Abstract
Effects of serum cholesterol on cholesterol content in the retina are currently unknown. It is also unclear how cholesterol levels are controlled in the retina. High-cholesterol diet and oral administrations of simvastatin were used to modulate serum cholesterol in mice. These treatments only modestly affected cholesterol content in the retina and had no significant effect on retinal expression of the major cholesterol- and vision-related genes; the sterol-regulatory element binding protein pathway of transcriptional regulation does not seem to be operative in the retina under the experimental conditions used. Evidence is obtained that posttranslational mechanisms play a role in the control of retinal cholesterol. Retinal genes were only upregulated by oral administrations of TO901317 activating liver X receptors. Three of the upregulated genes could be of particular importance (apoD, Idol, and Rpe65) and have not yet been considered in the context of cholesterol homeostasis in the retina. Collectively, the data obtained identify specific features of retinal cholesterol maintenance and suggest additional therapies for age-related macular degeneration, a blinding disease characterized by cholesterol and lipid accumulations in chorioretinal tissues.
Keywords: cytochrome P450, transcriptional regulation, posttranslational regulation, 3-hydroxy-3-methyl-glutaryl-CoA reductase, liver X receptor, sterol-regulatory element binding protein, RPE65, age-related macular degeneration
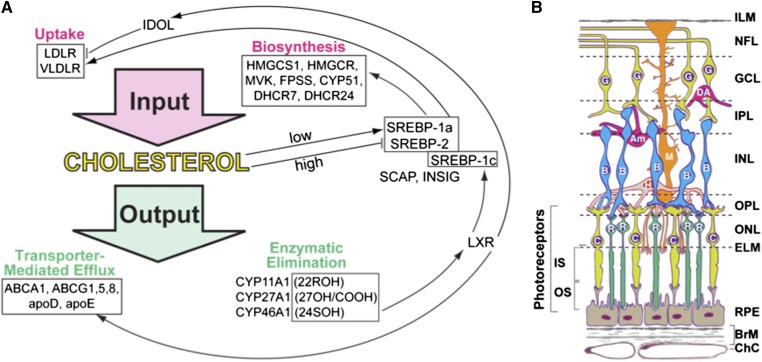
Cholesterol is an important endogenous compound, whose cellular and tissue levels are normally controlled by homeostatic mechanisms that balance the pathways of cholesterol input and output (1). The two most common sources of cellular cholesterol are in situ biosynthesis and receptor-mediated uptake of cholesterol-containing lipoprotein particles such as LDL and, to a lesser extent, VLDL (Fig. 1A; supplementary Table I). Pathways of input are regulated in large part by transcription factors of the sterol-regulatory element binding protein (SREBP) family, which stay, when cholesterol levels are high, as precursor proteins in the endoplasmic reticulum (ER) due to complex formation with cholesterol-sensing protein SCAP (SREBP cleavage-activating protein) and the ER-retention protein INSIG (insulin-induced gene) (1, 2). Cholesterol output is also mainly determined by the two major pathways: transporter (ABCA1)-mediated cholesterol efflux and metabolism to oxysterols by cytochrome P450 (CYP) enzymes. Once outside the cell or in the systemic circulation, cholesterol and oxysterols become associated with lipoproteins and are delivered to the liver for further degradation to bile acids. Depending on the tissue, cholesterol could be metabolized to 27-hydroxycholesterol or 5-cholestenoic acid by CYP27A1 or 24(S)-hydroxycholesterol by CYP46A1. In steroidogenic organs, cholesterol is utilized for steroid hormone production and is converted to pregnenolone by CYP11A1 via the intermediate 22R-hydroxycholesterol. Oxysterols as well as some of the cholesterol precursors in the pathway of cholesterol biosynthesis activate a family of transcription factors of the liver X receptor (LXR) family, which regulate the expression of the efflux transporter ABCA1 and apos involved in cholesterol removal (3–5). In addition, LXRs control the expression of the SREBP isoform SREBP-1c, which enhances transcription of the genes required for fatty acid synthesis (2). Thus, there is a coordinated regulation of the pathways of cholesterol input and output.
Fig. 1.
Cholesterol and retina essentials. A: Pathways of cholesterol input and output showing some of the proteins whose genes were investigated in the present work. SREBP-1a is a potent activator of all SREBP-responsive genes, whereas SREBP-1c preferentially acts on genes in the pathway of fatty acid synthesis. SREBP-2 preferentially enhances cholesterologenic genes. 22ROH, 22(R)-hydroxycholesterol, the product of CYP11A1; 27OH/COOH, 27-hydroxycholesterol and 5-cholestenoic acid, respectively, the products of CYP27A1; 24SOH, 24(S)-hydroxycholesterol, the product of CYP46A1. B: Chorioretinal layers and cells. The neurosensory retina has distinct layers (from top to bottom): ILM, inner limiting membrane; NFL, nerve fiber layer (ganglion cell axons); GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer (synapses); ONL, outer nuclear layer; ELM, external limiting membrane (junctional complexes); IS, PR inner segments; and OS, PR outer segments. RPE lies outside the neurosensory retina but is considered a part of the retina. BrM, Bruch’s membrane (vessel wall and RPE substratum); ChC, choriocapillaris (capillary bed for RPE and PRs). Non-PR layers of the retina are supplied by the retinal circulation (not shown). The major retinal cell types are ganglion cells (G), diffuse amacrine cells (DA), amacrine cells (Am), Müller cells (M), bipolar cells (B), horizontal cells (H), rods (R), and cones (C). Modified from Zheng et al. (11).
The neural retina (NR) is a light-sensitive tissue in the back of the eye mediating the transmission of the visual signal to the brain. This tissue is composed of multiple layers and cell types and is underlain by the retinal pigment epithelium (RPE), a polarized monolayer of cells serving as the interface between the NR and choroidal blood vessels (Fig. 1B). Although different systems, the NR and RPE are often considered as one structure called the retina because the RPE is a part of the NR embryologically and is closely associated with the NR via the strong interdigitation of its apical microvilli with the retinal photoreceptor (PR) outer segments (OS) (6). Different types and functions of the cells in the NR/RPE complex make understanding the overall cholesterol maintenance in the retina difficult. Nevertheless, progress has been made, and now we know that the retina obtains cholesterol via local biosynthesis and uptake of blood-borne cholesterol and contains all the genes necessary for independent cholesterol homeostasis (7–12). The means of cholesterol elimination have also been ascertained and comprise lipoprotein-mediated transport to the systemic circulation, metabolism to oxysterols by CYP27A1 and CYP46A1, and PR phagocytosis by the RPE, the retina-specific mechanism (13, 14). However, the quantitative importance of each of the retinal pathways of cholesterol input and output is currently unknown, and knowledge of their regulation is limited to immunohistochemistry studies showing protein expression and retinal localization of SREBPs, SCAP, INSIG, and LXRs in the retina of humans and other species (11, 15). Because in the retina transcriptional responsiveness of the SREBP and LXR target genes has not yet been comprehensively evaluated, herein, we investigated whether cholesterol content in mouse retina could be modulated by dietary and pharmacologic treatments and affect retinal expression of cholesterol-related genes. We used WT mice as well as Cyp27a1−/− mice (16) lacking the major enzyme, CYP27A1, metabolizing cholesterol in the retina (17–19). The Cyp27a1−/− retina has increased cholesterol content and vascular lesions associated with focal deposits of cholesterol (20). The pathways of cholesterol input and output are uncoupled in the Cyp27a1−/− retina (20); hence, Cyp27a1−/− mice were investigated as a model having dysregulated maintenance of retinal cholesterol and to obtain mechanistic insight(s) into the nature of this dysregulation. We established transcriptional responsiveness of the WT and Cyp27a1−/− retina to pharmacologic activation by LXRs and obtained evidence for the presence of nontranscriptional mechanisms regulating retinal cholesterol input. We identified apoD and Idol (inducible degrader of the LDL receptor), as potentially important for cholesterol maintenance in specific retinal cell types. Finally, we found that the levels of Rpe65 encoding an important protein of the visual cycle (21–24) are upregulated by an LXR agonist, TO901317 (TO9).
MATERIALS AND METHODS
Materials
Regular rodent chow (5P75-5P76-Prolab Isopro RMH 3000) was from LabDiet, and the investigational diets were custom-made by Research Diets. TO9 and simvastatin were purchased from Cayman Chemical. Trizol reagent was from Life Technologies. The source and dilution of primary and secondary antibodies for immunohistochemistry were as described (11) and also included rabbit polyclonal antibodies against ABCG1 (Novus Biologicals, diluted 1:100), APOD (LifeSpan BioSciences, diluted 1:100), and IDOL (GeneTex, diluted 1:100). Other chemicals were from Sigma-Aldrich unless otherwise indicated.
Animals
Cyp27a1+/− mice on the C57BL/6J background were obtained from the laboratory of Dr. Sandra Erickson (University of California, San Francisco) (16) and were bred to generate the Cyp27a1−/− line and Cyp27a1+/+ littermates (WT). Mice were housed in the Animal Resource Center at Case Western Reserve University and maintained in a standard 12 h light (∼10 lux)/12 h dark cycle environment. Water and food were provided ad libitum. All animal-handling procedures were approved by the Case Western Reserve University Institutional Animal Care and Use Committee and conformed to recommendations of the American Veterinary Association Panel on Euthanasia and the Association for Research in Vision and Ophthalmology.
Dietary studies
After weaning, WT and Cyp27a1−/− female mice were put on four different diets (Table 1) for 3 months. The investigational diet was a modified rodent chow containing a high content of fat (HF, 30% energy) and cholesterol (HC, 0.5%, w/w) and a small amount of cholic acid (CA, 0.05%, w/w). The three control diets were the regular rodent chow (5P75-5P76-Prolab Isopro RMH 3000) and modified rodent chows containing either 0.05% CA (CA diet) or HF (30% energy) plus 0.05% CA (HF/CA diet). CA was necessary because Cyp27a1−/− mice have reduced production of bile acids, and supplementation with CA improves their decreased intestinal absorption of dietary fat and cholesterol (16, 25–27). Accordingly, WT mice were also fed CA-supplemented diets to enable comparisons between genetic lines. Because CA is a ligand for the farnesoid X receptor (FXR), a transcription factor (28), the expression of Pltp, apoE, and Shp, the three FXR target genes (29, 30), was also monitored.
TABLE 1.
