Abstract
Cellular senescence is described to be a consequence of telomere erosion during the replicative life span of primary human cells. Quiescence should therefore not contribute to cellular aging but rather extend lifespan. Here we tested this hypothesis and demonstrate that cultured long-term quiescent human fibroblasts transit into senescence due to similar cellular mechanisms with similar dynamics and with a similar maximum life span as proliferating controls, even under physiological oxygen conditions. Both, long-term quiescent and senescent fibroblasts almost completely fail to undergo apoptosis. The transition of long-term quiescent fibroblasts into senescence is also independent of HES1 which protects short-term quiescent cells from becoming senescent. Most significantly, DNA damage accumulates during senescence as well as during long-term quiescence at physiological oxygen levels. We suggest that telomere-independent, potentially maintenance driven gradual induction of cellular senescence during quiescence is a counterbalance to tumor development.
Introduction
Many cells within our bodies, including fibroblasts, hepatocytes, lymphocytes, stem cells and germ cells, are in the state of quiescence, defined as a reversible cell cycle arrest with temporary absence of proliferation [1]. Pathologies associated with quiescence include auto-immune diseases, fibrosis, and chronic wounds. Some of these cells maintain a quiescent state for long periods of time, even years, and quiescent cells are defined to retain the ability to return into the cell cycle. In vivo, quiescence is considered to limit the uncontrolled proliferation of cells, especially stem cells, whose proliferation has to be controlled properly in order to maintain tissue function, therefore contributing to tissue homeostasis [2]–[6]. A number of functional changes have been associated with quiescence including modified metabolism [7]–[9] and altered chromatin conformation [10]–[12]. Quiescence is not a passive default state, but instead is actively maintained by specific molecular mechanisms [13], [14]. Human diploid fibroblasts can enter quiescence in response to signals including loss of adhesion, contact inhibition, and mitogen withdrawal. Each of these anti-proliferative signals induce a major induction-specific reprogramming of gene expression, either enforcing the non-dividing state by regulating genes involved in cell division, or ensuring the reversibility of quiescence by protecting cells from damage induced by free radicals, while other changes indicate the involvement in pathways protecting quiescent cells against transition into terminal differentiation [15]. Thus, quiescence is a collection of states determined by the initiating signal; however, a number of genes are universally characteristic of quiescence, implying the existence of a genetic program of quiescence common to the different quiescent states [15].
Quiescent cells show low expression of cyclins and cyclin dependent kinases (CDKs) [6], [16], [17] as well as of the CDK inhibitors (CDKIs) p19 or p16 [1], [18] but high expression of CDKIs p21, p27, p53 and p57 [2]. Up-regulation of p21 occurs during several cell cycle arrested states, including quiescence, senescence and terminal differentiation [19]–[22], and is mostly accompanied by expression of p27 [18], [23]–[26]. Quiescence can easily be reversed by depletion of p21 [1], and, vice versa, cells with depleted p21 show impaired entry into quiescence. Quiescence is not simply a downstream consequence of cell cycle exit. Specific inhibition of CDKs arrests the cell cycle, but this neither induces the quiescence-specific gene expression program nor resistance to terminal differentiation [15]. Thus, the quiescence program of gene expression, but not direct CDK inhibition, ensures the reversibility of the quiescence state. Due to the up-regulation of p21, quiescent cells are endangered to transit into senescence. Cells having been quiescent for 10 days are protected against this transition into senescence by the up-regulation of the transcriptional repressor HES1 [27].
In order to be reversible, quiescence must grant the return into the cell cycle. Consequently, quiescent cells repress transition into terminal differentiation in which cell cycle arrest is irreversible [15]. However, when transition into irreversible cell cycle arrest is suppressed, reversible non-dividing quiescent cells are less protected against cancer development and are subject to tumor development. While short-term quiescent cells were described to be protected against transition into senescence [27], long-term quiescent cells may protect themselves against malignant transformation by implementing a senescence-associated cell cycle arrest over longer periods of time. Indeed, most of a human foetal skin fibroblast cell population while being long-term quiescent, were observed to transit into senescence [28]. This however violates the definition of a quiescent cell population of being able, even after years of quiescence, to completely return into the cell cycle. Here we resolve this contradiction by showing that long-term quiescent primary human cultured fibroblast MRC-5 and WI-38 cells transit into senescence. It remains to be shown to what extent these findings, observed for cultured cells, also hold for cells in tissue (in vivo). After months of quiescence, MRC-5 and WI-38 cells still display HES1 up-regulation, but this is unable to protect the cells against transition into senescence. Only a few percent of the cells in the population are able to return into the cell cycle.
Cellular senescence is a cell cycle arrested state in which normal diploid primary cells have lost their proliferative potential [29]. Senescence is regarded as a tumor suppressor mechanism at young age but a contributor to tissue aging later in life (“antagonistic pleiotropy”), [30]. In vivo, cellular senescence is thought to significantly contribute to the aging process [31]–[35]. Senescence displays a specific phenotype, including a flattened and enlarged cellular morphology [36], [37], an increased activity of senescence-associated β-galactosidase (SA-β Gal) activity [38], and expression of additional, more or less specific, molecular signatures. Premature senescence is supposed to be mainly mediated by an increased and persistent DNA damage response resulting from genotoxic stress [34], [39]–[49]. In particular, replicative senescence can be mediated by telomere shortening and a subsequent persistent DNA damage response from unprotected telomeres [36], [50]–[53]. Permanent senescence-associated cell cycle arrest is initiated and maintained by the p53-p21 and p16-pRb pathways [54], [55]. Cellular senescence can be reversed when maintained only by p53-p21 induction [1], [19], [56], [57]. RNAi-mediated depletion of p21 but not p16 leads to cell cycle re-entry of senescent keratinocytes [1]. Suppression of the p16 pathway might lead to S-phase re-entry and replication but not cell division [57], [58]. Therefore, both p21 and p16 driven pathways constitute important mechanism to ensure the irreversibility of cellular senescence.
Telomere shortening as a basic concept for aging assumes that each successive cell division acts as a mitotic counting mechanism inducing replicative senescence [59]–[63]. According to this concept, induction of quiescence for a defined amount of time would be predicted to prolong the lifespan of fibroblasts in comparison to constantly proliferating cells. In contrast to this prediction, after long-term quiescence primary human foreskin fibroblasts (HFF) were observed to transit into senescence despite of negligible telomere shortening [28], questioning that cell division and telomeric attrition is necessarily required for senescence [64]–[67]. Here we detect that during long-term quiescence also other human fibroblasts enter senescence. Thus, other effects than telomere shortening, like oxidative stress induced DNA damage, may be responsible for this transition [67]. This is supported by the fact that mouse fibroblasts senesce in culture although mice have very long telomeres. We therefore analyzed the transition of quiescent human primary MRC-5 and WI-38 fibroblast cells into senescence and apoptosis. We reduced the oxidative stress and found that WI-38 cells did not respond at all to this stress reduction, and MRC-5 cells only to a small amount. Our results suggest that other mechanisms beyond telomere shortening and oxidative stress drive human fibroblasts into senescence.
Results
Short periods of quiescence delay fibroblast aging but do not extend life span
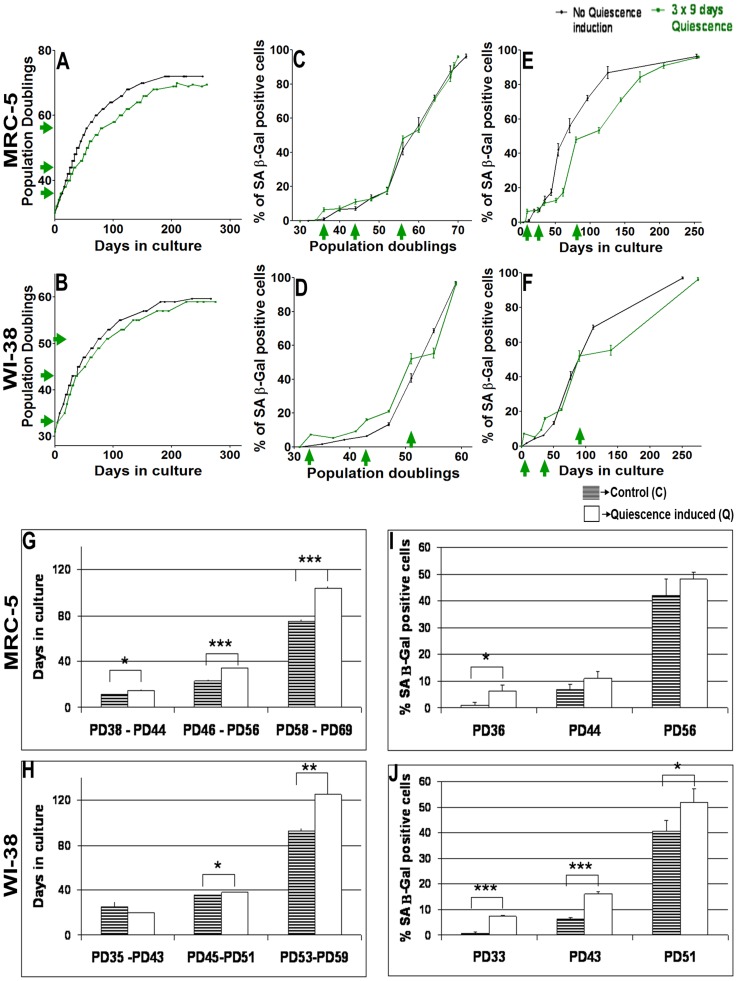
In a first set of experiments, quiescence was induced (three times for a period of 9 days each) by contact inhibition in replicatively aging MRC-5 fibroblasts (at population doublings (PDs) 36, 44 and 56) or WI-38 cells (at PDs 33, 43, and 51) (Fig. 1). Contact inhibition as the quiescence inducing signal was selected because it keeps the quiescent cells in the same serum condition as the proliferating control cells, and thus allows for a direct comparison of cellular and molecular signatures. Nevertheless, we performed similar quiescence induction experiments implying serum starvation over 8 days, which yielded similar results. Cells resumed proliferation after each release from quiescence (Fig. 1 A and B). Constantly proliferating control MRC-5 or WI-38 fibroblasts reached maximum PDs of 72 or 61, respectively, whereas cells subjected to pulses of quiescence reached 69 PDs (MRC-5) or 59 PDs (WI-38). Thus, repeated short-term quiescence induction did not result in an increased proliferative potential of MRC-5 or WI-38 cells; rather, we observed a slightly reduced number of cell divisions in quiescence-pulsed cell cultures. The periods of quiescence had an influence on cellular behavior: after periods of quiescence, between same PD values MRC-5 and WI-38 fibroblast cells grew detectably slower compared to constantly proliferating control fibroblast cells (Fig. 1 A and B). A quantitative analysis revealed, compared to control cells, a significantly decreased growth rate of MRC-5 or WI-38 cells having been quiescent for three 9-day periods, but not after one period (Fig. 1 G and H). The increase of the amount of SA-β Gal positive cells was similar in both populations when being plotted versus PDs (Fig. 1 C and D), with the time-limited exception of the single time points at the end of the quiescence periods. An increase of β Gal after confluency-induced quiescence has been observed before [38], [68]–[72]. This effect is stronger in WI-38 compared to MRC-5 cells (Fig. 1 C, D, I and J). Plotting the SA-β Gal activity versus time in culture revealed delayed aging in MRC-5 cells after two and three 9-day periods of quiescence (Fig. 1 E) while for WI-38 cells this effect was observed only after three periods of quiescence (Fig. 1 F). Short-term (9 days) quiescence induction also resulted in a significant increase of anti-apoptotic protein Bcl-2 in both MRC-5 and WI-38 fibroblasts (data not shown), in agreement with previous studies [73], [74].
Figure 1. Impact of short term quiescence induction (3×9 days) in MRC-5 and WI-38 fibroblasts.
