Abstract
Exposure of endothelial cells (ECs) to agents such as oxidized glycerophospholipids (oxGPs) and cytokines, known to accumulate in atherosclerotic lesions, perturbs the expression of hundreds of genes in ECs involved in inflammatory and other biological processes. We hypothesized that microRNAs (miRNAs) are involved in regulating the inflammatory response in human aortic endothelial cells (HAECs) in response to oxGPs and interleukin 1β (IL-1β). Using next-generation sequencing and RT-quantitative PCR, we characterized the profile of expressed miRNAs in HAECs pre- and postexposure to oxGPs. Using this data, we identified miR-21-3p and miR-27a-5p to be induced 3- to 4-fold in response to oxGP and IL-1β treatment compared with control treatment. Transient overexpression of miR-21-3p and miR-27a-5p resulted in the downregulation of 1,253 genes with 922 genes overlapping between the two miRNAs. Gene Ontology functional enrichment analysis predicted that the two miRNAs were involved in the regulation of nuclear factor κB (NF-κB) signaling. Overexpression of these two miRNAs leads to changes in p65 nuclear translocation. Using 3′ untranslated region luciferase assay, we identified 20 genes within the NF-κB signaling cascade as putative targets of miRs-21-3p and -27a-5p, implicating these two miRNAs as modulators of NF-κB signaling in ECs.
Keywords: cell signaling, cytokines, gene expression, inflammation oxidized lipids, microRNAs, nuclear factor κB, interleukin 1β
The endothelium transitions from a quiescent state to an active state during the initiation of the innate immune response. The active endothelium is characterized by an increased expression of leukocyte adhesion molecules and chemotactic factors as well as increased permeability of the vascular beds allowing for leukocyte recruitment into the periphery from the blood. The persistence of endothelial activation due to chronic exposure of inflammatory stimuli is critical to the development of many diseases, including atherosclerosis (1).
During the initiation of atherosclerosis, oxidized low density lipoprotein (oxLDL) trapped in the vessel wall contributes to the activation of ECs (2). In vitro treatment of human aortic endothelial cells (HAECs) with a component of oxLDL, oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphatidylcholine (Ox-PAPC), changes the expression of hundreds of genes involved in inflammation, unfolded protein response, coagulation, and sterol biosynthesis (3–8). Similar global effects of interleukin 1β (IL-1β) and TNFα on ECs have been observed (9). However, the regulation of the response to these stimuli has not been fully characterized. We hypothesized that miRNAs partially regulate the expression of genes that are involved in the response of ECs to Ox-PAPC.
MicroRNAs (miRNAs) are small noncoding RNAs that can affect the expression of hundreds to thousands of genes by binding to their target mRNAs causing translational repression or target degradation (10). miRNAs contribute to the regulation of many endothelial functions including angiogenesis, vasodilation, coagulation, and inflammation (11–14). Furthermore, expression of EC miRNAs is highly sensitive to environmental cues. Common risk factors for atherosclerosis including shear stress, high blood glucose, and oxidized lipids have been shown to alter EC miRNA expression suggesting a role for miRNAs during disease pathology (15–18).
In this study, we first characterized the repertoire of miRNAs expressed in HAECs using next-generation sequencing of small RNAs. We then identified miRs-21-3p and -27a-5p to be responsive to Ox-PAPC, IL-1β, and TNFα. We found that these two miRNAs act to regulate hundreds of genes, including genes associated with nuclear factor κB (NF-κB) signaling in ECs. We demonstrate that transient overexpression of miRs-21-3p and -27a-5p leads to decreased p65 nuclear translocation in response to proinflammatory stimulant IL-1β suggesting that miRs-21-3p and -27a-5p act to repress NF-κB signaling in HAECs. We show using 3′ untranslated region (UTR) luciferase assay that miRs-21-3p and -27a-5p alter the expression of both repressors and activators of the NF-κB signaling cascade suggesting that these two miRNAs act to regulate the extent of NF-κB signaling in ECs.
METHODS
Cell culture and treatments
HAECs were isolated from aortas of human heart transplant donors, and cultures were maintained in MCDB-131C complete medium (VEC Technologies, Rensselar, NY). Ox-PAPC was prepared as described previously (19). For some studies, cells were treated in 1% FBS medium containing 40 µg/ml Ox-PAPC or vehicle for 4 h. In other studies, cells were treated with 20 ng/ml or 2 ng/ml IL-1β (201-LB-005/CF) and 20 ng/ml TNFα (210-TA) purchased from R and D Systems in 1% FBS medium for 2 h.
miRNA overexpression experiments were performed using Dharmacon miRDIAN miRNA mimics (miR-21-3p: C-301023-01-0005; miR-27a-5p: C-301028-01-0005; negative control: CN-001000-01-05) at indicated concentrations for 24 h. miRNA knockdown experiments were performed using the Exiquon miRCURY LNA™ miRNA inhibitors (miR-21-3p: 410136-00; miR-27a-5p: 410168-00; negative control: 199004-00) at the indicated concentrations for 24 h. All transfections were performed with Invitrogen Lipofectamine 2000 reagent.
miRNA library preparation and sequencing
Following treatment, cells were lysed, and total RNA was isolated using the Qiagen miRNeasy Kit to retain the small RNA fraction. RNA integrity number (RIN) values were assessed with the Agilent Bioanalyzer 2100 instrument. Samples with RIN values > 9.0 were used for transcriptional profiling. Small RNA libraries were prepared using the Illumina Small RNA v1.5 protocol and sequenced for 76 base reads on the Illumina GAIIx platform.
Alignment and quantification of miRNA deep sequencing
The small RNA sequencing data analysis method has been described in detail elsewhere (20). Briefly, the sequencing files for the four HAEC libraries were converted to the FASTQ format using a custom Perl script. The reads were then aligned to the hg19 version of the genome using the Novoalign tool with the following settings: −l16 −t30 −h90 −rA −R 1 −m −g 200 −k. These settings allowed for a single read to map to multiple regions of the genome with up to one mismatch. We quantified the number of reads aligning to the genomic coordinates of known mature miRNAs downloaded from miRBase version 19 using the Bioconductor package GenomicRanges for R (v.2.14.0) (21). Whenever a read mapped to “x” genomic loci, the read would contribute a count of 1/x to those regions. To enable comparison of counts between samples, we normalized the expression values by dividing the counts for a given mature miRNA by the sum of all the miRNA counts for the corresponding library.
Whole genome transcript profiling and differential gene expression analysis
HAECs from two different donors were transfected with 1 nM miRNA mimics and negative control for 24 h in triplicate. Total RNA was hybridized to Illumina Human HT-12 v4 Expression BeadChips. Genome Studio software (2010.v3) was used for obtaining fluorescent intensities for each probe. The probes were processed using nonparametric background correction, followed by quantile normalization with control and expression probes using the neqc function in the limma package (R v2.14.0) (22). The probes with detection P values <0.01 were considered expressed and used for subsequent analysis. Differential gene expression analyses to compare overexpression of miR-21-3p or miR-27a-5p with negative control were performed using Patterns of Gene Expression (PaGE v5.1.6) (23). Genes were considered to be differentially expressed if their expression changed at least 25% with <5% false discovery rate (FDR). The list of differentially expressed genes was interrogated for statistically significant overrepresented biological themes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (24). Gene expression data have been deposited in Gene Expression Omnibus with the accession number GSE48006.
miRNA and mRNA RT-qPCR
miRNA reverse transcription was performed from total RNA samples that retained the small RNA fraction. The Applied Biosystems Taqman microRNA Reverse Transcription Kit was used to generate cDNA. miRNAs and small RNAs were quantified using the following Taqman assays: RNU44 (Assay ID 001095), hsa-miR-21-5p (Assay ID 000397), hsa-miR-21-3p (Assay ID 002438), hsa-miR-23a (Assay ID 000399), hsa-miR-24 (Assay ID 000402), hsa-miR-27a-3p (Assay ID 000408), and hsa-miR-27a-5p (Assay ID: 002445). mRNA reverse transcription was performed using the Taqman Reverse Transcription Kit. Quantitative PCR (qPCR) for miRNAs and mRNAs was performed using the Roche Probes 480 Master Mix or KAPA SYBRFast Master Mix, respectively, in a Roche LightCycler 480 instrument.
Western blots
Cytosolic and nuclear fractions of total cellular protein were isolated using the Thermo Fisher Scientific NE-PER Nuclear and Cytoplasmic Extraction Kit. Isolates were run on 4–12% Bis-Tris gels and transferred onto polyvinylidene difluoride membranes. Antibodies used were p65 (ab7970, Abcam) at 1:250, Lamin A/C (sc-6215 and sc-7292, Santa Cruz) at 1:500, and β-Actin (5125, Cell Signaling) at 1:250 dilutions.
3′ UTR assay
The 3′ UTRs for all indicated genes were cloned into a Promega psiCHECK-2 plasmid. Fifteen nanograms per well 3′ UTR plasmid and 100 ng/well of β-galactosidase plasmid as filler were transfected along with miRNA mimic into HEK293 cells grown in 48-well plates for 24 h. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System from Promega (E1960). The amount of Firefly luciferase activity was normalized to Renilla luciferase activity to account for transfection efficiency in each well.
