Abstract
Epithelial cells play an important role in host defense as sentinels for invading microbial pathogens. Chlamydia trachomatis is an intracellular bacterial pathogen that replicates in reproductive tract epithelium. Epithelial cells lining the reproductive tract likely play a key role in triggering inflammation and adaptive immunity during Chlamydia infections. For this report a murine oviduct epithelial cell line was derived in order to determine how epithelial cells influence innate and adaptive immune responses during Chlamydia infections. As expected, oviduct epithelial cells infected by Chlamydia muridarum produced a broad spectrum of chemokines, including CXCL16, and regulators of the acute-phase response, including interleukin-1α (IL-1α), IL-6, and tumor necrosis factor alpha. In addition, infected epithelial cells expressed cytokines that augment gamma interferon (IFN) production, including IFN-α/β and IL-12-p70. To my knowledge this is the first report of a non-myeloid/lymphoid cell type making IL-12-p70 in response to an infection. Equally interesting, infected epithelial cells significantly upregulated transforming growth factor alpha precursor expression, suggesting a mechanism by which they might play a direct role in the pathological scarring seen as a consequence of Chlamydia infections. Data from these in vitro studies predict that infected oviduct epithelium contributes significantly to host innate and adaptive defenses but may also participate in the immunopathology seen with Chlamydia infections.
Chlamydia trachomatis serovars D to K, referred to as the urogenital serovars, are sexually transmitted intracellular bacterial pathogens that cause acute urethritis, epididymitis, and pelvic inflammatory disease. C. trachomatis serovars A to C cause an eye infection called trachoma. Serovars L1 to L3 cause lymphogranuloma venereum (LGV), a disseminating sexually transmitted disease that begins with a primary small ulcerative lesion and progresses to fever, regional lymphadenopathy, and genital ulceration and/or lymphatic obstruction in later stages. Trachoma and LGV are uncommon in the developed world, while the urogenital serovars D to K are ubiquitous. With the exception of neonatal pneumonia and eye infections, urogenital serovars of C. trachomatis replicate exclusively in the epithelium lining the reproductive tract. The infectious particle is a metabolically inactive intermediate known as an elementary body. Elementary bodies deposited onto reproductive tract mucosa are endocytosed by epithelial cells, become metabolically active, and replicate in modified vacuoles, termed inclusions. The replicating form of C. trachomatis is a noninfectious form known as the reticulate body. As the developmental cycle progresses, progeny Chlamydia condense into infectious elementary bodies that are released by exocytosis or host cell lysis. Urogenital strains release apically, infecting adjacent mucosal epithelial cells, while disseminating serovars release basolaterally, penetrating beyond the mucosal epithelium (71).
During C. trachomatis infections with the urogenital serovars, progression of the infection into the upper female reproductive tract causes marked inflammation of the Fallopian tubes (47). Clinical data suggests that C. trachomatis evades immune-mediated elimination to cause persistent infections in some individuals (4, 11, 45, 54, 57, 64, 68). In women, chronic or recurrent C. trachomatis urogenital infections scar the Fallopian tubes, resulting in infertility and ectopic pregnancies, the two clinically important sequelae of infection (12, 13).
Epithelial cells lining mucosal surfaces serve as sentinels for invasion by microbial pathogens through the release of inflammatory cytokines (reviewed in reference 30). Because epithelial cells are the only cell type productively infected by C. trachomatis, they are the logical epicenter for the local inflammatory response. An increasing body of clinical data supports the existence of chronic Chlamydia infections and raises the possibility that infected epithelial cells directly contribute to Fallopian tube scarring, infertility, and ectopic pregnancies (reviewed in reference 65).
Studies of epithelial cell responses to C. trachomatis infection have utilized human epithelial tumors. HeLa cervical carcinoma cells infected with LGV secreted interleukin-1α (IL-1α), IL-6, IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF), and Groα (CXCL1) (58). Polarized HEC-1-B endometrial adenocarcinoma cells infected with C. trachomatis urogenital serovar E upregulated mRNA for neutrophil chemokines ENA-78 (CXCL5) and GCP-2 (CXCL6) (72). Microarray analysis of serovar D in nonpolarized HeLa cells showed upregulation of 18 genes. Of the 18, 4 were cytokines: IL-8, IL-11, leukemia inhibitory factor, and macrophage inhibitory protein 2α (MIP-2α; CXCL2) (25). Similarly, analysis of C. trachomatis serovar E-infected polarized HeLa cells showed upregulation of IL-11 mRNA and secretion of IL-6, IL-8, and IL-11 (20). HeLa cells infected by LGV, urogenital serovars, and C. muridarum secreted preformed IL-18 via a posttranslational mechanism dependent on caspase-1 (39). Recently, the effects of Chlamydia infection on HeLa cells were investigated by using high-density oligonucleotide microarrays (73). Nonpolarized HeLa cells infected with LGV altered the transcription of 130 genes on a 15,000-gene microarray. Upregulated cytokine transcription was seen for IL-1β, IL-6, and IL-16 (73). Analysis of the response of primary epithelial cells to Chlamydia infection is very limited (32). No studies to date have examined the response of oviduct epithelial cells to C. trachomatis infection.
Previous studies utilizing human epithelial tumors have identified a cadre of inflammatory cytokines induced by C. trachomatis infection. However, the existing data set was notable for the absence of some expected key inflammatory mediators, such as tumor necrosis factor alpha (TNF-α) (3), IP-10 gamma interferon (IFN-γ)-inducible 10-kDa protein (CXCL10), and RANTES (CCL5) (44). This suggested that existing in vitro models based on epithelial tumor cell lines might not fully mimic the immunobiology of Chlamydia-infected epithelium. To address this possibility, a nontransformed primary murine oviduct epithelial cell line was derived for experiments with C. muridarum, a murine pathogen closely related to C. trachomatis that is often used as an experimental surrogate for human infections (5, 14, 35). The cytokine data from those experiments, presented here, are consistent with the histopathology and cytokine polarization seen during natural infections with urogenital Chlamydia serovars.
MATERIALS AND METHODS
Mice.
Female B6.C-H2bm1/ByJ mice were purchased from The Jackson Laboratory (Bar Harbor, Maine) and were housed in Indiana University Purdue University—Indianapolis specific-pathogen-free facilities. The Institutional Animal Care and Utilization Committee approved all experimental protocols.
Cell lines.
McCoy mouse fibroblasts were generously provided by Barbara Van der Pol (Indiana University School of Medicine, Indianapolis). Murine B-cell lymphoma A20 (TIB-208) and normal mouse fibroblast CL.7 (TIB-80) were obtained from the American Type Culture Collection (Manassas, Va.). A murine oviduct cell line designated Bm1.11 was generated by resection of the proximal two-thirds of the oviducts, excluding the ovaries, from a female B6.C-H2bm1/ByJ mouse. Oviduct epithelial cells were released from oviduct lumens by canulating the lumens with a 0.5-ml syringe and insufflating them with pancreatin-hyaluronidase-collagenase cocktail (69) and then rinsing the lumens with the same cocktail after 15 min at 37°C. Released cells were recovered and grown in 1:1 Dulbecco's modified Eagle medium:F12K (Sigma Chemical Co., St. Louis, Mo.) supplemented with 10% characterized fetal bovine serum (HyClone, Logan, Utah), 2 mM l-alanyl-l-glutamine (Glutamax I; Gibco/Invitrogen, Carlsbad, Calif.), 5 μg of bovine insulin/ml, and 12.5 ng of recombinant human FGF-7 (KGF; Sigma Chemical Co.)/ml. Ciprofloxacin (10 μg/ml), gentamicin (25 μg/ml) (Sigma Chemical Co.), and 10 mM HEPES (Biowhittaker, Walkersville, Md.) were included in the culture medium for the first 4 weeks after explantation. Once a stable line was established, cloned cell lines were generated by dispersion at low density. Individual colonies were picked by using cloning rings. Resulting cell lines were screened for morphology, expression of cytokeratins, and IFN-γ-inducible expression of I-Ab. A cloned cell line designated Bm1.11 had an epithelial morphology, expressed cytokeratins, and had IFN-γ-inducible major histocompatibility complex (MHC) class II expression. The Bm1.11 cell line, maintained as described above, showed no evidence for senescence after more than 100 passages. All cell lines tested negative for mycoplasma contamination by either susceptibility to 6-methylpurine deoxyriboside (MycoTect kit; Invitrogen Corp.) or PCR (Mycoplasma detection kit; American Type Culture Collection).
Chlamydia.
C. muridarum, previously known as C. trachomatis strain MoPn, was generously provided by Barbara Van der Pol and was grown in McCoy cells. A mycoplasma-free stock was generated by exposure of existing stocks to 1% Triton X-100 as previously described (53), followed by detergent removal with Extracti-Gel D beads (Pierce Chemical, Rockford, Ill.) and limiting dilution on McCoy monolayers. Wells with visible cytopathic effect at the highest dilution (calculated to be ∼800 inclusion-forming units [IFU] prior to detergent treatment) after 5 days were harvested and expanded. Treated stocks were tested for the presence of mycoplasma by exposing nonpermissive A20 B cells to Chlamydia stocks, passing the A20 cells for more than 2 weeks in the absence of antibiotics, and then testing for A20 susceptibility to 6-methylpurine deoxyriboside (MycoTect kit; Invitrogen Corporation). The titers of mycoplasma-free C. muridarum stocks were determined on McCoy cells with centrifugation as previously described (61).
