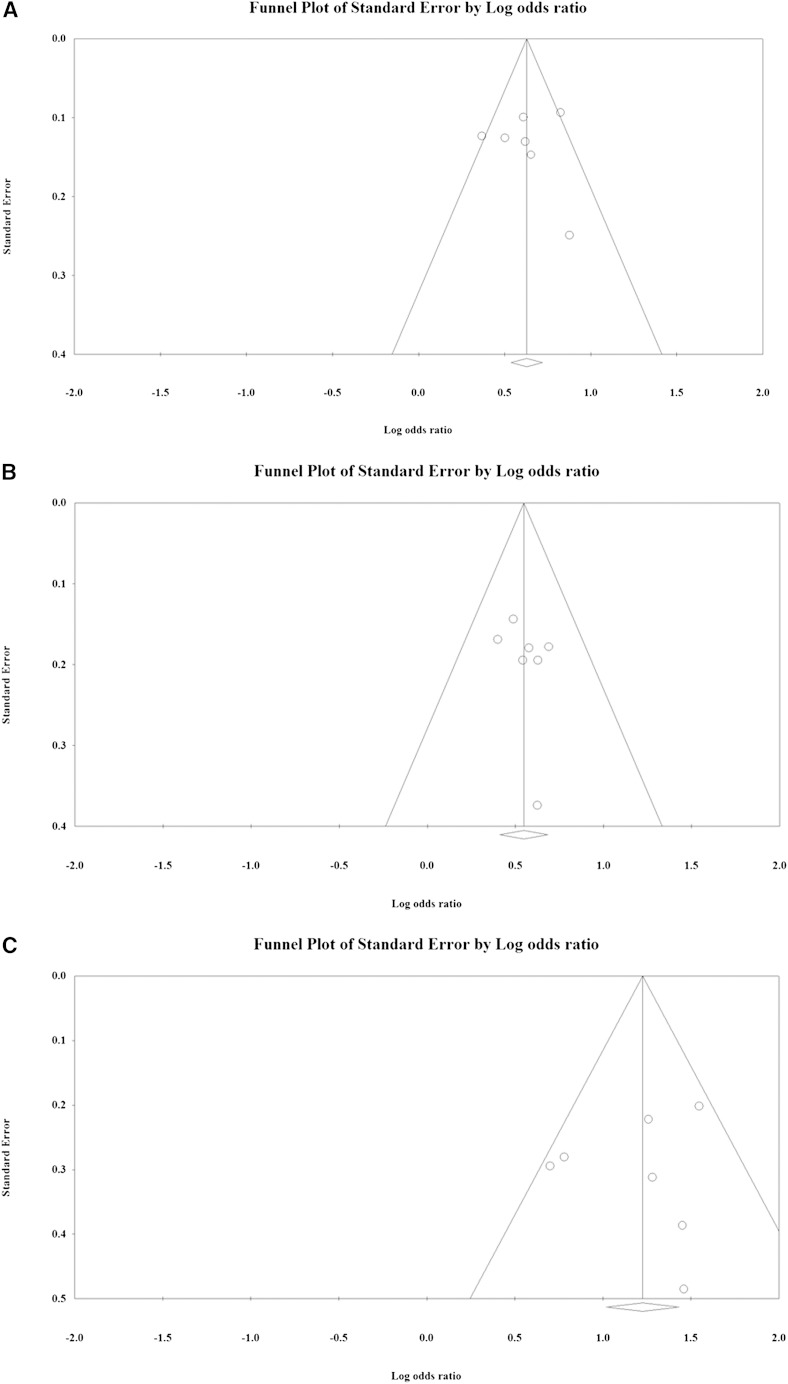
Sensitivity analysis using the leave-one-out approach for each of the different allele and genotypic analyses indicated that removal of any single study did not significantly influence the direction or magnitude of the pooled estimates (Fig. 3). These findings indicate that no one study dominated the findings. In addition, funnel plot analysis revealed that there was no significant evidence of bias for the G versus the C allele (t = 0.12, P = 0.456), the GC versus the CC genotype (t = 0.87, P = 0.212), or the GG versus the CC genotype (t = 0.40, P = 0.351) comparisons as determined using Egger’s test (Fig. 4).
Fig. 3.
Results of sensitivity analysis to examine the influence of individual studies on pooled estimates as determined using the leave-one-out approach: G versus C allele (A); GC versus CC genotype (B); and GG versus CC genotype (C).
Fig. 4.