Statistical analysis
Microarray differential expression was determined using a permutation-based method (23). In order to assess the statistical significance of changes in gene expression or luciferase activity, a two-sided Student’s t-test with unequal variance was used.
RESULTS
miRNAs expressed in HAECs
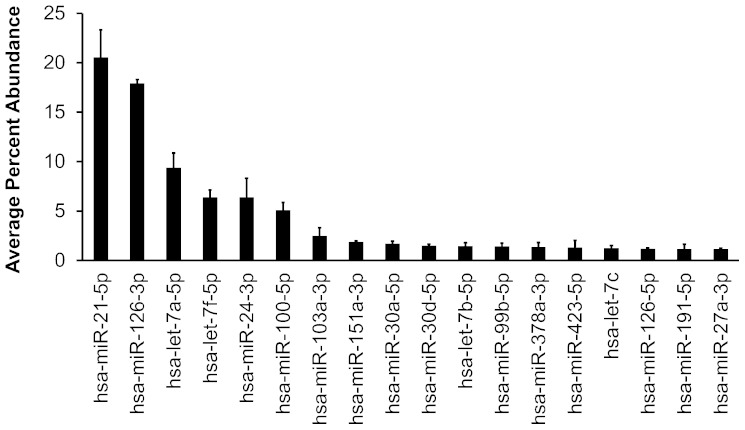
We sequenced four small RNA libraries prepared using total RNA from HAECs from two separate donors treated with media alone or media containing 40 μg/ml Ox-PAPC for 4 h. We obtained ∼15–26 million reads per library (supplementary Table I). Between 40% and 62% of the reads mapped to genomic regions that overlapped with known mature miRNA sequences (mirBase v19) (25). To account for differences between sequencing depth of the libraries, we normalized the miRNA expression levels by calculating the percentage of reads that mapped to a specific miRNA with respect to the total number of reads that mapped to all known miRNAs for each sample. We detected between 386 and 583 miRNAs expressed per library, representing a total of 618 unique miRNAs expressed in HAECs (supplementary Table I and supplementary File). Of the 618 miRNAs identified, we found that 18 miRNAs were expressed at an abundance >1% of total miRNA reads. miRs-21-5p and -126-3p consisted of 40% of all the mapped sequencing reads. These findings indicate that while many miRNAs are expressed, only a small fraction constitutes the majority of the miRNA pool in HAECs (Fig. 1; supplementary File).
Fig. 1.
Highly expressed HAEC miRNAs. miRNAs with average percent abundance >1% according to next-generation sequencing results are shown. Bars indicate average ± SEM in four libraries.
In order to validate the deep-sequencing results, we used miRNA-specific RT-qPCR to assay the expression levels of seven miRNAs whose abundance levels varied over a 10,000-fold range according to our deep-sequencing results. The correlation between the deep-sequencing and RT-qPCR results was highly significant (Pearson’s R = 0.91, P value = 4.39 × 10−6) suggesting that we were able to reliably quantify miRNA expression in HAECs using deep sequencing (supplementary Fig. I).
Effect of Ox-PAPC treatment on endothelial miRNAs
Oxidized glycerophospholipids (oxGPs) are known to contribute to numerous disease processes including atherosclerosis. Ox-PAPC is the major glycerophospholipid oxidized in minimally modified LDL that induces inflammatory and other responses in ECs. Treatment of cultured ECs with Ox-PAPC is known to change the expression of hundreds of genes in comparison to untreated ECs. Changes in miRNA expression levels as a response to environmental stimuli is known to modulate the expression of hundreds of genes; therefore, we hypothesized that Ox-PAPC may act to alter EC gene expression through changes in miRNA expression.
To assess the effects of Ox-PAPC treatment on EC miRNA expression, we calculated the fold change in abundance of miRNAs with respect to control cells. We observed that miRs-21-3p and -27a-5p were induced by 3.36- and 4.04-fold in response to Ox-PAPC treatment, respectively (Table 1). Mature miRNAs arise from Dicer processed pre-miRNA, a stem-loop precursor transcript that can produce two mature miRNAs, one from each arm (10). Although miRs-21-3p and -27a-5p are upregulated in response to Ox-PAPC, the expression of miRs-21-5p and -27a-3p, which arise from the same pre-miRNA transcript, was not altered by Ox-PAPC treatment (Table 1).
TABLE 1.
Abundance of miR-21 and miR-27a (percentage) in HAECs
| Donor 1 | Donor 2 | Fold Change | |||
| Control | Ox-PAPC | Control | Ox-PAPC | Ox-PAPC versus Control | |
| miR-21-5p | 1.64 × 101 | 1.56 × 101 | 2.35 × 101 | 2.68 × 101 | 1.04 |
| miR-21-3p | 5.15 × 10−3 | 1.80 × 10−2 | 1.32 × 10−2 | 4.28 × 10−2 | 3.37 |
| miR-27a-3p | 1.11 × 100 | 9.31 × 10−1 | 1.21 × 100 | 1.34 × 100 | 0.97 |
| miR-27a-5p | 2.43 × 10−2 | 1.06 × 10−1 | 2.36 × 10−2 | 8.77 × 10−2 | 4.03 |
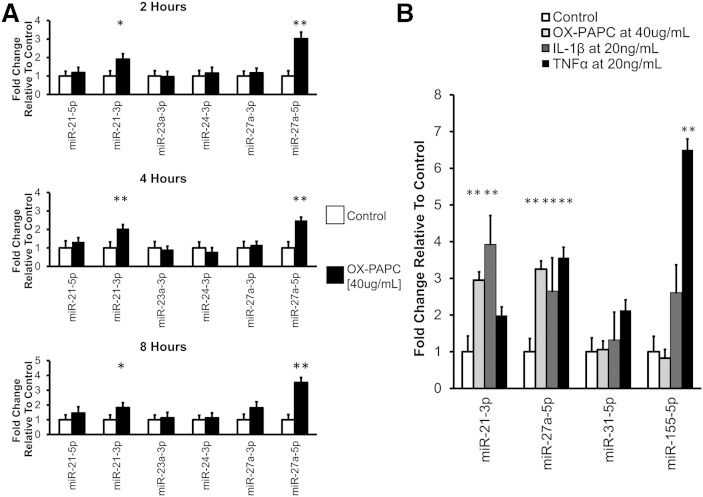
To determine the time course of miRNA induction, we measured the expression of miRs-21-3p and -27a-5p at 2, 4, and 8 h using miRNA-specific RT-qPCR. Consistent with the deep-sequencing results, we found that miRs-21-3p and -27a-5p were upregulated 2.0- and 2.5-fold, respectively at 4 h (Fig. 2A). In addition, we observed that miRs-21-3p and -27a-5p were upregulated 2- and 3-fold in response to Ox-PAPC as early as 2 h. By 8 h, miRs-21-3p and -27a-5p were still upregulated 2- and 3.5-fold, respectively.
Fig. 2.
Expression of select miRNAs in HAECs in response to various stimuli. A: miRNA expression in HAECs over time in response to Ox-PAPC treatment. HAECs were incubated with either control media or media containing Ox-PAPC for the indicated durations. miRNA levels were quantified using RT-qPCR. Plots show average ± SEM. N = 8–9. B: miRNA expression in HAECs treated with inflammatory stimuli. HAECs were treated with either control media or Ox-PAPC for 4 h, or IL-1β or TNFα for 2 h. miRNA levels were quantified using RT-qPCR. Plots show average ± SEM. N = 6. ** P < 0.01.
Several distinct miRNAs can be expressed as a cluster from a single primary transcript. miR-27a-5p is transcribed from an miRNA gene cluster including the genes MIR23A and MIR24-2. To determine whether Ox-PAPC specifically induced the expression of miRs-21-3p and -27a-5p, and rather than all miRNAs that arise from the same primary transcripts, we measured the expression of additional miRNAs at the three time points. Among the six miRNAs measured, only miRs-21-3p and -27a-5p showed induction upon treatment (Fig. 2A) suggesting that the regulation of miRs-21-3p and -27a-5p expression by Ox-PAPC occurs after these miRNAs are processed to mature form.
To determine whether induction of miRs-21-3p and -27a-5p in ECs was specific for Ox-PAPC, we treated HAECs with alternative inflammatory stimuli: IL-1β and TNFα. Exposure to TNFα is known to induce the expression of miRs-155-5p and -31-5p in human umbilical vein endothelial cells (HUVECs) (26). Therefore, to confirm TNFα activity, we measured the expression of these two miRNAs to serve as positive controls. We observed that miR-21-3p was induced ∼4- and 2-fold by IL-1β and TNFα, respectively, while miR-27a-5p was induced ∼2.5- and 3.5-fold by IL-1β and TNFα, respectively (Fig. 2B). These results suggest that miRs-21-3p and -27a-5p may be induced as a general response to inflammatory stimuli in HAECs.