Formulation of the diets used in the present study
| Content | Regular Chow | CA Diet | HF/CA Diet | HF/HC/CA Diet | ||||
| g (%) | kcal (%) | g (%) | kcal (%) | g (%) | kcal (%) | g (%) | kcal (%) | |
| Protein | 23 | 28 | 19 | 20 | 21.5 | 20 | 21.4 | 20 |
| Carbohydrates | 64.5 | 58 | 67 | 70 | 53.8 | 50 | 53.5 | 50 |
| Fat | 4.5 | 14 | 4 | 10 | 14.3 | 30 | 14.2 | 30 |
| Minerals | 8 | 10 | 0 | 10 | 0 | 10 | 0 | |
| Total | 100 | 100 | 100 | 100 | ||||
| kcal/g | 4.1 | 3.84 | 4.3 | 4.28 | ||||
| Cholesterol | 0 | 0 | 0 | 0 | 0 | 0 | 4.8 | 0 |
| CA | 0 | 0 | 0.52 | 0 | 0.48 | 0 | 0.48 | 0 |
Pharmacologic treatments
Simvastatin and TO9 were delivered orally by gavage to male mice 3 to 6 months of age fed regular chow (5P75-5P76-Prolab Isopro RMH 3000). Drug solutions were prepared from stocks in DMSO (187.5 mg/ml) diluted, depending on the treatment dose, either with PBS or DMSO and then PBS to have the final concentration of DMSO the same in all the solutions (4%). For all treatment doses, the volumes of administered drugs were approximately the same (from 0.28 ml to 0.35 ml) and equal to 10 ml/kg of body weight (BW). Control animals received the same volumes of 4% DMSO in PBS. Simvastatin, 30 mg/kg BW, was administered once a day for 7 days, and mice were euthanized 24 h after the administration of the last dose. TO9 was administered either once at the 4–75 mg/kg BW dose or daily for 3 consecutive days at the 20 mg/kg BW and 50 mg/kg BW doses. Mice treated with a single TO9 dose were euthanized at the indicated time, whereas the multidose treated mice were euthanized 24 h after the administration of the last drug dose.
Organ isolation
Mouse retina (the NR/RPE complex) was isolated as described (31), dipped quickly three times in cold PBS, and used either individually or combined with washed NR/RPE from a companion eye or eyes of other animals of the same gender and genotype. The brain and liver were excised and cut sagittally (the brain) or to separate the four lobes (the liver). Tissue pieces were rinsed several times in cold PBS, blotted to remove PBS, and used for homogenization.
RNA isolation and cDNA synthesis
One NR/RPE, a half of the brain, or combined pieces of the liver (four total, one ∼25 mg piece from each liver lobe) were used for homogenization in 10 vol (w/v) of Trizol reagent. Subsequent RNA extraction was according to the manufacturer’s protocol (Life Technologies). Contaminating genomic DNA was completely eliminated by treatment with the RNase-Free DNase Set (Qiagen), and RNA was considered DNA-free when 40 cycles of real-time PCR did not give an amplification signal with the primers for β-actin (supplementary Table II).
Gene profiling by PCR array
This was carried out as described (11) using the “Mouse Lipoprotein Signaling and Cholesterol metabolism” PCR array (Qiagen) and the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Gene expression was normalized to the mean threshold cycle (Ct) of the five housekeeping genes: β-glucuronidase (Gusb), hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1), β-heat shock protein 1 (Hspcb), Gapdh, and β-actin (Actb).
Quantitative real-time PCR (qRT-PCR)
These experiments were carried out in the ABI PRISM 7000 Sequence Detection System and utilized 1 μg of total RNA and high-capacity cDNA reverse transcription kit (Life Technologies) for reverse transcriptase reaction. The primers for gene quantifications (supplementary Table II) were designed by the Primer Express 3.0 software (Applied Biosystems) with the amplicon length being <130. The amplification products were analyzed by agarose gel electrophoresis to ensure that they were of the right size. Gene expression was normalized to the expression of Actb.
Immunohistochemistry
The preparation of retinal sections and immunostaining was as described (11), except that the last step, treatment with Sudan black to reduce autofluorescence, was omitted. Stained slides were imaged on a Leica DMI 6000 B inverted microscope (Leica Microsystems) equipped with a Retiga EXI camera (QImaging). Image analysis was performed using Metamorph Imaging Software (Molecular Devices).
Western blotting
SREBP-1 and SREBP-2 were detected in retinal lysates that were obtained by shredding a pooled retinal sample (10 NR/RPEs from five animals) with a 25 G needle in 10 vol (w/v) of 50 mM Tris-Cl buffer, pH 7.4, containing 1 mM EDTA, 0.5 mM EGTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40, 150 mM NaCl, 50 mM sodium fluoride, and a mixture of protease inhibitors (1 mM PMSF and 1× cOmplete EDTA-free from Roche). After solubilization for 30 min on ice, retinal lysate was subjected to centrifugation at 14,000 g for 15 min at 4°C, and the supernatant was used for SDS-PAGE (50 μg protein/lane) and Western blotting. HMGCR was detected in the microsomes from the retina and liver that were isolated from a pooled sample of 10 NR/RPEs from five mice or a pooled sample of five livers. Microsome isolation was as described (32) by differential centrifugation. After SDS-PAGE (10 μg protein/lane), microsomal proteins were transferred to a nitrocellulose membrane (Thermo Scientific), which was treated as described (33), except that the blocking buffer contained 0.25% Tween-20 instead of 0.1% Tween-20. Proteins were visualized with rabbit polyclonal primary antibodies against SREBP-1 (Santa Cruz Biotechnology Inc., dilution 1:200), SREBP-2 (Abcam, dilution 1:50), or HMGCR (United States Biological, dilution 1:250) and goat anti-rabbit secondary antibodies (IRDye 680LT from Li-Cor, dilution 1:20,000). Membranes were imaged by the Odyssey infrared imaging system (Li-Cor). Quantifications of HMGCR were performed by Metamorph software (Molecular Devices Corp.).
Sterol quantifications
Tissue and serum concentrations of sterols in diet-fed and simvastatin-treated mice were determined by isotope dilution gas chromatography-mass spectrometry as described (18) using saponified samples from animals fasted overnight. Serum total cholesterol, HDL, LDL, and triglycerides in the TO9 treatments were measured in nonfasting mice by Marshfield Labs (Marshfield Clinic, Marshfield, WI).
Computational predictions of the transcription factor binding sites
The analysis of the gene promoter regions was carried out by the MatInspector (34) and encompassed the 5 kb region upstream of the transcriptional start site.
Statistics
All data represent mean ± SD, and the statistics are indicated in each figure. A two-tailed, unpaired Student’s t-test was used to determine statistical significance, which is defined as * P < 0.05, ** P < 0.01, and *** P < 0.001.
RESULTS
Retinal expression of cholesterol-related genes
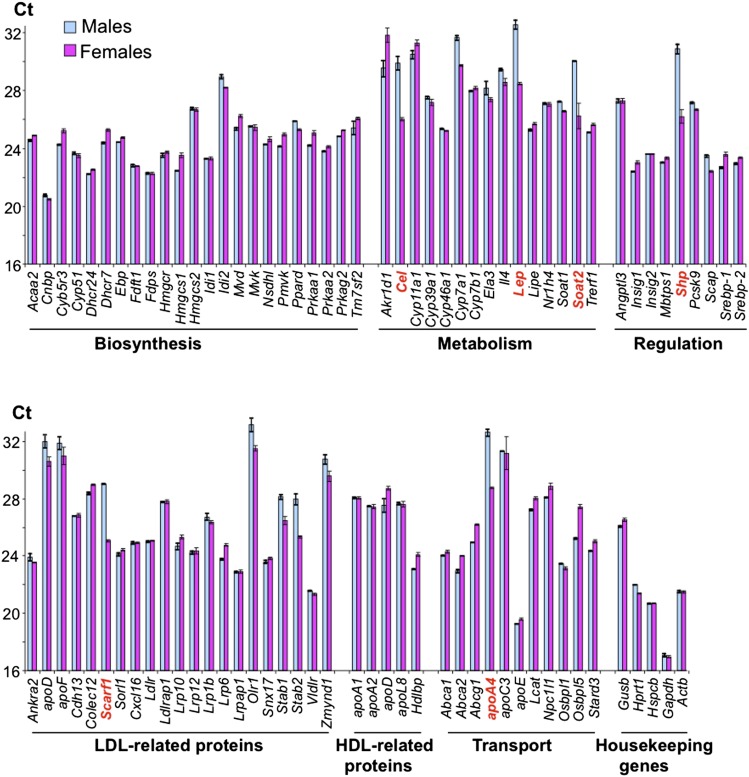
Eighty-four major cholesterol-related genes were profiled by PCR array in mouse retina (Fig. 2). These studies detected the mRNA transcripts for every gene in the array and showed a comparable, within an order of magnitude, gene expression in males and females. Significant gender differences were in the expression of only six genes (Cel, Lep, Soat2, Shp, Scarf1, and Apoa4), whose levels varied >10-fold between males and females.
Fig. 2.
Retinal expression of cholesterol-related genes in WT mice as assessed by PCR array. Each bar represents the mean of duplicate measurements in a pooled sample of three NR/RPEs from three WT mice 3 to 6 months of age. The Ct number reflects the gene expression level with the lower number corresponding to the higher gene expression. Genes in bold red are those whose expression varies >10-fold between males and females.
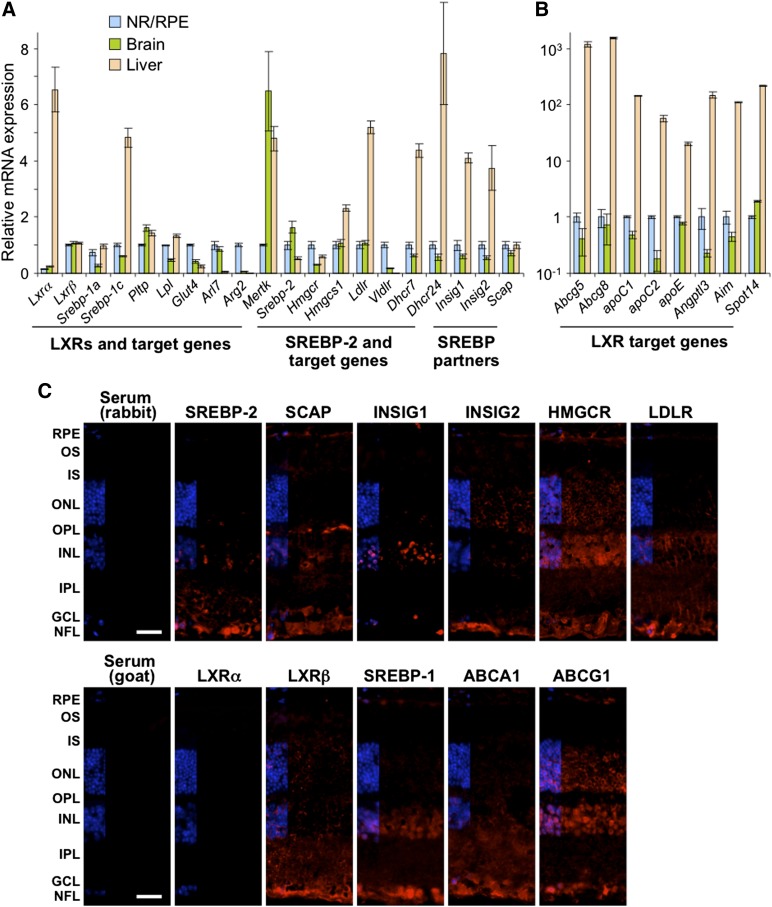
The transcript amounts for 28 genes were then quantified by qRT-PCR. These genes encoded the master regulators of cellular cholesterol and fatty acid/triglyceride content (LXRα, LXRβ, SREBP-1a, SREBP-1c, and SREBP-2) and proteins that are under transcriptional regulation of LXRs and SREBPs, or their partners in the SREBP-SCAP-INSIG complex (Fig. 3A, B). Mice of only one gender (males) were used, and the retina was compared with the brain as an example of another neural organ and the liver as the organ playing a central role in the maintenance of whole body cholesterol homeostasis. The measurements by qRT-PCR confirmed retinal expression of the selected genes and revealed that, in the brain, the expression of these genes was comparable, within an order of magnitude, to that in the retina. In the liver, however, only about two-thirds of the studied genes showed expression comparable to that in the retina including Lxrβ (expressed in almost all tissues and organs), Srebp-1a (a potent activator of all SREBP-responsive genes), Srebp-2 (activates Ldlr encoding the receptor for LDL and many cholesterogenic genes), Hmgcr (the rate-limiting enzyme in cholesterol biosynthesis), and Scap (the escort protein for SREBPs). The expression of the remaining one-third of the genes was from 100- to 1,000-fold higher in the liver than in the retina (Fig. 3B), and these genes pertained to cellular cholesterol efflux (Abcg5 and Abcg8), lipoprotein-mediated cholesterol transport (apoC1, apoC2, and apoE), and processes unrelated to cholesterol maintenance (Angptl3, AIM, and Spot14).