Infections.
Bm1.11 cells were plated in 6-well tissue culture plates and were used when confluent to minimize changes in cell number over the time course of individual experiments. For all experiments, Bm1.11 cells were infected with 10 IFU of C. muridarum/cell in 2 ml of culture medium without centrifugation or subsequent change of medium. Mock-infected wells (0 h) received an inoculum-equivalent volume of sucrose-phosphate-glutamic acid buffer (SPG buffer) but lacked C. muridarum in the medium.
Immunohistochemistry.
Bm1.11 cells grown on glass chamber slides (Labtek, Naperville, Ill.) were fixed for 2 min at room temperature with 1:1 methanol-acetone, blocked with 5% normal goat serum-phosphate-buffered saline for 45 min, and then stained with 36-7-5, a monoclonal antibody specific for H-2Kk (an epitope not present on Bm1.11 cells) (BD Pharmingen, San Diego, Calif.) or monoclonal antibody cocktail AE1/AE3 specific for acidic and basic cytokeratins (Cappel/ICN, Irvine, Calif.). Detection was accomplished with a fluorescein isothiocyanate-coupled goat anti-mouse immunoglobulin G F(ab′)2 affinity-purified antiserum (Cappel/ICN).
Flow cytometry.
Bm1.11 cells were dislodged from tissue culture plastic by using a Hank's salt-based enzyme-free cell dissociation buffer (Gibco/Invitrogen). Cells were fixed with StabilCyte (Ebio, St. Paul, Minn.) and then were washed and stained for 20 min on ice in phosphate-buffered saline-2% bovine serum albumin with phycoerythrin-coupled M5/114.15.2 (I-Ab) (Ebioscience, San Diego, Calif.). IFN-γ induction was accomplished by addition of 10 ng of murine recombinant IFN-γ (Sigma Chemical Co.)/ml to the culture medium for 14 h prior to fixation and staining. Cells were analyzed with a FACS Calibur cytometer (BD Biosciences, San Diego, Calif.).
RPA.
Standard template sets mCK-1b, mCK-3b, and mCK-5c were purchased from a commercial vendor. In addition, a custom template was designed and purchased that included probe templates for IL-12-p35, IL-12-p40, IL-1β, IL-11, IL-7, IL-1α, transforming growth factor α (TGFα), and IFNα-4. 32P-multiprobe RNase protection assays (RPA) were done with a commercial kit according to the manufacturer's protocol (Riboquant; BD Pharmingen). In all experiments 2 μg of yeast tRNA served as the control for hybridization; L32 and GAPDH probes served as loading and RNA quality controls. Untreated 32P probe (2,000 cpm) was loaded on each denaturing polyacrylamide gel to generate the standard curves used to identify protected RNA fragments. Experimental probe hybridizations contained total RNA from 5 × 105 Bm1.11 cell equivalents. Total RNA was isolated with a commercial kit according to manufacturer protocols (RNeasy; QIAGEN, Valencia, Calif.).
Generation of mature peritoneal macrophages.
Peritoneal macrophages were isolated from three female C57BL/6 mice by peritoneal lavage. Recovered cells were suspended at 106 cells/ml in RPMI 1640 complete medium containing 20 ng of recombinant murine IL-3 (R&D Systems, Minneapolis, Minn.)/ml and were plated at 2 ml per well in a 12-well tissue culture-treated plate. On day 3, 1 ml of the medium was removed and replaced with 1 ml of fresh medium containing IL-3 as described above. On day 6 the cultures were activated by adding 10 ng of E. coli LPS/ml and 10 ng of recombinant murine IFN-γ (Sigma Chemical Co.)/ml. After 24 h total RNA from each well was isolated with the RNeasy commercial kit as described above.
RT-PCR.
Optimized primer pairs were designed using Vector NTI Suite (Infomax Inc., Frederick, Md.). Primer pairs (Amitof, Alston, Mass.) are described in Table 1. Reverse transcriptase PCR (RT-PCR) was accomplished by using a single-tube avian myeloblastosis virus RT-Tfl polymerase kit (Access RT-PCR; Promega, Madison, Wis.) with 32 cycles. RT-negative reactions were included to rule out DNA artifacts. ABCD-1 (CCL22) and TARC (CCL17) primer pairs did not generate specific RT-PCR products in infected epithelial cells. The ABCD-1 (CCL22) primer pair detected ABCD-1 (CCL22) mRNA in LPS-treated splenocyte and activated peritoneal macrophage total RNA by RT-PCR. A 280-bp spurious PCR product representing mouse mitochondria RNA was also amplified by the CCL22 primer pair based on 10 out of 11 and 8 out of 9 base identities with mitochondria RNA in the primer termini. I do not have an adequate positive control to confirm the TARC RT-PCR assay. TARC (CCL17) expression is limited to activated Langerhan and myeloid dendritic cells. It is not expressed in the spleen even during systemic microbial infection (2).
TABLE 1.
Primers used for RT-PCR
| Chemokine | Sense primer | Antisense primer | PCR product size (bp) |
|---|---|---|---|
| I-Ab alpha chain | 5′ATGCTCAGCCTCTGTGGAGGTG3′ | 5′CGTCTGCGACTGACTTGCTATT3′ | 424 |
| Mig (CXCL9) | 5′TGCCATGAAGTCCGCTGTTC3′ | 5′GGGGTGTTTTGGGTTTTCTG3′ | 353 |
| CX3CL1 | 5′TCACCAGGCTGCTGCCCTCACTAA3′ | 5′TGGCCTTCGGACCCAACATT3′ | 465 |
| ABCD-1 (CCL22) | 5′ACTCCTGGTGGCTCTCGTCCTT3′ | 5′CCTGGGATCGGCACAGATAT3′ | 220 |
| TARC (CCL17) | 5′ATGAGGTCACTTCAGATGCT3′ | 5′ATGTTTGTCTTTGGGGTCTG3′ | 240 |
| MCP-5 (CCL12) | 5′ATGAAGATTTCCACACTTCT3′ | 5′CTTATCCAGTATGGTCCTGA3′ | 213 |
| CXCL16 | 5′TTGTTCTTGTGATCGTACCA3′ | 5′TGAAGGGAGAGTGGTCATCT3′ | 400 |
| MARC (CCL7) | 5′ATGAGGATCTCTGCCACGCT3′ | 5′GGAGTTGGGGTTTTCATGTC3′ | 284 |
Northern blots.
RT-PCR products from monokine induced by IFN-γ (Mig; CXCL9), CX3CL1, CXCL16, and MARC (CCL7) primer pairs were gel purified and cloned into pCRII-TOPO (Invitrogen). Insert identity was confirmed by sequencing each cloned chemokine fragment. A mouse β-actin control amplimer was purchased from a commercial vendor (Clontech/BD Biosciences). In all experiments total RNA from 5 × 105 Bm1.11 cells was loaded into each lane of a 1% agarose formaldehyde gel. Digoxigenin-labeled RNA molecular size markers were purchased from a commercial vendor (Roche Diagnostic Corp., Indianapolis, Ind.). Neutral transfer to positively charged nylon membranes was accomplished by using a downward transfer system (Schleicher & Schuell, Keene, N.H.). After transfer to the membrane, RNA was UV cross-linked to the membrane. Membranes were probed with PCR-generated digoxigenin-labeled DNA probes (Roche Diagnostic Corp.). Approximately 500 ng of labeled probe was denatured at 95°C for 3 min and then was hybridized with the membrane for 1 h at 68°C in ExpressHyb following the manufacturer's protocol (Clontech/BD Biosciences). Digoxigenin-labeled probes retained on the membrane were detected by using chemiluminescence with a digoxigenin-specific monoclonal antibody coupled to alkaline phosphatase and disodium 3-(4-methoxyspiro{1,2-dioetane-3,2-(5-chloro)tricyclo[3,3,1,13,7]decon}-4 yl)phenylphosphate (CSPD) substrate according to the manufacturer's protocol (Roche Diagnostic Corp.). All film exposures were for 1 h.
Multiplex ELISA determination of cytokine production.