(A & B) Growth curve of 2 independent MRC-5 (A) and WI-38 (B) fibroblast cell lines (control with no quiescence induction and a cell line where quiescence was induced 3 times separately for a span of 9 days by contact inhibition) maintained in culture at 20% O2 as triplicates from an early PD until senescence at late PDs. Each growth curve is measured in triplicate. Data points of all measurements are displayed (not the mean). (B, C, D, E & F) Percentage of SA-β gal positive cells at different time points of their growth in culture in the control MRC-5 (C & E) and WI-38 (D & F) fibroblast cell line and in the cell line where quiescence was induced 3 times separately. Fig. 1 C and D are plotted with PDs, whereas Fig. 1 E and F are plotted with days in culture in the y-axis. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). (G & H) Quiescence was induced by contact inhibition in short periods of 9 days at 3 stages of the lifespan of MRC-5 (at PDs = 36, 44, 56) and WI-38 (at PDs = 33, 43, 51) fibroblasts maintained in culture at 20% O2. The plot shows the number of days spent by MRC-5 (G) and WI-38 (H) fibroblasts in culture between PDs 38 and 44, 46 and 56, and 58 and 69 for MRC-5 (G) and between PDs 35 and 43, 45 and 51, and 53 and 59 for WI-38 (H) for cells having been repeatedly quiescent compared to the control fibroblasts. (I & J) Percentage of SA-β gal positive cells at PD immediately after quiescence induction compared to their respective non-quiescence induced MRC-5 (I) and WI-38 (J) controls. The bars indicate the mean ± S.D. Values statistically different from their controls (t-test) are indicated with an asterix: * p<0.05, ** p<0.01, *** p<0.001. n = 3
In cells having experienced periods of quiescence, p21 was up-regulated (S1 and S2 Figs.) [27] while p16 was reduced (S1 and S2 Figs., with the exception of WI-38 at PD 33) compared to proliferating controls. We detected only insignificant differences in expression levels of cyclin D1 (S1 and S2 Figs.) and D2 (S1 and S2 Figs.) and the DNA damage marker γH2A.X (S1 and S2 Figs.) between cells having experienced periods of quiescence and control cells (S1 and S2 Figs.).
Taken together, we observed that the longer times in culture of the cells having been in quiescence (3 times 9 days) compared to control cells, influence cellular properties. Next, we studied MRC-5 and WI-38 cells during and after longer periods of quiescence.
Long-term quiescence of human fibroblasts resulted in senescence induction
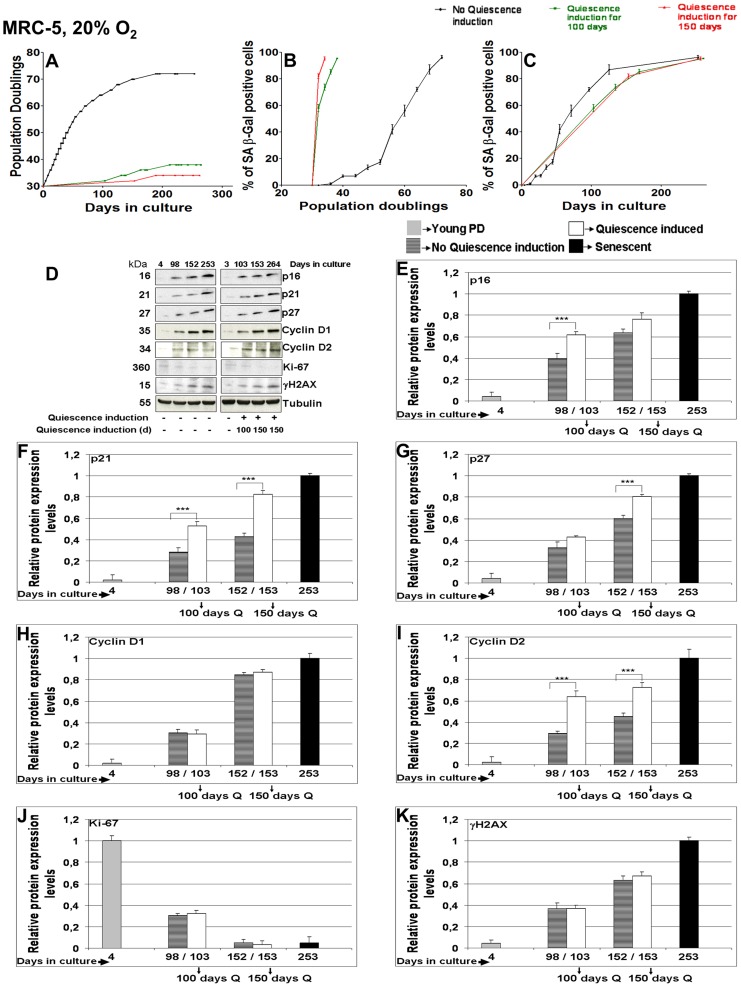
MRC-5 and WI-38 fibroblasts were subjected to long-term quiescence induction by contact inhibition for 100 or 150 consecutive days at 20% O2. After release from long-term quiescence, fibroblasts were able to undergo only three (after 100 days of quiescence) or one (after 150 days) further PD(s) (Figs. 2 A and S3). Thus, the replicative life span of long-term quiescence fibroblasts is severely reduced consistent with previous observations in fibroblasts [28] and in yeast [75]. WI-38 fibroblasts released from long term quiescence showed >70% SA-β Gal positive cells after 100 and 150 days of quiescence (S3 Fig.), whereas corresponding MRC-5 cells showed >55% and >80% SA-β Gal positive cells after 100 and 150 days of quiescence induction, respectively (Fig. 2 B and C).
Figure 2. Effect of long term quiescence induction (100 or 150 days) in MRC-5 fibroblasts maintained at 20% O2.
(A) Growth curve of 3 independent MRC-5 fibroblast cell lines (control with no quiescence induction, and cell lines where quiescence was induced for 100 or 150 days respectively by contact inhibition and then maintained in culture till they approached senescence) maintained in culture at 20% O2 as triplicates from an early PD until senescence at late PDs. Each growth curve is measured in triplicate. Data points of all measurements are displayed (not the mean). (B & C) Percentage of SA-β gal positive cells at different time points of their growth in culture in the control MRC-5 fibroblast cell line and in the cell lines where quiescence was induced for 100 or 150 days respectively. Fig. 2 B and C are plotted with PDs and days in the y-axis respectively. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). (D) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2, Ki-67 and γH2A.X in MRC-5 fibroblast cell lines (subjected to different culture conditions of 100 or 150 days quiescence by contact inhibition and no quiescence induction) maintained in culture at 20% O2 until they approached senescence at late PD. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (E, F, G, H, I, J, K) Comparison of mean fold change of protein expression levels of p16 (E), p21 (F), p27 (G), Cyclin D1 (H), Cyclin D2 (I), Ki-67 (J) and γH2A.X (K) in MRC-5 cell lines where quiescence was induced for 100 or 150 days by contact inhibition respectively compared to controls at corresponding span of time in culture. Cell lines were maintained at 20% O2 as triplicates. The bars indicate the mean ± S.D. *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
The influence of long-term quiescence on WI-38 and MRC-5 cells was further analyzed by measuring the levels of markers for proliferation and cell cycle arrest. For both cell types, the proliferation marker Ki-67 strongly decreased reaching lowest values in the senescent state (Figs. 2 D, J and S3). The levels of all three cell cycle arrest markers p21, p16, and p27 were increased after release from long-term quiescence (100 and 150 days) compared to controls at corresponding PDs (Figs. 2 D, E, F, G and S3). This behavior of p21 and p16 in quiescent cells is in agreement with published observations [1], [2], [18]. We noticed that, compared to controls, the increase of p21 levels, particularly after 150 days of quiescence, was higher than the increase of p16 and p27 levels (Figs. 2 D, F and S3). After long-term quiescence as well as in normally proliferating control cells, in both fibroblast cell lines Cyclin D1 and D2 increased, as described earlier [76]–[78]. In MRC-5 cells, Cyclin D1 levels were very similar to their control levels (Fig. 2 D and H) while Cyclin D2 showed a strong increase at the end of long-term quiescence (Fig. 2 D and I). In contrast, in WI-38 fibroblasts, Cyclin D1 expression levels strongly increased (S3 Fig.) whereas Cyclin D2 levels were significantly higher only in cells at the end of long-term quiescence after 100 days of quiescence but hardly any increase after 150 days (S3 Fig.). For these markers (p16, p21, p27, Cyclin D1 and D2), highest values were measured for MRC-5 and WI-38 cells when being senescent (Figs. 2 D, E, F, G, H, I, J, K and S3). While both fibroblast cell lines, MRC-5 and WI-38, showed a similar overall behavior and comparable marker increases for p16, p21, p27, we observed a clear difference between these two cell lines for Cyclin D1 (compare Fig. 2 H with S3 Fig.), indicating that both cells lines respond individually different to environmental cues, as proposed before [79].
After long-term quiescence induction, the percentage of SA-β Gal positive cells, when plotted versus time in culture, were found to be similar to controls for WI-38 cells, and only slightly delayed for intermediate time periods for MRC-5 cells (Figs. 2 C and S3). We asked if this similar transition into senescence is driven by a similar level of DNA damage. DNA damage foci can be identified by the marker γH2A.X which is recruited to DNA repair sites [80]–[82]. Consistent with published results [20], [83], [84], in both normally proliferating fibroblast cell lines γH2A.X levels increased with age (Figs. 2 K and S3). Indeed, after long-term quiescence an increase of γH2A.X levels, very similar to controls, was observed in MRC-5 as well as in WI-38 fibroblasts (both 100 and 150 days; Figs. 2 D, K and S3).
High-throughput RNA sequencing of MRC-5 cells at PD 32 showed up-regulation of p16, p27, Cyclin D2, Ki-67 and γH2A.X mRNAs after 150 days compared to 9 days of quiescence (unpublished data), correlating with corresponding protein levels. This suggests that up-regulation of the abundance of these proteins during quiescence is due to transcription. We observed that during long-term quiescence MRC-5 or WI-38 fibroblasts cells do not suffer from telomere shorting (data not shown), in agreement with previous reports [28], [85]. Taken together, our observations show that DNA damage accumulation can occur in non-cycling fibroblasts over time independent of the telomere status.
Long-term quiescence induction at 3% O2
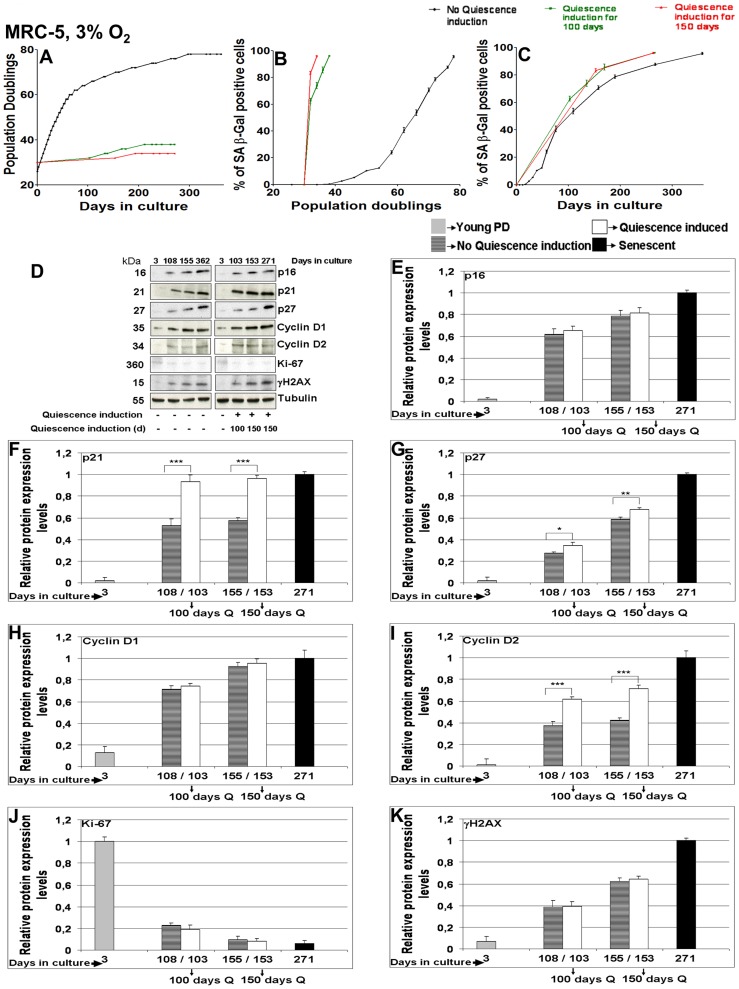
A main source of DNA damage accumulation may be ambient oxygen concentration. We reasoned that reducing O2 levels from 20% down to physiological 3% [86] may reduce cellular DNA damage, resulting in lower γH2A.X levels and a delayed transition into senescence [87], [88]. Therefore, MRC-5 and WI-38 were now subjected to long-term quiescence induction by contact inhibition for 100 or 150 consecutive days at 3% O2 (Figs. 3 A and S4). After release from long-term quiescence at 3% O2, MRC-5 and WI-38 fibroblasts were able to undergo only three or one more PD(s), respectively (Figs. 3 A and S4). We detected >60% and >80% SA-β Gal positive cells in MRC-5 and WI-38 fibroblasts maintained at 3% O2 after 100 and 150 days of quiescence, respectively (Figs. 3 B, C and S4). The proliferation marker Ki-67 strongly decreased with time for MRC-5 and WI-38 cells being released from long-term quiescence as well as for proliferating control cells (Figs. 3 D, J and S4). In MRC-5 fibroblasts maintained at 3% O2, p16 (Fig. 3 D and E) and Cyclin D1 (Fig. 3 D and H) values were similar to control values and p27 (Fig. 3 D and G) values only slightly increased for cells released from long-term quiescence. In contrast, p21 (Fig. 3 D and F) and Cyclin D2 (Fig. 3 D and I) values strongly increased in cells released from long-term quiescence compared to proliferating control cells. In WI-38 fibroblasts maintained at 3% O2, the expression levels of all cell cycle markers analyzed (p16, p21, p27, Cyclin D1 and D2) increased after release from 100 or 150 days of quiescence compared to proliferating controls of corresponding PDs (S4 Fig.). Furthermore, the levels of DNA damage marker γH2A.X revealed no differences between MRC-5 and WI-38 fibroblasts cells after release from long-term quiescence (both 100 and 150 days) compared to normally proliferating controls (Figs. 3 D, K and S4). Importantly, after about 100 or 150 days in culture, γH2A.X levels were very similar in MRC-5 cells either kept under 20% or 3% O2 (Figs. 3 K and S4). We therefore conclude that a non-physiological increased oxygen level, at least the one applied here, seems not to be the direct linear source of DNA damage accumulation during quiescence or senescence. The cellular effect of oxygen may be complex: oxygen levels were found to influence the transition into senescence in a non-proportional way with low oxygen having a protective effect only at the end of the cellular lifespan [89], [90].