Fig. 3.
Retinal gene and protein expression of transcriptional regulators of cholesterol homeostasis and some of their targets. A: Genes expressed at comparable levels in the NR/RPE, brain, and liver. B: Genes expressed at significantly different levels. mRNA was quantified by qRT-PCR. Bars represent mean ± SD of duplicate measurements in individual samples from five WT male mice 3 to 6 months of age. The expression of Lxrα and Srebp-1a is relative to that of Lxrβ and Srebp-1c, respectively. In the retina, the Ct numbers for Lxrα and Lxrβ are 26.2 and 24.0, respectively, and those for Srebp-1a and Srebp-1c are 25.6 and 25.1, respectively. Retinal Ct for Actb is 20.4. C: Immunohistochemical localization of protein expression in the retina. All images are representative of stainings in multiple sections from four animals. Scale bars: 15 μm.
Immunohistochemistry was used next to evaluate retinal expression and localization of SREBPs, LXRs, and related proteins (Fig. 3B). Overall, SREBP-2 and SREBP-1 had a similar pattern of retinal expression, which was mainly confined to the layers of the inner retina: the GCL, IPL, and INL. The GCL, IPL, and INL also seemed to express the SREBP partner protein SCAP and either INSIG1 (GCL and INL) or INSIG2 (GCL and to a lesser extent INL). In addition, signals for the individual proteins from the SREBP-SCAP-INSIG complex were detected in the OPL, which showed immunostaining for SCAP; ONL, which had immunofluorescence for INSIGs; PR IS, which had a very faint staining for SCAP; and the RPE containing immunosignals for SREBP-1, SCAP, and INSIG2. The signal for the SREBP-2 target HMGCR was present throughout the retina, except the PR IS and OS, suggesting that in the layers where SREBP, SCAP, and INSIG do not colocalize with HMGCR (ONL, IS, and OS), HMGCR could be regulated by the mechanisms other than gene transcription. The expression pattern of LDLR, another SPEBP-2 target, was more restricted and was mainly associated with the layers expressing the proteins of the SREBP-SCAP-INSIG complex. The signal for transcription factor LXRα was not detectable, in agreement with expression of this isoform mostly in visceral organs (3), whereas that for LXRβ was seen in almost every retinal layer, although at different intensities. Thus, most of the studied genes seem to be expressed as proteins in the retina. Mouse retina can now be compared with human retina (11), the retina of other species (10, 14), and immunohistochemistry characterizations by others (35). Quantitative methodologies should be used to confirm immunolocalizations.
Gene expression in mice in response to altered cholesterol content
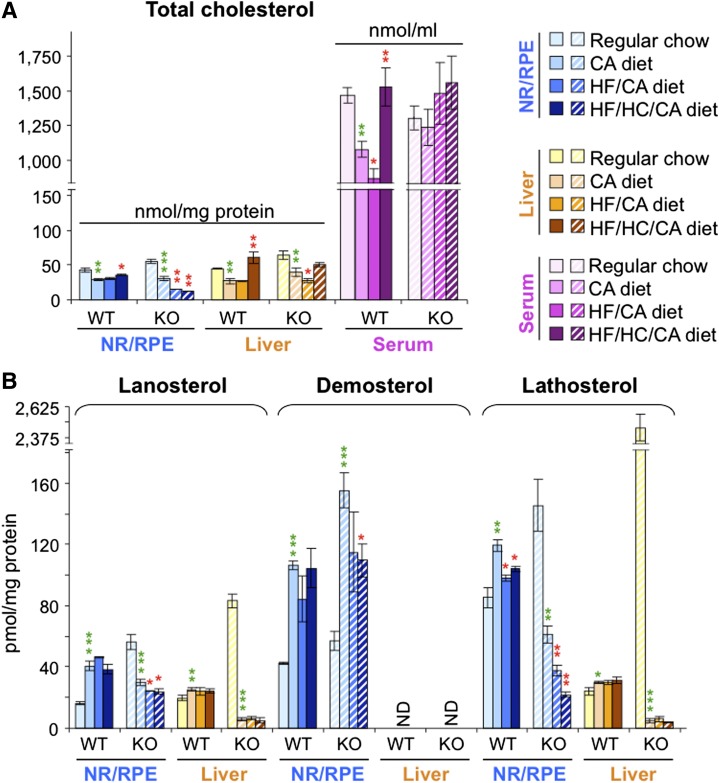
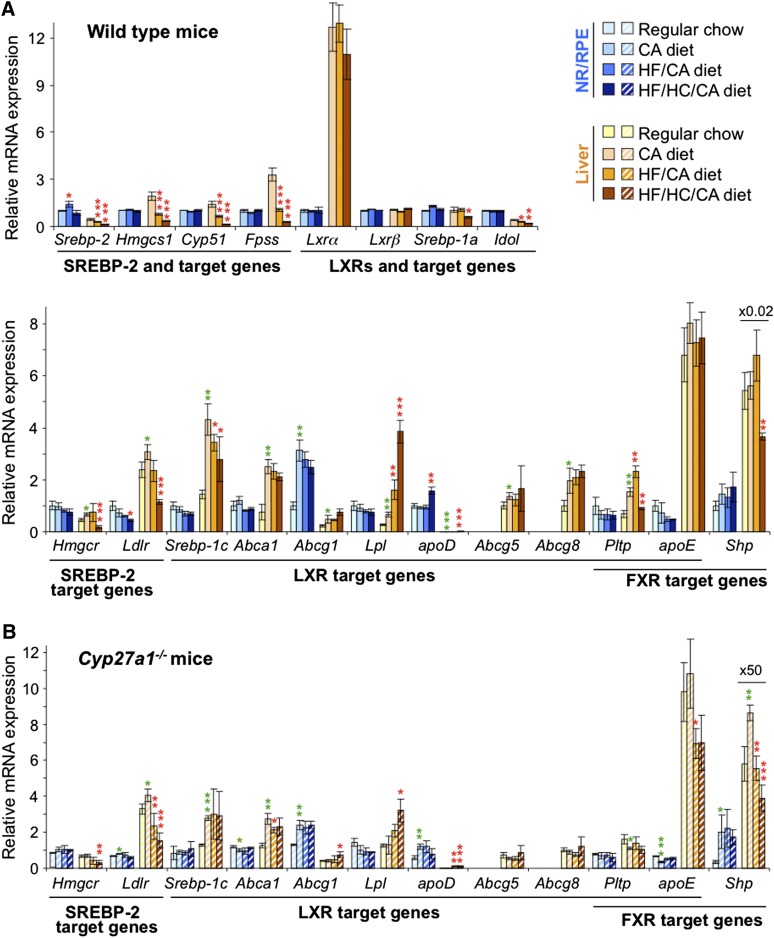
Feeding the WT mice the HF/HC/CA diet for 3 months led to a 1.4-fold increase in serum cholesterol and a smaller 1.2-fold increase in retinal cholesterol as compared with the control CA diet (Fig. 4A). There was also a decrease in retinal levels of cholesterol precursor lathosterol (Fig. 4B). Retinal expression of cholesterogenic genes (Hmgcs, Cyp51, Fpps, and Hmgcr) and Srebp-2 regulating the expression of these genes was, nevertheless, unchanged in WT mice on the HF/HC/CA versus the CA diet (Fig. 5A) indicating a nontranscriptional mechanism of downregulation of cholesterol biosynthesis. Transcriptional response was observed in a different pathway of cholesterol input, cellular uptake of LDL-bound cholesterol, as the HF/HC/CA diet decreased retinal levels of Ldlr (Fig. 5A). An increase in retinal cholesterol in WT mice on the HF/HC/CA diet versus the CA diet did not seem lead to a compensatory upregulation of a pathway of cholesterol output as the levels of retinal genes controlling lipoprotein-mediated cholesterol elimination (Lxrα and Lxrβ) and their targets in this pathways (Abca1, Abcg1, Lpl, Abcg5, Abcg8, and apoE) were not altered, except the upregulation of apoD. This is in contrast to the liver, where transcriptional regulation was fully operative on the HF/HC/CA and HF/CA diets as compared with the CA diet affecting most of the genes involved in the pathways of cholesterol input (Srebp-2, Hmgcs1, Cyp51, Fpps, Srebp-1a, Idol, Hmgcr, Ldlr, and Srebp-1c) and some of the genes mediating cholesterol output (Lpl, apoD, and Pltp). Despite this regulation, cholesterol accumulation was more significant in the WT liver than in the WT retina of mice on the HF/HC/CA diet versus the CA diet (2.3-fold versus 1.2-fold) (Fig. 4A). The WT liver was also the organ sensitive to the supplementation with CA as compared with the regular chow as the transcription of the FXR/LXR target Pltp and some of the LXR targets (Srebp-1c, Abca1, Abcg1, Lpl, apoD, and Abcg8) was altered in mice on the CA diet as compared with the regular chow (Fig. 5A). Conversely, in the WT retina, the CA diet did not affect the expression of any of the studied FXR targets and most other genes, except Abcg1, as compared with the regular chow. Despite differential effects on the gene transcription, the CA diet had similar effects on retinal and hepatic cholesterol levels, which were lower than those in WT mice on the regular chow (Fig. 4A).
Fig. 4.
Effect of different diets on sterol content in the retina and liver of WT and Cyp27a1−/− (KO) mice. A: The content of total cholesterol including the measurements in the serum. B: The content of cholesterol precursors lanosterol, desmosterol, and lathosterol. Dietary treatments of female mice started after their weaning (about 2 weeks of age) and continued for 3 months; n = 7 per dietary group. Bars pertaining to the NR/RPE are in bluish colors, and those pertaining to the liver and serum are in yellow-brown and magenta-purple colors, respectively. The results are mean ± SD of the measurements in individual animals. CHO, cholesterol; ND, not detectable (the limit of detection is ≥10 pmol/mg protein). Green asterisks are significant changes versus regular chow. Red asterisks are significant changes versus the CA-containing diet. * P < 0.05; ** P < 0.01; *** P < 0.001.
Fig. 5.
Effect of different diets on gene expression in the retina and liver of WT (A) and Cyp27a1−/− (B) mice. The dietary treatments, mouse gender, bar color, code, and abbreviations are as in Fig. 4. mRNA was measured by qRT-PCR. Bars represent mean ± SD of individual measurements in five mice. Green asterisks are significant changes versus regular chow. Red asterisks are significant changes versus the CA-containing diet. * P < 0.05; ** P < 0.01; *** P < 0.001.