Confluent Bm1.11 monolayers in 12-well tissue culture-treated plates were infected with 10 IFU of C. muridarum/cell in 2 ml of fresh medium (to minimize concentrations of cytokines made constitutively) without centrifugation or subsequent change of the medium. Mock-infected wells (0 h) received an inoculum-equivalent volume of SPG buffer but did not have C. muridarum in the medium. At indicated time points supernatants were harvested, spun for 2 min at 16,000 × g in a microcentrifuge to remove debris, brought to 0.5% (vol/vol) NP-40 (to neutralize C. muridarum), and frozen at −80°C. Samples representing two independent experiments were shipped to a commercial vendor to be analyzed for cytokine content by multiplex enzyme-linked immunosorbent assay (ELISA) technology (Searchlight; Pierce Chemical Co.). Cytokines tested included murine GM-CSF, TNF-α, IL-6, IL-12-p70, KC (CXCL1), MIP-2 (CXCL2), MCP-1 (CCL2), RANTES (CCL5), and MCP-5 (CCL12). Basal-level secretion of chemokine MCP-1 (CCL2) by Bm1.11 cells exceeded the capacity of the multiplex ELISA (data not shown). Per the manufacturer, sensitivity of the assay for individual cytokines ranged from 0.2 to 2.7 pg/ml.
Analysis of cytokine data.
To permit semiquantitative comparisons of cytokine mRNA levels, all cytokines detected by RPA were quantified by densitometry (Quantiscan software; Biosoft, Ferguson, Mo.). The resulting values were normalized to the untreated GAPDH probe band on each gel, thereby adjusting for exposure differences between films. Northern blots were also quantified by densitometry. Determination that a cytokine had been upregulated by infection was defined as a more than threefold increase in intensity after normalizing data for slight differences in loading as determined by a parallel β-actin blot (Fig. 3D). Like the RPA, all Northern blots were done with 5 × 105 Bm1.11 cell equivalents per lane, permitting at least crude comparisons of cytokine mRNA levels between the RPA and Northern blots. Multiplex ELISA data are quantitative. Every cytokine upregulated by C. muridarum infection was assigned a relative expression value of 1+ to 4+ (low level to high level). The breakpoints for ELISA concentrations (in picograms/milliliter) and densitometry values (in arbitrary units) are depicted in Table 2.
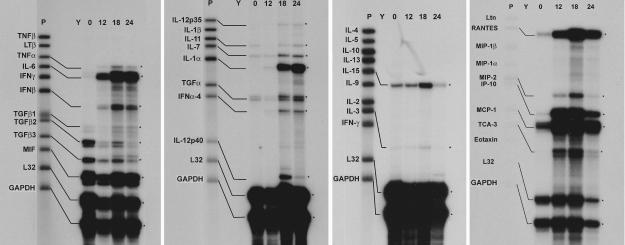
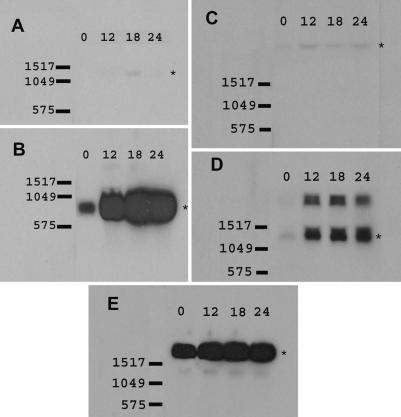
FIG. 3.
RPA analysis of Bm1.11 cell cytokine responses to C. muridarum infection. On all gels the leftmost lane (P) is the untreated probe set. 32P RNA probes that detected mRNA transcripts are further labeled with diagrammatic bars. Asterisks on the right of each panel highlight the protected RNA fragments indicated by the bars on the left. Y, control hybridization with yeast tRNA. 0, 12, 18, and 24 represent the time course of C. muridarum infection in hours. Data presented are representative of three independent experiments with each probe set showing the same results.
TABLE 2.
ELISA break points and densitometry values
| Assaya | Expression levels and assay breakpoint values
|
|||
|---|---|---|---|---|
| 1+ | 2+ | 3+ | 4+ | |
| Multiplex ELISA (pg/ml) | <100 | >100<1,000 | >1,000<15,000 | >15,000 |
| RPA—cytokines (AU) | <10 | >10<100 | >100<200 | >200 |
| RPA—chemokines (AU) | <100 | >100<1,000 | >1,000<5,000 | >5,000 |
| Northern blots (AU) | <100 | >100<1,000 | >1,000<5,000 | >5,000 |
AU, arbitrary units.
Cytokines analyzed by both ELISA and RPA were used to crudely standardize the results of the two techniques. ELISA breakpoints were empirically chosen based on the desire to have an expression scale range of 1+ to 4+. The ELISA breakpoints were then used to set the breakpoints for the RPA such that the RPA and ELISA data gave the same 1+ to 4+ score for each cytokine analyzed by both techniques. RPA chemokine breakpoints differ from the cytokine breakpoints, because chemokine mRNA levels were 10 to 20 times higher than that of any nonchemokine cytokine. A different set of chemokine breakpoints was therefore needed to reflect the spectrum of chemokine expression levels. As in any large survey of cytokines based on mRNA quantification, the results presented here are open to the criticism that mRNA levels do not necessarily reflect relative levels of secreted cytokines due to differences in mRNA stability (62), RNA translation efficiency (23), and efficiency of secretion. Furthermore, relative protein levels of individual cytokines do not address their relative biologic potency, much less how cytokine biologic activities may be altered in a complex cytokine milieu. These considerations aside, it is useful to apply reasonable assumptions and attempt to analyze cytokine survey data for the purpose of generating hypotheses to drive future experimentation.
RESULTS
Characterization of an oviduct epithelial cell line.
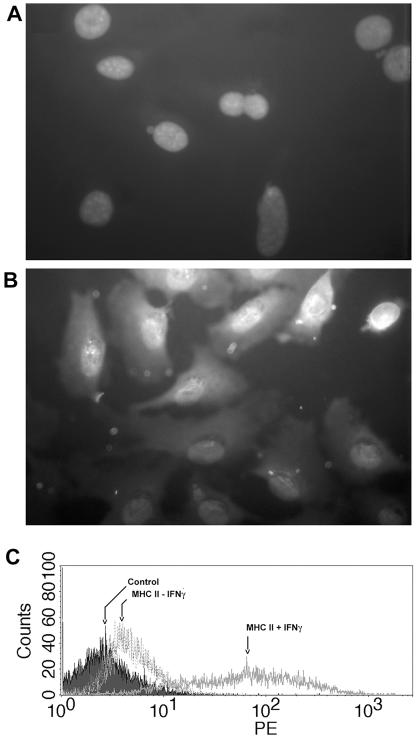
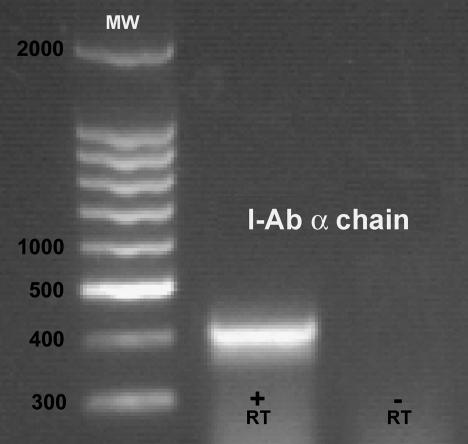
A primary murine oviduct cell line designated Bm1.11 was derived to study the epithelial cytokine response to infection in the absence of leukocytes and nonepithelial stroma. Murine epithelial cells have several properties that are not shared by other murine stromal cell types, including expression of cytokeratins and IFN-γ-inducible expression of MHC class II genes. Figure 1 shows Bm1.11 expression of cytokeratins by immunohistochemistry and marked upregulation of MHC II expression with IFN-γ treatment. Expression of the I-Ab α chain was confirmed by RT-PCR (Fig. 2). IFN-γ treatment of CL.7, a murine fibroblast cell line, did not induce MHC II expression (data not shown).
FIG. 1.
Characterization of oviduct epithelial cell line Bm1.11. (A) Bm1.11 cells grown in chamber slides were fixed and stained with irrelevant monoclonal antibody 36-7-5 specific for H-2Kk (Bm1.11 cells are H-2Kbm1). (B) Bm1.11 cells stained with the monoclonal antibody cocktail AE1/AE3 specific for acidic and basic cytokeratins. Cell nuclei in panels A and B were visualized by staining with 4′,6′-diamidino-2-phenylindole. (C) Flow cytometry analysis of cell surface levels of MHC II without treatment and after 14 h of exposure to 10 ng of recombinant murine IFN-γ/ml. Control refers to autofluorescence. PE, phycoerythrin.
FIG. 2.
Detection of I-Ab α chain mRNA in Bm1.11 cells exposed to 10 ng of IFN-γ/ml for 14 h. MW designates the lane of the 100-bp molecular size ladder. +RT, RT-PCR containing RT; −RT, control reaction lacking RT.
Chlamydia infection induces multiple inflammatory cytokines.