Identification of biological pathways regulated miRs-21-3p and -27a-5p in ECs
A small number of studies have characterized mRNA targets of miRs-21-3p and -27a-5p in immune cells such as eosinophils and natural killer (NK) cells and cancers such as hepatocellular carcinoma and head and neck squamous cell carcinoma (27–30). However, how miRs-21-3p and -27a-5p regulate gene expression in ECs is not known. Therefore, we sought to identify the targets and biological pathways modulated by these two miRNAs in ECs using an unbiased approach. Based on previous dose response experiments with miR-21-3p and miR-27a-5p miRNA mimics, we transfected HAECs from two additional donors with 1 nM mimic leading to 5- to 30-fold induction of miR-21-5p and 15- to 30-fold induction of miR-27a-5p, respectively (supplementary Figs. II and III). Twenty-four hours after transfection, we isolated the total RNA and performed whole genome transcript profiling. Because miRNAs often function to degrade mRNAs, we focused on the transcripts that were downregulated in response to the transient overexpression.
We found that miRs-21-3p and -27a-5p decreased the expression of 1,037 and 1,138 genes >25% at 5% FDR, respectively. Of the 1,253 genes that were downregulated by either of the miRNAs, 922 of them were common, suggesting the regulation of similar biological pathways by these two miRNAs in ECs. While a single miRNA can target several hundred mRNAs, it is likely that most of the downregulation of the genes in HAECs is due to secondary effects. miRNAs typically bind to the 3′ UTRs of their mRNA targets leading to transcript degradation (10). We examined all 1,253 genes downregulated by either miRNA using the miRNA target prediction algorithm miRANDA (31). We found that 471 and 411 genes contained predicted binding sites for miRs-21-3p or -27a-5p, respectively. Of those genes with predicted miRNA binding sites, 206 genes were predicted targets for both miRNAs.
Using DAVID, we searched for Gene Ontology enrichment in the genes downregulated by miRNA overexpression. We observed significant enrichment for genes involved in the defense response, inflammatory response, and response to wounding functional categories (Table 2). Within the defense response category, genes belonging to the NF-κB signaling pathway were highly represented comprising ∼30% and 26% percent of genes downregulated by miRs-21-3p and -27a-5p, respectively. Therefore, we hypothesized that modulation of miRs-21-3p and -27a-5p expression in ECs would lead to functional consequences for NF-κB signaling.
TABLE 2.
Functional enrichment of Gene Ontology categories of genes downregulated by miR-21-3p and miR-27a-5p
| miR-21-3p Overexpression | miR-27a-5p Overexpression | |||||
| Gene Ontology Category | Number of Genes | Enrichment P | Bonferroni-Corrected Enrichment P | Number of Genes | Enrichment P | Bonferroni-Corrected Enrichment P |
| Defense response | 119 | 6.04 × 10−15 | 2.58 × 10−11 | 130 | 4.99 × 10−15 | 2.32 × 10−11 |
| Inflammatory response | 74 | 5.77 × 10−12 | 2.49 × 10−8 | 80 | 7.43 × 10−12 | 3.45 × 10−8 |
| Response to wounding | 107 | 1.21 × 10−10 | 5.20 × 10−7 | 119 | 3.01 × 10−11 | 1.40 × 10−7 |
| Response to lipopolysaccharide | 27 | 8.33 × 10−5 | 3.01 × 10−1 | 28 | 3.28 × 10−4 | 7.82 × 10−1 |
| Regulation of cell death | 173 | 3.65 × 10−5 | 1.46 × 10−1 | 193 | 5.41 × 10−5 | 2.22 × 10−1 |
| Regulation of IκB kinase/NF-κB cascade | 36 | 4.53 × 10−4 | 8.58 × 10−1 | 34 | 1.94 × 10−2 | 1.00 × 100 |
miRs-21-3p and -27a-5p regulate p65 nuclear translocation
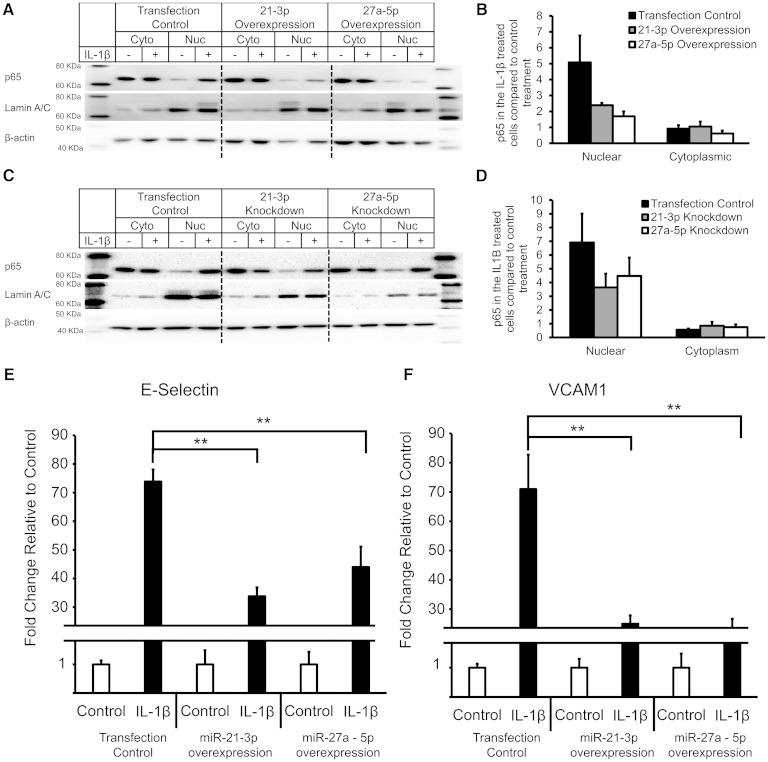
The p65, one of the NF-κB subunits, is sequestered in the cytoplasm by the IκB proteins in an inactive state (32). Once the signaling cascade is activated, the IκB proteins are phosphorylated by the Iκκ family of kinases, and p65 is free to translocate to the nucleus. Based on the significant decrease in NF-κB signaling related genes in response to miRNA overexpression, we hypothesized that overexpression of miRs-21-3p and -27a-5p would impair p65 nuclear translocation. Previous work in HeLa cells using a luciferase reporter construct containing three tandem NF-κB binding sites has strongly suggested that Ox-PAPC does not activate NF-κB signaling (33). We observed that treatment of HAECs with Ox-PAPC did not induce p65 nuclear translocation but appeared to decrease basal nuclear translocation (supplementary Fig. IV). Consequently, to determine the effect of miRs-21-3p and -27a-5p on p65 nuclear translocation, we chose to treat HAECs overexpressing miRs-21-3p and -27a-5p with IL-1β, a known activator of NF-κB signaling.
As expected, p65 translocated to the nucleus in response to IL-1β treatment in cells transfected with control miRNA (Fig. 3A). We observed a significant decrease in p65 nuclear translocation following overexpression of miRs-21-3p and -27a-5p (Fig. 3A, B). We observed a lesser decrease in p65 nuclear translocation with IL-1β treatment of HAECs following knockdown of miRs-21-3p and -27a-5p expression with miRNA inhibitors (Fig. 3C, D).
Fig. 3.
miR-21-3p and miR-27a-5p inhibit NF-κB signaling in HAECs. A: HAECs were transfected with 1 nM control or miR-21-3p or miR-27a-5p mimic for 24 h and then treated with 20 ng/ml IL-1β for 2 h. Representative Western blot of both cytosolic and nuclear protein fractions from HAECs. B: Quantification of p65 nuclear translocation in three donors. Bars show average ± SD. N = 3. C: HAECs were transfected with 50 nM control or miR-21-3p or miR-27a-5p inhibitor for 24 h and then treated with 20 ng/ml IL-1β for 2 h. Representative Western blot of both cytosolic and nuclear protein fractions from HAECs. D: Quantification of p65 nuclear translocation in three donors. Bars show average ± SD. N = 3. E and F: After 24 h transfection with control and mimics, cells were treated with 2 ng/ml of IL-1β for 2 h; gene expression was quantified using RT-qPCR. Bars represent the average ± SEM. N = 6. ** P < 0.01.
Altered p65 nuclear translocation leads to changes in the expression of downstream targets of NF-κB signaling. We measured the expression of E-selectin and vascular cell adhesion molecule-1 (VCAM-1), direct targets of NF-κB, in HAECs overexpressing the two miRNAs. In control cells, E-selectin and VCAM-1 expression were induced >70-fold in response to IL-1β treatment. As expected, we observed impaired induction of E-selectin and VCAM-1 following miRNA overexpression (Fig. 3E, F).
As knockdown of the two miRNAs appeared to also alter p65 nuclear translocation, we measured the expression of E-selectin and VCAM-1 in ECs transfected with miRNA inhibitors. We found that in comparison to control cells, knockdown of miR-27a-5p had no significant change on the expression of E-selectin or VCAM-1 (supplementary Fig. V). In comparison, knockdown of miR-21-3p led to an increased expression of both E-selectin and VCAM-1, despite impaired p65 nuclear translocation (supplementary Fig. V).