Nutritional studies on Cyp27a1−/− mice, which have a dysregulated maintenance of retinal cholesterol (20), led to unexpected results. Retinal cholesterol was decreased on the HF/HC/CA diet 2.4-fold as compared with the CA diet (Fig. 4A), and this decrease was observed under essentially unchanged serum cholesterol levels (Fig. 4A). Retinal lathosterol was decreased as well (2.9-fold) on the HF/HC/CA diet versus the CA diet with changes being larger than those in WT mice on the same diets (Fig. 4B). In addition, there was a decrease in retinal desmosterol in Cyp27a1−/− mice on the HF/HC/CA diet versus the CA diet. Lathosterol and desmosterol may reflect changes in cholesterol biosynthesis in different cell types in the retina because in the brain the two sterols are considered to be the markers for cholesterol biosynthesis in neurons (lathosterol) and astrocytes (desmosterol) (36). Despite significant decreases in the levels of retinal sterols (cholesterol, lanosterol, desmosterol, and lathosterol) in Cyp27a1−/− mice on the HF/HC/CA diet versus the CA diet, retinal expression of cholesterol-related genes was not affected in this line (Fig. 5B). The Cyp27a1−/− retina, however, showed some sensitivity to dietary CA as the retinal (and hepatic) expression of Shp was increased, while the levels of apoD and apoE were up- and downregulated, respectively (Fig. 5B), in mice on the CA diet versus the regular chow. In the Cyp27a1−/− liver, the levels of cholesterol and lathosterol were not changed significantly on the HF/HC/CA diet versus the CA diet (Fig. 4B), possibly because of the compensatory changes in the expression of Hmgcr, Ldlr, Lpl, and apoD.
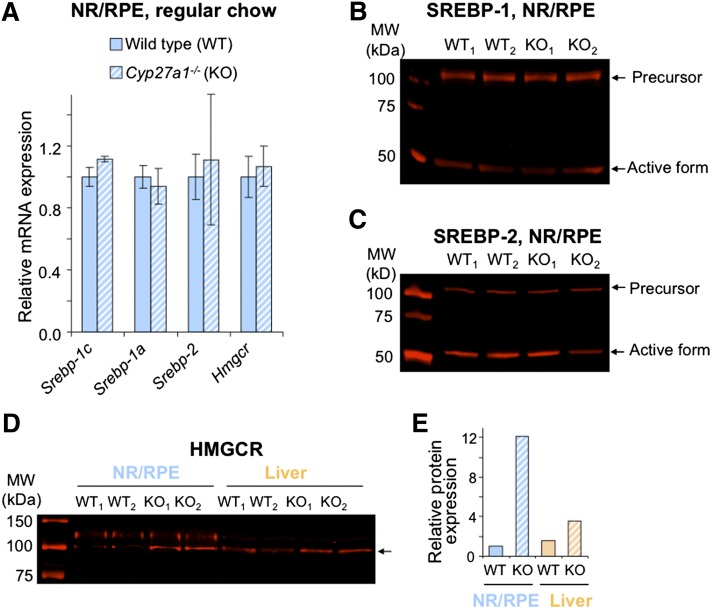
Nutritional studies on WT and Cyp27a1−/− mice suggested that altered levels of retinal cholesterol do not significantly affect the transcription of retinal cholesterol-related genes. These studies also showed that the expression of Srebp-2 and Hmgcr, the two key players in cholesterol biosynthesis, is similar between the WT and Cyp27a1−/− mice when they are fed regular chow (Fig. 6A), despite retinal cholesterol biosynthesis in Cyp27a1−/− mice being upregulated as indicated by an increase in retinal lathosterol. Hence, we evaluated protein expression of SREBPs and HMGCR in WT and Cyp27a1−/− mice on regular chow (Fig. 6B–D). In the retina, protein levels of the SREBP precursors (125 kDa) and active forms (∼50 kDa) were similar in WT and Cyp27a1−/− mice, whereas the protein expression of HMGCR was higher in the Cyp27a1−/− retina than in the WT retina (12.0-fold) consistent with an increase in retinal lathosterol. An increase in protein levels of HMGCR was also observed in the Cyp27a1−/− liver as compared with the WT liver (2.3-fold), although it was smaller than in the retina. Thus, posttranslational regulation of HMGCR (37–39) seems to contribute to cholesterol biosynthesis in the retina and plays a larger role in the retina than in the liver.
Fig. 6.
Gene (A) and protein (B–E) expression of SREBPs and HMGCR in the retina or liver of mice on regular chow. A: Relative mRNA expression as assessed by qRT-PCR. Bars represent mean ± SD of individual measurements in five mice. B–D: protein detection by Western blot. Retinal lysates (B and C) or microsomes (D) were individually prepared from the four different samples (WT1, WT2, KO1, and KO2), each representing a pooled sample of the NR/RPE or liver from 5 mice, a total of 10 mice per genotype. Black arrow in D corresponds to the molecular weight of HMGCR. E: The quantification of HMGCR expression in D. Results are the mean of the signal intensity of the two WT samples (WT1 and WT2) and two KO (Cyp27a1−/−, KO1 and KO2) samples. Male mice 3 to 6 months of age were used in all experiments.
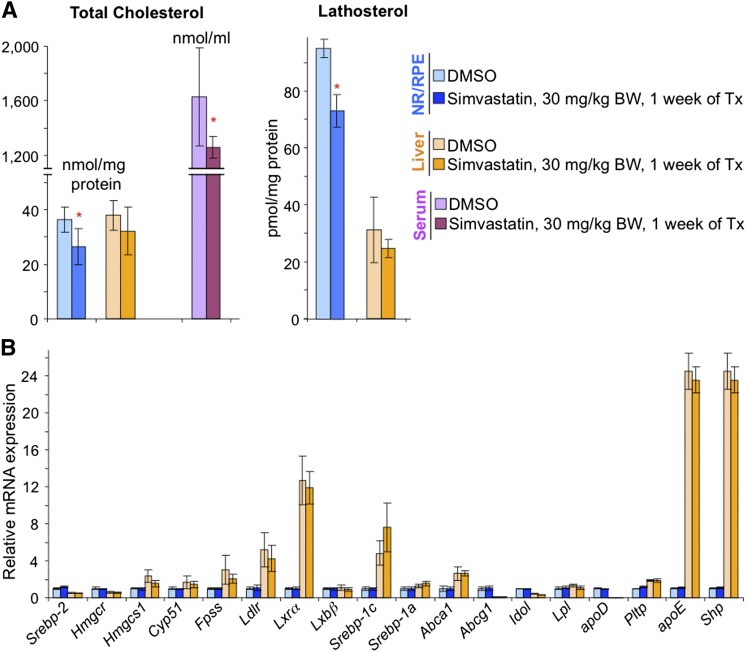
To confirm that a decrease in retinal cholesterol does not lead to changes in retinal expression of cholesterol related-genes as indicated by Cyp27a1−/− mice on the HF/HC/CA diet versus the CA diet, we treated WT mice on a regular chow with a cholesterol-lowering drug simvastatin, which inhibits enzymatic activity of HMGCR. At a daily drug dose of 30 mg/kg body BW and drug administration for 7 days, mice showed a 1.3-fold decrease in serum cholesterol and a 1.4-fold decrease in retinal cholesterol; however, their retinal expression of the tested genes was unchanged (Fig. 7). Hepatic expression of these genes was not affected as well, but unlike the retina, the liver cholesterol was not altered as a result of the simvastatin treatment.
Fig. 7.
Effects of the simvastatin treatment on sterol content (A) and gene expression (B) in mouse retina and liver. Male mice 3 to 6 months of age were euthanized 24 h after the administration of the last drug dose. Bars represent mean ± SD of individual measurements in six mice. The measurements, bar color code, and abbreviations are as in Fig. 4. Red asterisks are significant changes versus control mice that received the vehicle (DMSO). * P < 0.05. Tx, treatment.