Multiprobe RPA were chosen for initial analysis of the epithelial cytokine response to infection by C. muridarum, because the technique yields quantitative data on multiple gene transcripts without risk of amplification artifacts. Wells of Bm1.11 cells were infected with 10 IFU of C. muridarum/cell in a staggered fashion such that simultaneous harvest of multiple wells gave time points for mock infection (0 h) and for infection at 12, 18, and 24 h (Fig. 3). RPA analysis revealed infection-associated induction of mRNA for TNF-α, IL-6, IFN-β, IL-12-p35, IL-12-p40, IL-1α, IFN-α, MIP-2 (CXCL2), and TCA-3 (CCL1) and infection-associated upregulation of IL-7, TGFα, IL-15, IL-3, RANTES (CCL5), IP-10 (CXCL10), and MCP-1 (CCL2). Transcription of housekeeping genes L32 and GAPDH was not affected by infection, nor were mRNA levels of TGFβ1, MIF, and IL-11. Interestingly, levels of TGFβ2 and TGFβ3 mRNA were reproducibly decreased in response to C. muridarum infection.
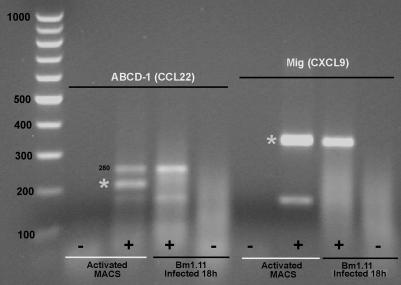
Commercially available RPA probe template sets for murine cytokines are relatively limited; therefore, RT-PCR was used to broaden the analysis of the chemokine response of oviduct epithelial cells to infection by C. muridarum. Primers pairs were designed for the inducible chemokines MARC (CCL7), MCP-5 (CCL12), ABCD-1 (CCL22), Mig (CXCL9), CXCL16, and CX3CL1. RT-PCR products for MARC (CCL7), MCP-5 (CCL12), Mig (CXCL9), CXCL16, and CX3CL1 were detected in Bm1.11 cells infected for 18 h with C. muridarum at 10 IFU/cell (data not shown). No RT-PCR products were detected for ABCD-1 (CCL22) (Fig. 4).
FIG. 4.
Absence of ABCD-1 (CCL22) mRNA in infected Bm1.11 cells. RT-PCR assays for ABCD-1 and Mig (CXCL9) were performed on total RNA isolated from Bm1.11 cells infected with C. muridarum for 18 h and activated peritoneal macrophages (Activated MACS) as a positive control. Asterisks mark the specific RT-PCR products for each primer pair. There is a spurious PCR band (280 bp) in the ABCD-1 (220 bp) RT-PCR that is present in both activated peritoneal macrophages and infected Bm1.11 cells (see Materials and Methods). Mig, a low-level transcript in infected epithelial cells, is readily detected in infected Bm1.11 cells by using the same RNA samples. Data are representative of three independent experiments showing the same result.
To get quantitative data on the levels of MARC (CCL7), Mig (CXCL9), CXCL16, and CX3CL1, Northern blot analysis was performed on Bm1.11 cells mock infected or infected with C. muridarum for 12, 18, and 24 h. RT-PCR products for MARC (CCL7), Mig (CXCL9), CXCL16, and CX3CL1 were gel purified, cloned, and sequenced to confirm identity. Cloned gene fragments for each chemokine were then used as templates to make digoxigenin-labeled probes for Northern blot analysis. Representative Northern blots from two independent time course experiments are shown in Fig. 5. C. muridarum infection of Bm1.11 cells induced very modest levels of Mig (CXCL9) mRNA and robust levels of MARC (CCL7) and CXCL16 mRNA. Very low levels of CX3CL1 mRNA are present constitutively and were not upregulated by Chlamydia infection. MCP-5 (CCL12) levels were determined by multiplex ELISA (see below).
FIG. 5.
Northern blot analysis of additional chemokines induced by C. muridarum infection. (A) Mig (CXCL9), 1,284 bp; (B) MARC (CCL7), 777 bp; (C) CX3CL1, 3,095 bp; (D) CXCL16, 1,375 bp. The higher-molecular-mass band is likely a splicing precursor of CXCL16. (E) β-actin, 1,892 bp. 0, 12, 18, and 24 represent the time course of C. muridarum infection in hours. Data presented are representative of two independent experiments for each chemokine showing the same result.
To further characterize the cytokine response and confirm secretion of cytokines detected by RPA, time course experiments were repeated to generate cell culture supernatants for analysis by multiplex ELISA. Cell culture supernatants from Bm1.11 cells mock infected or infected with C. muridarum at 10 IFU/cell for 6, 12, 18, 24, and 36 h were collected and analyzed for GM-CSF, TNF-α, IL-6, IL-12-p70, KC (CXCL1), MIP-2 (CXCL2), RANTES (CCL5), and MCP-5 (CCL12). Results of two independent experiments are presented in Table 3.
TABLE 3.
Results of time course experiments
| Cytokine | Expt no. | Time course supernatant analysis (pg/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| 0 h | 6 h | 12 h | 18 h | 24 h | 36 h | ||
| GM-CSF | 1 | 46.0 | 55.7 | 256.7 | 3,030.0 | 10,112.7 | 16,063.4 |
| 2 | 32.9 | 73.7 | 240.6 | 2,760.0 | 9,258.6 | 22,647.3 | |
| TNFα | 1 | 20.2 | 38.8 | 26.3 | 94.9 | 316.0 | 500.3 |
| 2 | 15.7 | 15.5 | 23.0 | 93.7 | 321.4 | 575.0 | |
| IL-6 | 1 | 1,847.3 | 5,496.1 | 25,785.6 | 279,505.4 | >280,000a | >280,000 |
| 2 | 2,625.0 | 5,423.3 | 25,890.2 | 237,723.6 | >280,000a | >280,000 | |
| IL-12-p70 | 1 | 39.4 | 45.6 | 48.0 | 78.5 | 94.5 | 155.1 |
| 2 | 31.9 | 35.8 | 44.5 | 85.6 | 135.9 | 516.0 | |
| KC | 1 | 11,904.9 | 8,730.8 | 22,958.7 | >40,000a | >40,000 | >40,000 |
| 2 | 13,502.3 | 10,007.4 | 22,606.0 | >40,000a | >40,000 | >40,000 | |
| MIP-2 | 1 | 30.9 | 31.2 | 147.8 | 1,586.2 | 4,975.9 | 11,748.4 |
| 2 | 37.6 | 53.0 | 165.9 | 1753.2 | 5,124.3 | 9,853.1 | |
| RANTES | 1 | 7,590.0 | 3,235.0 | 8,305.0 | >20,000a | >20,000 | >20,000 |
| 2 | 8,930.0 | 3,560.0 | 8,425.0 | >20,000a | >20,000 | >20,000 | |
| MCP-5 | 1 | 4.6 | 6.3 | 8.6 | 27.0 | 33.6 | 27.6 |
| 2 | 6.7 | 8.0 | 10.1 | 29.9 | 39.5 | 30.0 | |
Concentration in supernatant exceeded the capacity of the ELISA.
Summary of cytokine data.
RNA levels determined by RPA have previously been shown to correlate well with protein levels in other experimental systems (26, 38). In this report, there was good agreement between mRNA transcript levels detected by RPA and the amount of protein detected in culture supernatants by ELISA for all cytokines [TNF-α, IL-6, IL-12-p70, RANTES (CCL5), and MIP-2 (CXCL2)] analyzed by both techniques (compare Fig. 3 and Table 3). There was also good agreement between the time course of mRNA induction and the appearance of a specific cytokine in the culture medium. Early transcription of IL-6 mRNA correlated with detectable IL-6 in the ELISA by 6 h postinfection. In contrast, later induction of transcripts for TNF-α and IL-12 were reflected by delays in detection of these cytokines out to 18 and 24 h in the ELISA. No mRNA was detected for TNF-β, lymphotoxin β, IFN-γ, IL-1β, IL-4, IL-5, IL-10, IL-13, IL-9, IL-2, lymphotactin, MIP-1α, MIP-1β, eotaxin (CCL11), and ABCD-1 (CCL22). Figure 6 is a semiquantitative summary of all cytokines upregulated by oviduct epithelial cells in response to infection by C. trachomatis based on the algorithm described in Materials and Methods. This type of analysis ignores potential differences in mRNA stability, efficiency of translation, and cytokine potency but provides a useful starting point for discussing results of a large cytokine survey.
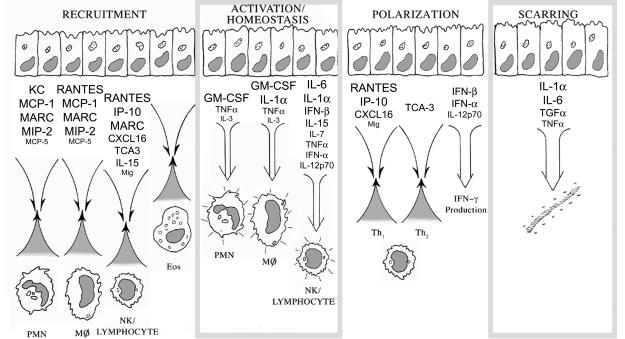
FIG. 6.