Figure 3. Effect of long term quiescence induction (100 or 150 days) in MRC-5 fibroblasts maintained at 3% O2.
(A) Growth curve of 3 independent MRC-5 fibroblast cell lines (control with no quiescence induction, and cell lines where quiescence was induced for 100 or 150 days respectively by contact inhibition and then maintained in culture till they approached senescence) maintained in culture at 3% O2 as triplicates from an early PD until senescence at late PDs. Each growth curve is measured in triplicate. Data points of all measurements are displayed (not the mean). (B & C) Percentage of SA-β gal positive cells at different time points of their growth in culture in the control MRC-5 fibroblast cell line and in the cell lines where quiescence was induced for 100 or 150 days respectively. Fig. 3 B and C are plotted with PDs and days in the y-axis respectively. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). (D) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2, Ki-67 and γH2A.X in MRC-5 fibroblast cell lines (subjected to different culture conditions of 100 or 150 days quiescence by contact inhibition and no quiescence induction) maintained in culture at 3% O2 until they approached senescence at late PD. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (E, F, G, H, I, J, K) Comparison of mean fold change of protein expression levels of p16 (E), p21 (F), p27 (G), Cyclin D1 (H), Cyclin D2 (I), Ki-67 (J) and γH2A.X (K) in MRC-5 cell lines where quiescence was induced for 100 or 150 days by contact inhibition respectively compared to controls at corresponding span of time in culture. Cell lines were maintained at 3% O2 as triplicates. The bars indicate the mean ± S.D. * p<0.05, ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
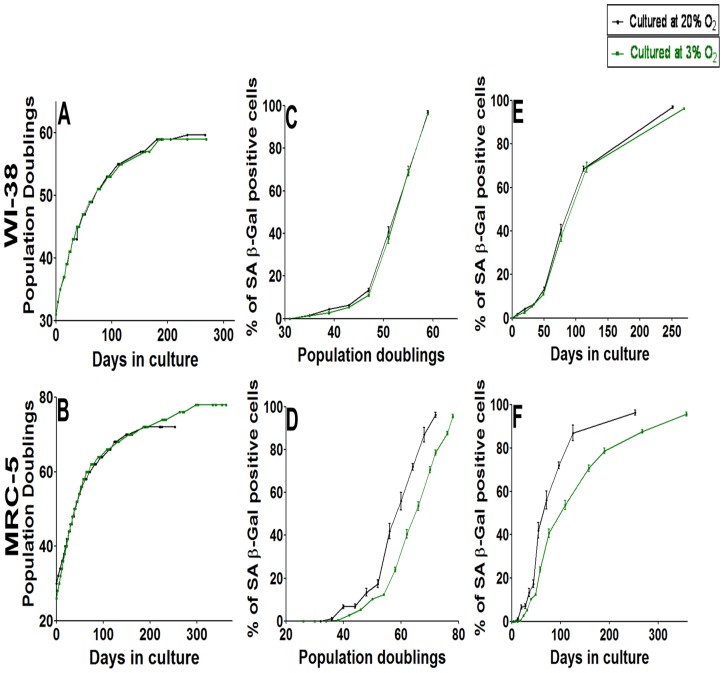
We then asked if the low oxygen level had an influence on the aging of these fibroblast cells. We found that WI-38 fibroblasts, maintained at 3% O2, approached senescence quantitatively in the same way as those maintained at 20% O2, as shown by the growth curve (Fig. 4 A) and the percentage of SA-β Gal positive cells (Fig. 4 C and E). Instead, MRC-5 fibroblasts, maintained at 3% O2, had a longer lifespan compared to cells kept at 20% O2 (Fig. 4 B). Under low O2 levels, MRC-5 cells were able to divide even after 300 days in culture while cells under 20% O2 underwent their last division after ca. 185 days, and MRC-5 cells at 3% O2 were able to undergo considerably more PDs (cumulative PD = 78) compared to those at 20% O2 (cumulative PD = 72). Parallel to the MRC-5 growth curve (Fig. 4 B), the transition into senescence is delayed for MRC-5 maintained at 3% O2 (Fig. 4 D and F). The results indicate a clear difference in the behavior of WI-38 compared to MRC-5 cells: reducing the oxygen level from 20% to 3% did not change the aging of WI-38 cells but delayed aging in MRC-5 cells. Considerable cell-strain variability in response to oxygen had been observed before [87].
Figure 4. Comparison of the effect of O2 levels (3 or 20%) in culture on the growth curve and induction of senescence revealed by β gal in MRC-5 and WI-38 fibroblasts.
(A & B) Growth curve of 2 independent WI-38 (A) and MRC-5 (B) fibroblast cell lines maintained in culture at 20% or 3% O2 till they achieved senescence at late PD. Data points of all measurements are displayed (not the mean). (C & D) Percentage of SA-β gal positive cells at different time points of their growth in culture in WI-38 (C) and MRC-5 (D) fibroblast cell lines maintained at 20% or 3% O2. The figures were plotted with PDs on the x-axis (E & F) Percentage of SA-β gal positive cells at different time points of their growth in culture in WI-38 (E) and MRC-5 (F) fibroblast cell lines maintained at 20% or 3% O2. The figures were plotted with days in culture on the x-axis. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). n = 3.
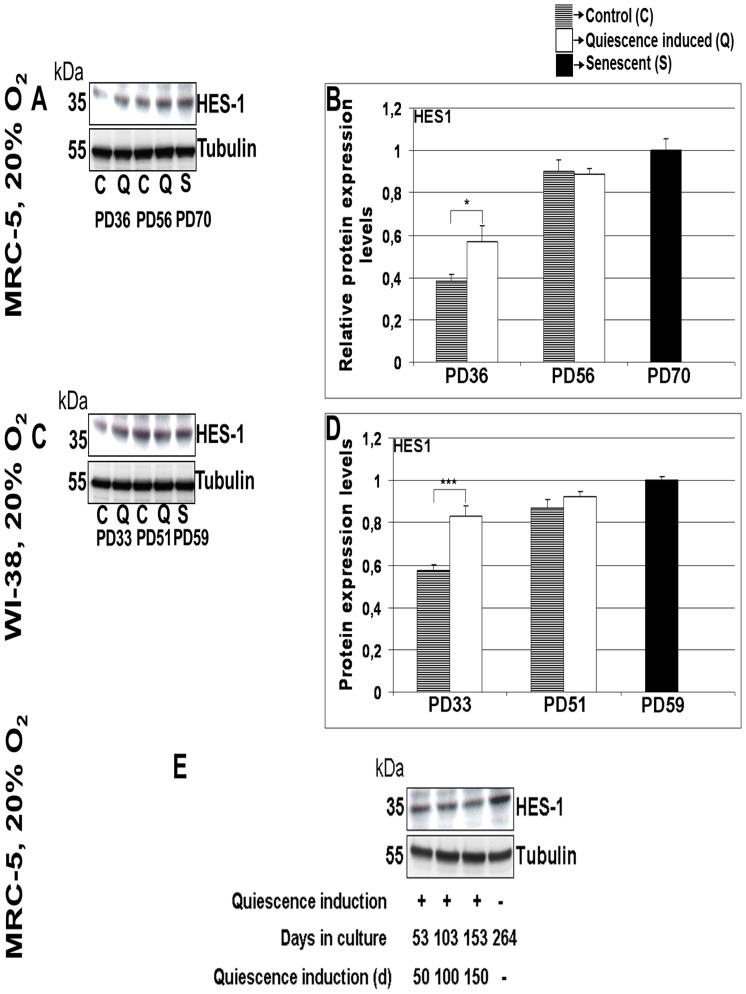
HES1 is up-regulated in long-term quiescent and senescent fibroblast cells
In quiescent cells, the transcriptional repressor HES1 is up-regulated and, during a period of several days, prevents transition into senescence although during quiescence p21 is up-regulated [27]. Thus, HES1 guarantees reversibility of quiescent cells into the cell cycle. Consistent with these observations, we detected only minor influences of short term quiescence on the number of SA-β Gal positive cells compared to normally proliferating control cells (Fig. 1) despite detecting p21 up-regulation (S1 and S2 Figs.). After short-term (9 days) quiescence of MRC-5 and WI-38 cells, we observed an up-regulation of HES1 compared to the HES1 level in normally proliferating cells (Fig. 5 A, B, C and D) while after 3 times 9 days of quiescence, the HES1 levels had further increased and now are similar to the HES1 levels in control cells (Fig. 5 A, B, C and D). We then asked if HES1 remains up-regulated also after long-term quiescence. We found high levels of HES1 in MRC-5 (Fig. 5 E) as well as WI-38 cells after 100 and 150 days of quiescence independent of 3% or 20% oxygen levels. Surprisingly, we find high levels of HES1 in both cells types also when being senescent (Fig. 5). In order to check that the antibody indeed recognizes HES1, we cloned HES1 in fusion with GFP and verified that the HES1 antibody recognizes the fusion protein which in parallel is identified by an anti-GFP antibody (data not shown). Thus, HES1 remains up-regulated; however, after long-term quiescence it is no longer able to prevent transition into senescence (as indicated by our data above).
Figure 5. Expression levels of HES1 in MRC-5 and WI-38 fibroblasts subjected to short term or long term quiescence induction.
(A) The blot show the protein expression levels of HES1 in two MRC-5 fibroblast cell lines (control with no quiescence induction and a cell line where quiescence was induced 3 times separately for a span of 9 days) maintained at 20% O2 at different stages of their span in culture. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (B) Comparison of mean fold change of protein expression levels of HES1 in 3 times quiescence induced MRC-5 cell lines and control MRC-5 cell lines maintained in culture as triplicates. (C) The protein expression levels of HES1 in two WI-38 fibroblast cell lines (control with no quiescence induction and a cell line where quiescence was induced 3 times separately for a span of 9 days) maintained at 20% O2 at different stages of their span in culture. (D) Comparison of mean fold change of protein expression levels of HES1 in 3 times quiescence induced WI-38 cell lines and control WI-38 cell lines maintained in culture as triplicates. The bars indicate the mean ± S.D. * p<0.05, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3 (E) The protein expression levels of HES1 in MRC-5 fibroblast cell lines maintained at 20% subjected to 50, 100 or 150 days of quiescence by contact inhibition and in senescent state. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. n = 2.