Regulation of NF-κB signaling through 3′ UTR targeting of NF-κB genes
We used two distinct approaches to identify candidate target genes for miRs-21-3p and -27a-5p (supplementary Fig. VI). In our first approach, we focused on the differentially downregulated genes in our microarrays studies. There were 1,037 and 1,138 downregulated genes in response to miRs-21-3p and -27a-5p overexpression, respectively. 119 and 130 of these genes belonged to the defense response Gene Ontology functional category, respectively. The two gene lists along with known NF-κB pathway genes that were found to be downregulated by miRs-21-3p or -27a-5p overexpression were then analyzed for putative binding sites for either miRs-21-3p or -27a-5p as predicted by the miRANDA algorithm (supplementary Fig. VI). This approach generated 26 preliminary candidate genes that were then assayed using RT-qPCR in an additional HAEC donor overexpressing miRs-21-3p or -27a-5p. We were able to confirm downregulation for 15 of the 26 genes that contained putative binding sites for either of the two miRNAs (P ≤ 0.1) (supplementary Fig. VII).
In the second approach, we queried the whole genome for predicted targets of miRs-21-3p and -27a-5p using miRWALK, a database of miRNA prediction algorithms (36). Genes that showed predicted target sites for either miRs-21-3p or -27a-5p by a minimum of three algorithms were then searched in the literature for known associations with NF-κB signaling or known relevance to EC biology. This resulted in 12 putative targets for miR-21-3p and 8 putative targets for miR-27a-5p. Five genes out of the 20 genes were selected at random to be validated as miRNA targets using the 3′ UTR assay. The combination of these two approaches resulted in 20 candidate target genes representing three distinct miRs-21-3p and -27a-5p predicted target types: joint miRs-21-3p and -27a-5p target, miR-21-3p target only, and miR-27a-5p target only (Table 3; supplementary Fig. VI).
TABLE 3.
Putative gene targets of miR-21-3p and miR-27a-5p for validation
| Gene Symbol | Gene Name | miR-21-3p Binding Site? | miR-27a-5p Binding Site? |
| BCL2 | B-cell CLL/lymphoma 2 | Yes | Yes |
| HMOX1 | Heme oxygenase (decycling) 1 | Yes | Yes |
| ICOSLG | Inducible T-cell costimulator ligand | Yes | Yes |
| IL1R1 | Interleukin 1 receptor, type I | Yes | Yes |
| TICAM2 | Toll-like receptor adaptor molecule 2 | Yes | Yes |
| UBE2N | Ubiquitin-conjugating enzyme E2N | Yes | Yes |
| BCL10 | B-cell CLL/lymphoma 10 | Yes | No |
| CBX4 | Chromobox homolog 4 | Yes | No |
| CEBPB | CCAAT/enhancer binding protein (C/EBP) beta | Yes | No |
| CX3CL1 | Chemokine (C-X3-C motif) ligand 1 | Yes | No |
| GCH1 | GTP cyclohydrolase 1 | Yes | No |
| HMGB1 | High mobility group box 1 | Yes | No |
| IL12A | Interleukin 12A | Yes | No |
| TAP1 | Transporter 1, ATP-binding cassette, subfamily B | Yes | No |
| TLR3 | Toll-like receptor 3 | Yes | No |
| TNFAIP3 | Tumor necrosis factor, alpha-induced protein 3 | Yes | No |
| RELA | V-rel reticuloendotheliosis viral oncogene homolog A/p65 | No | Yes |
| RFC1 | Replication factor C (activator 1) 1, 145 kDa | No | Yes |
| TNIP1 | TNFAIP3 interacting protein 1 | No | Yes |
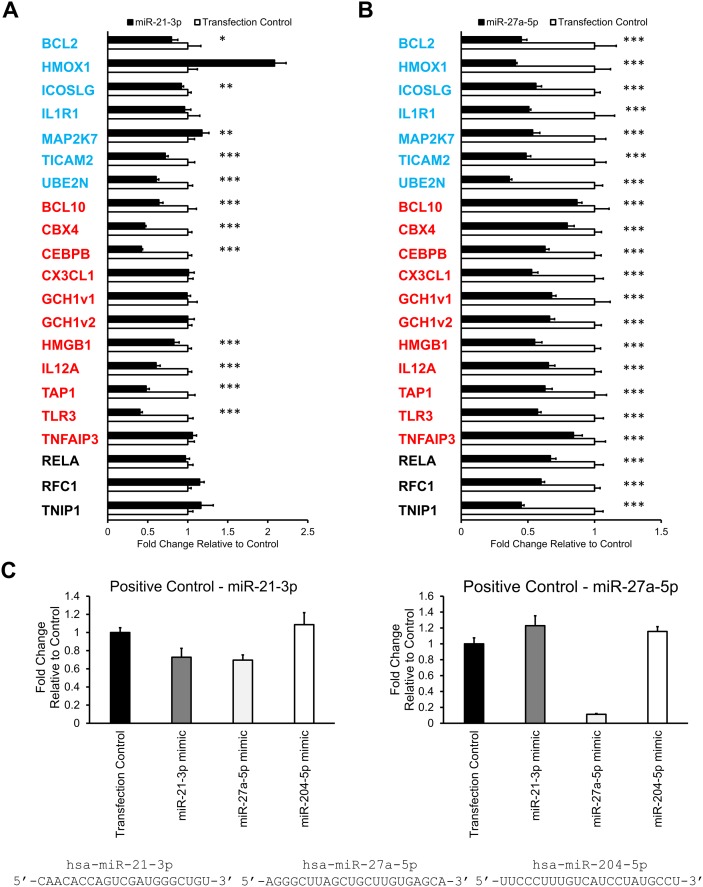
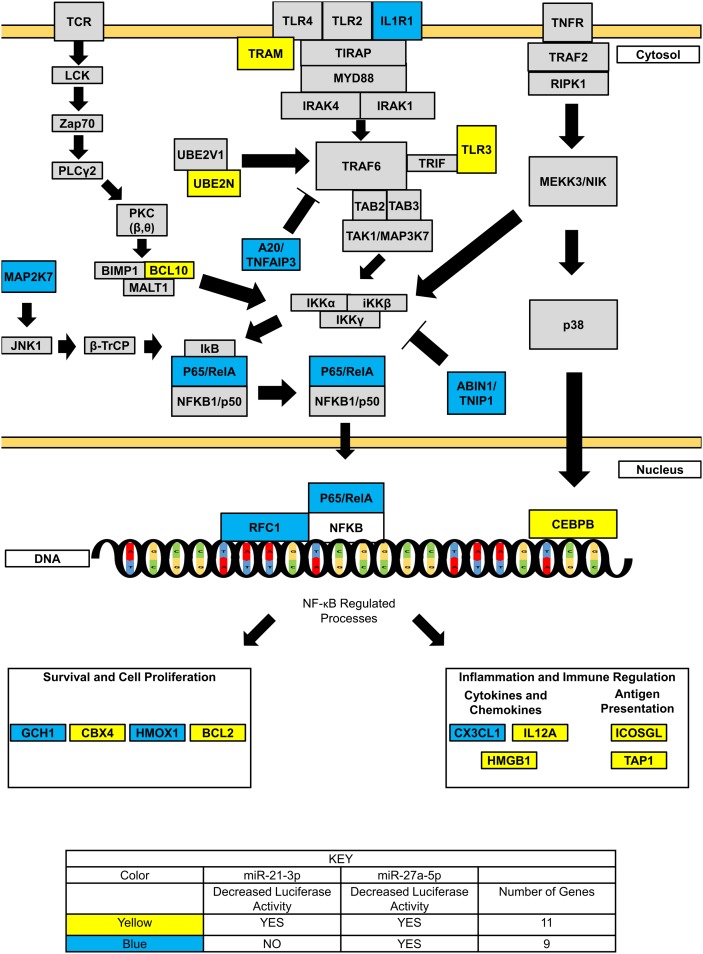
Upon close inspection of these 20 candidate target genes, we found they could be classified into three major subgroups related to their location/role in the NF-κB signaling cascade. Eight genes (IL1R1, TICAM2, TLR3, UBE2N, BCL10, MAP2K7, TNFAIP3, and TNIP1) have been shown to act upstream of p65 nuclear translocation. The remaining 12 genes involved processes downstream of p65 nuclear translocation and could be broken up into two additional groups. Three genes (RELA, RFC1, and CEBPB) were found to act at the site of NF-κB transcriptional binding sites. The remaining nine genes (GCH1, CBX4, HMOX1, BCL2, CX3CL1, IL12A, HMGB1, ICOSLG, and TAP1) were known downstream targets of NF-κB. These nine genes based on known biological function could be classified into two main subcategories: cell survival and proliferation (GCH1, CBX4, HMOX1, and BCL2) and immune response/inflammation (CX3CL1, IL12A, HMGB1, ICOSLG, and TAP1). Thus, it is likely that miRs-21-3p and -27a-5p act at many points in the signaling cascade to regulate the extent of NF-κB signaling.
miRNAs regulate gene expression through direct interactions with the 3′ UTR of the target mRNA. Therefore, to determine direct miRNA/mRNA interactions by miRs-21-3p and -27a-5p with their putative target genes, we utilized 3′ UTR luciferase reporter assay. The 3′ UTRs of the 20 genes were cloned to create fusion transcripts with the firefly luciferase in the psiCHECK-2 plasmid. The plasmids containing the candidate gene 3′ UTRs were then cotransfected in HEK293 cells with 5 nM mimics of miR-21-3p, miR-27a-5p, or cel-miR-67 (a negative control) to overexpress the miRNAs of interest.