Summary of Bm1.11 oviduct epithelial cell cytokine responses to infection by C. muridarum. Expression levels of cytokines upregulated by C. muridarum infection were graded on a scale of 1+ to 4+ (low to high) as described in Materials and Methods. Relative expression levels are reflected by type sizes within the figure, with 4+ represented by the largest type size [e.g., MARC (CCL7)], 3+ represented by the second-largest type size (e.g., CXCL16), 2+ represented by the second-smallest type size (e.g., TNF-α), and 1+ indicated by the smallest type size (e.g., IL-3). PMN, polymorphonuclear cell; Eos, eosinophil.
DISCUSSION
To facilitate understanding of the multifaceted oviduct epithelial response to C. muridarum infection, cytokines induced by infection with respect to their predicted biological effects are discussed below. The first panel of Fig. 6 depicts the predicted role of Chlamydia-infected epithelial cells in leukocyte recruitment based on their cytokine response. Chemokines are critical cytokines for leukocyte recruitment (52). Bm1.11 cells infected by C. muridarum transcribed a plethora of chemokines, including myeloid-active chemokines KC (CXCL1), MIP-2 (CXCL2), MCP-1 (CCL2), MARC (CCL7), and MCP-5 (CCL12) that are important in neutrophil, monocyte/macrophage, and dendritic cell recruitment, and RANTES (CCL5), a chemokine involved in recruitment of monocyte/macrophage and dendritic cells. NK cell and lymphocyte-active chemokines upregulated by infection included TCA-3 (CCL1), RANTES (CCL5), MARC (CCL7), Mig (CXCL9), IP-10 (CXCL10), and CXCL16. Eotaxin (CCL11), the dominant chemokine for eosinophil recruitment (29), was not detected. Additional cytokines detected that have roles in leukocyte recruitment included IL-15, an important T-cell growth factor that also serves as a chemoattractant for T lymphocytes (70), and IL-1α and TNF-α, which play indirect roles in leukocyte recruitment by upregulating ICAM-1, E-selectin, and VCAM-1 on adjacent endothelia (41). Recruitment of neutrophils, monocytes, and lymphocytes without recruitment of eosinophils is consistent with the histology seen in humans with C. trachomatis salpingitis (17, 47).
Cytokines released at sites of infection trigger antimicrobial activity in NK cells, T cells, and phagocytes. The second panel of Fig. 6 depicts the predicted effects of infected epithelial cells on local leukocyte activation and homeostasis. Bm1.11 cells infected by C. muridarum secreted TNF-α, a major activator of neutrophils within infected tissues. TNF-α, IL-1α, and GM-CSF made by infected epithelial cells would activate macrophages and trigger dendritic cell maturation. IFN-α/β increase the cytolytic function of NK cells and T cells (10). IL-15 can trigger NK killing (34). IL-6 is an important cytokine in cytotoxic T-cell differentiation (46, 51).
Growth factors for T cells and macrophages released at the sites of infection may affect homeostasis by counteracting apoptotic or anergizing stimuli, thereby prolonging functional immune responses at sites of infection (21, 49, 59, 75) and possibly contributing to local expansion of effector cells (1). C. muridarum-infected epithelial cells expressed or secreted IL-3 and GM-CSF, growth factors for neutrophils and monocytes/macrophages, and upregulated transcription of IL-7 and IL-15, growth factors for T lymphocytes and NK cells. Anergy is a major theoretical barrier for T lymphocytes responding to infection, because epithelial cells do not express the CD28 ligands CD80 and CD86 (reviewed in reference 42). However, cytokines have been demonstrated to provide alternative costimulatory signals capable of substituting for the classic costimulatory signal delivered through CD28. Naïve CD4 T cells use IL-1 as an alternative costimulatory signal, while naïve CD8 T cells use IFN-α/β (15). C. muridarum-infected oviduct epithelial cells make IL-1α and IFN-α/β, thereby potentially making up for their perceived antigen presentation deficiencies related to lack of CD80 and CD86 expression.
The host immune response to urogenital infections by C. trachomatis is dominated by IFN-γ in animal models and human infections (48). The third panel of Fig. 6 shows the predicted effects of infected epithelial cells on T-lymphocyte cytokine polarization. The data presented in this report suggest that Chlamydia-infected oviduct epithelial cells could facilitate IFN-γ production through three different mechanisms. The first would be through expression of chemokines that preferentially recruit Th1 over Th2 lymphocytes. C. muridarum-infected Bm1.11 cells induced or upregulated transcription of chemokines Mig (CXCL9), IP-10 (CXCL10), and CXCL16 that selectively recruit Th1 lymphocytes bearing CXCR3 and CXCR6 receptors (8, 33, 60). CXCR6, the receptor for CXCL16, is highly expressed on intraepithelial lymphocytes (43), consistent with a role in the reproductive tract mucosal immune compartment. To my knowledge this is the first report suggesting a role for CXCL16 in host defense against Chlamydia infections. RANTES (CCL5), the chemokine ligand for CCR5, also preferentially favors Th1 recruitment based on its receptors' relative distribution on Th1 and Th2 lymphocytes (8, 60). The only Th2-selective chemokine transcribed by infected Bm1.11 cells was TCA-3 (CCL1). TCA-3 (CCL1) is the ligand for CCR8 on Th2 cells, and Th2-selective chemotaxis has been demonstrated in vitro (76). However, the role of CCR8 in recruitment of Th2 cells within a complex of in vivo Th2 cytokine milieu is unclear (7). The other important Th2 chemokine receptors are CCR3 and CCR4. Infected Bm1.11 cells did not transcribe mRNA for the CCR3 ligand eotaxin (CCL11) or the CCR4 ligand ABCD-1 (CCL22). The other CCR4 ligand is TARC. We did not detect mRNA for TARC in infected oviduct epithelial cells by RT-PCR but have not rigorously excluded its presence, lacking an adequate positive control (data not shown). TARC is not likely to be made by infected oviduct epithelial cells, as it is selectively produced by activated myeloid dendritic cells and Langerhans cells (2). The chemokine data presented here would predict that Chlamydia-infected epithelium in vivo would favor recruitment of Th1 over Th2 lymphocytes.
A second mechanism for facilitating IFN-γ production would be through IFN-α and IFN-β. Infected Bm1.11 cells expressed IFN-α and IFN-β. IFN-α/β facilitate IFN-γ production by activating CD8 T cells through an IL-12 independent pathway (50). Conversely, IFN-α/β are known to downmodulate IL-12 production by dendritic cells (6). The latter effect would presumably be counterbalanced by epithelial secretion of IL-12.
The third and potentially most important mechanism is IL-12-p70 secretion by infected oviduct epithelial cells. Surprisingly, C. muridarum-infected Bm1.11 cells transcribed both chains, IL-12-p35 and IL-12-p40, of the IL-12-p70 heterodimer and secreted IL-12-p70 into culture supernatants. IL-12-p70 is the principle mediator of cytokine polarization in vitro and in vivo, acting directly on T cells and NK cells to enhance IFN-γ production. While IL-12-p35 is made constitutively by many cell types, IL-12-p70 secretion is classically the exclusive domain of macrophages and dendritic cells (67). The finding of IL-12-p70 secretion by infected epithelial cells was surprising and suggests there may be redundant sources of IL-12-p70 locally during genital tract Chlamydia infections. The biology of IL-12 is complex, as the binding of the bioactive IL-12-p70 heterodimer by the IL-12 receptor is antagonized by IL-12-p40 homodimers (22, 31). At the same time, there is evidence that IL-12-p40 plays an important role in host defense against microbes that is independent of IL-12-p70 (18, 27, 36). In this report, RPA analysis showed roughly balanced mRNA levels for IL-12-p35 and IL-12-p40 at 24 h postinfection. Assessment of IL-12-p40 homodimer protein levels in infected cell supernatants was not done.
Immunopathologic scarring of the Fallopian tubes is a clinically important aspect of human urogenital infections caused by C. trachomatis. The fourth panel of Fig. 6 illustrates the predicted role of infected epithelial cells in scarring. C. muridarum-infected oviduct epithelial cells transcribed TGFα, TNF-α, and large quantities of IL-1α and IL-6. These cytokines have been associated with scarring and fibrosis in animal models and humans (37, 40, 55, 74), raising the possibility that Chlamydia-infected epithelial cells directly contribute to scarring of the Fallopian tubes. TGFβ family members have been strongly associated with scarring (28). Interestingly, in Bm1.11 cells TGFβ2 and TGFβ3 were reproducibly downregulated by C. muridarum infection. The data presented here raise the possibility that TGFα rather than TGFβ is the cytokine driving the scarring seen during C. trachomatis infections. TGFα is translated as a transmembrane glycoprotein known as a TGFα precursor. TGFα precursor RNA levels at 24 h postinfection were upregulated fourfold (normalized to GAPDH housekeeping gene transcript) compared to that for the mock-infected epithelial cell control (Fig. 3). TGFα precursor is cleaved by cell surface proteases, releasing TGFα. Both TGFα precursor and TGFα are biologically active. Selective overexpression of TGFα in the lungs of transgenic mice causes fibrosis and dilation of the airways (24). There are no commercial sources for recombinant murine TGFα or antibodies to murine TGFα. Within the 50 amino acids of TGFα there are 4 amino acid differences between human and mouse TGFα. To our knowledge, there are no published studies successfully using anti-human TGFα immunoreagents to study murine TGFα. We are generating mouse TGFα immunoreagents to address the role of TGFα precursor and TGFα in scarring. Other cell types, such as Th3 lymphocytes, tissue macrophages, or nonepithelial stromal components, could still serve as sources of TGFβ.