Quiescent as well as senescent fibroblast cells are protected from transition into apoptosis
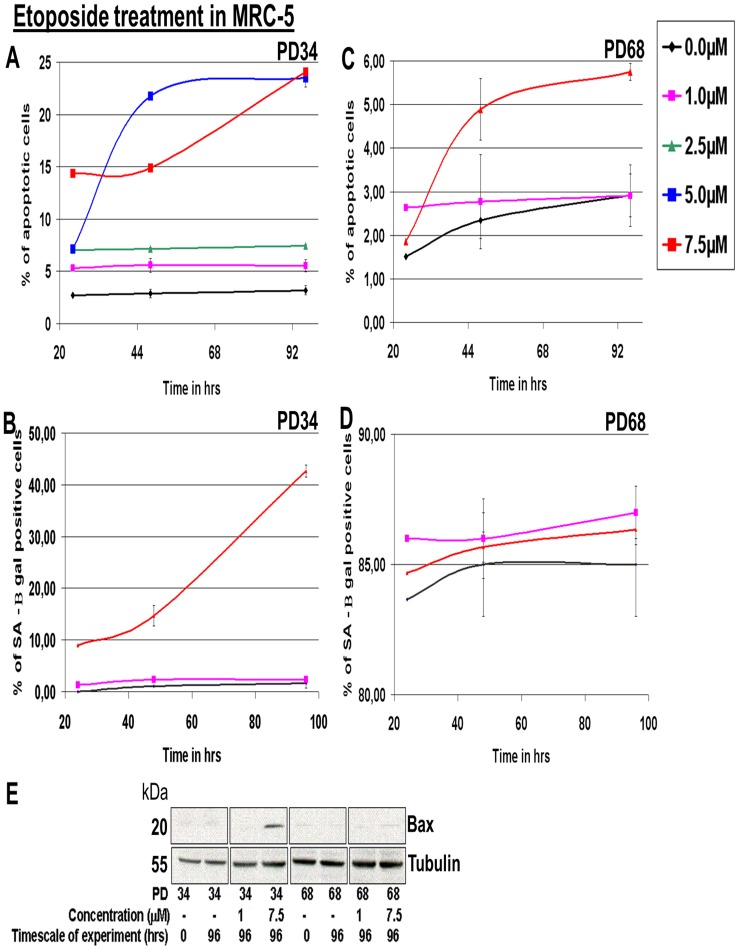
To avoid danger to the cell and its framing tissue, not only normally proliferating but also quiescent cells must be able to react to harmful stress by inducing apoptosis. We therefore analyzed the potential of fibroblast cells for apoptosis induction. As inducing agents, we applied Etoposide [91] as well as Staurosporine [92] and detected the apoptotic state by immunostaining of Bax phosphorylation [93], [94]. Young MRC-5 fibroblasts at PD 34 were exposed to increasing concentrations of Etoposide (0.0 to 7.5 µM). As detected by immunostaining in Western blots, 5.0 µM or 7.5 µM of Etoposide resulted in 23% apoptotic MRC-5 fibroblasts after 96 hrs of treatment in culture compared to 3% apoptotic control cells at 0.0 µM Etoposide (Fig. 6 A). Treatment of young MRC-5 fibroblasts with 7.5 µM Etoposide also resulted in the induction of pro-apoptotic Bax protein (Fig. 6 E). Exposure to 7.5 µM Etoposide for 96 hrs revealed an induction of ca. 43% SA-β Gal positive cells while lower Etoposide concentrations did not induce senescence (Fig. 6 B). In contrast, when old MRC-5 fibroblasts at PD 68 were exposed to Etoposide for 96 hrs, 2–3% induction of apoptosis was observed for 1 µM and only a slight increase from 3–6% at 7.5 µM (Fig. 6 C). This minor induction could be explained by the small amount of non-senescent cells in the population being induced to apoptosis. Consistent with these results, Bax proteins were not induced as revealed by Western blots (Fig. 6 E). Treatment of old MRC-5 fibroblasts at PD 68 displayed about 85% SA-β Gal positive cells independent of Etoposide concentrations (0.0, 1.0 or 7.5 µM; Fig. 6 D). A similar behaviour was observed when the cells were treated with Staurosporine: increasing concentrations of 0.0, 0.1, 0.5, 1.0, and 2.0 µM Staurosporine in the serum of young MRC-5 fibroblasts at PD 34 resulted in an increase in the percentage of apoptotic cells. 1.0 and 2.0 µM Staurosporine were able to induce apoptosis in 20–25% of cells. The presence of apoptotic cells was also detected by Bax protein induction. 2.0 µM Staurosporine induced senescence in more than 50% of the cells. When old MRC-5 fibroblasts (PD 68) were exposed to 2.0 µM Staurosporine for 96 hrs, an only minor induction of apoptosis was detected (from 2% to 4%) while about 85% of the cells were SA-β Gal positive (Fig. S5). In summary, apoptosis can be induced in young fibroblast cells; however, even at high concentrations of inducing agents, apoptosis is induced only to a minor extent in a senescent fibroblast cell population, consistent with [95].
Figure 6. Impact of Etoposide treatment in young and old PD MRC-5 fibroblasts.
(A) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (young PD = 34) treated with different concentrations of Etoposide for different time spans (B) Percentage of SA-β gal positive cells in MRC-5 fibroblast cell lines (young PD = 34) treated with different concentrations of Etoposide for different time spans (C) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (old PD = 68) treated with different concentrations of Etoposide for different time spans (D) Percentage of SA-β gal positive cells in MRC-5 fibroblast cell lines (old PD = 68) treated with different concentrations of Etoposide for different time spans. In each instance, MRC-5 fibroblasts were maintained in culture at 20% O2 as triplicates. In A & C, the bars indicate the mean ± S.D. In B & D error bars indicate ± S.E). (E) The blots show absence or induction of apoptotic protein Bax in MRC-5 fibroblast cell lines (young PD = 34 & old PD = 68) treated with 1.0 or 7.5 µM of Etoposide for 96 hrs compared to controls (0 hrs). The MRC-5 fibroblasts were maintained in culture at 20% O2. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. n = 3.
Then we asked if high concentrations of these agents can induce apoptosis in quiescent cells. Treating short-term (9 day) quiescent MRC-5 fibroblasts (PD 35) for 96 hours with 7.5 µM Etoposide or 2.0 µM Staurosporine revealed 16% and 13% apoptotic cells, respectively (S6 Fig.), as indicated by Bax phosphorylation. Thus, apoptosis can be induced in short-term quiescent cells, however, to a reduced amount compared to normally proliferating cells. In long-term (150 days) quiescent MRC-5 fibroblasts (PD 32), 7.5 µM Etoposide or 2.0 µM Staurosporine were unable to induce significant levels of apoptosis (S6 Fig.): the percentage of apoptotic cells of 4–5% in young (PD 32) fibroblasts subjected to 150 days of quiescence were similar to the percentage of apoptotic cells observed in senescent fibroblasts (PD 68) (S6 Fig.). Supporting this result, the treatment of long-term quiescence fibroblasts (150 days) with either Etoposide or Staurosporine did not induce the pro-apoptotic protein Bax (S6 Fig.). Thus, consistent with earlier findings [96], during quiescence the cells lose their potential for apoptosis induction, as do normally proliferating cells during their life span.
Differential regulation of mRNA expression in long-term quiescent fibroblasts is similar to that in senescent cells
We observed that long-term quiescent fibroblasts transit into senescence by a similar rate as do normally proliferating cells (Figs. 3 C and S4). We speculated that this similarity in rates may be due to similar cellular mechanisms ruling this transition in both cellular states. We therefore compared gene expression profiles, measured by RNA-seq, of young MRC-5 cells (PD 32) with long-term quiescent MRC-5 fibroblasts (subjected to 150 days contact inhibition). We identified 1,499 up- and 1,216 down-regulated differentially expressed genes (DEGs). As a next step, we intersected these DEGs with senescence-regulated genes identified by [97] which resulted in 60 up- and 194 down-regulated DEGs (S7 Fig.). Identification of the common significantly enriched GO terms and KEGG pathways between the two data sets (150 days quiescent versus senescent fibroblasts) revealed that “cell cycle (GO:0007049)”, “ATP binding (GO:0005524)”, and “DNA repair (GO:0006281)” were among the most significantly down-regulated pathways (data not shown). A number of similarly regulated KEGG pathways found by gene set enrichment analysis, were obtained for both datasets (S8 Fig.), again identifying “repair” as one of the commonly down-regulated pathways. Thus, both, quiescent and normally proliferating cells, transit into senescence by modifying the same pathways (in particular down-regulating the DNA repair pathway) by similar amounts, i.e. they transit into senescence due to similar cellular processes.
Discussion
Several independent signals (like mitogen withdrawal, contact inhibition, loss of adhesion, or RAS induction) induce cellular quiescence, a reversible cell cycle arrest, by up-regulation of the CDK inhibitor p21. Each of these signals causes regulation of a unique set of genes known to terminate growth and division [15]. Nevertheless, underlying this diversity of states, the expression of a set of genes was found to be independent of the quiescence induction signal, and thus describing a general quiescent state, regardless of the signal that induced it [15]. Reversibility of quiescence into the cell cycle is insured by suppressing the transition into terminal differentiation [15]. Furthermore, short-term quiescent cells are described to be protected against transition into senescence by up-regulation of the transcriptional repressor HES1 [27]. Considered to being permanently reversible, long-term quiescent cells would be cancer-prone: although not dividing, the quiescent cells accumulate DNA damage (as shown here), due to their re-directed but still high metabolic activity [9]. Low level DNA damage is considered to be routinely repaired (maintenance, see [79]) while, in normally proliferating cells, harmful damage induces apoptosis. However, after long-term quiescence, as we show here in agreement with others [96] apoptosis induction is strongly suppressed which, after return into the cell cycle, immediately increases the potential for tumor development. In order to avoid this dangerous fate, while being quiescent for longer periods of time, cells might transit into senescence. Indeed, quiescent HFF fibroblast cells were found to become senescent [28]. This however, when being of general nature, would contradict the HES1 protection mechanism and the definition of quiescence, i.e. that all quiescent cells can return into the cell cycle.
We resolved this contradiction in cultured cells by analyzing further human primary fibroblasts (to what extent these results hold in vivo, remains to be shown). We induced short-term and long-term quiescence in MRC-5 and WI-38 fibroblasts. After short-term (9 day) quiescence we observed p21 and p27 up-regulation but no induction of p16 (S1 and S2 Figs.), in agreement with earlier findings [1], [18]. In these cells, HES1 was up-regulated (Fig. 5 A, B, C, D); DNA damage was not induced as indicated by low γH2A.X levels (S1 and S2 Figs.), and the growth curve was similar to that of normally proliferating control cells. These and the SA-β Gal results show that, after short-term quiescence, these cells were in a reversible cell cycle arrested state with no indications for transition into senescence. This marker pattern strongly changed after long-term quiescence. Still, after 100 and 150 days of quiescence, p21 and p27 were up-regulated, but now also p16, while the proliferation marker Ki-67 decreased, the DNA damage marker γH2A.X increased, and the growth curve strongly changed (Fig. 2). The number of SA-β Gal positive cells, when plotted versus population doublings (PDs), showed a markedly different behavior compared to that of normally proliferating control cells. We observed that during long-term quiescence, DNA damage accumulates at comparable amounts as in control cells (as detected by γH2A.X levels), and most of the quiescent cells transit into senescence (as indicated by a similar increase of SA-β Gal positive cells), with only a small part of the population still being proliferative active (Ki-67 positive).
Thus, not all quiescent cells are able to return into the cell cycle but, after release from quiescence, the cell population is re-established out of the small non-senescent cell fraction. When indeed during long-term quiescence the proliferative cell fraction of the population decreases, then, after release from quiescence, re-establishing a proliferative population must take the longer, the longer the cells have been quiescent. This prediction agrees with our observations and published findings: cells deprived of serum for weeks rather than days, took longer to re-enter the cell cycle [98]–[100], correlating with cell size [8]. We observed that cells having experienced quiescence subsequently grow slower (Fig. 1 G and H). Furthermore, the time required to grow to confluency (time required for 1 PD) is clearly longer after 100 or 150 days of quiescence than required by young proliferating cells, instead it relates to the time required by old cell populations close to senescence.
When plotting the number of SA-β Gal positive cells versus number of days in culture, during and after long-term quiescence the cells behaved rather similar to control cells. This similar transition rate into senescence made us speculate that with time (days in culture) during quiescence as well as during normal proliferation, similar cellular processes within these cells might rule this transition. We therefore compared the gene expression profile of young with that of long-term quiescent fibroblasts, and determined which genes are either up- or down-regulated when cells are in long-term quiescence. Interestingly, we found DNA repair pathway genes down-regulated, explaining accumulation of DNA damage in long-term quiescent cells. These differentially expressed genes were compared to those recently found when comparing young and senescent fibroblast cells [97]. In both data sets, we identified a number of pathways which are differentially regulated in a similar way. For example [97] found that also in senescent cells, the DNA repair pathway is down-regulated. Thus, quiescent and normally proliferating cells transit into senescence not only in similar time frames but also due to similar cellular mechanisms. As observed here for fibroblasts, during quiescence also hematopoietic stem cells (HSCs) were recently found to accumulate DNA strand breaks [101], and also these cells transcriptionally attenuated the DNA damage response and repair (DDR) pathway.