Gene expression in mice in response to the TO9 treatment
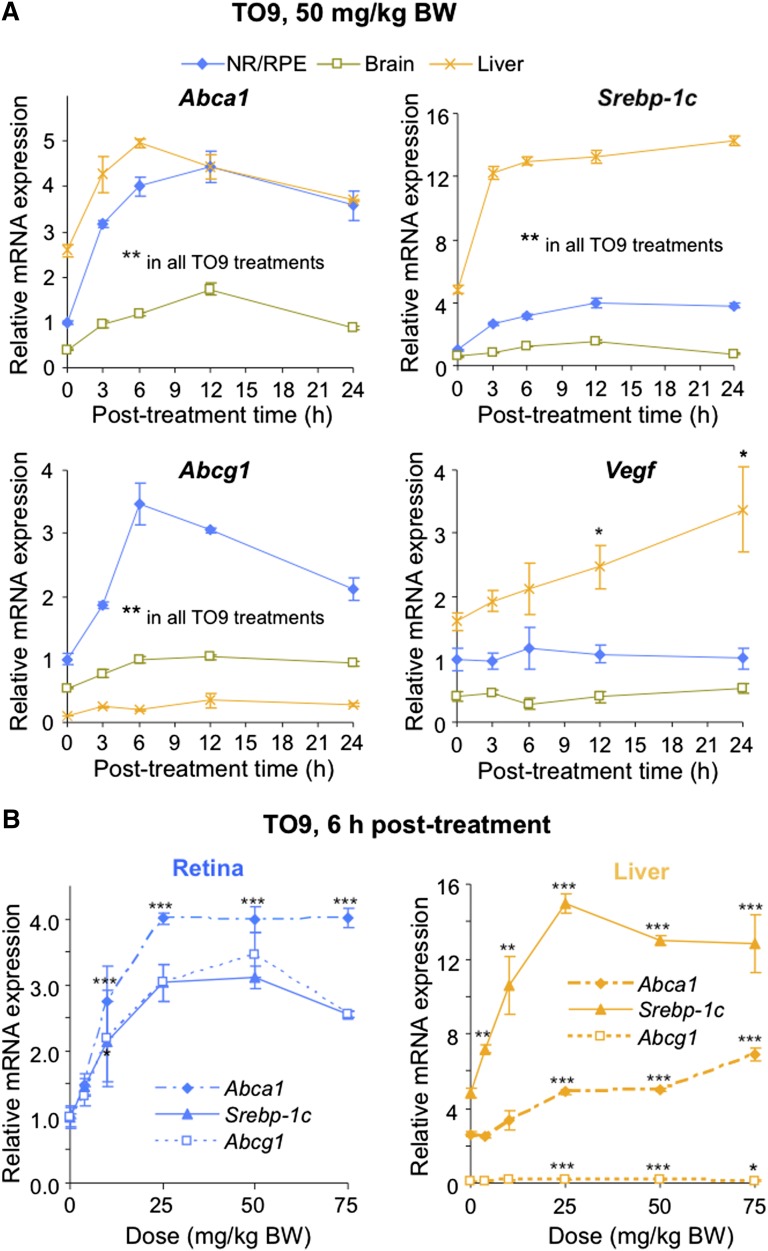
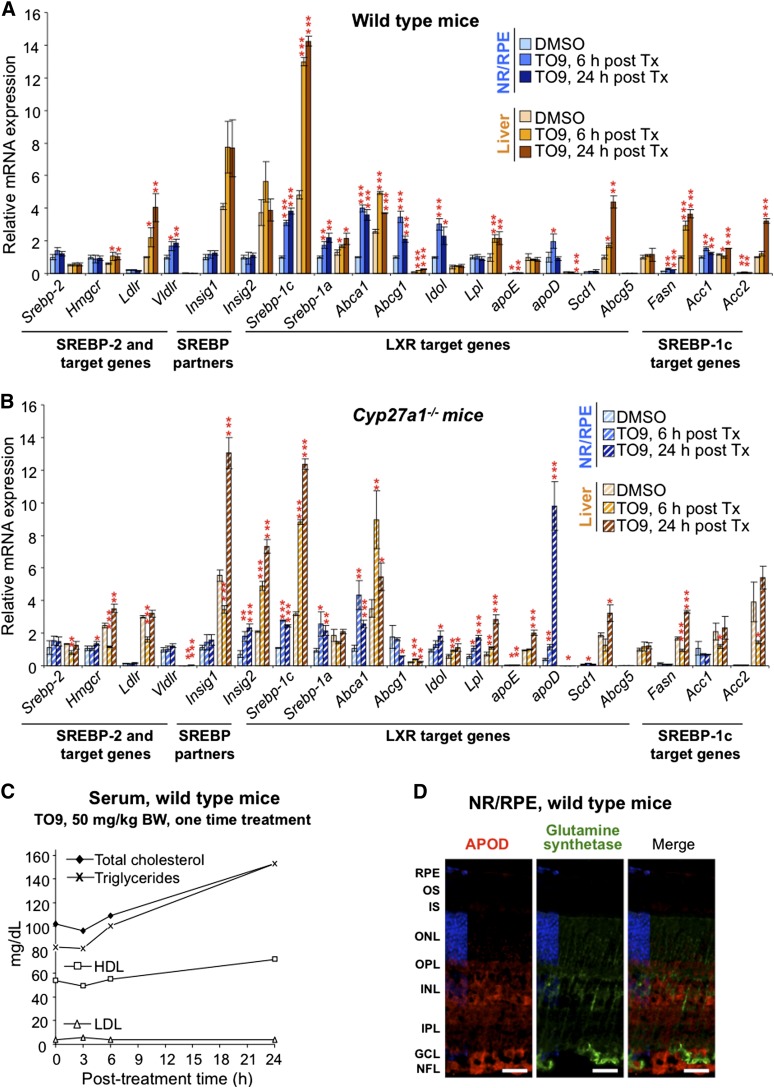
Because the transcription of retinal cholesterol-related genes was not significantly affected by the changes in the levels of retinal cholesterol, we next tested whether the retina is transcriptionally responsive to mouse treatment with TO9, a potent synthetic agonist of LXRs and modulator of the LXR target genes in nonocular tissues (40). Mice were first given by gavage a single dose of TO9 (50 mg/kg BW), and transcriptional response was assessed at different time points (3–24 h posttreatment) in the retina, brain, and liver (Fig. 8A). The retina was the primary organ of investigation, whereas the liver served as a positive control, and the brain as an indicator of the ability of TO9 to cross the barriers separating the brain (and retina) from the systemic circulation. Four LXR target genes were analyzed: three (Abca1, Srebp-1c, and Abcg1) regulated primarily by LXRs and one (Vegf) regulated by multiple transcription factors including LXRs (28, 41). Vegf was selected because of its deleterious contribution to the progression of several eye diseases such as neovascular age-related macular degeneration (AMD) and diabetic retinopathy (42, 43). After the TO9 treatment, we also analyzed mouse serum (Fig. 9C) because hypertriglyceridemia is a known side effect of LXR agonists including TO9 (40).
Fig. 8.
Time course (A) and TO901307 (TO9) dose dependence (B) of retinal and hepatic expression of some of the LXR target genes. Each point represents mean ± SD of individual measurements in five retinas or livers from five male mice 3 to 6 months of age. Black asterisks are significant changes versus control animals that received the vehicle (DMSO). * P < 0.05; ** P < 0.01; *** P < 0.001.
Fig. 9.
Effect of TO9 treatment on gene expression in mouse retina and liver (A and B) and lipid profile in the serum (C). A–C: A single dose of TO9 (50 mg/kg BW) was administered by gavage to male mice 3 to 6 months of age, and animals were euthanized at the indicated time. The bar color code is as in Fig. 4. Bars represent mean ± SD of individual measurements in five retinas or livers from five mice. * P < 0.05; ** P < 0.01; *** P < 0.001 compared with animals that received the vehicle (DMSO). The data on the serum lipid content represent single measurements in a pooled sample from three mice. D: Immunohistochemical localization of APOD (in red) and glutamine synthetase (in green, a marker for Müller cells) in mouse retina. All images are representative of stainings in multiple sections from four male mice 3 to 6 months of age. Scale bars: 15 μm.
A single TO9 dose (50 mg/kg BW) upregulated Abca1, Srebp-1c, and Abcg1 in all studied tissues and during all posttreatment times (Fig. 8A). Vegf was upregulated only in the liver and only 12 h and 24 h posttreatment. The kinetics of the TO9-induced upregulation were gene- and tissue-specific and were either of a parabolic or hyperbolic shape for Abca1, Srebp-1c, and Abcg1 or linear for Vegf. At ∼6 h posttreatment, the upregulation of Abca1, Srebp-1c, and Abcg1 was maximal (or close to maximal), and the elevation of serum cholesterol and triglycerides was modest (<25%). Hence, this time point was selected for the next experiment evaluating gene responses to different doses of TO9 (Fig. 8B). At 6 h posttreatment, retinal upregulation of Abca1, Srebp-1c, and Abcg1 was dose dependent at the 4–25 mg/kg BW TO9 doses, reached the plateau (Abca1 and Srebp-1c) or further increased (Abcg1) at the 25–50 mg/kg BW doses, and leveled off (Abca1) or began to decline (Abcg1 and Srebp-1c) at the 75 mg/kg BW dose. Hepatic upregulation of Abca1, Srebp-1c, and Abcg1 was also dose dependent at the 4–25 mg/kg BW doses, started to decline (Srebp-1c) or plateaued (Spebp-1c and Abcg1) after the 25 mg/kg BW dose, and showed no change (Srebp-1c), an increase (Abca1), or decrease (Abcga1) in gene expression at the 75 mg/kg BW dose.
We then expanded the number of the analyzed genes in WT mice and added gene analysis in Cyp27a1−/− mice (Fig. 9A, B). Animals received a single 50 mg/kg BW TO9 dose and were euthanized 6 h and 24 h posttreatment. The 24 h time point was used to test the genes (Fasn, Acc1, and Acc2) that are regulated by LXRs indirectly through the LXR target SREBP-1c and hence may require a longer time for transcriptional response. In addition, we assessed the key genes of cholesterol input (Srebp-2, Hmgcr, Ldlr, Vldr, Insig1, and Insig 2) that are not regulated by LXRs because serum cholesterol was increased significantly 24 h post TO9 treatment (Fig. 9C). Most of the LXR targets, direct and indirect, were upregulated in the retina and liver of WT mice at least at one postadministration time point. The intertissue differences were only in the expression of Idol upregulated in the retina but not the liver. Conversely, Lpl was upregulated in the liver but not the retina. Retinal expression of the genes of cholesterol input (Srebp-2, Hmgcr, Ldlr, Insig1, and Insig 2) was not altered at either 6 h or 24 h time points with the exception of Vldlr. In contrast, in the liver, some of the genes of cholesterol input were upregulated either slightly (Hmgcr, up to 1.8-fold) or more significantly (Ldlr, up to 4-fold). Because of the role in posttranslational regulation of receptor-mediated cholesterol input (44–46) and upregulation by TO9 in the retina, we attempted to immunolocalize IDOL. Effective anti-IDOL antibodies have not yet been reported (47), and the assessment of new commercial antibodies is a challenge because IDOL is a rare and unstable protein (47). We used the retina from Idol−/− mice (Hong et al., unpublished observations) as an additional control and obtained a staining pattern similar to that in the WT retina, while control sections treated with nonimmune serum showed no signal (not shown). Thus, of all anti-IDOL antibodies that are currently on the market, none are of a good quality as tested by others (47) and us.
Similar to the WT retina, the Cyp27a1−/− retina was also responsive to TO9 treatments (Fig. 9B) but had several differences as compared with the response of the WT retina. The expression of the Srebp-1c targets Fasn, Acc1, and Acc2 was unchanged at both posttreatment times, and the expression of Hmgcr and Lpl was upregulated at least one posttreatment time. Remarkably, there was up to a 25-fold upregulation of apoD encoding a lipoprotein responsible for transport of lipids and other small hydrophobic molecules in several tissues including the brain and liver (48). Little is currently known about APOD in the retina. Therefore, retinal immunolocalization of APOD was carried out (Fig. 9D). The protein was found to be mainly localized in the inner retina and OPL but not in Müller cells spanning these layers as indicated by staining with glutamine synthetase, a marker of Müller cells.
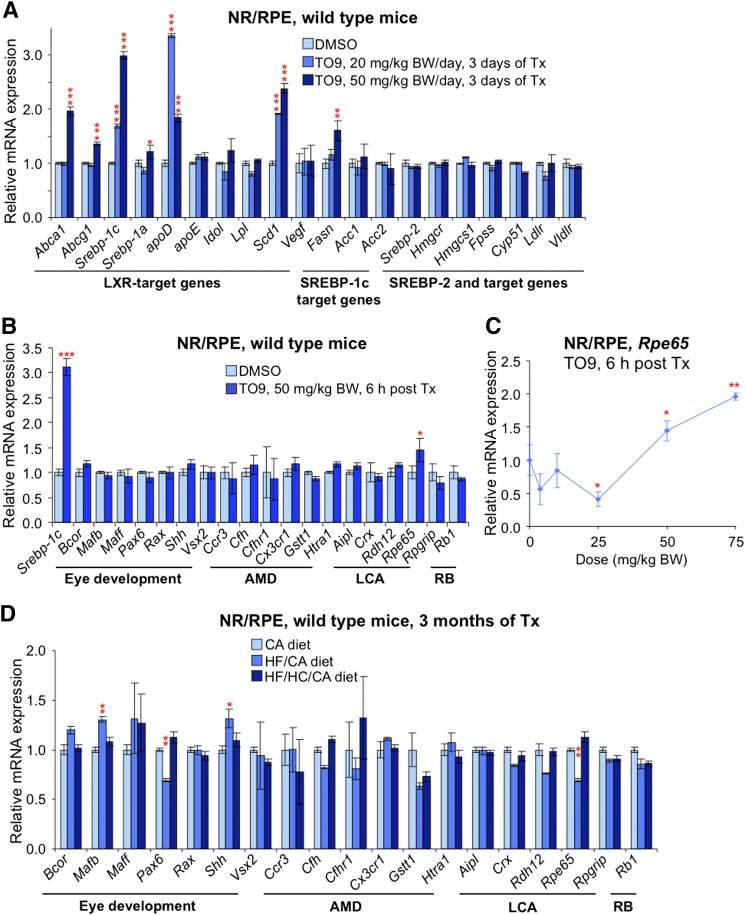
To test whether multiple TO9 treatments lead to stable upregulation of retinal genes, we assessed gene expression after a daily treatment of WT mice either with 20 mg/kg BW or 50 mg/kg BW TO9 dose for 3 consecutive days (Fig. 10A). Six of 10 LXR targets and one of three SREBP-1c targets showed upregulation in this treatment paradigm. Importantly, the expression of Vegf was not upregulated as was the expression of Srebp-2 and target cholesterogenic genes.