Infected Bm1.11 oviduct epithelial cells have a unique cytokine signature. Analysis of cytokines within genital secretions of infected mice reflects all cell types participating in the inflammatory response. Genital secretions of mice infected with C. muridarum have detectable levels of IL-1β, IL-6, GM-CSF, and MIP-2 (CXCL2) by ELISA (16). Infected Bm1.11 oviduct epithelial cells in this report secreted IL-6, GM-CSF, and MIP-2 (CXCL2), but no mRNA for IL-1β was detected by RPA analysis, suggesting that leukocytes or other stromal components are the source of IL-1β. Semiquantitative RT-PCR has shown increased IL-11 mRNA levels in genital tract homogenates from mice infected with C. muridarum (19). A less than twofold increase in basal transcription of IL-11 was seen in infected Bm1.11 cells (Fig. 3B), suggesting that other cell types or epithelial cells lower in the reproductive tract are the source of IL-11 mRNA in vivo. IL-11 is an antiinflammatory cytokine that antagonizes IL-12, TNF-α, and IL-1 production by macrophages (66) and influences CD4 T-cell polarization toward a Th2 phenotype (9). Chemokine analyses of the upper and lower reproductive tracts of mice infected by C. muridarum have been performed. IP-10 (CXCL10), Mig (CXCL9), and RANTES (CCL5) were the predominant chemokines induced in the upper reproductive tract by infection; modest amounts of eotaxin (CCL11) were seen during the resolution phase of the infection (44). Bm1.11 oviduct epithelial cells in this report upregulated expression of RANTES (CCL5), Mig (CXCL9), and IP-10 (CXCL10) during infection, though Mig (CXCL9) induction was modest. Maxion and Kelly (44) defined the sources of IP-10 (CXCL10) within the upper tract by immunohistochemistry showing staining of columnar epithelial cells, endothelial cells, and stromal cells.
Cytokine profiles of dendritic cells pulsed with heat-inactivated C. muridarum elementary bodies have been investigated with RPA. C. muridarum-pulsed dendritic cells upregulated several cytokines not seen in C. muridarum-infected oviduct epithelial cells, including MIP-1α (CCL3), MIP-1β (CCL4), and IL-1β. Conversely, C. muridarum-infected Bm1.11 cells expressed TCA-3 (CCL1), a chemokine not detected in C. muridarum-pulsed dendritic cells. Among the cytokines investigated for both cell types, infected oviduct epithelial cells and Chlamydia-pulsed dendritic cells shared IL-1α, IL-6, IL-12-p40, TNF-α, RANTES (CCL5), MIP-2 (CXCL2), MCP-1 (CCL2), and IP-10 (CXCL10) (63).
The spectrum of inflammatory cytokines made by the Bm1.11 oviduct epithelial cell line was somewhat surprising in light of previous work with human HeLa and HEC-1-B cell lines. Those studies identified the principle cytokines induced by Chlamydia infection to be IL-1α, IL-1β, IL-6, IL-8, IL-11, IL-16, IL-18, GM-CSF, Groα (CXCL1), ENA-78 (CXCL5), and GCP-2 (CXCL6). Murine Bm1.11 cells infected by C. muridarum initiated or enhanced transcription and/or secretion of IL-1α, IL-3, IL-6, IL-7, IL-12, IL-15, IFN-α, IFN-β, TNF-α, TGFα, GM-CSF, KC (CXCL1), MIP-2 (CXCL2), TCA-3 (CCL1), MCP-1 (CCL2), RANTES (CCL5), MARC (CCL7), MCP-5 (CCL12), Mig (CXCL9), IP-10 (CXCL10), and CXCL16. The spectrum of cytokines made by Bm1.11 cells infected with C. muridarum encompassed all mutually investigated cytokines previously documented in the human HeLa and HEC-1-B cells, with the exception of IL-1β and IL-11.
The broader spectrum of cytokines made by the Bm1.11 oviduct cell line may reflect differences in the Chlamydia serovars tested, differences between mice and humans, or differences between cell and tissue types. HeLa cells and HEC-1-B cells are carcinomas that originated in the cervix and uterus, respectively. Epithelium lining the cervix and uterus may have a different biology than epithelium lining the oviducts. This could explain differences such as IL-11 upregulation in infected HeLa cells but not in infected Bm1.11 cells. An additional possibility is that the process of transformation and evasion of host tumor immunosurveillance has changed the biology of HeLa and HEC-1-B cells. HeLa cells contain multiple integrated copies of human papillomavirus 18 and actively transcribe human papillomavirus genes (56).
The oviduct epithelial data presented here support the hypothesis that oviduct epithelial cells contribute directly to innate and adaptive immunity during C. trachomatis infections. The epithelial contributions to host defense include recruiting leukocytes and facilitating production of IFN-γ critical for controlling Chlamydia infections, but epithelial cytokines such as TGFα may contribute to detrimental scarring associated with the infection.
Acknowledgments
I gratefully acknowledge Roger Rank and Stanley Spinola for their thoughtful comments during preparation of the manuscript, Darron Brown for assistance with immunohistochemistry, Thomas Weinzerl for graphics assistance with Fig. 6, and Barbara Van der Pol at the Indiana University Chlamydia Laboratory for McCoy cells and C. muridarum.
This work was supported by grant 1K08AI52128-01 from the National Institute of Allergy and Infectious Diseases.
Editor: J. D. Clements
REFERENCES
- 1.Agostini, C., L. Trentin, R. Sancetta, M. Facco, C. Tassinari, A. Cerutti, M. Bortolin, A. Milani, M. Siviero, R. Zambello, and G. Semenzato. 1997. Interleukin-15 triggers activation and growth of the CD8 T-cell pool in extravascular tissues of patients with acquired immunodeficiency syndrome. Blood 90:1115-1123. [PubMed] [Google Scholar]
- 2.Alferink, J., I. Lieberam, W. Reindl, A. Behrens, S. Weiss, N. Huser, K. Gerauer, R. Ross, A. B. Reske-Kunz, P. Ahmad-Nejad, H. Wagner, and I. Forster. 2003. Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J. Exp. Med. 197:585-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ault, K. A., O. W. Tawfik, M. M. Smith-King, J. Gunter, and P. F. Terranova. 1996. Tumor necrosis factor-alpha response to infection with Chlamydia trachomatis in human fallopian tube organ culture. Am. J. Obstet. Gynecol. 175:1242-1245. [DOI] [PubMed] [Google Scholar]
- 4.Barlow, R. E., I. D. Cooke, O. Odukoya, M. K. Heatley, J. Jenkins, G. Narayansingh, S. S. Ramsewak, and A. Eley. 2001. The prevalence of Chlamydia trachomatis in fresh tissue specimens from patients with ectopic pregnancy or tubal factor infertility as determined by PCR and in-situ hybridisation. J. Med. Microbiol. 50:902-908. [DOI] [PubMed] [Google Scholar]
- 5.Barron, A. L., H. J. White, R. G. Rank, B. L. Soloff, and E. B. Moses. 1981. A new animal model for the study of Chlamydia trachomatis genital infections: infection of mice with the agent of mouse pneumonitis. J. Infect. Dis. 143:63-66. [DOI] [PubMed] [Google Scholar]
- 6.Biron, C. A. 2001. Interferons alpha and beta as immune regulators-a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, B., and C. M. Lloyd. 2003. CC chemokine ligand 1 promotes recruitment of eosinophils but not Th2 cells during the development of allergic airways disease. J. Immunol. 170:4810-4817. [DOI] [PubMed] [Google Scholar]
- 8.Bonecchi, R., G. Bianchi, P. P. Bordignon, D. D'Ambrosio, R. Lang, A. Borsatti, S. Sozzani, P. Allavena, P. A. Gray, A. Mantovani, and F. Sinigaglia. 1998. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 187:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozza, M., J. L. Bliss, A. J. Dorner, and W. L. Trepicchio. 2001. Interleukin-11 modulates Th1/Th2 cytokine production from activated CD4+ T cells. J. Interferon Cytokine Res. 21:21-30. [DOI] [PubMed] [Google Scholar]
- 10.Brooks, C. G., M. Holscher, and D. Urdal. 1985. Natural killer activity in cloned cytotoxic T lymphocytes: regulation by interleukin 2, interferon, and specific antigen. J. Immunol. 135:1145-1152. [PubMed] [Google Scholar]
- 11.Campbell, L. A., D. L. Patton, D. E. Moore, A. L. Cappuccio, B. A. Mueller, and S. P. Wang. 1993. Detection of Chlamydia trachomatis deoxyribonucleic acid in women with tubal infertility. Fertil. Steril. 59:45-50. [PubMed] [Google Scholar]
- 12.Cates, W., Jr., R. T. Rolfs, Jr., and S. O. Aral. 1990. Sexually transmitted diseases, pelvic inflammatory disease, and infertility: an epidemiologic update. Epidemiol. Rev. 12:199-220. [DOI] [PubMed] [Google Scholar]
- 13.Coste, J., B. Laumon, A. Bremond, P. Collet, and N. Job-Spira. 1994. Sexually transmitted diseases as major causes of ectopic pregnancy: results from a large case-control study in France. Fertil. Steril. 62:289-295. [DOI] [PubMed] [Google Scholar]
- 14.Cotter, T. W., and G. I. Byrne. 1996. Immunity to Chlamydia: comparison of human infections and murine models. Res. Immunol. 147:587-595. [DOI] [PubMed] [Google Scholar]
- 15.Curtsinger, J. M., C. S. Schmidt, A. Mondino, D. C. Lins, R. M. Kedl, M. K. Jenkins, and M. F. Mescher. 1999. Inflammatory cytokines provide a third signal for activation of naive CD4+ and CD8+ T cells. J. Immunol. 162:3256-3262. [PubMed] [Google Scholar]
- 16.Darville, T., C. W. Andrews, Jr., J. D. Sikes, P. L. Fraley, and R. G. Rank. 2001. Early local cytokine profiles in strains of mice with different outcomes from chlamydial genital tract infection. Infect. Immun. 69:3556-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean, D. 1997. Chlamydia infections of Chlamydia trachomatis, p. 473-497. In D. H. Connor, F. W. Chandler, D. A. Schwartz, H. J. Manz, and E. E. Lack (ed.), Pathology of infectious diseases. Appleton & Lange, Stamford, Conn.