Our observations suggest the following model for fibroblasts in cell culture. Cells proliferate with the rate “r” [79]. Whenever repair is required, the cells exit the cell cycle for a short period of time by up-regulating the CDK inhibitor p21, carry out the repair and return into the cell cycle. This “maintenance” reduces the rate of cell growth [79]. When cells are unable to complete the repair process, the cells transit into senescence, with only a very minor part of the population still being proliferatively active (as indicated by the low but detectable Ki-67 levels). This transition correlates with the up-regulation of the CDK inhibitor p16. The p53-p21 pathway is connected to the p16-pRB pathway [55], [59], with p16 being up-regulated after some time delay (estimated from our data: ca. 30–40 days). We suggest that this delayed response of the p16 pathway is the time frame the cells allow for cell cycle reversible DNA damage response, the process of normal cellular maintenance. Once this time is used up, p16 up-regulation induces a permanent cell cycle arrest, which in human cells is not necessarily irreversible [57]. Consistent with this view, (i) murine p16 knock-out cells do not become senescent [102], [103], (ii) p16 is inactivated in many human tumors [104], [105], and (iii) ectopic expression of p16 in human cancer cell lines being negative in p16 expression, induced growth arrest and senescence [106], [107]. Here we show that a corresponding process also takes place during long-term quiescence; indeed, quiescent and cells in maintenance transit into senescence due to similar cellular mechanisms. Although we detected quantitative differences between cell types, the same qualitative behavior was observed between MRC-5 and WI-38 cells. It remains to be shown if this model also holds true for cells in tissue. We observed that apoptosis could be induced in young but hardly in senescent cells [91], [95] in contrast to observations in HS74 [108] and PAEC cells [109], discussed in [37]. Correspondingly, after short-term (9 days) but hardly after long-term (150 days) quiescence, apoptosis could be induced, again indicating a complementary development over time in proliferating and quiescent cells. Interestingly, senescent cells cannot transit into quiescence [100].
Fibroblast cells can divide up to about 70 times before stably arresting and becoming senescent [29]. Senescent cells, being arrested in the cell cycle, remain metabolically active but prevent cancer development. Shortening of telomeres is considered to be an origin for this transition into senescence: telomere shortening might act as a counting mechanism (“replicometer”) triggering replicative senescence in normal diploid cells since these cells do not contain telomerase [110], [111]. Telomere shortening triggers a p53/p21-dependent cell cycle arrest through accumulation of G-rich single stranded DNA fragments. This prevents replication of damaged DNA, allowing sufficient time for repair [112], [113]. If cells are unable to repair the DNA damage, p16 is up-regulated and the arrest becomes permanent [57]. If telomere shortening would be the main mechanism triggering senescence, during non-replicating quiescence, cells should not transit into senescence and time in the quiescent state would not count for their replicative life span since telomeric attrition strongly depends on cell division, both in vitro [85], [114] and in vivo [114]. In contrast to this and consistent with [28], we find that quiescent human fibroblast cells age with a rate similar to normally proliferating cells. Thus, in human fibroblasts, telomere shortening cannot be the dominant mechanism triggering senescence but factors other than mere telomere length, or different mechanisms must drive this transition [67]. Consistently, in human cells expression of telomerase did not reverse the senescence arrest [57]. Further arguments support this notion: individual cells from clonally derived populations show heterogeneous division potential [115], and the fraction of senescent cells, present in a large population, increases progressively with PDs, and not most of them together at a given PD. This indicates that the lifespan of an individual cell lineage is not controlled simply by telomere shortening during each round of cell division, but instead also by sensing genotoxic stresses [116] or other independent mechanisms upstream of telomere shortening [37], like for example: enhanced cellular ability to bind DNA-ends may be important for longevity [66], and telomerase might down-regulate the p16 pathway [103], [117].
We speculated that high oxygen levels might induce the increasing amount of DNA damage. We reduced the extracellular oxygen levels from 20% to physiological 3% and detected no or only minor quantitative differences. Thus, the increasing amounts of DNA damage and the increase of SA-β Gal positive cells with time in culture are not mainly due to high (20%) oxygen levels; instead we consider internal cellular processes like metabolic effects responsible for this. Furthermore, when high oxygen stress (inducing DNA damage and not telomere shortening) would trigger senescence in quiescent cells, lower oxygen concentrations should delay the transition into senescence [87], [88]. However, in quiescent WI-38 cells the transition into senescence was not altered when reducing the oxygen level from 20% to 3%; thus, WI-38 cells do not respond to this difference in stress. MRC-5 did sense the reduction in stress level: during quiescence, the increase of the number of SA-β Gal positive MRC-5 cells was slightly delayed. Nevertheless, MRC-5 cells became senescent also under low oxygen levels. These results clearly identify quantitative differences in the behavior of these human cell lines, as detected before [79], [87]. Thus, other cellular processes must be involved in senescence induction [49], [118] which must be active in non-replicating quiescent cells also at low oxygen levels [119]. We speculate that cellular maintenance is the common basic mechanism driving normally proliferating as well as quiescent cells into senescence.
Materials and Methods
Cell culture
Primary human lung fibroblasts MRC-5 (derived from normal lung tissue of a 14 week old male fetus) and WI-38 (derived from 12 week old female fetus) were obtained from ATCC (LGC Promochem GmbH, Wesel, Germany) at young population doublings (PD) of 26-28. Cells were cultured in Dulbeccos modified Eagles medium (DMEM) with L-Glutamine, low glucose (PAA Laboratories, Pasching, Austria), supplemented with 10% fetal bovine serum (FBS) (PAA Laboratories, Pasching, Austria). Cell culture was carried out under normal air conditions in a 9.5% CO2 atmosphere at 37°C. For subculturing procedures, 1x PBS (pH 7.4) (PAA Laboratories, Pasching, Austria) and trypsine/EDTA (PAA Laboratories, Pasching, Austria) were used. Fibroblast cultures approaching confluency were splitted at a ratio of 1∶4. For replicative senescence, fibroblasts were maintained until the end of their lifespan (>95% of cells SA-β Gal positive). Quiescence induction was performed by contact inhibition or serum starvation. For contact inhibition, quiescence was induced either repetitively (3 times for 9 days during the culture span of the fibroblasts) or in long-term (100 or 150 days of consecutive quiescence induction). In MRC-5 fibroblasts, short-term quiescence was induced during PDs 36, 44 and 56 whereas in WI-38 fibroblasts, short-term quiescence was induced at PDs 33, 43 and 51. Once the cells were confluent at their respective PDs, they were left in the confluent state for 9 days with media change (containing 10% FBS) every 3 days (contact inhibition). Then the cells were split and transferred back to normal culture conditions with subsequent canonical splitting. With respect to the long-term quiescence induction, the cells were allowed to reach confluence at early PDs and were left in a confluent state for 100 or 150 days. In this instance, media (containing 10% FBS) was changed every 3 days. Otherwise, quiescence was induced for 8 days by incubation of cells with serum-deprived medium, DMEM supplemented with 0.5% FBS (serum starvation). After addition of normal growth medium, cells resumed proliferation.
SA-β Gal activity assay
SA-β Gal activity was determined as described by [38]. SA-β Gal was measured for fibroblasts in culture at every four PDs, analyzing mean values ± standard deviation of 3×60 cells, each.
Western blotting
For Western blotting, 104 cells/µl were used per lane. Immunodetection was performed using 5%-powdered milk in PBS-T (1xPBS, pH 7.4 and 1% Tween20) for blocking (Roth, Germany). Primary antibodies, anti-p21 mouse antibody (OP64; Calbiochem; dilution 1∶200), anti-p16 mouse antibody (550834; BD Pharmingen; 1∶200), anti-p27 rabbit antibody (sc-528; Santa Cruz; 1∶200), anti-γH2AX (07-164; Millipore; 1∶50), anti-Cyclin D1 rabbit antibody (ab16663; Abcam; 1∶500), anti-Cyclin D2 mouse antibody (ab3805; Abcam; 1∶500), anti-Ki-67 mouse antibody (ab6526; Abcam; 1∶200), anti-HES1 rabbit antibody (sc-25392; Santa Cruz; 1∶200), anti-Bcl-2 (IMG-80093; IMGENEX; 1∶200), anti-Bax (IMG-80165; 1∶250) and anti-tubulin mouse antibody (T-9026; SIGMA-Aldrich; 1∶5000) were diluted as indicated in 5%-powdered milk (in PBS-T) and incubated for one hour at room temperature. Washing steps were performed 3×10 min in 1×PBS-T. The secondary horseradish peroxidase-labeled antibodies (Jackson Immuno Research Lab) were incubated for 1 hr at room temperature. Detection of horseradish peroxidase was performed using ECL-detection system and radiographic film (GE Healthcare, Germany). After film development, quantification of signal intensities of the bands in the Western blots was carried out using Metamorph software.
Treatment of human embryonic lung fibroblasts with apoptotic agents
Two well established apoptotic inducers; Staurosporine [92], [120]–[122] and Etoposide [91], [123] were used in this study. Young (PD = 34) and aged (PD = 68) MRC-5 fibroblasts were treated with different concentrations of Staurosporine (0.1, 0.5, 1.0, 2.0 µM) or Etoposide (1.0, 2.5, 5.0, 7.5 µM) for different time spans (24, 48, 96 hrs) and maintained in culture at 20% O2. The fibroblasts were subjected to Etoposide or Staurosporine treatment after every 24 hrs of their span in culture. The percentage of SA-β Gal positive and apoptotic cells were investigated in the MRC-5 fibroblasts after different spans of treatment with Staurosporine and Etoposide.
Detection of apoptotic cells by Hoechst staining using flow cytometry
A BD FACS Canto II was used for flow cytometry. Ca. 106/ml of MRC-5 fibroblasts cells treated with different concentrations of apoptotic agents were stained with Hoechst 33342 and propidium iodide staining to detect the percentage of apoptotic cells [120]. Hoechst 33342 blue fluorescence dye (excited by 405 nm laser line, emission detected using a 450±50 nm band pass filter) stains the condensed chromatin of apoptotic cells brighter than the chromatin of non-apoptotic cells, while the red fluorescence dye propidium iodide (excited by 488 nm laser line, emission detected using a 670 nm long pass filter) permeates only into dead cells, enabling the differentiation of dead from apoptotic cells. The percentage of apoptotic cells was then retrieved by performing flow cytometry [124]. Experiments were performed according to the protocols of the manufacturer (BD Bioscience) and fluorescence signals were analyzed using the FACS Diva software 6.1.7 (BD Bioscience).
High-throughput RNA sequencing
For quality check, total RNA was analyzed using Agilent Bioanalyzer 2100 (Agilent Technologies) and RNA 6000 Nano Kit (Agilent) to ensure appropriate RNA quality in terms of degradation. Total RNA was used for Illumina library preparation and next-generation sequencing. Around 2.5 µg total RNA was used for indexed library preparation using Illumina's TruSeq RNA Sample Prep Kit v2 following the manufacturer's instruction. Libraries were pooled and sequenced using Illumina HiSeq2000 and HiSeq2500 sequencing machines in single read mode with 50 cycles using sequencing chemistry v3. Reads were extracted in FastQ format using CASAVA v1.8.2 (Illumina).
RNA-seq data analysis
Raw data sequencing results were received in FastQ format. Read mapping was performed using Tophat 2.0.6 [125] and the human genome references assembly GRCh37.66 obtained from Ensembl. Uniquely mapped reads were counted for all genes using featureCounts [126]. RPKM values were computed using exon lengths provided by featureCounts and the sum of all mapped reads per sample. Differentially expressed genes (DEGs) were identified using the DESeq [127], edgeR [128] and baySeq [129] statistical software analysis tools. The resulting p-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate (FDR) [130]. For comparing DEGs from this study with data by [97] the same statistical cutoffs have been used (FDR of all three tests <0.05 and absolute log2 fold-change >1). P-values for the overlap of gene lists were calculated using the web tool at http://nemates.org/MA/progs/overlap_stats.html with a total number of genes of 16,035 representing the number of comparable genes between both datasets. Singular enrichment analysis was performed for the intersection of either up- and down-regulated DEGs found in both studies using DAVID [131]. Generally applicable gene set enrichment for pathway analysis (GAGE) [132] was used in order to detect significantly regulated KEGG pathways (FDR adjusted p<0.01) while log2 fold-changes and normalized gene counts have been used for data by [97] and data of this study, respectively.