Of the 20 candidate genes, 16 genes contained predicted miR-21-3p sites in their 3′ UTR. For 11 of 16 genes (BCL2, ICOSLG, TICAM2, UBE2N, BCL10, CBX4, CEBPB, HMGB1, IL12A, TAP1, and TLR3), we observed significant downregulation of luciferase activity following cotransfection with miR-21-3p mimic (Fig. 4A). Seven 3′ UTR plasmid constructs representing the three miR-21-3p candidate target genes (GCH1, TNFAIP3, and CX3CL1) and three joint miR-21-3p and miR-27a-5p candidate target genes (HMOX1, MAP2K7, and IL1R) did not show decreased luciferase activity following miR-21-3p overexpression. Numerous factors can contribute to the strength of miRNA-3′ UTR interactions including location of predicted miRNA binding site in the 3′ UTR, extent of complementarity between miRNA seed sequence and target 3′ UTR, and total extent complementarity between the full-length miRNA sequence and target 3′ UTR (10). We hypothesized that variation in one of these three factors was shared by the six candidate genes leading to a lack of response following miR-21-3p overexpression. However, upon closer examination, we were unable to find significant similarity among the three factors in the six candidate genes.
Fig. 4.
Identification of candidate target genes of miRs-21-3p and -27a-5p. A and B: For each gene, a plasmid containing the 3′ UTR was cotransfected with miRNA mimic at 5 nM concentration into HEK293 cells. Results for the miR-21-3p mimic transfected cells (A) and for the miR-27a-5p mimic transfected cells (B). Bars represent average ± SD. N = 6. A and B: Blue color indicates genes that are predicted to be miR-21-3p and miR-27a-5p targets. Red color indicates genes that are predicted to be miR-21-3p targets. Black color indicates genes that are predicted to be miR-27a-5p targets. GCH1v1 and GCH1v2 represent two different 3′ UTR isoforms of the same gene GCH1. C and D: 3′ UTR luciferase assay positive controls for miRs-21-3p and -27a-5p. For each miRNA, a positive control plasmid containing the full-length complementary sequence of the miRNA in triplicate was generated. These positive control plasmids were cotransfected with either mimic control (transfection control), miR-21-3p, miR-27a-5p, or miR-204-5p (an unrelated miRNA) into HEK293 cells. C: Results from the miR-21-3p positive control and from the miR-27a-5p positive control. Bars represent average ± SD. N = 6. Asterisks indicate significant downregulation following miRNA overexpression. * P < 0.1, ** P < 0.05, *** P < 0.01.
In contrast, 10 out of 20 candidate genes contained predicted miR-27a-5p interaction sites as determined by miRANDA. Using the luciferase reporter assay, we found that all 10 predicted candidate genes showed decreased luciferase activity in response to miR-27a-5p overexpression (Fig. 4B). Furthermore, we observed that all 10 candidate genes predicted by miRANDA to contain only miR-21-3p binding sites also showed downregulation in response to miR-27a-5p, independent of their response to miR-21-3p overexpression (Fig. 4A, B).
miRNA recognition of its target 3′ UTRs is primarily mediated by complementary in the target 3′ UTR to miRNA seed sequence, positions 2–8 of the mature miRNA. However, studies using Argonaute (Ago)-bound miRNA/mRNA pairings suggest that up to 45% miRNA targets lack a perfect seed sequence match (37). Therefore, we tested the hypothesis that there was a nonspecific interaction between miR-27a-5p and miR-21-3p target 3′ UTR sites. We generated positive control luciferase plasmids for both miR-21-3p and miR-27a-5p by inserting sequences that were complementary to the full sequence of miRs-21-3p or -27a-5p into the luciferase plasmid. We then cotransfected the positive control miRNA target plasmids with the miRNA mimics and then measured luciferase activity 48 h after transfection. As a negative control we cotransfected the positive control miRNA target plasmids with miR-204-5p mimic, an unrelated miRNA that shows no sequence similarity to either miR-21-3p or miR-27a-5p (Fig. 4C). As expected, both the miRs-21-3p and -27a-5p positive controls showed no downregulation of luciferase activity in response to miR-204-5p overexpression in addition to downregulation of luciferase activity in response to overexpression of their respective miRNA (Fig. 4C).
There was no effect of miR-21-3p or miR-204-5p overexpression on the luciferase construct harboring the miR-27a-5p complimentary sequence (Fig. 4C). However, we observed a decrease in luciferase activity of the miR-21-3p positive control plasmid with miR-27a-5p overexpression and not with miR-204-5p overexpression (Fig. 4C). This finding suggested that there is possible direct interaction between miR-21-3p target sites and miR-27a-5p that is not predicted by in silico prediction methods (supplementary Table II). Therefore, to confirm that miR-27a-5p can indeed interact with predicted miR-21-3p sites in 3′ UTRs of genes, we mutated the predicted miR-21-3p site and measured luciferase activity in response to miR-21-3p and miR-27a-5p overexpression. At random we selected the 3′ UTR of the CBEPB gene and mutated the miR-21-3p predicted target site (supplementary Fig. VIII B). Upon overexpression of the two miRNAs, we found that luciferase activity decreased ∼60% and 40% in response to miRs-21-3p and -27a-5p overexpression, respectively (supplementary Fig. VIII A). In comparison, mutation of the predicted miR-21-3p in the CBEPB 3′ UTR abolished the effects of miRs-21-3p and -27a-5p overexpression on luciferase activity. These findings strongly suggest that miR-27a-5p can interact with predicted miR-21-3p sites in 3′ UTR leading to downregulation of mRNA expression (supplementary Fig. VIII).
DISCUSSION
The relevance of the effects of oxGPs on endothelial function in various pathologies, such as acute inflammation, lung injury, and atherosclerosis, has been extensively documented (38). Here, we identified a role for two miRNAs, miRs-21-3p and -27a-5p, in regulating the response of HAECs to oxGPs. Using an unbiased global approach, we determined that the two miRNAs regulate the NF-κB signaling pathway in ECs in response to additional inflammatory stimuli including IL-1β. Using the 3′ UTR luciferase assay system, we show interactions between the two miRNAs with multiple transcripts in the NF-κB signaling cascade suggesting that these miRNAs act as fine-tuning regulators of NF-κB signaling.
Profile of miRNA-expressed in ECs
The endothelium contributes to the regulation of numerous physiological functions including the vasomotor tone, hemostatic balance, and inflammation. Here we present a catalog of miRNAs expressed in HAECs as identified by next-generation sequencing of the small RNAs. We identified a total of 618 unique miRNAs expressed in HAECs. Previous studies characterizing miRNA expression in ECs from seven different vascular beds expression using miRNA microarrays identified 166 EC miRNAs (39). Our study identified 474 HAEC miRNAs in addition to replicating 144 of the 166 previously reported EC miRNAs. The greater number of miRNAs detected in our study compared with the previous study can in part be attributed to the use of deep sequencing for detection. In contrast to microarray methods, deep sequencing contains a greater dynamic range for quantification of miRNA expression as well as increased accuracy in distinguishing between similar miRNAs and allowing for identification of novel miRNAs (40).
Previously reported deep-sequencing results identified 427 known and novel miRNAs expressed in HUVECs (41). Our study identified 369 of these 427 miRNAs. While we cannot rule out the differences in sequencing depth and data analysis between the two studies, these results suggest that although the majority of miRNAs are common between HAECs and HUVECs, differences in miRNA expression may exist based on the vascular bed.
Characterization of miRNA-regulated biological processes in ECs through identification of miRNA/mRNA interactions has shown that miRNAs can act as key regulators of numerous physiological processes in ECs including angiogenesis, lineage commitment, and inflammation (11, 42, 43). Of the 18 highly expressed EC miRNAs identified in our study, 11 have been previously studied in ECs. Seven of the 11 previously expressed miRNAs have been implicated as regulators of angiogenesis. miRs-126-5p, -126-3p, -27a-3p, -103a-3p, -30a-5p, and -let-7f are proangiogenic and target known inhibitors of angiogenesis, including the genes SPRED1, DLK1, SEMA6, TSP-1, and DLL4 to promote vessel development (11, 44–47). miR-24-3p, through regulation of the transcription factor of GATA2 and PAK4, acts to inhibit angiogenesis (48).
EC miRNAs in the immune response
In addition to angiogenesis, highly expressed EC miRNAs have also been shown to act as regulators of the inflammatory process. miR-126-3p, in addition to its role as a modulator of angiogenesis, is a known anti-inflammatory miRNA through direct repression of the leukocyte adhesion molecule VCAM-1 (49). In addition, many miRNAs that are expressed at moderate to low levels in our data set have also been shown to regulate inflammation in ECs. The moderately expressed miRs-17-5p and -31-5p directly inhibit the transcription of leukocyte adhesion molecules intracellular adhesion molecule-1 and E-selectin, respectively (26). Along with miR-126-3p, this provides evidence for miRNAs as direct suppressors of inflammation in ECs (26, 49). In addition to direct suppression of the expression of inflammatory molecules, anti-inflammatory EC miRNAs also act to regulate major signaling pathways. For example, miRs-181b-5p, -155-5p, and -10a-5p act to suppress inflammation through inhibition of NF-κB signaling (15, 43). Our studies identified two EC miRNAs that act as inhibitors of inflammation in ECs, miRs-21-3-p and -27a-5p, through regulation of NF-κB signaling.