Fig. 10.
Effects of TO9 (A–C) and dietary (D) treatments on gene expression in mouse retina. A: A single dose of TO9 (either 20 mg/kg BW or 50 mg/kg BW) was administered by gavage to mice for 3 consecutive days, and animals were euthanized 24 h after the last treatment. B: Onetime TO9 administration. Animals were euthanized 6 h post treatment (Tx). C: Response of retinal Rpe65 expression to varied doses of TO9. Mice were euthanized 6 h after the Tx. D: Effect of diets (see Fig. 4 for details of the dietary treatments) on vision-related genes. Male mice 3 to 6 months of age were used in A–C, and female mice 3 to 6 months of age were used in D. LCA, Leber’s congenital amaurosis; RB, retinoblastoma. All data represent mean ± SD of individual measurements in five retinas from five mice. Red asterisks are significant changes versus control mice that received the vehicle (A–C) or versus mice on the CA-containing diet (D). * P < 0.05; ** P < 0.01; *** P < 0.001.
Studies with TO9 were concluded by the computational analysis of the vision-related genes for the presence in their promoter regions of LXRE, the LXR binding sequence. Such genes were found (supplementary Table III) and evaluated for retinal expression after a onetime administration of TO9 (50 mg/kg BW) to WT mice (Fig. 10B). Only one gene (Rpe65) was upregulated, and this upregulation was confirmed in subsequent studies in TO9 dose dependence (Fig. 10C). Because LXRs are sensors of intracellular cholesterol/oxysterols, the expression of vision-related genes was then assessed in mice on different diets. Consistent with the upregulation by TO9, Rpe65 was among the four genes (Rpe65, Maff, Cfhr1, and Pax6) that showed a trend toward upregulation in response to the HF/HC/CA diet versus the CA diet (Fig. 10D).
DISCUSSION
The present work is a part of the ongoing investigation in this laboratory aimed at the identification of the major mechanisms that control cholesterol levels in the retina, a tissue of unique functions and cell types. These studies are necessary for development of therapeutics for AMD, a complex blinding disease linked, among other risk factors, to disturbances in chorioretinal cholesterol (13). Previously, we studied spatial distribution of the pathways of cholesterol input, output, and homeostatic regulation in human retina (11). Herein we used mice and assessed the sensitivity of these pathways to dietary and pharmacologic treatments. Our most important findings are as follows.
First, the diet-induced changes in the levels of serum cholesterol were larger than the changes in the levels of retinal cholesterol: WT mice on the HF/HC/CA diet versus the CA diet had a 1.4-fold increase in serum cholesterol and only a 1.2-fold increase in retinal cholesterol (Fig. 4A). A possible explanation for this difference is that the blood-retina barrier protects in part the retina from the fluctuations of serum cholesterol. Animal studies, however, do not support this mechanism: when fluorescently labeled lipoprotein particles were injected into the systemic circulation of monkeys and rats, they were taken up by the retina rapidly and avidly (9, 10). A more plausible scenario is that the retina has the means to counteract and diminish the effects of systemic circulation on retinal cholesterol. This scenario is in agreement with human studies showing inconsistent associations between serum cholesterol and the incidence of AMD (49). Conversely, the sterol profile of Cyp27a1−/− mice on the HF/HC/CA diet, which had unchanged serum cholesterol but a 2.4-fold reduction in retinal cholesterol (Fig. 4A), indicates that serum cholesterol could be normal but that retinal cholesterol maintenance is dysregulated.
Second, only lowering serum cholesterol by the simvastatin treatment (a 1.3-fold decrease) led to a similar (1.4-fold) decrease in retinal cholesterol (Fig. 7A). There was also a decrease in the retina in the levels of lathosterol (1.3-fold), a general marker of cholesterol biosynthesis and the activity of HMGCR, the rate-limiting enzyme in this pathway (50–52). Retinal lathosterol, however, has not yet been proved to be a suitable marker of cholesterol biosynthesis and the activity of HMGCR in the retina. Also, it is unclear whether simvastatin crosses the blood-retina barrier as it does the blood-brain barrier (53, 54). Accordingly, a 1.3-fold decrease in retinal lathosterol suggests but does not unambiguously prove that systemic administration of simvastatin directly inhibits retinal HMGCR and retinal biosynthesis of cholesterol. It is also possible that retinal cholesterol is decreased due to a decrease of serum cholesterol without the inhibition of retinal HMGCR. Regardless of the mechanism, the most important outcome of these studies is the demonstration that simavastatin treatment does decrease retinal cholesterol when the drug is delivered through the systemic circulation. Perhaps simvastatin and other lipophilic statins should be considered for the effect on the formation, reduction, or removal of drusen, cholesterol-containing deposits (>40%, v/v) appearing with age under the RPE and being a pathologic hallmark of AMD (55). Combined with the recent investigation showing significant improvements in the ultrastructure and function of the PR/RPE/Bruch’s membrane region in the simvastatin-treated mice fed high-fat atherogenic diet (56), the present work supports the idea of revisiting statins for the treatment or prevention of AMD (13, 56, 57). Testing of statins is also warranted by their cholesterol-independent effects (58, 59), some of which (e.g., improvement of endothelial function, reduction of inflammation and angiogenesis) could be of therapeutic value for AMD (57).
Third, unexpectedly, the modulations of retinal cholesterol achieved by dietary and simvastatin treatments did not affect (with the two exceptions, Ldlr and apoD; Fig. 5A) retinal expression of the key cholesterol-related genes (Figs. 5, 7). This lack of transcriptional responsiveness indicates that either changes in retinal cholesterol were not large enough to activate SREBPs and LXRs or that nontranscriptional mechanisms play a role in the regulation of cholesterol homeostasis in the retina. The latter was investigated and supported by a 12-fold increase in protein, but not mRNA, levels of HMGCR in the Cyp27a1−/− retina versus WT retina (Fig. 6A, D, E). Indeed, the Cyp27a1−/− retina lacks the CYP27A1 metabolite 27-hydroxycholesterol shown to bind to INSIG and accelerate through this binding the degradation of HMGCR (60). Also, the difference in the HMGCR protein levels is larger between the Cyp27a1−/−- WT retina (12.0-fold) and the Cyp27a1−/−- WT liver (2.3-fold) likely reflecting substrate availability for CYP27A1 in the two organs: cholesterol in the retina and mostly bile acid intermediates in the liver (17, 18, 61). Thus, of transcriptional, translational, and posttranslational mechanisms controlling the levels of HMGCR in nonocular organs (38), the oxysterol-dependent HMGCR degradation seems to be of particular importance in the retina. Such a regulation of retinal HMGCR provides an explanation for a paradoxical increase of cholesterol biosynthesis in the Cyp27a1−/− retina and the dysregulation in this line of the retinal cholesterol maintenance (20). Besides the degradation of HMGCR, the retina seems to have another mechanism of posttranslational regulation of cholesterol input. IDOL is an E3 ubiquitin ligase mediating the ubiquitination and degradation of the cell surface receptors for LDL, VLDL, and apoE and leading to rapid receptor elimination from the cell surface (44–46). Idol was not upregulated by TO9 in the liver but showed an increased expression in the retina (Fig. 9A) consistent with previous studies demonstrating that the mechanism governing IDOL responsiveness to LXR is tissue specific and present in the intestine, macrophages, and fat but not the liver (44). The retina appears to be another organ with the active LXR-IDOL axis.
Fourth, in the retina, not only Idol but also 10 other cholesterol-related genes were upregulated by TO9 (Figs. 9A, B and 10AFigs. 9A, B and 10A). These were eight direct LXR targets (Srebp-1c, Srebp-1a, Idol, Abca1, Abcg1, apoE, apoD, and Scd1) and three indirect targets regulated via SREBP-1c (Fasn, Acc1, and Acc2). Only two of these genes (Abca1 and Abcg1) were shown to be responsive to TO9 previously (62, 63). When combined, the data on gene responsiveness to dietary and pharmacologic treatments enable two conclusions. First, of the two major pathways controlling the transcription of cholesterol-related genes in nonocular organs (SREBP and LXR dependent) and operative in the liver, only one (LXR dependent) is of regulatory importance for the retina. Second, to compensate for a weak transcriptional control of cholesterol biosynthesis by the SREBP pathway, the retina seemed to develop strong reliance on posttranslational regulation of cholesterol input, both biosynthesis (HMGCR) and receptor-mediated uptake of cholesterol-containing lipoproteins (IDOL).
LXR agonists were suggested for the treatment of AMD and diabetic retinopathy (62, 63). Our results support this approach by demonstrating that while a number of LXR targets were upregulated by TO9 in the retina, Vegf and cholesterogenic genes were not among these genes (Figs. 8–10). Agonists of LXRs are currently not in clinical use because of their side effects including a marked increase in hepatic and serum triglyceride content (40). Therefore, partial, isoform- (LXRα or LXRβ), tissue-, promoter-, or pathway-specific LXR modulators are in development to address the undesired effects of the pan LXR agonists such as TO9 (64, 65). Our work justifies further investigation of synthetic LXR agonists for retinal diseases and their testing in humans once safer compounds are developed. Of drugs that are already on the market and therefore are available for immediate testing are those that increase the levels of endogenous LXR ligands. For example, the levels of 24-hydroxycholesterol, a high-affinity LXR ligand (66), could be increased pharmacologically (67) by stimulating the activity of CYP46A1, the brain- and retina-specific enzyme, which produces this oxysterol (17, 68, 69).
Fifth, the present work suggested new proteins that should be evaluated for retinal significance. Besides Idol, our study highlighted apoD, the only gene upregulated in the WT retina when mice were fed the HF/HC/CA diet (Fig. 5B) or treated with TO9 (Figs. 9A, 10AFigs. 9A, 10A). ApoD was also the retinal gene showing the highest level of modulation, >10-fold upregulation, upon the TO9 treatment of Cyp27a1−/− mice (Fig. 9B). APOD is a member of the lipocalin superfamily of proteins known to facilitate extra- and intercellular transfer of fatty acids and retinoids (70, 71), and an atypical lipoprotein present in the systemic circulation mainly in the HDL. Unlike other lipoproteins, APOD is expressed not only in the liver and intestine but also in a variety of other tissues including the brain, where it is normally found in the glial cells and the wall of blood vessels. Of importance for the retina could be the finding that systemic ablation of apoE leads to upregulation of APOD in the brain (72, 73). We did not detect APOD expression in Müller cells (Fig. 9D), the major glial cells of the retina and cells that express APOE (74–77). It is conceivable that the function of APOD in the retina is similar to and complements that of APOE. Both apos could be the constituents of the HDL-like lipoprotein particles proposed to circulate in the intraretinal space and facilitate lipid exchange between different cell types (78).