Statistical analysis of the samples
Where appropriate, quantitative results were examined for statistical significance using paired two-sample type 2 Student's t-tests assuming equal variances; p values are presented where appropriate.
Ethics Statement
The human fibroblast cell lines (MRC-5 and WI-38) used in this investigation was ordered from ATCC. A number of senescence related studies has been undertaken using MRC-5 and WI-38 [133], [134], [135] fibroblast cell lines.
Supporting Information
Effect of short term quiescence induction on protein expression levels of a number of cell cycle associated genes and a marker for DNA damage in MRC-5 fibroblasts. (A) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2 and γH2A.X in two MRC-5 fibroblast cell lines (control with no quiescence induction and a cell line where quiescence was induced 3 times separately for a span of 9 days) maintained at 20% O2 at different stages of their span in culture. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (B, C, D, E, F, G) Comparison of mean fold change of protein expression levels of p16 (B), p21 (C), p27 (D), Cyclin D1 (E), Cyclin D2 (F) and γH2A.X (G) in 3 times quiescence induced MRC-5 cell lines and control MRC-5 cell lines maintained in culture as triplicates. The bars indicate the mean ± S.D. ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
(TIF)
Effect of short term quiescence induction on protein expression levels of a number of cell cycle associated genes and a marker for DNA damage in WI-38 fibroblasts. (A) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2 and γH2A.X in two WI-38 fibroblast cell lines (control with no quiescence induction and a cell line where quiescence was induced 3 times separately for a span of 9 days) maintained at 20% O2 at different stages of their span in culture. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (B, C, D, E, F, G) Comparison of mean fold change of protein expression levels of p16 (B), p21 (C), p27 (D), Cyclin D1 (E), Cyclin D2 (F) and γH2A.X (G) in 3 times quiescence induced WI-38 cell lines and control WI-38 cell lines maintained in culture as triplicates. The bars indicate the mean ± S.D. ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
(TIF)
Effect of long term quiescence induction (100 or 150 days) in WI-38 fibroblasts maintained at 20% O2. (A) Growth curve of 3 independent WI-38 fibroblast cell lines (control with no quiescence induction, and cell lines where quiescence was induced for 100 or 150 days respectively by contact inhibition and then maintained in culture till they approached senescence) maintained in culture at 20% O2 as triplicates from an early PD until senescence at late PDs. Each growth curve is measured in triplicate. Data points of all measurements are displayed (not the mean). (B & C) Percentage of SA-β gal positive cells at different time points of their growth in culture in the control WI-38 fibroblast cell line and in the cell lines where quiescence was induced for 100 or 150 days respectively. S3B and S3C Figs. are plotted with PDs and days in the y-axis respectively. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). (D) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2, Ki-67 and γH2A.X in WI-38 fibroblast cell lines (subjected to different culture conditions of 100 or 150 days quiescence by contact inhibition and no quiescence induction) maintained in culture at 20% O2 until they approached senescence at late PD. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (E, F, G, H, I, J, K) Comparison of mean fold change of protein expression levels of p16 (E), p21 (F), p27 (G), Cyclin D1 (H), Cyclin D2 (I), Ki-67 (J) and γH2A.X (K) in WI-38 cell lines where quiescence was induced for 100 or 150 days by contact inhibition respectively compared to controls at corresponding span of time in culture. Cell lines were maintained at 20% O2 as triplicates. The bars indicate the mean ± S.D. ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3 specifies the number of samples except for γH2A.X (S3K Fig. where n = 2).
(TIF)
Effect of long term quiescence induction (100 or 150 days) in WI-38 fibroblasts maintained at 3% O2. (A) Growth curve of 3 independent WI-38 fibroblast cell lines (control with no quiescence induction, and cell lines where quiescence was induced for 100 or 150 days respectively by contact inhibition and then maintained in culture till they approached senescence) maintained in culture at 3% O2 as triplicates from an early PD until senescence at late PDs. Each growth curve is measured in triplicate. Data points of all measurements are displayed (not the mean). (B & C) Percentage of SA-β gal positive cells at different time points of their growth in culture in the control WI-38 fibroblast cell line and in the cell lines where quiescence was induced for 100 or 150 days respectively. S4B and S4C Figs. are plotted with PDs and days in the y-axis respectively. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). (D) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2, Ki-67 and γH2A.X in WI-38 fibroblast cell lines (subjected to different culture conditions of 100 or 150 days quiescence by contact inhibition and no quiescence induction) maintained in culture at 3% O2 until they approached senescence at late PD. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (E, F, G, H, I, J, K) Comparison of mean fold change of protein expression levels of p16 (E), p21 (F), p27 (G), Cyclin D1 (H), Cyclin D2 (I), Ki-67 (J) and γH2A.X (K) in WI-38 cell lines where quiescence was induced for 100 or 150 days by contact inhibition respectively compared to controls at corresponding span of time in culture. Cell lines were maintained at 3% O2 as triplicates. The bars indicate the mean ± S.D. * p<0.05, ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
(TIF)
Impact of Staurosporine treatment in young and old PD MRC-5 fibroblasts. (A) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (young PD = 34) treated with different concentrations of Staurosporine for different time spans (B) Percentage of SA-β gal positive cells in MRC-5 fibroblast cell lines (young PD = 34) treated with different concentrations of Staurosporine for different time spans (C) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (old PD = 68) treated with different concentrations of Staurosporine for different time spans (D) Percentage of SA-β gal positive cells in MRC-5 fibroblast cell lines (old PD = 68) treated with different concentrations of Staurosporine for different time spans. In each instance, MRC-5 fibroblasts were maintained in culture at 20% O2 as triplicates. In A & C, the bars indicate the mean ± S.D. The mean values in B & D is displayed with error bar (± S.E). (E) The blots show the protein expression levels apoptotic protein Bax in MRC-5 fibroblast cell lines (young PD = 34 & old PD = 68) treated with 0.1 or 2.0 µM of Staurosporine for 96 hrs compared to controls (0 hrs). The MRC-5 fibroblasts were maintained in culture at 20% O2. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. n = 3.
(TIF)
Impact of Etoposide or Staurosporine treatment in MRC-5 fibroblasts subjected to short or long term quiescence. (A) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (untreated and treated with 7.5 µM Etoposide or 2.0 µM Staurosporine for 96 hrs) maintained at different culture conditions - after 9 (MRC-5 PD 35) or 150 days (MRC-5 PD 32) of quiescence induction by contact inhibition, fibroblasts of young PD = 34 and in fibroblasts of old PD = 68. (B) The blots show the protein expression levels of apoptotic protein Bax in MRC-5 fibroblast cell lines (young PD = 34, MRC-5 cell lines subjected to 150 consecutive days of quiescence induction by contact inhibition) treated with 2.0 (Staurosporine) or 7.5 µM (Etoposide) of apoptotic agents compared to controls (0 hrs). The MRC-5 fibroblasts were maintained in culture at 20% O2. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. Values statistically different from their controls (t-test) are indicated with an asterix: * p<0.05, ** p<0.01, *** p<0.001. n = 3.
(TIF)
Intersection of the most commonly differentially regulated genes with age (both up and down) in IMR-90 fibroblasts subjected to replicative senescence [97] compared to genes retrieved from MRC-5 fibroblasts subjected to long term quiescence induction compared to their controls. Venn plots of DEGs in young vs. senescent IMR-90 fibroblasts (red circles, data of [97]) and young (MRC-5, PD 32) vs. quiescent MRC-5 fibroblasts (PD 32+150 days quiescent; blue circles, data determined here), (A) up-, (B) down-regulated. Both numbers of DEGs in the intersection are significant with regards to the expected overlap of two independent groups.
(TIF)
Heatmap showing scaled gene-set level changes for KEGG pathways identified using gene set enrichment analysis. The left column represents the comparison of young and senescent IMR-90 cells [97] while the right column is based on expression changes between young and long-term quiescent MRC-5 fibroblasts. Each KEGG pathway was found to be significantly regulated in IMR-90, MRC-5 or both datasets. The histogram on the left explains the color coding (e.g. red boxes encode down-regulation in senescent IMR-90 and quiescent MRC-5 fibroblasts).
(TIF)
Acknowledgments
The work described here is part of the research programme of the Jena Centre for Systems Biology of Ageing - JenAge. We acknowledge the German Ministry for Education and Research (Bundesministerium für Bildung und Forschung – BMBF) for enabling us to undertake this research study. We are grateful to Dr. Marco Groth and Dr. Mathias Platzer from the genome analysis group in Fritz Lipmann Institute, Jena for undertaking the high-throughput RNA sequencing. We would also like to thank Sabine Ohndorf from our research group for her technical assistance.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All the data except sequencing data are available in the manuscript and the supplement section. Sequencing results can be found at: NCBI GEO under the accession number GSE60883.
Funding Statement
This work was supported by Bundesministerium für Bildung und Forschung (BMBF), Grant code: 0315581. http://www.bmbf.de/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pajalunga D, Mazzola A, Salzano AM, Biferi MG, De Luca G, et al. (2007) Critical requirement for cell cycle inhibitors in sustaining nonproliferative states. Journal of Cell Biology 176:807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes & Development 13(12):1501–1512. [DOI] [PubMed] [Google Scholar]
- 3. Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, et al. (2009) p53 Regulates Hematopoietic Stem Cell Quiescence. Cell Stem Cell 4(1):37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Os R, de Haan G, Dykstra BJ (2009) Hematopoietic Stem Cell Quiescence: Yet another Role for p53. Cell Stem Cell 4(1):7–8. [DOI] [PubMed] [Google Scholar]
- 5. Asai T, Liu Y, Bae N, Nimer SD (2011) The p53 tumor suppressor protein regulates hematopoietic stem cell fate. Journal of Cellular Physiology 226(9):2215–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dulic V (2011) Be quiet and you'll keep young: does mTOR underlie p53 action in protecting against senescence by favoring quiescence? Aging (Albany NY) 3(1):3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, et al. (2004) Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. The FASEB Journal 18(11):1303–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lum JJ, Bauer DE, Kong M, Harris MH, Li C, et al. (2005) Growth Factor Regulation of Autophagy and Cell Survival in the Absence of Apoptosis. Cell 120(2):237–248. [DOI] [PubMed] [Google Scholar]
- 9. Lemons JMS, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, et al. (2010) Quiescent fibroblasts exhibit high metabolic activity. PLoS Biology 8(10):e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grigoryev SA, Nikitina T, Pehrson JR, Singh PB, Woodcock CL (2004) Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. Journal of Cell Science 117(25):6153–6162. [DOI] [PubMed] [Google Scholar]
- 11. Laporte D, Courtout F, Salin B, Ceschin J, Sagot I (2013) An array of nuclear microtubules reorganizes the budding yeast nucleus during quiescence. The Journal of Cell Biology 203(4):585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, et al. (2013) H4K20 methylation regulates quiescence and chromatin compaction. Molecular Biology of the Cell 24(19):3025–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yusuf I, Fruman DA (2003) Regulation of quiescence in lymphocytes. Trends in Immunology 24(7):380–386. [DOI] [PubMed] [Google Scholar]