Regulation of NF-κB signaling by miRs-21-3p and -27a-5p in ECs
NF-κB signaling is one of the key pathways in the activation of the immune response in ECs. Various environmental cues, including cytokines, bacterial infections, and hemodynamic forces, can activate NF-κB signaling in ECs leading to the increased expression of leukocyte adhesion molecules, cytokines, and chemokines (50–52). In ECs, miRNAs have been shown to suppress the activation of NF-κB at various points in the signaling cascade. A key site of miRNA-mediated suppression of NF-κB signaling is through degradation of the kinases responsible for phosphorylating IκB repressor proteins. miRs-10a-5p, -155-5p, -199a-5p, -223-3p, -15a-5p, and -16-5p all function in this manner to repress NF-κB signaling (15, 53–55). In addition, EC miRNAs can also repress NF-κB signaling by inhibiting the translocation of the NF-κB subunits to the nucleus, such as miR-181b-5p-mediated transcriptional repression of importin-α3, an adaptor protein functioning in nuclear protein import (43).
Based on our microarray data, we predicted that Ox-PAPC induced an increase in the levels of miRs-21-3p and -27a-5p to suppress NF-κB signaling in ECs. Previous studies from several groups have demonstrated that in HAECs and HUVECs, Ox-PAPC minimally activates or does not activate the expression of many NF-κB targets strongly induced by lipopolysaccharide (LPS), TNFα, or IL-1β such as E-selectin and VCAM-1 (34, 35). Using a luciferase construct containing three copies of the NF-κB responsive element, it has previously shown that unlike TNFα, Ox-PAPC treatment does not induce activation of NF-κB (33). Rather, these and other studies indicate that Ox-PAPC is a strong inhibitor of activation of NF-κB targets by LPS in all EC types (35). Our studies suggest that the induction of miRs-21-3p and -27a-5p by Ox-PAPC can be a possible mechanism that suppresses the activation of NF-κB signaling in ECs.
In contrast to Ox-PAPC, IL-1β and TNFα have also been shown to strongly induce synthesis of proinflammatory molecules by the NF-κB pathway but have well established feedback mechanisms to block long-term activation, including miRNAs. Our studies demonstrate that miRs-21-3p and -27a-5p are used to regulate NF-κB activation by these divergent activators of inflammation.
Identifying miRNA targets
In an attempt to identify direct targets of miRs-21-3p and -27a-5p, we used two in silico approaches. In our first approach, we identified candidate genes by focusing on genes whose expression was downregulated following miRNA overexpression. While this approach has been well established for the identification of miRNA targets, the large number of genes whose expression was altered by either miRs-21-3p or -27a-5p overexpression in our data set, even at a highly conservative statistical cutoff, necessitated candidate prioritization. In our second method, we utilized an in silico-based approach for identifying miRNA target genes by using multiple miRNA prediction programs to query the whole genome for predicted targets of miRs-21-3p and -27a-5p. In comparison to direct measurements of miRNA/mRNA interactions, the utilization of miRNA prediction algorithms such as miRANDA, which rely on base complementarity of the mature miRNA sequence in the 3′ UTR, biased our final candidate gene list to genes that contained sequences in the 3′ UTR complementary to miRs-21-3p and -27a-5p seed sequence (56).
While numerous methods have been developed for the identification of direct miRNA-mRNA interactions, the application of these methods is highly dependent on the miRNA-mRNA interaction being investigated. cross-linking immunoprecipitation-based methods, which utilize immunoprecipitation of Ago proteins in the RNA-induced silencing complex complex in combination with cross-linking of bound direct miRNA-mRNA interactions, have the advantage of direct determination of in vivo miRNA-mRNA interactions. However, it should be noted that low-abundance target miRNA-mRNA interactions may not be captured regardless of their affinity of interaction with Ago (57).To validate our list of 20 candidate genes, we chose to use the well-established 3′ UTR luciferase assay. One of the drawbacks of this method is its reliance on excessively high overexpression of both the target UTR and miRNA to determine affinity between the miRNA and its target.
Upon testing of these 20 candidate genes using the 3′ UTR luciferase assay, we made two distinct observations. First, in testing miR-21-3p target sites, we were only able to confirm downregulation for 11 of the 16 predicted target genes, representing an ∼69% concordance of miRNA/mRNA interaction predictions for miR-21-3p by miRANDA with experimental observation. In contrast, previous studies on the accuracy of miRNA prediction programs for predicting true miRNA/mRNA interactions have reported 20–49% concordance between experimental validation and in silico prediction for miRANDA (58, 59).
Second, we observed downregulation of all 10 putative miR-21-3p targets containing no predicted miR-27a-5p targets with miR-27a-5p overexpression. We further confirm this observation using a positive control luciferase plasmid containing only the complementary sequence of miR-21-3p. In addition, we do not observe this effect with miR-21-3p overexpression and miR-27a-5p target sites. Upon visual inspection of the mature sequences of miRs-21-3p and -27a-5p, we have identified two regions of limited sequence similarity that may contribute to the interaction of miR-27a-5p to miR-21-3p target sites. In mammals, canonical miRNA/mRNA interactions involve complementarity between the 3′ UTR and the given miRNA’s 5′ end. The first region of limited region of sequence similarity involves bases 13 and 15–19 of miR-21-3p being identical to bases 1–5 of miR-27a-5p, which represents a large fraction of the miR-27a-5p seed sequence suggesting a possible mechanism for downregulation of miR-21-3p targets by miR-27a-5p.
Complementarity in other regions beyond the 2–8 base pair seed sequence at the 5′ UTR has also been reported to mediate miRNA/mRNA interactions (10). One of these alternate miRNA/mRNA interactions, centered miRNA binding, involves the presence of 11 base perfect complementary starting at base 3, 4, or 5 of the miRNA sequence to 3′ UTR (60). In addition to similarity to miR-27a-5p seed sequence, between bases 8 and 17 of both miRNAs, bases 8, 9, 12, 14, and 16 are identical between the two miRNAs, suggesting an alternate interaction site between miR-21-3p sites and miR-27a-5p.
We show further evidence that miR-27a-5p can interact with predicted miR-21-3p sites as mutation of the predicted miR-21-3p site in the 3′ UTR of CBEPB abolished the effects of both miRs-21-3p and -27a-5p overexpression on luciferase activity. These observations strongly suggest that these two miRNAs act to regulate gene expression in the NF-κB signaling pathway through both through downregulation miRNA-specific target genes and, in a synergistic manner, through direct targeting of the same genes.
miRs-21-3p and -27a-5p modulate the extent of NF-κB signaling in ECs
miRNAs are biological rheostats that act to fine-tune gene expression to provide robustness and modulate the extent of activation of the signaling cascade (61). One mechanism of modulation of a signaling cascade is through direct repression of both activators and repressors, as seen in zebrafish miR-430 targeting both transforming growth factor β nodal agonist squinty and nodal antagonist lefty (62). In total, we found evidence for direct interactions between miRs-21-3p and/or -27a-5p for 20 candidate genes (Fig. 4, 5). Upon closer inspection of these 20 candidate genes, we found that several of these genes are either known repressors of NF-κB signaling, such as TNFAIP3/A20 and TNIP1/ABIN-1, or activators of NF-κB signaling, such as MAP2K7 (63). In addition, we found that 9 of the 20 candidate genes lie downstream of NF-κB signaling (5). These two findings, taken in consideration with the experimental observations that both overexpression and knockdown of miRs-21-3p and -27a-5p impairs p65 nuclear translocation following IL-1β treatment, suggest a dual role for miRs-21-3p and -27a-5p interacting with both the activators and regulators of NF-κB signaling.
Fig. 5.
miRs-21-3p and -27a-5p affect the expression of multiple genes in the NF-κB signaling cascade. The 20 candidate genes tested in the 3′ UTR luciferase assay were overlaid onto the NF-κB signaling cascade. Yellow color indicates genes that were shown to be regulated by both miRs-21-3p and -27a-5p overexpression using 3′ UTR luciferase assay. Blue color indicates genes that were shown to be regulated by miR-27a-5p overexpression, but not miR-21-3p overexpression using 3′ UTR luciferase assay.
Activation of gene expression in response to IL-1β is complex and involves the activation of numerous signaling cascades including c-Jun N-terminal kinase, MAPK, and NF-κB (64). Following inhibition of miRs-21-3p and -27a-5p with miRNA inhibitors, we did not observe the expected decrease in expression of E-selectin and VCAM-1 given that loss of the miRNAs led to a decrease in p65 nuclear translocation. Treatment of HAECs with IL-1β following knockdown of miR-21-3p leads to an increase in the expression of E-selectin and VCAM-1 despite impaired p65 nuclear translocation. Promoters of these two genes contain binding sites for the activator protein 1 (AP-1) family of transcription factors (65, 66). Therefore, despite impaired p65 nuclear translocation in response to knockdown of miR-21-3p, IL-1β could act through AP-1 to induce the expression of E-selectin and VCAM-1.