Finally, we found that the expression of Rpe65 catalyzing the rate-limiting step in the visual cycle (79) could be enhanced by TO9 in vivo (Fig. 10B, C). The Rpe65 upregulation, however, required high doses of TO9 (50 and 75 mg/kg BW) suggesting indirect regulation by LXRs. Because LXRs serve as cholesterol/oxysterol sensors, and increased cholesterol content is known to inhibit the activation of rhodopsin, another essential protein in the visual cycle (80, 81), the TO9 upregulation of Rpe65 could be a mechanism protecting the function of rhodopsin in response to the elevation of retinal cholesterol. Indeed, cholesterol inhibits rhodopsin indirectly by ordering the lipid bilayer and limiting the ability of this membrane-bound enzyme to undergo conformational changes in the lipid bilayer (80). Accordingly, when retinal cholesterol homeostasis is disturbed and cholesterol content is elevated throughout the retina including the OS and RPE, increased production of a bulky 11-cis retinal by RPE65 could reduce increased ordering of the lipid bilayer due to the increased cholesterol content and thereby rescue rhodopsin inhibition.
In summary, the evaluation of transcriptional responsiveness of cholesterol- and vision-related genes in mouse retina to modulations of retinal cholesterol provided insights of relevance both clinically and to basic science, identified new proteins of potential importance to retinal cholesterol maintenance, and suggested that cholesterol could regulate the expression of an important protein of the visual cycle.
Supplementary Material
Acknowledgments
The authors thank Dr. P. Tontonoz (University of California, Los Angeles) for providing eyes from Idol−/− mice, Dr. Philip Kiser for helpful discussions, and the Visual Sciences Research Center Core Facility for assistance with mouse breeding (Kathryn Franke), animal genotyping (Dr. Ming-Jin Chang), tissue sectioning (Cathy Doller), and microscopy (Dr. Scott Howell).
Footnotes
Abbreviations:
- AMD
- age-related macular degeneration
- BW
- body weight
- CA
- cholic acid
- CYP
- cytochrome P450
- FXR
- farnesoid X receptor
- GCL
- ganglion cell layer
- HC
- high cholesterol
- HF
- high fat
- IDOL
- inducible degrader of the LDL receptor
- INL
- inner nuclear layer
- HMGCR
- 3-hydroxy-3-methyl-glutaryl-CoA reductase
- INSIG
- insulin-induced gene
- IPL
- the inner plexiform layer
- IS
- inner segments
- LDLR
- LDL receptor
- LXR
- liver X receptor
- NR
- neural retina
- ONL
- outer nuclear layer
- OPL
- outer plexiform layer
- OS
- outer segments
- PR
- photoreceptor
- qRT-PCR
- quantitative real-time PCR
- RPE
- retinal pigment epithelium
- SCAP
- SREBP cleavage-activating protein
- SREBP
- sterol-regulatory element binding protein
- TO9
- TO901317
This work was supported in part by National Institutes of Health Grants EY018383 (I.A.P.) and Core grant EY11373, funds from the Ohio Lions Eye Research Foundation, and unrestricted grants from Research to Prevent Blindness. I.A.P. is a recipient of the Jules and Doris Stein Professorship from the Research to Prevent Blindness Foundation.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
REFERENCES
- 1.Brown M. S., Goldstein J. L. 2009. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J. Lipid Res. 50 (Suppl.): S15–S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Repa J. J., Mangelsdorf D. J. 2000. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu. Rev. Cell Dev. Biol. 16: 459–481. [DOI] [PubMed] [Google Scholar]
- 4.Yang C., McDonald J. G., Patel A., Zhang Y., Umetani M., Xu F., Westover E. J., Covey D. F., Mangelsdorf D. J., Cohen J. C., et al. 2006. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 281: 27816–27826. [DOI] [PubMed] [Google Scholar]
- 5.Spann N. J., Garmire L. X., McDonald J. G., Myers D. S., Milne S. B., Shibata N., Reichart D., Fox J. N., Shaked I., Heudobler D., et al. 2012. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 151: 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang N., Koutz C. A., Anderson R. E. 1993. A method for the isolation of retinal pigment epithelial cells from adult rats. Invest. Ophthalmol. Vis. Sci. 34: 101–107. [PubMed] [Google Scholar]
- 7.Fliesler S. J., Florman R., Rapp L. M., Pittler S. J., Keller R. K. 1993. In vivo biosynthesis of cholesterol in the rat retina. FEBS Lett. 335: 234–238. [DOI] [PubMed] [Google Scholar]
- 8.Fliesler S. J., Keller R. K. 1995. Metabolism of [3H]farnesol to cholesterol and cholesterogenic intermediates in the living rat eye. Biochem. Biophys. Res. Commun. 210: 695–702. [DOI] [PubMed] [Google Scholar]
- 9.Elner V. M. 2002. Retinal pigment epithelial acid lipase activity and lipoprotein receptors: effects of dietary omega-3 fatty acids. Trans. Am. Ophthalmol. Soc. 100: 301–338. [PMC free article] [PubMed] [Google Scholar]
- 10.Tserentsoodol N., Sztein J., Campos M., Gordiyenko N. V., Fariss R. N., Lee J. W., Fliesler S. J., Rodriguez I. R. 2006. Uptake of cholesterol by the retina occurs primarily via a low density lipoprotein receptor-mediated process. Mol. Vis. 12: 1306–1318. [PubMed] [Google Scholar]
- 11.Zheng W., Reem R. E., Omarova S., Huang S., DiPatre P. L., Charvet C. D., Curcio C. A., Pikuleva I. A. 2012. Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS ONE. 7: e37926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., Jia C., Kazmierkiewicz K. L., Bowman A. S., Tian L., Liu Y., Gupta N. A., Gudiseva H. V., Yee S. S., Kim M., et al. 2014. Comprehensive analysis of gene expression in human retina and supporting tissues. Hum. Mol. Genet. 23: 4001–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pikuleva I. A., Curcio C. A. 2014. Cholesterol in the retina: the best is yet to come. Prog. Retin. Eye Res. 41: 64–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fliesler S. J., Bretillon L. 2010. The ins and outs of cholesterol in the vertebrate retina. J. Lipid Res. 51: 3399–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H., Zheng S., Qiu Y., Yang Y., Wang C., Yang P., Li Q., Lei B. 2014. Activation of liver X receptor alleviates ocular inflammation in experimental autoimmune uveitis. Invest. Ophthalmol. Vis. Sci. 55: 2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrac S., Lear S. R., Ananthanarayanan M., Balasubramaniyan N., Bollineni J., Shefer S., Hyogo H., Cohen D. E., Blanche P. J., Krauss R. M., et al. 2005. Role of CYP27A in cholesterol and bile acid metabolism. J. Lipid Res. 46: 76–85. [DOI] [PubMed] [Google Scholar]
- 17.Liao W-L., Heo G-Y., Dodder N. G., Reem R. E., Mast N., Huang S., DiPatre P. L., Turko I. V., Pikuleva I. A. 2011. Quantification of cholesterol-metabolizing P450s CYP27A1 and CYP46A1 in neural tissues reveals a lack of enzyme-product correlations in human retina but not human brain. J. Proteome Res. 10: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mast N., Reem R., Bederman I., Huang S., DiPatre P. L., Bjorkhem I., Pikuleva I. A. 2011. Cholestenoic acid is an important elimination product of cholesterol in the retina: comparison of retinal cholesterol metabolism with that in the brain. Invest. Ophthalmol. Vis. Sci. 52: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M., Heo G. Y., Omarova S., Pikuleva I. A., Turko I. V. 2012. Sample prefractionation for mass spectrometry quantification of low-abundance membrane proteins. Anal. Chem. 84: 5186–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omarova S., Charvet C. D., Reem R. E., Mast N., Zheng W., Huang S., Peachey N. S., Pikuleva I. A. 2012. Abnormal vascularization in mouse retina with dysregulated retinal cholesterol homeostasis. J. Clin. Invest. 122: 3012–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamel C. P., Tsilou E., Pfeffer B. A., Hooks J. J., Detrick B., Redmond T. M. 1993. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 268: 15751–15757. [PubMed] [Google Scholar]
- 22.Jin M., Li S., Moghrabi W. N., Sun H., Travis G. H. 2005. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 122: 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moiseyev G., Chen Y., Takahashi Y., Wu B. X., Ma J. X. 2005. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad. Sci. USA. 102: 12413–12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redmond T. M., Poliakov E., Yu S., Tsai J. Y., Lu Z., Gentleman S. 2005. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. USA. 102: 13658–13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen H., Reshef A., Maeda N., Lippoldt A., Shpizen S., Triger L., Eggertsen G., Bjorkhem I., Leitersdorf E. 1998. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J. Biol. Chem. 273: 14805–14812. [DOI] [PubMed] [Google Scholar]
- 26.Repa J. J., Lund E. G., Horton J. D., Leitersdorf E., Russell D. W., Dietschy J. M., Turley S. D. 2000. Disruption of the sterol 27-hydroxylase gene in mice results in hepatomegaly and hypertriglyceridemia. Reversal by cholic acid feeding. J. Biol. Chem. 275: 39685–39692. [DOI] [PubMed] [Google Scholar]
- 27.Båvner A., Shafaati M., Hansson M., Olin M., Shpitzen S., Meiner V., Leitersdorf E., Björkhem I. 2010. On the mechanism of accumulation of cholestanol in the brain of mice with a disruption of sterol 27-hydroxylase. J. Lipid Res. 51: 2722–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalaany N. Y., Mangelsdorf D. J. 2006. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 68: 159–191. [DOI] [PubMed] [Google Scholar]
- 29.Mak P. A., Kast-Woelbern H. R., Anisfeld A. M., Edwards P. A. 2002. Identification of PLTP as an LXR target gene and apoE as an FXR target gene reveals overlapping targets for the two nuclear receptors. J. Lipid Res. 43: 2037–2041. [DOI] [PubMed] [Google Scholar]
- 30.Hoeke M. O., Heegsma J., Hoekstra M., Moshage H., Faber K. N. 2014. Human FXR regulates SHP expression through direct binding to an LRH-1 binding site, independent of an IR-1 and LRH-1. PLoS ONE. 9: e88011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saadane A., Mast N., Charvet C., Omarova S., Zheng W., Huang S. S., Kern T. S., Peachey N. S., Pikuleva I. A. 2014. Retinal and non-ocular abnormalities in Cyp27a1−/− Cyp64a1−/− mice with dysfunctional metabolism of cholesterol. Am. J. Pathol. 184: 2403–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mast N., Linger M., Clark M., Wiseman J., Stout C. D., Pikuleva I. A. 2012. In silico and intuitive predictions of CYP46A1 inhibition by marketed drugs with subsequent enzyme crystallization in complex with fluvoxamine. Mol. Pharmacol. 82: 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heo G. Y., Liao W. L., Turko I. V., Pikuleva I. A. 2012. Features of the retinal environment which affect the activities and product profile of cholesterol-metabolizing cytochromes P450 CYP27A1 and CYP11A1. Arch. Biochem. Biophys. 518: 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 21: 2933–2942. [DOI] [PubMed] [Google Scholar]