- 18.Decken, K., G. Kohler, K. Palmer-Lehmann, A. Wunderlin, F. Mattner, J. Magram, M. K. Gately, and G. Alber. 1998. Interleukin-12 is essential for a protective Th1 response in mice infected with Cryptococcus neoformans. Infect. Immun. 66:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dessus-Babus, S., T. L. Darville, F. P. Cuozzo, K. Ferguson, and P. B. Wyrick. 2002. Differences in innate immune responses (in vitro) to HeLa cells infected with nondisseminating serovar E and disseminating serovar L2 of Chlamydia trachomatis. Infect. Immun. 70:3234-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dessus-Babus, S., S. T. Knight, and P. B. Wyrick. 2000. Chlamydial infection of polarized HeLa cells induces PMN chemotaxis but the cytokine profile varies between disseminating and non-disseminating strains. Cell Microbiol. 2:317-327. [DOI] [PubMed] [Google Scholar]
- 21.Flad, H. D., E. Grage-Griebenow, F. Petersen, B. Scheuerer, E. Brandt, J. Baran, J. Pryjma, and M. Ernst. 1999. The role of cytokines in monocyte apoptosis. Pathobiology 67:291-293. [DOI] [PubMed] [Google Scholar]
- 22.Germann, T., and E. Rude. 1995. Interleukin-12. Int. Arch. Allergy Immunol. 108:103-112. [DOI] [PubMed] [Google Scholar]
- 23.Grafi, G., I. Sela, and G. Galili. 1993. Translational regulation of human beta interferon mRNA: association of the 3′ AU-rich sequence with the poly(A) tail reduces translation efficiency in vitro. Mol. Cell. Biol. 13:3487-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardie, W. D., M. D. Bruno, K. M. Huelsman, H. S. Iwamoto, P. E. Carrigan, G. D. Leikauf, J. A. Whitsett, and T. R. Korfhagen. 1997. Postnatal lung function and morphology in transgenic mice expressing transforming growth factor-alpha. Am. J. Pathol. 151:1075-1083. [PMC free article] [PubMed] [Google Scholar]
- 25.Hess, S., C. Rheinheimer, F. Tidow, G. Bartling, C. Kaps, J. Lauber, J. Buer, and A. Klos. 2001. The reprogrammed host: Chlamydia trachomatis-induced up-regulation of glycoprotein 130 cytokines, transcription factors, and antiapoptotic genes. Arthritis Rheum. 44:2392-2401. [DOI] [PubMed] [Google Scholar]
- 26.Hobbs, M. V., W. O. Weigle, D. J. Noonan, B. E. Torbett, R. J. McEvilly, R. J. Koch, G. J. Cardenas, and D. N. Ernst. 1993. Patterns of cytokine gene expression by CD4+ T cells from young and old mice. J. Immunol. 150:3602-3614. [PubMed] [Google Scholar]
- 27.Holscher, C., R. A. Atkinson, B. Arendse, N. Brown, E. Myburgh, G. Alber, and F. Brombacher. 2001. A protective and agonistic function of IL-12p40 in mycobacterial infection. J. Immunol. 167:6957-6966. [DOI] [PubMed] [Google Scholar]
- 28.Huang, J. S., Y. H. Wang, T. Y. Ling, S. S. Chuang, F. E. Johnson, and S. S. Huang. 2002. Synthetic TGF-beta antagonist accelerates wound healing and reduces scarring. FASEB J. 16:1269-1270. [DOI] [PubMed] [Google Scholar]
- 29.Humbles, A. A., D. M. Conroy, S. Marleau, S. M. Rankin, R. T. Palframan, A. E. Proudfoot, T. N. Wells, D. Li, P. K. Jeffery, D. A. Griffiths-Johnson, T. J. Williams, and P. J. Jose. 1997. Kinetics of eotaxin generation and its relationship to eosinophil accumulation in allergic airways disease: analysis in a guinea pig model in vivo. J. Exp. Med. 186:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagnoff, M. F., and L. Eckmann. 1997. Epithelial cells as sensors for microbial infection. J. Clin. Investig. 100:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalinski, P., P. L. Vieira, J. H. Schuitemaker, E. C. de Jong, and M. L. Kapsenberg. 2001. Prostaglandin E(2) is a selective inducer of interleukin-12 p40 (IL-12p40) production and an inhibitor of bioactive IL-12p70 heterodimer. Blood 97:3466-3469. [DOI] [PubMed] [Google Scholar]
- 32.Kaushic, C., K. Grant, M. Crane, and C. R. Wira. 2000. Infection of polarized primary epithelial cells from rat uterus with Chlamydia trachomatis: cell-cell interaction and cytokine secretion. Am. J. Reprod. Immunol. 44:73-79. [DOI] [PubMed] [Google Scholar]
- 33.Kim, C. H., E. J. Kunkel, J. Boisvert, B. Johnston, J. J. Campbell, M. C. Genovese, H. B. Greenberg, and E. C. Butcher. 2001. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J. Clin. Investig. 107:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinoshita, N., T. Hiroi, N. Ohta, S. Fukuyama, E. J. Park, and H. Kiyono. 2002. Autocrine IL-15 mediates intestinal epithelial cell death via the activation of neighboring intraepithelial NK cells. J. Immunol. 169:6187-6192. [DOI] [PubMed] [Google Scholar]
- 35.Kuo, C., and W. J. Chen. 1980. A mouse model of Chlamydia trachomatis pneumonitis. J. Infect. Dis. 141:198-202. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann, J., S. Bellmann, C. Werner, R. Schroder, N. Schutze, and G. Alber. 2001. IL-12p40-dependent agonistic effects on the development of protective innate and adaptive immunity against Salmonella enteritidis. J. Immunol. 167:5304-5315. [DOI] [PubMed] [Google Scholar]
- 37.Lin, Z. Q., T. Kondo, Y. Ishida, T. Takayasu, and N. Mukaida. 2003. Essential involvement of IL-6 in the skin wound-healing process as evidenced by delayed wound healing in IL-6-deficient mice. J. Leukoc. Biol. 73:713-721. [DOI] [PubMed] [Google Scholar]
- 38.Liu, W., and R. J. Kurlander. 1995. Analysis of the interrelationship between IL-12, TNF-alpha, and IFN-gamma production during murine listeriosis. Cell Immunol. 163:260-267. [DOI] [PubMed] [Google Scholar]
- 39.Lu, H., C. Shen, and R. C. Brunham. 2000. Chlamydia trachomatis infection of epithelial cells induces the activation of caspase-1 and release of mature IL-18. J. Immunol. 165:1463-1469. [DOI] [PubMed] [Google Scholar]
- 40.Madtes, D. K., A. L. Elston, R. C. Hackman, A. R. Dunn, and J. G. Clark. 1999. Transforming growth factor-alpha deficiency reduces pulmonary fibrosis in transgenic mice. Am. J. Respir. Cell. Mol. Biol. 20:924-934. [DOI] [PubMed] [Google Scholar]
- 41.Mantovani, A., and E. Dejana. 1989. Cytokines as communication signals between leukocytes and endothelial cells. Immunol. Today. 10:370-375. [DOI] [PubMed] [Google Scholar]
- 42.Marelli-Berg, F. M., and R. I. Lechler. 1999. Antigen presentation by parenchymal cells: a route to peripheral tolerance? Immunol. Rev. 172:297-314. [DOI] [PubMed] [Google Scholar]
- 43.Matloubian, M., A. David, S. Engel, J. E. Ryan, and J. G. Cyster. 2000. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat. Immunol. 1:298-304. [DOI] [PubMed] [Google Scholar]
- 44.Maxion, H. K., and K. A. Kelly. 2002. Chemokine expression patterns differ within anatomically distinct regions of the genital tract during Chlamydia trachomatis infection. Infect. Immun. 70:1538-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCormack, W. M., S. Alpert, D. E. McComb, R. L. Nichols, D. Z. Semine, and S. H. Zinner. 1979. Fifteen-month follow-up study of women infected with Chlamydia trachomatis. N. Engl. J. Med. 300:123-125. [DOI] [PubMed] [Google Scholar]