The role of tissue specificity in the identification of miRs-21-3p and -27a-5p target genes
Recently published reports have begun to characterize gene targets of miRs-21-3p and -27a-5p in different tissue types and diseases. Activation of epidermal growth factor receptor signaling is key in the development and progression of numerous cancers such as head and neck squamous cell carcinoma. Recently published work has shown that miR-27a-5p, whose expression is decreased in head and neck squamous cell carcinoma cell lines, acts to regulate epidermal growth factor receptor signaling through the repression of v-akt murine thymoma viral oncogene homolog 1 (AKT1) and mechanistic target of rapamycin (30). NK-cells cytotoxicity is mediated through release of granules containing proteins such as perforin and granzymes to induce apoptosis. Recent work has shown that expression of miR-27a-5p leads to loss of cytoxicity of NK-cells through direct transcriptional repression of perforin 1 (PRF1) and granzyme B (GZMB), suggesting miR-27a-5p as repressor of apoptosis (28).
Recent work has also implicated miR-21-3p as a regulator of apoptosis and cell growth in allergic inflammation; the continued presence of eosinophils at the site of inflammation is highly dependent on the secretion of granulocyte macrophage-colony-stimulating factor (GM-CSF), which acts to suppress apoptosis (67). In culture, GM-CSF upregulates the expression of miR-21-3p, which enhances ERK activation leading to a suppression of apoptosis in eosinophils, suggesting that miR-21-3p is a key component in GM-CSF-mediated survival (27). In hepatocellular carcinoma, increased expression of MAT2A and MAT2B, which are targets of miR-21-3p, contributes to progression of the tumor growth (29). In our data, we found downregulation of the reported targets that are expressed in ECs (AKT1, MAT2A, and MAT2B) following 24 h overexpression (data not shown). Given the known role of NF-κB signaling in the promotion of cell survival and proliferation, confirmation of the downregulation of these genes in our own data is suggestive of modulation of other key processes by these two miRNAs in ECs.
In summary, our study catalogs the profile of expressed miRNAs in HAECs both pre- and postexposure to Ox-PAPC. Furthermore, we identified the miRNAs, miRs-21-3p and -27a-5p, to be induced in ECs in response to OxPAPC, IL-1β, and TNFα treatment. We find these two miRNAs act to regulate NF-κB signaling through suppression of p65 nuclear translocation. We present evidence using the 3′ UTR luciferase assay that these two miRNAs modulate the extent of activation NF-κB signaling by regulating genes involved in both the suppression and activation of NF-κB signaling. Taken together, our studies suggest an important and complex role for miRs-21-3p and -27a-5p in the regulation of NF-κB activation in HAECs.
Supplementary Material
Footnotes
Abbreviations:
- Ago
- Argonaute, AKT1, v-akt murine thymoma viral oncogene homolog 1
- AP-1
- activator protein 1
- EC
- endothelial cell
- GM-CSF
- granulocyte macrophage-colony-stimulating factor
- HAEC
- human aortic endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- IL-1β
- interleukin 1β miRNA, microRNA
- NF-κB
- nuclear factor κB
- NK
- natural killer
- oxGP
- oxidized glycerophospholipid
- Ox-PAPC
- oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphatidylcholine
- UTR
- untranslated region
- VCAM-1
- vascular cell adhesion molecule-1
This work was supported by National Institutes of Health Grants HL-30568 and HL-28481 (A.J.L. and J.A.B.); Ruth L. Kirschstein National Research Service Award, Grant T32GM718539 (M.C.R.); American Heart Association postdoctoral fellowship, Grant 10POST3660048; National Institutes of Health Ruth L. Kirschstein National Research Service Award, Grant T32HL69766; and National Institutes of Health Pathway to Independence Award, Grant K99HL121172 (M.C.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of eight figures, two tables, and a supplementary file.
REFERENCES
- 1.Endemann D. H., Schiffrin E. L. 2004. Endothelial dysfunction. J. Am. Soc. Nephrol. 15: 1983–1992. [DOI] [PubMed] [Google Scholar]
- 2.Libby P., Ridker P. M., Hansson G. K. 2009. Inflammation in atherosclerosis: from pathophysiology to practice. J. Am. Coll. Cardiol. 54: 2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gargalovic P. S., Imura M., Zhang B., Gharavi N. M., Clark M. J., Pagnon J., Yang W-P., He A., Truong A., Patel S., et al. 2006. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc. Natl. Acad. Sci. USA. 103: 12741–12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banfi C., Colli S., Eligini S. 2002. Oxidized LDLs influence thrombotic response and cyclooxygenase 2. Prostaglandins Leukot. Essent. Fatty Acids. 67: 169–173. [DOI] [PubMed] [Google Scholar]
- 5.Gargalovic P. S., Gharavi N. M., Clark M. J., Pagnon J., Yang W-P., He A., Truong A., Baruch-Oren T., Berliner J. A., Kirchgessner T. G., et al. 2006. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 26: 2490–2496. [DOI] [PubMed] [Google Scholar]
- 6.Reddy S. T., Grijalva V., Ng C., Hassan K., Hama S., Mottahedeh R., Wadleigh D. J., Navab M., Fogelman A. M. 2002. Identification of genes induced by oxidized phospholipids in human aortic endothelial cells. Vascul. Pharmacol. 38: 211–218. [DOI] [PubMed] [Google Scholar]
- 7.Yeh M., Cole A. L., Choi J., Liu Y., Tulchinsky D., Qiao J-H., Fishbein M. C., Dooley A. N., Hovnanian T., Mouilleseaux K., et al. 2004. Role for sterol regulatory element-binding protein in activation of endothelial cells by phospholipid oxidation products. Circ. Res. 95: 780–788. [DOI] [PubMed] [Google Scholar]
- 8.Watson A. D., Navab M., Hama S. Y., Sevanian A., Prescott S. M., Stafforini D. M., McIntyre T. M., Du B. N., Fogelman A. M., Berliner J. A. 1995. Effect of platelet activating factor-acetylhydrolase on the formation and action of minimally oxidized low density lipoprotein. J. Clin. Invest. 95: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao B., Stavchansky S. A., Bowden R. A., Bowman P. D. 2003. Effect of interleukin-1β and tumor necrosis factor-α on gene expression in human endothelial cells. Am. J. Physiol. Cell Physiol. 284: C1577–C1583. [DOI] [PubMed] [Google Scholar]
- 10.Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuehbacher A., Urbich C., Zeiher A. M., Dimmeler S. 2007. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 101: 59–68. [DOI] [PubMed] [Google Scholar]
- 12.Suárez Y., Fernández-Hernando C., Pober J. S., Sessa W. C. 2007. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 100: 1164–1173. [DOI] [PubMed] [Google Scholar]
- 13.Sun H-X., Zeng D-Y., Li R-T., Pang R-P., Yang H., Hu Y-L., Zhang Q., Jiang Y., Huang L-Y., Tang Y-B., et al. 2012. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension. 60: 1407–1414. [DOI] [PubMed] [Google Scholar]
- 14.Marchand A., Proust C., Morange P-E., Lompré A-M., Trégouët D-A. 2012. miR-421 and miR-30c inhibit SERPINE 1 gene expression in human endothelial cells. PLoS ONE. 7: e44532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang Y., Shi C., Manduchi E., Civelek M., Davies P. F. 2010. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 107: 13450–13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng B., Chen S., McArthur K., Wu Y., Sen S., Ding Q., Feldman R. D., Chakrabarti S. 2011. miR-146a-mediated extracellular matrix protein production in chronic diabetes complications. Diabetes. 60: 2975–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afonyushkin T., Oskolkova O. V., Bochkov V. N. 2012. Permissive role of miR-663 in induction of VEGF and activation of the ATF4 branch of unfolded protein response in endothelial cells by oxidized phospholipids. Atherosclerosis. 225: 50–55. [DOI] [PubMed] [Google Scholar]
- 18.Fang Y., Davies P. F. 2012. Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium. Arterioscler. Thromb. Vasc. Biol. 32: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson A. D., Leitinger N., Navab M., Faull K. F., Hörkkö S., Witztum J. L., Palinski W., Schwenke D., Salomon R. G., Sha W., et al. 1997. Structural identification by mass spectrometry of oxidized phospholipids in minimally oxidized low density lipoprotein that induce monocyte/endothelial interactions and evidence for their presence in vivo. J. Biol. Chem. 272: 13597–13607. [DOI] [PubMed] [Google Scholar]
- 20.Civelek M., Hagopian R., Pan C., Che N., Yang W-P., Kayne P. S., Saleem N. K., Cederberg H., Kuusisto J., Gargalovic P. S., et al. 2013. Genetic regulation of human adipose microRNA expression and its consequences for metabolic traits. Hum. Mol. Genet. 22: 3023–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi W., Oshlack A., Smyth G. K. 2010. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 38: e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant G. R., Liu J., Stoeckert C. J. 2005. A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 21: 2684–2690. [DOI] [PubMed] [Google Scholar]