Funnel plot for publication bias: G versus C alleles (A); GC versus CC genotype (B); and GG versus CC genotype (C). White circles represent published articles. White rhombuses represent the actual combined effect sizes.
DISCUSSION
In this meta-analysis, we examined the risk of liver cirrhosis on the basis of PNPLA3 rs738409 C>G polymorphism allele frequency and genotype. A total of seven studies, involving more than 2,000 patients with cirrhosis, were found to be eligible for inclusion in our study. Notably, subsequent analysis of data extracted from these studies revealed that the rs738409 C>G polymorphism was associated with an increased risk of liver cirrhosis.
Our meta-analysis revealed that the G allele was associated with a significantly increased risk of cirrhosis than the C allele, and, likewise, that both the GG and GC genotypes were associated with a significantly increased risk of cirrhosis than the CC genotype. These findings strongly support the notion that the PNPLA3 rs738409 C>G polymorphism increases the risk of liver cirrhosis. Interestingly, our findings differ from that reported in a study that was not eligible for inclusion in our meta-analysis. Specifically, in a study of Italian patients with hereditary hemochromatosis, Valenti et al. (33) did not find any association between cirrhosis and the G allele or the GC or GG genotype. Subanalysis, however, did reveal a significant association between cirrhosis and the G allele in patients with a body mass index <25. Presumably, the disparity in the overall results between our analysis and that reported by Valenti et al. (33) is a reflection of the fact that all patients in Valenti’s study had hereditary hemochromatosis, whereas none of the studies in our meta-analysis were restricted to including patients with this condition.
Other reports have examined the association between the PNPLA3 rs738409 C>G polymorphism and other forms of liver disease, most notably NAFLD, which leads to cirrhosis in ∼6% of cases (38). Indeed, there is extensive evidence of a link between NAFLD and this polymorphism. Specifically, in the first published genome-wide association study, Romeo et al. (11) reported that there was strong genetic signal for the rs738409 variant and NAFLD in a population comprising Hispanic, African American, and European American individuals. More recently, the rs738409 allele was found to be associated with NAFLD in a number of studies that included patients from the United States, Taiwan, China, Japan, or Malaysia (39–43). Indeed, in a Japanese study, Hotta et al. (44) found that the risk of carrying the G allele was 0.44 in control patients and 0.60 in patients with NAFLD. In a meta-analysis published in 2011, Sookoian and Pirola (28) found that the rs738409 variant was associated with increased hepatic triglyceride content and heightened severity of NAFLD.
Unsurprisingly, the rs738409 variant has also been found to be a risk factor for progression in patients with alcoholic fatty liver disease (AFLD) (45). In addition to NAFLD and AFLD, the results of a recent meta-analysis reported by Trépo et al. (46) indicate that the PNPLA3 rs738409 C>G polymorphism is significantly associated with hepatocellular carcinoma in patients of European descent with cirrhosis. The GG genotype was also found to be associated with a higher risk of hepatocellular carcinoma in a US patient population (41). Taken together, our findings and those from previously published reports demonstrate that the PNPLA3 rs738409 C>G polymorphism increases the risk of liver disease. The mechanism through which the PNPLA3 rs738409 C >G polymorphism might contribute to cirrhosis and liver disease is unclear. Recent evidence suggests that this polymorphism may result in a number of potentially important changes that may precede liver disease, including increased accumulation of triacylglycerols (47) in hepatocytes and decreased lipase activity (48). Effects of inflammation and fibrogenesis have also been proposed (29).
Our analyses have several limitations that warrant mention. First, most studies included Caucasian patients only; hence, the generalizability of our results to other racial/ethnic groups may be limited. Race/ethnicity is also a potential confounding factor as indicated by findings from several studies, which reported that individuals of Hispanic, black, or Indian race/ethnicity may be more likely to develop cirrhosis compared with Caucasians (37, 49). Additional studies involving participants of different ethnic backgrounds are necessary to further whether differences in the frequency of cirrhosis among different ethnic groups is or is not associated with the rs738409 variant. Second, the control groups differed between studies, with some including healthy subjects and others including noncirrhotic patients from the same patient population as the patients with cirrhosis. This lack of homogeneity between studies may have affected the results of our analyses. Finally, due to the lack of consistently available data, we were not able to perform analyses controlling for other factors that may affect the risk of cirrhosis, such as alcohol consumption, age, sex, and so forth, or indeed the underlying cause of cirrhosis. Clearly, these factors may also have had some impact on the results of our analyses. Despite these limitations, the findings across the studies were, in general, similar suggesting that differences in study design and patient characteristics did not significantly affect our results. Our findings are also strengthened by the fact that all studies were considered to be of high quality (all had Newcastle-Ottawa scores ≥7) and the lack of publication bias.