- 35.Duncan K. G., Hosseini K., Bailey K. R., Yang H., Lowe R. J., Matthes M. T., Kane J. P., LaVail M. M., Schwartz D. M., Duncan J. L. 2009. Expression of reverse cholesterol transport proteins ATP-binding cassette A1 (ABCA1) and scavenger receptor BI (SR-BI) in the retina and retinal pigment epithelium. Br. J. Ophthalmol. 93: 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfrieger F. W., Ungerer N. 2011. Cholesterol metabolism in neurons and astrocytes. Prog. Lipid Res. 50: 357–371. [DOI] [PubMed] [Google Scholar]
- 37.DeBose-Boyd R. A. 2008. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 18: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo Y., Debose-Boyd R. A. 2010. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit. Rev. Biochem. Mol. Biol. 45: 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burg J. S., Espenshade P. J. 2011. Regulation of HMG-CoA reductase in mammals and yeast. Prog. Lipid Res. 50: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walczak R., Joseph S. B., Laffitte B. A., Castrillo A., Pei L., Tontonoz P. 2004. Transcription of the vascular endothelial growth factor gene in macrophages is regulated by liver X receptors. J. Biol. Chem. 279: 9905–9911. [DOI] [PubMed] [Google Scholar]
- 42.Jager R. D., Mieler W. F., Miller J. W. 2008. Age-related macular degeneration. N. Engl. J. Med. 358: 2606–2617. [DOI] [PubMed] [Google Scholar]
- 43.Tang J., Kern T. S. 2011. Inflammation in diabetic retinopathy. Prog. Retin. Eye Res. 30: 343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelcer N., Hong C., Boyadjian R., Tontonoz P. 2009. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 325: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong C., Duit S., Jalonen P., Out R., Scheer L., Sorrentino V., Boyadjian R., Rodenburg K. W., Foley E., Korhonen L., et al. 2010. The E3 ubiquitin ligase IDOL induces the degradation of the low density lipoprotein receptor family members VLDLR and ApoER2. J. Biol. Chem. 285: 19720–19726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L., Reue K., Fong L. G., Young S. G., Tontonoz P. 2012. Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis. Arterioscler. Thromb. Vasc. Biol. 32: 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scotti E., Calamai M., Goulbourne C. N., Zhang L., Hong C., Lin R. R., Choi J., Pilch P. F., Fong L. G., Zou P., et al. 2013. IDOL stimulates clathrin-independent endocytosis and multivesicular body-mediated lysosomal degradation of the low-density lipoprotein receptor. Mol. Cell. Biol. 33: 1503–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perdomo G., Henry Dong H. 2009. Apolipoprotein D in lipid metabolism and its functional implication in atherosclerosis and aging. Aging (Albany NY). 1: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakravarthy U., Wong T. Y., Fletcher A., Piault E., Evans C., Zlateva G., Buggage R., Pleil A., Mitchell P. 2010. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 10: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Björkhem I., Miettinen T., Reihnér E., Ewerth S., Angelin B., Einarsson K. 1987. Correlation between serum levels of some cholesterol precursors and activity of HMG-CoA reductase in human liver. J. Lipid Res. 28: 1137–1143. [PubMed] [Google Scholar]
- 51.Kempen H. J., Glatz J. F., Gevers Leuven J. A., van der Voort H. A., Katan M. B. 1988. Serum lathosterol concentration is an indicator of whole-body cholesterol synthesis in humans. J. Lipid Res. 29: 1149–1155. [PubMed] [Google Scholar]
- 52.Lund E., Sisfontes L., Reihner E., Bjorkhem I. 1989. Determination of serum levels of unesterified lathosterol by isotope dilution-mass spectrometry. Scand. J. Clin. Lab. Invest. 49: 165–171. [DOI] [PubMed] [Google Scholar]
- 53.Sierra S., Ramos M. C., Molina P., Esteo C., Vazquez J. A., Burgos J. S. 2011. Statins as neuroprotectants: a comparative in vitro study of lipophilicity, blood-brain-barrier penetration, lowering of brain cholesterol, and decrease of neuron cell death. J. Alzheimers Dis. 23: 307–318. [DOI] [PubMed] [Google Scholar]
- 54.Evans B. A., Evans J. E., Baker S. P., Kane K., Swearer J., Hinerfeld D., Caselli R., Rogaeva E., St George-Hyslop P., Moonis M., et al. 2009. Long-term statin therapy and CSF cholesterol levels: implications for Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 27: 519–524. [DOI] [PubMed] [Google Scholar]
- 55.Curcio C. A., Johnson M., Rudolf M., Huang J. D. 2011. The oil spill in ageing Bruch membrane. Br. J. Ophthalmol. 95: 1638–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barathi V. A., Yeo S. W., Guymer R. H., Wong T. Y., Luu C. D. 2014. Effects of simvastatin on retinal structure and function of a high-fat atherogenic mouse model of thickened Bruch’s membrane. Invest. Ophthalmol. Vis. Sci. 55: 460–468. [DOI] [PubMed] [Google Scholar]
- 57.Guymer R. H., Baird P. N., Varsamidis M., Busija L., Dimitrov P. N., Aung K. Z., Makeyeva G. A., Richardson A. J., Lim L., Robman L. D. 2013. Proof of concept, randomized, placebo-controlled study of the effect of simvastatin on the course of age-related macular degeneration. PLoS ONE. 8: e83759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao J. K., Laufs U. 2005. Pleiotropic effects of statins. Annu. Rev. Pharmacol. Toxicol. 45: 89–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calabrò P., Yeh E. T. 2005. The pleiotropic effects of statins. Curr. Opin. Cardiol. 20: 541–546. [DOI] [PubMed] [Google Scholar]
- 60.Song B. L., DeBose-Boyd R. A. 2004. Ubiquitination of 3-hydroxy-3-methylglutaryl-CoA reductase in permeabilized cells mediated by cytosolic E1 and a putative membrane-bound ubiquitin ligase. J. Biol. Chem. 279: 28798–28806. [DOI] [PubMed] [Google Scholar]
- 61.Wikvall K. 1984. Hydroxylations in biosynthesis of bile acids. Isolation of a cytochrome P-450 from rabbit liver mitochondria catalyzing 26-hydroxylation of C27-steroids. J. Biol. Chem. 259: 3800–3804. [PubMed] [Google Scholar]
- 62.Sene A., Khan A. A., Cox D., Nakamura R. E., Santeford A., Kim B. M., Sidhu R., Onken M. D., Harbour J. W., Hagbi-Levi S., et al. 2013. Impaired cholesterol efflux in senescent macrophages promotes age-related macular degeneration. Cell Metab. 17: 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hazra S., Rasheed A., Bhatwadekar A., Wang X., Shaw L. C., Patel M., Caballero S., Magomedova L., Solis N., Yan Y., et al. 2012. Liver X receptor modulates diabetic retinopathy outcome in a mouse model of streptozotocin-induced diabetes. Diabetes. 61: 3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X., Yeh V., Molteni V. 2010. Liver X receptor modulators: a review of recently patented compounds (2007–2009). Expert Opin. Ther. Pat. 20: 535–562. [DOI] [PubMed] [Google Scholar]
- 65.Jakobsson T., Treuter E., Gustafsson J. A., Steffensen K. R. 2012. Liver X receptor biology and pharmacology: new pathways, challenges and opportunities. Trends Pharmacol. Sci. 33: 394–404. [DOI] [PubMed] [Google Scholar]
- 66.Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. 1999. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. USA. 96: 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mast N., Li Y., Linger M., Clark M., Wiseman J., Pikuleva I. A. 2014. Pharmacologic stimulation of cytochrome P450 46A1 and cerebral cholesterol turnover in mice. J. Biol. Chem. 289: 3529–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lund E. G., Guileyardo J. M., Russell D. W. 1999. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc. Natl. Acad. Sci. USA. 96: 7238–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bretillon L., Diczfalusy U., Bjorkhem I., Maire M. A., Martine L., Joffre C., Acar N., Bron A., Creuzot-Garcher C. 2007. Cholesterol-24S-hydroxylase (CYP46A1) is specifically expressed in neurons of the neural retina. Curr. Eye Res. 32: 361–366. [DOI] [PubMed] [Google Scholar]
- 70.Skerra A. 2000. Lipocalins as a scaffold. Biochim. Biophys. Acta. 1482: 337–350. [DOI] [PubMed] [Google Scholar]
- 71.Eichinger A., Nasreen A., Kim H. J., Skerra A. 2007. Structural insight into the dual ligand specificity and mode of high density lipoprotein association of apolipoprotein D. J. Biol. Chem. 282: 31068–31075. [DOI] [PubMed] [Google Scholar]
- 72.Hu C. Y., Ong W. Y., Sundaram R. K., Chan C., Patel S. C. 2001. Immunocytochemical localization of apolipoprotein D in oligodendrocyte precursor-like cells, perivascular cells, and pericytes in the human cerebral cortex. J. Neurocytol. 30: 209–218. [DOI] [PubMed] [Google Scholar]
- 73.Jansen P. J., Lutjohann D., Thelen K. M., von Bergmann K., van Leuven F., Ramaekers F. C., Monique M. 2009. Absence of ApoE upregulates murine brain ApoD and ABCA1 levels, but does not affect brain sterol levels, while human ApoE3 and human ApoE4 upregulate brain cholesterol precursor levels. J. Alzheimers Dis. 18: 319–329. [DOI] [PubMed] [Google Scholar]
- 74.Amaratunga A., Abraham C. R., Edwards R. B., Sandell J. H., Schreiber B. M., Fine R. E. 1996. Apolipoprotein E is synthesized in the retina by Muller glial cells, secreted into the vitreous, and rapidly transported into the optic nerve by retinal ganglion cells. J. Biol. Chem. 271: 5628–5632. [DOI] [PubMed] [Google Scholar]
- 75.Kuhrt H., Hartig W., Grimm D., Faude F., Kasper M., Reichenbach A. 1997. Changes in CD44 and ApoE immunoreactivities due to retinal pathology of man and rat. J. Hirnforsch. 38: 223–229. [PubMed] [Google Scholar]
- 76.Shanmugaratnam J., Berg E., Kimerer L., Johnson R. J., Amaratunga A., Schreiber B. M., Fine R. E. 1997. Retinal Muller glia secrete apolipoproteins E and J which are efficiently assembled into lipoprotein particles. Brain Res. Mol. Brain Res. 50: 113–120. [DOI] [PubMed] [Google Scholar]
- 77.Anderson D. H., Ozaki S., Nealon M., Neitz J., Mullins R. F., Hageman G. S., Johnson L. V. 2001. Local cellular sources of apolipoprotein E in the human retina and retinal pigmented epithelium: implications for the process of drusen formation. Am. J. Ophthalmol. 131: 767–781. [DOI] [PubMed] [Google Scholar]
- 78.Tserentsoodol N., Gordiyenko N. V., Pascual I., Lee J. W., Fliesler S. J., Rodriguez I. R. 2006. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol. Vis. 12: 1319–1333. [PubMed] [Google Scholar]
- 79.Kiser P. D., Palczewski K. 2010. Membrane-binding and enzymatic properties of RPE65. Prog. Retin. Eye Res. 29: 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albert A. D., Boesze-Battaglia K. 2005. The role of cholesterol in rod outer segment membranes. Prog. Lipid Res. 44: 99–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park P. S. 2014. Constitutively active rhodopsin and retinal disease. Adv. Pharmacol. 70: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.