- 14. Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, et al. (2004) “Sleeping Beauty”: Quiescence in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews 68(2):187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coller HA, Sang L, Roberts JM (2006) A New Description of Cellular Quiescence. PLoS Biol 4(3):e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ekholm SV, Reed SI (2000) Regulation of G1 cyclin-dependent kinases in the mammalian cell cycle. Current Opinion in Cell Biology 12(6):676–684. [DOI] [PubMed] [Google Scholar]
- 17. Sherr CJ, Roberts JM (2004) Living with or without cyclins and cyclin-dependent kinases. Genes & Development 18(22):2699–2711. [DOI] [PubMed] [Google Scholar]
- 18. Perucca P, Cazzalini O, Madine M, Savio M, Laskey RA, et al. (2009) Loss of p21(CDKN1A) impairs entry to quiescence and activates a DNA damage response in normal fibroblasts induced to quiescence. Cell Cycle 8(1):105–114. [DOI] [PubMed] [Google Scholar]
- 19. Brown JP, Wei W, Sedivy JM (1997) Bypass of Senescence After Disruption of p21CIP1/WAF1 Gene in Normal Diploid Human Fibroblasts. Science 277(5327):831–834. [DOI] [PubMed] [Google Scholar]
- 20. Herbig U, Jobling WA, Chen BPC, Chen DJ, Sedivy JM (2004) Telomere Shortening Triggers Senescence of Human Cells through a Pathway Involving ATM, p53, and p21CIP1, but Not p16INK4a. Molecular Cell 14(4):501–513. [DOI] [PubMed] [Google Scholar]
- 21. Jiang H, Lin J, Su Z, Collart F, Huberman E, et al. (1994) Induction of differentiation in human promyelocytic HL-60 leukemia cells activates p21waf1cip1 expression in the absence of p53. Oncogene 9:3397–3406. [PubMed] [Google Scholar]
- 22.Zezula J, Casaccia-Bonnefil P, Ezhevsky SA, Osterhout DJ, Levine JM, et al. (2001) p21cip1 is required for the differentiation of oligodendrocytes independently of cell cycle withdrawal pp 27–34. [DOI] [PMC free article] [PubMed]
- 23. Rivard N, L'Allemain G, Bartek J, Pouysségur J (1996) Abrogation of p27Kip1 by cDNA Antisense Suppresses Quiescence (G0 State) in Fibroblasts. Journal of Biological Chemistry 271(31):18337–18341. [DOI] [PubMed] [Google Scholar]
- 24. Ladha MH, Lee KY, Upton TM, Reed MF, Ewen ME (1998) Regulation of Exit from Quiescence by p27 and Cyclin D1-CDK4. Molecular and Cellular Biology 18(11):6605–6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sgambato A, Cittadini A, Faraglia B, Weinstein IB (2000) Multiple functions of p27Kip1 and its alterations in tumor cells: a review. Journal of Cellular Physiology 183(1):18–27. [DOI] [PubMed] [Google Scholar]
- 26. Chassot AA, Lossaint G, Turchi L, Meneguzzi G, Fisher D, et al. (2008) Confluence-induced cell cycle exit involves pre-mitotic CDK inhibition by p27Kip1 and cyclin D1 downregulation. Cell Cycle 7(13):2038–2046. [DOI] [PubMed] [Google Scholar]
- 27. Sang L, Coller HA, Roberts JM (2008) Control of the Reversibility of Cellular Quiescence by the Transcriptional Repressor HES1. Science 321(5892):1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Munro J, Steeghs K, Morrison V, Ireland H, Parkinson EK (2001) Human fibroblast replicative senescence can occur in the absence of extensive cell division and short telomeres. Oncogene 20:3541–3552. [DOI] [PubMed] [Google Scholar]
- 29. Hayflick L, Moorhead P (1961) The serial cultivation of human diploid cell strains. Experimental Cell Research 25:585–621. [DOI] [PubMed] [Google Scholar]
- 30. Krtolica A, Campisi J (2002) Cancer and aging: a model for the cancer promoting effects of the aging stroma. The International Journal of Biochemistry & Cell Biology 34:1401–1414. [DOI] [PubMed] [Google Scholar]
- 31. Hayflick L (1985) Theories of biological aging. Experimental Gerontology 20:145–159. [DOI] [PubMed] [Google Scholar]
- 32. Sager R (1991) Senescence as a mode of tumor suppression. Environ. Health Perspect 93:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hornsby PJ (2002) Cellular Senescence and Tissue Aging In Vivo. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences 57(7):B251–B256. [DOI] [PubMed] [Google Scholar]
- 34. Campisi J (2005) Suppressing Cancer: The Importance of Being Senescent. Science 309(5736):886–887. [DOI] [PubMed] [Google Scholar]
- 35. Adams PD (2009) Healing and Hurting: Molecular Mechanisms, Functions, and Pathologies of Cellular Senescence. Molecular Cell 36(1):2–14. [DOI] [PubMed] [Google Scholar]
- 36. Ben-Porath I, Weinberg RA (2004) When cells get stressed: an integrative view of cellular senescence. The Journal of Clinical Investigation 113(1):8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cristofalo VJ, Lorenzini A, Allen RG, Torres C, Tresini M (2004) Replicative senescence: a critical review. Mechanisms of Ageing and Development 125(10–11):827–848. [DOI] [PubMed] [Google Scholar]
- 38. Dimri GP, Lee X, Basile G, Acosta M, Scott G, et al. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proceedings of the National Academy of Sciences 92(20):9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Di Leonardo A, Linke SP, Clarkin K, Wahl GM (1994) DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes & Development 8(21):2540–2551. [DOI] [PubMed] [Google Scholar]
- 40. Bladier C, Wolvetang E, Hutchinson P, de Haan J, Kola I (1997) Response of a primary human fibroblast cell line to H2O2: senescence-like growth arrest or apoptosis? Cell Growth Differ 8(5):589–598. [PubMed] [Google Scholar]
- 41. Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras Provokes Premature Cell Senescence Associated with Accumulation of p53 and p16INK4a. Cell 88(5):593–602. [DOI] [PubMed] [Google Scholar]
- 42. Toussaint O, Medrano EE, von Zglinicki T (2000) Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Experimental Gerontology 35(8):927–945. [DOI] [PubMed] [Google Scholar]
- 43. Debacq-Chainiaux F, Borlon C, Pascal T, Royer V, Eliaers F, et al. (2005) Repeated exposure of human skin fibroblasts to UVB at subcytotoxic level triggers premature senescence through the TGF-β1 signaling pathway. Journal of Cell Science 118(4):743–758. [DOI] [PubMed] [Google Scholar]
- 44. Mallette FA, Gaumont-Leclerc MF, Ferbeyre G (2007) The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes & Development 21(1):43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dimri GP, Itahana K, Acosta M, Campisi J (2000) Regulation of a Senescence Checkpoint Response by the E2F1 Transcription Factor and p14ARF Tumor Suppressor. Molecular and Cellular Biology 20(1):273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pearson M, Carbone R, Sebastiani C, Cioc M, Fagioli M, et al. (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406(6792):207–210. [DOI] [PubMed] [Google Scholar]
- 47. Ben-Porath I, Weinberg RA (2005) The signals and pathways activating cellular senescence. The International Journal of Biochemistry & Cell Biology 37(5):961–976. [DOI] [PubMed] [Google Scholar]
- 48. Zglinicki Tv, Saretzki G, Ladhoff J, Fagagna FdAd, Jackson SP (2005) Human cell senescence as a DNA damage response. Mechanisms of Ageing and Development 126(1):111–117. [DOI] [PubMed] [Google Scholar]
- 49. Campisi J, d'Adda di Fagagna F (2007) Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8(9):729–740. [DOI] [PubMed] [Google Scholar]
- 50. Steinert S, Shay JW, Wright WE (2000) Transient Expression of Human Telomerase Extends the Life Span of Normal Human Fibroblasts. Biochemical and Biophysical Research Communications 273(3):1095–1098. [DOI] [PubMed] [Google Scholar]
- 51. Hemann MT, Strong MA, Hao LY, Greider CW (2001) The Shortest Telomere, Not Average Telomere Length, Is Critical for Cell Viability and Chromosome Stability. Cell 107(1):67–77. [DOI] [PubMed] [Google Scholar]
- 52. de Lange T (2002) Protection of mammalian telomeres. Oncogene 21:532–540. [DOI] [PubMed] [Google Scholar]
- 53. Campisi J (2008) Aging and cancer cell biology, 2008. Aging Cell 7(3):281–284. [DOI] [PubMed] [Google Scholar]
- 54. Sedelnikova OA, Horikawa I, Zimonjic D, Popescu N, Bonner W, et al. (2004) Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol 6(2):168–170. [DOI] [PubMed] [Google Scholar]
- 55. Hwang ES (2002) Replicative senescence and senescence-like state induced in cancer-derived cells. Mechanisms of Ageing and Development 123(12):1681–1694. [DOI] [PubMed] [Google Scholar]
- 56. Gire V, Wynford-Thomas D (1998) Reinitiation of DNA Synthesis and Cell Division in Senescent Human Fibroblasts by Microinjection of Anti-p53 Antibodies. Molecular and Cellular Biology 18(3):1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beauséjour CM, Krtolica A, Galimi F, Narita M, Lowe SW, et al. (2003) Reversal of human cellular senescence: roles of the p53 and p16 pathways pp 4212–4222. [DOI] [PMC free article] [PubMed]
- 58. Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, et al. (2010) Dissecting the Unique Role of the Retinoblastoma Tumor Suppressor during Cellular Senescence. Cancer Cell 17(4):376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hara E, Smith R, Parry D, Tahara H, Stone S, et al. (1996) Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Molecular and Cellular Biology 16(3):859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mazars GR, Jat PS (1997) Expression of p24, a novel p21Waf1/Cip1/Sdi1-related protein, correlates with measurement of the finite proliferative potential of rodent embryo fibroblasts. Proceedings of the National Academy of Sciences 94(1):151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jones CJ, Kipling D, Morris M, Hepburn P, Skinner J, et al. (2000) Evidence for a Telomere-Independent “Clock” Limiting RAS Oncogene-Driven Proliferation of Human Thyroid Epithelial Cells. Molecular and Cellular Biology 20(15):5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durand B, Gao FB, Raff M (1997) Accumulation of the cyclin-dependent kinase inhibitor p27/Kip1 and the timing of oligodendrocyte differentiation pp 306–317. [DOI] [PMC free article] [PubMed]
- 63. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The Hallmarks of Aging. Cell 153(6):1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harley CB (1991) Telomere loss: mitotic clock or genetic time bomb? Mutation Research/DNAging 256(2–6):271–282. [DOI] [PubMed] [Google Scholar]
- 65. Cristofalo VJ, Pignolo RJ (1993) Replicative senescence of human fibroblast like cells in culture. Physiological Reviews 73(3):617–638. [DOI] [PubMed] [Google Scholar]
- 66. Lorenzini A, Johnson FB, Oliver A, Tresini M, Smith JS, et al. (2009) Significant correlation of species longevity with DNA double strand break-recognition but not with telomere length. Mech. Ageing Dev 130:784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wright WE, Shay JW (2001) Cellular senescence as a tumor-protection mechanism: the essential role of counting. Current Opinion in Genetics & Development 11:98–103. [DOI] [PubMed] [Google Scholar]
- 68. Severino J, Allen RG, Balin S, Balin A, Cristofalo VJ (2000) Is β Galactosidase Staining a Marker of Senescence in Vitro and in Vivo? Experimental Cell Research 257(1):162–171. [DOI] [PubMed] [Google Scholar]
- 69. Yang NC, Hu ML (2005) The limitations and validities of senescence associated-β galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Experimental Gerontology 40(10):813–819. [DOI] [PubMed] [Google Scholar]
- 70. Cristofalo VJ (2005) SA β Gal staining: Biomarker or delusion. Experimental Gerontology 40(10):836–838. [DOI] [PubMed] [Google Scholar]
- 71. Cho S, Hwang E (2012) Status of mTOR activity may phenotypically differentiate senescence and quiescence. Mol Cells 33(6):597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Imai Y, Takahashi A, Hanyu A, Hori S, Sato S, et al. (2014) Crosstalk between the Rb Pathway and AKT Signaling Forms a Quiescence-Senescence Switch. Cell Reports 7(1):194–207. [DOI] [PubMed] [Google Scholar]
- 73. Lane AA, Scadden DT (2010) Stem Cells and DNA Damage: Persist or Perish? Cell 142(3):360–362. [DOI] [PubMed] [Google Scholar]
- 74. Crescenzi E, Palumbo G, Brady HJM (2003) Bcl-2 activates a programme of premature senescence in human carcinoma cells. Biochem J 375(2):263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ashrafi K, Sinclair D, Gordon JI, Guarente L (1999) Passage through stationary phase advances replicative ageing in S. cerevisiae . Proceedings of the National Academy of Sciences USA 96:9100–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lucibello FC, Sewing A, Brusselbach S, Burger C, Muller R (1993) Deregulation of cyclins D1 and E and suppression of cdk2 and cdk4 in senescent human fibroblasts. Journal of Cell Science 105(1):123–133. [DOI] [PubMed] [Google Scholar]
- 77. Atadja P, Wong H, Veillete C, Riabowol K (1995) Overexpression of Cyclin D1 Blocks Proliferation of Normal Diploid Fibroblasts. Experimental Cell Research 217(2):205–216. [DOI] [PubMed] [Google Scholar]