- 24.Huang D. W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 25.Kozomara A., Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suárez Y., Wang C., Manes T. D., Pober J. S. 2010. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J. Immunol. 184: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong C. K., Lau K. M., Chan I. H. S., Hu S., Lam Y. Y. O., Choi A. O. K., Lam C. W. K. 2013. MicroRNA-21* regulates the prosurvival effect of GM-CSF on human eosinophils. Immunobiology. 218: 255–262. [DOI] [PubMed] [Google Scholar]
- 28.Kim T-D., Lee S. U., Yun S., Sun H-N., Lee S. H., Kim J. W., Kim H. M., Park S-K., Lee C. W., Yoon S. R., et al. 2011. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood. 118: 5476–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo T-F., Tsai W-C., Chen S-T. 2013. MicroRNA-21–3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS ONE. 8: e75628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X., Bhayani M. K., Dodge C. T., Nicoloso M. S., Chen Y., Yan X., Adachi M., Thomas L., Galer C. E., Jiffar T., et al. 2013. Coordinated targeting of the EGFR signaling axis by microRNA-27a*. Oncotarget. 4: 1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betel D., Wilson M., Gabow A., Marks D. S., Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36: D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann A., Baltimore D. 2006. Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 210: 171–186. [DOI] [PubMed] [Google Scholar]
- 33.Yeh M., Leitinger N., de Martin R., Onai N., Matsushima K., Vora D. K., Berliner J. A., Reddy S. T. 2001. Increased transcription of IL-8 in endothelial cells is differentially regulated by TNF-alpha and oxidized phospholipids. Arterioscler. Thromb. Vasc. Biol. 21: 1585–1591. [DOI] [PubMed] [Google Scholar]
- 34.Romanoski C. E., Che N., Yin F., Mai N., Pouldar D., Civelek M., Pan C., Lee S., Vakili L., Yang W-P., et al. 2011. Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ. Res. 109: e27–e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochkov V. N., Oskolkova O. V., Birukov K. G., Levonen A-L., Binder C. J., Stöckl J. 2010. Generation and biological activities of oxidized phospholipids. Antioxid. Redox Signal. 12: 1009–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dweep H., Sticht C., Pandey P., Gretz N. 2011. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44: 839–847. [DOI] [PubMed] [Google Scholar]
- 37.Thomas M., Lieberman J., Lal A. 2010. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17: 1169–1174. [DOI] [PubMed] [Google Scholar]
- 38.Lee S., Birukov K. G., Romanoski C. E., Springstead J. R., Lusis A. J., Berliner J. A. 2012. Role of phospholipid oxidation products in atherosclerosis. Circ. Res. 111: 778–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCall M. N., Kent O. A., Yu J., Fox-Talbot K., Zaiman A. L., Halushka M. K. 2011. MicroRNA profiling of diverse endothelial cell types. BMC Med. Genomics. 4: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard C. C., Cheng H. H., Tewari M. 2012. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 13: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voellenkle C., Van Rooij J., Guffanti A., Brini E., Fasanaro P., Isaia E., Croft L., David M., Capogrossi M. C., Moles A., et al. 2012. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA. 18: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedrioli D. M. L., Karpanen T., Dabouras V., Jurisic G., van de Hoek G., Shin J. W., Marino D., Kälin R. E., Leidel S., Cinelli P., et al. 2010. miR-31 functions as a negative regulator of lymphatic vascular lineage-specific differentiation in vitro and vascular development in vivo. Mol. Cell. Biol. 30: 3620–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun X., Icli B., Wara A. K., Belkin N., He S., Kobzik L., Hunninghake G. M., Vera M. P., Registry M., Blackwell T. S., et al. 2012. MicroRNA-181b regulates NF-κB – mediated vascular inflammation. J. Clin. Invest. 122: 1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S., Aurora A. B., Johnson B. A., Qi X., McAnally J., Hill J. A., Richardson J. A., Bassel-Duby R., Olson E. N. 2008. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 15: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schober A., Nazari-Jahantigh M., Wei Y., Bidzhekov K., Gremse F., Grommes J., Megens R. T. A., Heyll K., Noels H., Hristov M., et al. 2014. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat. Med. 20: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urbich C., Kaluza D., Frömel T., Knau A., Bennewitz K., Boon R. A., Bonauer A., Doebele C., Boeckel J-N., Hergenreider E., et al. 2012. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood. 119: 1607–1616. [DOI] [PubMed] [Google Scholar]
- 47.Bridge G., Monteiro R., Henderson S., Emuss V., Lagos D., Georgopoulou D., Patient R., Boshoff C. 2012. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood. 120: 5063–5072. [DOI] [PubMed] [Google Scholar]
- 48.Fiedler J., Jazbutyte V., Kirchmaier B. C., Gupta S. K., Lorenzen J., Hartmann D., Galuppo P., Kneitz S., Pena J. T. G., Sohn-Lee C., et al. 2011. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 124: 720–730. [DOI] [PubMed] [Google Scholar]
- 49.Harris T. A., Yamakuchi M., Ferlito M., Mendell J. T., Lowenstein C. J. 2008. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA. 105: 1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohan S., Mohan N., Sprague E. A. 1997. Differential endothelial activation of NF-kappa B in human aortic cells conditioned to specific flow environments. Am. J. Physiol. Cell Physiol. 273: C572–C578. [DOI] [PubMed] [Google Scholar]
- 51.True A. L., Rahman A., Malik A. B. 2000. Activation of NF-kappaB induced by H(2)O(2) and TNF-alpha and its effects on ICAM-1 expression in endothelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 279: L302–L311. [DOI] [PubMed] [Google Scholar]
- 52.Faure E., Equils O., Sieling P. A., Thomas L., Zhang F. X., Kirschning C. J., Polentarutti N., Muzio M., Arditi M. 2000. Bacterial lipopolysaccharide activates NF-kappa B through Toll-like receptor 4 (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J. Biol. Chem. 275: 11058–11063. [DOI] [PubMed] [Google Scholar]
- 53.Tili E., Michaille J-J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., et al. 2007. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179: 5082–5089. [DOI] [PubMed] [Google Scholar]
- 54.Chen R., Alvero A. B., Silasi D. A., Kelly M. G., Fest S., Visintin I., Leiser A., Schwartz P. E., Rutherford T., Mor G. 2008. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 27: 4712–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li T., Morgan M. J., Choksi S., Zhang Y., Kim Y-S., Liu Z. 2010. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol. 11: 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Min H., Yoon S. 2010. Got target? Computational methods for microRNA target prediction and their extension. Exp. Mol. Med. 42: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hausser J., Zavolan M. 2014. Identification and consequences of miRNA-target interactions - beyond repression of gene expression. Nat. Rev. Genet. 15: 599–612. [DOI] [PubMed] [Google Scholar]
- 58.Sethupathy P., Megraw M., Hatzigeorgiou A. 2006. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 3: 881–886. [DOI] [PubMed] [Google Scholar]
- 59.Alexiou P., Maragkakis M., Papadopoulos G. L., Reczko M., Hatzigeorgiou A. G. 2009. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 25: 3049–3055. [DOI] [PubMed] [Google Scholar]
- 60.Martin H. C., Wani S., Steptoe A. L., Krishnan K., Nones K., Nourbakhsh E., Vlassov A., Grimmond S. M., Cloonan N. 2014. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 15: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ebert M. S., Sharp P. A. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell. 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi W-Y., Giraldez A. J., Schier A. F. 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 318: 271–274. [DOI] [PubMed] [Google Scholar]
- 63.Mauro C., Pacifico F., Lavorgna A., Mellone S., Iannetti A., Acquaviva R., Formisano S., Vito P., Leonardi A. 2006. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J. Biol. Chem. 281: 18482–18488. [DOI] [PubMed] [Google Scholar]
- 64.O’Neill L. A., Greene C. 1998. Signal transduction pathways activated by the IL-1 receptor family: ancient signaling machinery in mammals, insects, and plants. J. Leukoc. Biol. 63: 650–657. [PubMed] [Google Scholar]
- 65.Jensen L. E., Whitehead A. S. 2003. ELAM-1/E-selectin promoter contains an inducible AP-1/CREB site and is not NF-kappa B-specific. Biotechniques. 35: 54–56, 58. [DOI] [PubMed] [Google Scholar]
- 66.Cybulsky M. I., Fries J. W., Williams A. J., Sultan P., Eddy R., Byers M., Shows T., Gimbrone M. A., Collins T. 1991. Gene structure, chromosomal location, and basis for alternative mRNA splicing of the human VCAM1 gene. Proc. Natl. Acad. Sci. USA. 88: 7859–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon H. U., Yousefi S., Dibbert B., Levi-Schaffer F., Blaser K. 1997. Anti-apoptotic signals of granulocyte-macrophage colony-stimulating factor are transduced via Jak2 tyrosine kinase in eosinophils. Eur. J. Immunol. 27: 3536–3539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.