In summary, our findings show that the G allele of the PNPLA3 gene is a risk factor for cirrhosis and that the homozygous GG and GC genotype had a greater effect on this risk than the heterozygous CC genotype. Patients at risk for liver cirrhosis (and indeed other liver diseases) may benefit from PNPLA3 genotyping and, as a consequence, more intensive monitoring if the rs738409 C>G polymorphism is identified.
Footnotes
Abbreviations:
- df
- degrees of freedom
- NAFLD
- nonalcoholic fatty liver disease
- OR
- odds ratio
- PNPLA3
- patatin-like phospholipase domain-containing-3
This work was supported by LiaoNing Technology and Science Fund (Grant No. 2011225015) and ShenYang Technology and Science Fund Grant (No. F13-220-9-61).
REFERENCES
- 1.National Institutes of Health. Surveillance report #93: liver cirrhosis mortality in the United States, 1970–2009. Accessed September 10, 2013, at http://pubs.niaaa.nih.gov/publications/Surveillance93/Cirr09.htm.
- 2.Blachier M., Leleu H., Peck-Radosavljevic M., Valla D. C., Roudot-Thoraval F. 2013. The burden of liver disease in Europe: a review of available epidemiological data. J. Hepatol. 58: 593–608. [DOI] [PubMed] [Google Scholar]
- 3.Teli M. R., Day C. P., Burt A. D., Bennett M. K., James O. F. 1995. Determinants of progression to cirrhosis or fibrosis in pure alcoholic fatty liver. Lancet. 346: 987–990. [DOI] [PubMed] [Google Scholar]
- 4.Becker U., Deis A., Sorensen T. I., Gronbaek M., Borch-Johnsen K., Muller C. F., Schnohr P., Jensen G. 1996. Prediction of risk of liver disease by alcohol intake, sex, and age: a prospective population study. Hepatology. 23: 1025–1029. [DOI] [PubMed] [Google Scholar]
- 5.Naveau S., Giraud V., Borotto E., Aubert A., Capron F., Chaput J. C. 1997. Excess weight risk factor for alcoholic liver disease. Hepatology. 25: 108–111. [DOI] [PubMed] [Google Scholar]
- 6.Raynard B., Balian A., Fallik D., Capron F., Bedossa P., Chaput J. C., Naveau S. 2002. Risk factors of fibrosis in alcohol-induced liver disease. Hepatology. 35: 635–638. [DOI] [PubMed] [Google Scholar]
- 7.Mathurin P., Beuzin F., Louvet A., Carrie-Ganne N., Balian A., Trinchet J. C., Dalsoglio D., Prevot S., Naveau S. 2007. Fibrosis progression occurs in a subgroup of heavy drinkers with typical histological features. Aliment. Pharmacol. Ther. 25: 1047–1054. [DOI] [PubMed] [Google Scholar]
- 8.Nishiguchi S., Kuroki T., Yabusako T., Seki S., Kobayashi K., Monna T., Otani S., Sakurai M., Shikata T., Yamamoto S. 1991. Detection of hepatitis C virus antibodies and hepatitis C virus RNA in patients with alcoholic liver disease. Hepatology. 14: 985–989. [PubMed] [Google Scholar]
- 9.Mann R. E., Smart R. G., Govoni R. 2003. The epidemiology of alcoholic liver disease. Alcohol Res. Health. 27: 209–219. [PMC free article] [PubMed] [Google Scholar]
- 10.Stickel F., Osterreicher C. H. 2006. The role of genetic polymorphisms in alcoholic liver disease. Alcohol Alcohol. 41: 209–224. [DOI] [PubMed] [Google Scholar]
- 11.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L. A., Boerwinkle E., Cohen J. C., Hobbs H. H. 2008. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40: 1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan X., Waterworth D., Perry J. R., Lim N., Song K., Chambers J. C., Zhang W., Vollenweider P., Stirnadel H., Johnson T., et al. 2008. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 83: 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dongiovanni P., Donati B., Fares R., Lombardi R., Mancina R. M., Romeo S., Valenti L. 2013. PNPLA3 I148M polymorphism and progressive liver disease. World J. Gastroenterol. 19: 6969–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson P. A., Gardner S. D., Lambie N. M., Commans S. A., Crowther D. J. 2006. Characterization of the human patatin-like phospholipase family. J. Lipid Res. 47: 1940–1949. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y., Cohen J. C., Hobbs H. H. 2011. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J. Biol. Chem. 286: 37085–37093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kantartzis K., Peter A., Machicao F., Machann J., Wagner S., Konigsrainer I., Konigsrainer A., Schick F., Fritsche A., Haring H. U., et al. 2009. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes. 58: 2616–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotronen A., Johansson L. E., Johansson L. M., Roos C., Westerbacka J., Hamsten A., Bergholm R., Arkkila P., Arola J., Kiviluoto T., et al. 2009. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 52: 1056–1060. [DOI] [PubMed] [Google Scholar]