- 46.Ming, J. E., C. Cernetti, R. M. Steinman, and A. Granelli-Piperno. 1989. Interleukin 6 is the principal cytolytic T lymphocyte differentiation factor for thymocytes in human leukocyte conditioned medium. J. Mol. Cell. Immunol. 4:203-211. [PubMed] [Google Scholar]
- 47.Moller, B. R., L. Westrom, S. Ahrons, K. T. Ripa, L. Svensson, C. von Mecklenburg, H. Henrikson, and P. A. Mardh. 1979. Chlamydia trachomatis infection of the Fallopian tubes. Histological findings in two patients. Br J. Vener. Dis. 55:422-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison, R. P., and H. D. Caldwell. 2002. Immunity to murine chlamydial genital infection. Infect. Immun. 70:2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller, Y. M., P. M. Bojczuk, E. S. Halstead, A. H. Kim, J. Witek, J. D. Altman, and P. D. Katsikis. 2003. IL-15 enhances survival and function of HIV-specific CD8+ T cells. Blood 101:1024-1029. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen, K. B., W. T. Watford, R. Salomon, S. R. Hofmann, G. C. Pien, A. Morinobu, M. Gadina, J. J. O'Shea, and C. A. Biron. 2002. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 297:2063-2066. [DOI] [PubMed] [Google Scholar]
- 51.Okada, M., M. Kitahara, S. Kishimoto, T. Matsuda, T. Hirano, and T. Kishimoto. 1988. IL-6/BSF-2 functions as a killer helper factor in the in vitro induction of cytotoxic T cells. J. Immunol. 141:1543-1549. [PubMed] [Google Scholar]
- 52.Olson, T. S., and K. Ley. 2002. Chemokines and chemokine receptors in leukocyte trafficking. Am. J. Physiol. Regul. Integr. Comp. Physiol. 283:R7-R28. [DOI] [PubMed] [Google Scholar]
- 53.Ossewaarde, J. M., A. de Vries, T. Bestebroer, and A. F. Angulo. 1996. Application of a Mycoplasma group-specific PCR for monitoring decontamination of Mycoplasma-infected Chlamydia sp. strains. Appl. Environ. Microbiol. 62:328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patton, D. L., M. Askienazy-Elbhar, J. Henry-Suchet, L. A. Campbell, A. Cappuccio, W. Tannous, S. P. Wang, and C. C. Kuo. 1994. Detection of Chlamydia trachomatis in fallopian tube tissue in women with postinfectious tubal infertility. Am. J. Obstet. Gynecol. 171:95-101. [DOI] [PubMed] [Google Scholar]
- 55.Polo, M., F. Ko, F. Busillo, C. W. Cruse, T. J. Krizek, and M. C. Robson. 1997. Cytokine production in patients with hypertrophic burn scars. J. Burn Care Rehabil. 18:477-482. [DOI] [PubMed] [Google Scholar]
- 56.Popescu, N. C., J. A. DiPaolo, and S. C. Amsbaugh. 1987. Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet. Cell Genet. 44:58-62. [DOI] [PubMed] [Google Scholar]
- 57.Rahm, V. A., H. Gnarpe, and V. Odlind. 1988. Chlamydia trachomatis among sexually active teenage girls. Lack of correlation between chlamydial infection, history of the patient and clinical signs of infection. Br J. Obstet. Gynaecol. 95:916-919. [DOI] [PubMed] [Google Scholar]
- 58.Rasmussen, S. J., L. Eckmann, A. J. Quayle, L. Shen, Y. X. Zhang, D. J. Anderson, J. Fierer, R. S. Stephens, and M. F. Kagnoff. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Investig. 99:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saba, S., G. Soong, S. Greenberg, and A. Prince. 2002. Bacterial stimulation of epithelial G-CSF and GM-CSF expression promotes PMN survival in CF airways. Am. J. Respir. Cell. Mol. Biol. 27:561-567. [DOI] [PubMed] [Google Scholar]
- 60.Sallusto, F., D. Lenig, C. R. Mackay, and A. Lanzavecchia. 1998. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 187:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schachter, J. (ed.). 1980. Chlamydiae (Psittacosis-lymphogranuloma venereum-trachoma group), 3rd ed. American Society for Microbiology, Washington, D.C.
- 62.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 63.Shaw, J. H., V. R. Grund, L. Durling, and H. D. Caldwell. 2001. Expression of genes encoding Th1 cell-activating cytokines and lymphoid homing chemokines by chlamydia-pulsed dendritic cells correlates with protective immunizing efficacy. Infect. Immun. 69:4667-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shepard, M. K., and R. B. Jones. 1989. Recovery of Chlamydia trachomatis from endometrial and fallopian tube biopsies in women with infertility of tubal origin. Fertil. Steril. 52:232-238. [DOI] [PubMed] [Google Scholar]
- 65.Stephens, R. S. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 11:44-51. [DOI] [PubMed] [Google Scholar]
- 66.Trepicchio, W. L., M. Bozza, G. Pedneault, and A. J. Dorner. 1996. Recombinant human IL-11 attenuates the inflammatory response through down-regulation of proinflammatory cytokine release and nitric oxide production. J. Immunol. 157:3627-3634. [PubMed] [Google Scholar]
- 67.Trinchieri, G. 1998. Interleukin-12: a cytokine at the interface of inflammation and immunity. Adv. Immunol. 70:83-243. [DOI] [PubMed] [Google Scholar]
- 68.van den Brule, A. J., C. Munk, J. F. Winther, S. K. Kjaer, H. O. Jorgensen, C. J. Meijer, and S. A. Morre. 2002. Prevalence and persistence of asymptomatic Chlamydia trachomatis infections in urine specimens from Danish male military recruits. Int. J. STD AIDS (Suppl. 13) 2:19-22. [DOI] [PubMed] [Google Scholar]
- 69.White, H. D., R. H. Prabhala, S. L. Humphrey, K. M. Crassi, J. M. Richardson, and C. R. Wira. 2000. A method for the dispersal and characterization of leukocytes from the human female reproductive tract. Am. J. Reprod. Immunol. 44:96-103. [DOI] [PubMed] [Google Scholar]
- 70.Wilkinson, P. C., and F. Y. Liew. 1995. Chemoattraction of human blood T lymphocytes by interleukin-15. J. Exp. Med. 181:1255-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wyrick, P. B. 2000. Intracellular survival by Chlamydia. Cell Microbiol. 2:275-282. [DOI] [PubMed] [Google Scholar]
- 72.Wyrick, P. B., S. T. Knight, T. R. Paul, R. G. Rank, and C. S. Barbier. 1999. Persistent chlamydial envelope antigens in antibiotic-exposed infected cells trigger neutrophil chemotaxis. J. Infect. Dis. 179:954-966. [DOI] [PubMed] [Google Scholar]
- 73.Xia, M., R. E. Bumgarner, M. F. Lampe, and W. E. Stamm. 2003. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J. Infect. Dis. 187:424-434. [DOI] [PubMed] [Google Scholar]
- 74.Xue, H., R. L. McCauley, and W. Zhang. 2000. Elevated interleukin-6 expression in keloid fibroblasts. J. Surg. Res. 89:74-77. [DOI] [PubMed] [Google Scholar]
- 75.Yajima, T., H. Nishimura, R. Ishimitsu, T. Watase, D. H. Busch, E. G. Pamer, H. Kuwano, and Y. Yoshikai. 2002. Overexpression of IL-15 in vivo increases antigen-driven memory CD8+ T cells following a microbe exposure. J. Immunol. 168:1198-1203. [DOI] [PubMed] [Google Scholar]
- 76.Zingoni, A., H. Soto, J. A. Hedrick, A. Stoppacciaro, C. T. Storlazzi, F. Sinigaglia, D. D'Ambrosio, A. O'Garra, D. Robinson, M. Rocchi, A. Santoni, A. Zlotnik, and M. Napolitano. 1998. The chemokine receptor CCR8 is preferentially expressed in Th2 but not Th1 cells. J. Immunol. 161:547-551. [PubMed] [Google Scholar]