- 18.Romeo S., Sentinelli F., Dash S., Yeo G. S., Savage D. B., Leonetti F., Capoccia D., Incani M., Maglio C., Iacovino M., et al. 2010. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int. J. Obes. (Lond.). 34: 190–194. [DOI] [PubMed] [Google Scholar]
- 19.Sookoian S., Castano G. O., Burgueno A. L., Gianotti T. F., Rosselli M. S., Pirola C. J. 2009. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 50: 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krawczyk M., Grunhage F., Zimmer V., Lammert F. 2011. Variant adiponutrin (PNPLA3) represents a common fibrosis risk gene: non-invasive elastography-based study in chronic liver disease. J. Hepatol. 55: 299–306. [DOI] [PubMed] [Google Scholar]
- 21.Trépo E., Gustot T., Degré D., Lemmers A., Verset L., Demetter P., Ouziel R., Quertinmont E., Vercruysse V., Amininejad L., et al. 2011. Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J. Hepatol. 55: 906–912. [DOI] [PubMed] [Google Scholar]
- 22.Tian C., Stokowski R. P., Kershenobich D., Ballinger D. G., Hinds D. A. 2010. Variant in PNPLA3 is associated with alcoholic liver disease. Nat. Genet. 42: 21–23. [DOI] [PubMed] [Google Scholar]
- 23.Nischalke H. D., Berger C., Luda C., Berg T., Muller T., Grunhage F., Lammert F., Coenen M., Kramer B., Korner C., et al. 2011. The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS ONE. 6: e27087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stickel F., Buch S., Lau K., Meyer zu Schwabedissen H., Berg T., Ridinger M., Rietschel M., Schafmayer C., Braun F., Hinrichsen H., et al. 2011. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in Caucasians. Hepatology. 53: 86–95. [DOI] [PubMed] [Google Scholar]
- 25.Falleti E., Fabris C., Cmet S., Cussigh A., Bitetto D., Fontanini E., Fornasiere E., Bignulin S., Fumolo E., Bignulin E., et al. 2011. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 31: 1137–1143. [DOI] [PubMed] [Google Scholar]
- 26.Valenti L., Nobili V. 2011. PNPLA3 I148M polymorphism and liver damage in obese children. J. Pediatr. 159: 876. [DOI] [PubMed] [Google Scholar]
- 27.Valenti L., Rumi M., Galmozzi E., Aghemo A., Del Menico B., De Nicola S., Dongiovanni P., Maggioni M., Fracanzani A. L., Rametta R., et al. 2011. Patatin-like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 53: 791–799. [DOI] [PubMed] [Google Scholar]
- 28.Sookoian S., Pirola C. J. 2011. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 53: 1883–1894. [DOI] [PubMed] [Google Scholar]
- 29.Valenti L., Al-Serri A., Daly A. K., Galmozzi E., Rametta R., Dongiovanni P., Nobili V., Mozzi E., Roviaro G., Vanni E., et al. 2010. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 51: 1209–1217. [DOI] [PubMed] [Google Scholar]
- 30.Rotman Y., Koh C., Zmuda J. M., Kleiner D. E., Liang T. J. 2010. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 52: 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speliotes E. K., Butler J. L., Palmer C. D., Voight B. F., Hirschhorn J. N. 2010. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 52: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valenti L., Alisi A., Galmozzi E., Bartuli A., Del Menico B., Alterio A., Dongiovanni P., Fargion S., Nobili V. 2010. I148M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology. 52: 1274–1280. [DOI] [PubMed] [Google Scholar]
- 33.Valenti L., Maggioni P., Piperno A., Rametta R., Pelucchi S., Mariani R., Dongiovanni P., Fracanzani A. L., Fargion S. 2012. Patatin-like phospholipase domain containing-3 gene I148M polymorphism, steatosis, and liver damage in hereditary hemochromatosis. World J. Gastroenterol. 18: 2813–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moher D., Liberati A., Tetzlaff J., Altman D. G. 2010. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. 8: 336–341. [DOI] [PubMed] [Google Scholar]
- 35.Wells G. A., Shea B., O’Connell D., Peterson J., Welch V., Losos M., Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Accessed September 10, 2013, at http://www.medicine.mcgill.ca/rtamblyn/Readings/The%20Newcastle%20-%20Scale%20for%20assessing%20the%20quality%20of%20nonrandomised%20studies%20in%20meta-analyses.pdf.
- 36.DerSimonian R., Laird N. 1986. Meta-analysis in clinical trials. Control. Clin. Trials. 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 37.Dutta A. K. 2013. Genetic factors affecting susceptibility to alcoholic liver disease in an Indian population. Ann. Hepatol. 12: 901–907. [PubMed] [Google Scholar]
- 38.Lazo M., Clark J. M. 2008. The epidemiology of nonalcoholic fatty liver disease: a global perspective. Semin. Liver Dis. 28: 339–350. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y. C., Chang P. F., Chang M. H., Ni Y. H. 2014. Genetic variants in GCKR and PNPLA3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. Am. J. Clin. Nutr. 99: 869–874. [DOI] [PubMed] [Google Scholar]
- 40.Tan H. L., Zain S. M., Mohamed R., Rampal S., Chin K. F., Basu R. C., Cheah P. L., Mahadeva S., Mohamed Z. 2014. Association of glucokinase regulatory gene polymorphisms with risk and severity of non-alcoholic fatty liver disease: an interaction study with adiponutrin gene. J. Gastroenterol. 49: 1056–1064. [DOI] [PubMed] [Google Scholar]
- 41.Hassan M. M., Kaseb A., Etzel C. J., El-Serag H., Spitz M. R., Chang P., Hale K. S., Liu M., Rashid A., Shama M., et al. 2013. Genetic variation in the PNPLA3 gene and hepatocellular carcinoma in USA: risk and prognosis prediction. Mol. Carcinog. 52 (Suppl. 1): E139–E147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitamoto T., Kitamoto A., Yoneda M., Hyogo H., Ochi H., Nakamura T., Teranishi H., Mizusawa S., Ueno T., Chayama K., et al. 2013. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum. Genet. 132: 783–792. [DOI] [PubMed] [Google Scholar]
- 43.Peng X. E., Wu Y. L., Lin S. W., Lu Q. Q., Hu Z. J., Lin X. 2012. Genetic variants in PNPLA3 and risk of non-alcoholic fatty liver disease in a Han Chinese population. PLoS ONE. 7: e50256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotta K., Yoneda M., Hyogo H., Ochi H., Mizusawa S., Ueno T., Chayama K., Nakajima A., Nakao K., Sekine A. 2010. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med. Genet. 11: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stickel F., Hampe J. 2012. Genetic determinants of alcoholic liver disease. Gut. 61: 150–159. [DOI] [PubMed] [Google Scholar]
- 46.Trépo E., Nahon P., Bontempi G., Valenti L., Falleti E., Nischalke H. D., Hamza S., Corradini S. G., Burza M. A., Guyot E., et al. 2014. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: evidence from a meta-analysis of individual participant data. Hepatology. 59: 2170–2177. [DOI] [PubMed] [Google Scholar]
- 47.Ruhanen H., Perttila J., Holtta-Vuori M., Zhou Y., Yki-Jarvinen H., Ikonen E., Kakela R., Olkkonen V. M. 2014. PNPLA3 mediates hepatocyte triacylglycerol remodeling. J. Lipid Res. 55: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pingitore P., Pirazzi C., Mancina R. M., Motta B. M., Indiveri C., Pujia A., Montalcini T., Hedfalk K., Romeo S. 2014. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim. Biophys. Acta. 1841: 574–580. [DOI] [PubMed] [Google Scholar]
- 49.Flores Y. N., Yee H. F., Jr, Leng M., Escarce J. J., Bastani R., Salmeron J., Morales L. S. 2008. Risk factors for chronic liver disease in blacks, Mexican Americans, and whites in the United States: results from NHANES IV, 1999–2004. Am. J. Gastroenterol. 103: 2231–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]