- 78. Meyyappan M, Wong H, Hull C, Riabowol KT (1998) Increased Expression of Cyclin D2 during Multiple States of Growth Arrest in Primary and Established Cells. Molecular and Cellular Biology 18(6):3163–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schäuble S, Klement K, Marthandan S, Münch S, Heiland I, et al. (2012) Quantitative Model of Cell Cycle Arrest and Cellular Senescence in Primary Human Fibroblasts. Plos One 7:e42150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, et al. (2000) A critical role for histone H2A.X in recruitment of repair factors to nuclear foci after DNA damage. Current Biology 10(15):886–895. [DOI] [PubMed] [Google Scholar]
- 81. Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ (2001) ATM Phosphorylates Histone H2A.X in Response to DNA Double-strand Breaks. Journal of Biological Chemistry 276(45):42462–42467. [DOI] [PubMed] [Google Scholar]
- 82. Lowndes NF, Toh GWL (2005) DNA Repair: The Importance of Phosphorylating Histone H2A.X. Current Biology. 15(3):R99–R102. [DOI] [PubMed] [Google Scholar]
- 83. Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U (2007) Accumulation of senescent cells in mitotic tissue of aging primates. Mechanisms of Ageing and Development 128(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kouzarides T (2007) Chromatin Modifications and Their Function. Cell 128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 85. Sitte N, Saretzki G, von Zglinicki T (1998) Accelerated Telomere Shortening in Fibroblasts After Extended Periods of Confluency. Free Radical Biology and Medicine 24(6):885–893. [DOI] [PubMed] [Google Scholar]
- 86. Parrinello S, Samper E, Krtolica A, Goildstein J, Melov S, et al. (2003) Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 5(8):741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Forsyth NR, Evans AP, Shay JW, Wright WE (2003) Developmental differences in the immortalization of lung fibroblasts by telomerase. Aging Cell 2:235–243. [DOI] [PubMed] [Google Scholar]
- 88. Pitiyage GN, Slijepcevic P, Gabrani A, Chianea YG, Lim KP, et al. (2011) Senescent mesenchymal cells accumulate in human fibrosis by a telomere-independent mechanism and ameliorate fibrosis through matrix metalloproteinases. Journal of Pathology 223:604–617. [DOI] [PubMed] [Google Scholar]
- 89. Atamna H, Paler-Martinez A, Ames BN (2000) N-t-butyl hydroxylamine, a hydrolysis product of alpha-phenyl-N-t-butyl nitrone, is more potent in delaying senescence in human lung fibroblasts. Journal of Biologiocal Chemistry 275:6741–6748. [DOI] [PubMed] [Google Scholar]
- 90. Forsyth NR, Wright WE, Shay JW (2002) Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation 69(4-5):188–197. [DOI] [PubMed] [Google Scholar]
- 91. Seluanov A, Gorbunova V, Falcovitz A, Siqal A, Milyavsky M, et al. (2001) Change of the Death Pathway in Senescent Human Fibroblasts in Response to DNA Damage Is Caused by an Inability To Stabilize p53. Molecular and Cellular Biology 21(5):1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Manns J, Daubrawa M, Driessen S, Paasch F, Hoffmann N, et al. (2011) Triggering of a novel intrinsic apoptosis pathway by the kinase inhibitor staurosporine: activation of caspase-9 in the absence of Apaf-1. The FASEB Journal 25(9):3250–3261. [DOI] [PubMed] [Google Scholar]
- 93. Finucane DM, Bossy-Wetzel E, Waterhouse NJ, Cotter TG, Green DR (1999) Bax-induced Caspase Activation and Apoptosis via Cytochromec Release from Mitochondria Is Inhibitable by Bcl-xL. Journal of Biological Chemistry 274(4):2225–2233. [DOI] [PubMed] [Google Scholar]
- 94. Pawlowski J, Kraft AS (2000) Bax-induced apoptotic cell death. Proceedings of the National Academy of Sciences 97(2):529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang E (1995) Senescent human fibroblasts resist programmed cell death, and failure to suppress bcl2 is involved. Cancer Res 55:2284–2292. [PubMed] [Google Scholar]
- 96. Naderi J, Hung M, Pandey S (2003) Oxidative stress-induced apoptosis in dividing fibroblasts involves activation of p38 MAP kinase and over-expression of Bax: resistance of quiescent cells to oxidative stress. Apoptosis 8:91–100. [DOI] [PubMed] [Google Scholar]
- 97.Lackner DH, Hayashi MT, Cesare AJ, Karlseder J (2014) A genomics approach identifies senescence-specific gene expression regulation. Aging Cell 2014 May 23. doi: 10.1111/acel.12234. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 98. Augenlicht LH, Baserga R (1974) Changes in the Go state of WI-38 fibroblasts at different times after confluence. Exp Cell Res 89:255–262. [DOI] [PubMed] [Google Scholar]
- 99. Soprano KJ (1994) WI-38 cell long term quiescence model system. A valuable tool to study molecular events that regulate growth. Journal of Cellular Biochemistry 54(4):405–414. [DOI] [PubMed] [Google Scholar]
- 100. Pignolo RJ, Martin BG, Horton JH, Kalbach AN, Cristofalo VJ (1998) The Pathway of Cell Senescence: WI-38 Cells Arrest in Late G1 and Are Unable to Traverse the Cell Cycle from a True G0 State. Experimental Gerontology 33(1–2):67–80. [DOI] [PubMed] [Google Scholar]
- 101. Beerman I, Seita J, Inlay Matthew A, Weissman Irving L, Rossi Derrick J (2014) Quiescent Hematopoietic Stem Cells Accumulate DNA Damage during Aging that Is Repaired upon Entry into Cell Cycle. Cell Stem Cell 15(1):37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wolstein JM, Lee DH, Michaud J, Buot V, Stefanchik B, et al. (2010) INK4a knockout mice exhibit increased fibrosis under normal conditions and in response to unilateral ureteral obstruction pp F1486-F1495. [DOI] [PMC free article] [PubMed]
- 103. Zhang X, Wu X, Tang W, Luo Y (2012) Loss of p16 (Ink4a) function rescues cellular senescence induced by telomere dysfunction. Int J Mol Sci 13:5866–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Okamoto A, Demetrick DJ, Spillare EA, Hagiwara K, Hussain SP, et al. (1994) Mutations and altered expression of p16INK4 in human cancer. Proceedings of the National Academy of Sciences 91(23):11045–11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Serrano M, Lee HW, Chin L, Carlos-Cardo C, Bleach D, et al. (1996) Role of the INK4a Locus in Tumor Suppression and Cell Mortality. Cell 85(1):27–37. [DOI] [PubMed] [Google Scholar]
- 106. Uhrbom L, Nister M, Westermark B (1997) Induction of senescence in human malignant glioma cells by p16INK4a. Oncogene 15:505–514. [DOI] [PubMed] [Google Scholar]
- 107. Dai CY, Enders GH (2000) p16 INK4a can initiate an autonomous senescence program. Oncogene 19:1613–1622. [DOI] [PubMed] [Google Scholar]
- 108. DeJesus V, Rios I, Davis C, Chen Y, Calhoun D, et al. (2002) Induction of apoptosis in human replicative senescent fibroblasts. Exp. Cell Res 274:92–99. [DOI] [PubMed] [Google Scholar]
- 109. Zhang J, Patel JM, Block ER (2002) Enhanced apoptosis in prolonged cultures of senescent porcine pulmonary artery endothelial cells. Mech. Ageing Dev 123:613–625. [DOI] [PubMed] [Google Scholar]
- 110. Olovnikov AM (1971) Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk. SSSR 201:1496–1499. [PubMed] [Google Scholar]
- 111. Watson JD (1972) Origin of concatemeric T7 DNA. Nat New Biol 239:197–201. [DOI] [PubMed] [Google Scholar]
- 112. Saretzki G, Sitte N, Merkel U, Wurm RE (1999) Telomere shortening triggers a p53-dependent cell cycle arrest via accumulation of G-rich single stranded DNA fragments. Oncogene 18:5148–5158. [DOI] [PubMed] [Google Scholar]
- 113. Shiloh Y (2006) The ATM-mediated DNA-damage response: taking shape. Trends in Biochemical Sciences 31(7):402–410. [DOI] [PubMed] [Google Scholar]
- 114. Allsopp RC, Chang E, Kashefi-Aazam M, Rogaev EI, Piatyszek MA, et al. (1995) Telomere Shortening Is Associated with Cell Division in Vitro and in Vivo. Experimental Cell Research 220(1):194–200. [DOI] [PubMed] [Google Scholar]
- 115. Smith JR, Whitney RG (1980) Intraclonal variation in proliferative potential of human diploid fibroblasts: stochastic mechanism for cellular aging. Science 207:82–84. [DOI] [PubMed] [Google Scholar]
- 116. Suram A, Herbig U (2014) The replicometer is broken: telomeres activate cellular senescence in response to genotoxic stresses. Aging Cell 13:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bodnar AG, Quellette M, Frolkis M, Holt SE, Chiu CP, et al. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352. [DOI] [PubMed] [Google Scholar]
- 118. Kuilman T, Peeper DS (2009) Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer 9(2):81–94. [DOI] [PubMed] [Google Scholar]
- 119. Passos J, Simillion C, Hallinan J, Wipat A, von Zglinicki T (2009) Cellular senescence: unravelling complexity. AGE 31(4):353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Thuret G, Chiquet C, Herrag S, Dumollard JM, Boudard D, et al. (2003) Mechanisms of staurosporine induced apoptosis in a human corneal endothelial cell line. British Journal of Ophthalmology 87(3):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Johansson AC, Steen H, Ollinger K, Roberg K (2003) Cathepsin D mediates cytochrome c release and caspase activation in human fibroblast apoptosis induced by staurosporine. Cell Death Differ 10(11):1253–1259. [DOI] [PubMed] [Google Scholar]
- 122. Kim S, Ryu S, Kang H, Choi H, Park S (2011) Defective nuclear translocation of stress-activated signaling in senescent diploid human fibroblasts: a possible explanation for aging-associated apoptosis resistance. Apoptosis 16(8):795–807. [DOI] [PubMed] [Google Scholar]
- 123. Soldani C, Bottone MG, Pellicciari C, Scovassi AI (2001) Two-color fluorescence detection of Poly (ADP-Ribose) Polymerase-1 (PARP-1) cleavage and DNA strand breaks in etoposide-induced apoptotic cells. Eur J Histochem 45:389–392. [DOI] [PubMed] [Google Scholar]
- 124. Mazzini G, Ferrari C, Erba E (2003) Dual excitation multi-fluorescence flow cytometry for detailed analyses of viability and apoptotic cell transition. Europ J Histochem 47:289–298. [PubMed] [Google Scholar]
- 125. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features.”. Bioinformatics (Oxford, England) 30(7):923–930. [DOI] [PubMed] [Google Scholar]
- 127. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hardcastle TJ, Kelly KA (2010) baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B 57(1):289–300. [Google Scholar]
- 131. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 132. Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ (2009) GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Holliday R (2014) The commitment of human cells to senescence. Interdiscip Top Gerontol. 39:1–7. [DOI] [PubMed] [Google Scholar]
- 134. Jacobs JP, Jones CM, Baille JP (1970) Characteristics of a human diploid cell designated MRC-5. Nature 227(5254):168–170. [DOI] [PubMed] [Google Scholar]
- 135. Trlifjová J, Strízová V, Trlifaj L, Budsínsky´ Z, Frühbauer Z (1968) A prolonged cultivation of the human diploid cell strain WI-38. J Hyg Epidemiol Microbiol Immunol 12(2):212–226. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of short term quiescence induction on protein expression levels of a number of cell cycle associated genes and a marker for DNA damage in MRC-5 fibroblasts. (A) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2 and γH2A.X in two MRC-5 fibroblast cell lines (control with no quiescence induction and a cell line where quiescence was induced 3 times separately for a span of 9 days) maintained at 20% O2 at different stages of their span in culture. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (B, C, D, E, F, G) Comparison of mean fold change of protein expression levels of p16 (B), p21 (C), p27 (D), Cyclin D1 (E), Cyclin D2 (F) and γH2A.X (G) in 3 times quiescence induced MRC-5 cell lines and control MRC-5 cell lines maintained in culture as triplicates. The bars indicate the mean ± S.D. ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
(TIF)
Effect of short term quiescence induction on protein expression levels of a number of cell cycle associated genes and a marker for DNA damage in WI-38 fibroblasts. (A) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2 and γH2A.X in two WI-38 fibroblast cell lines (control with no quiescence induction and a cell line where quiescence was induced 3 times separately for a span of 9 days) maintained at 20% O2 at different stages of their span in culture. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (B, C, D, E, F, G) Comparison of mean fold change of protein expression levels of p16 (B), p21 (C), p27 (D), Cyclin D1 (E), Cyclin D2 (F) and γH2A.X (G) in 3 times quiescence induced WI-38 cell lines and control WI-38 cell lines maintained in culture as triplicates. The bars indicate the mean ± S.D. ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
(TIF)
Effect of long term quiescence induction (100 or 150 days) in WI-38 fibroblasts maintained at 20% O2. (A) Growth curve of 3 independent WI-38 fibroblast cell lines (control with no quiescence induction, and cell lines where quiescence was induced for 100 or 150 days respectively by contact inhibition and then maintained in culture till they approached senescence) maintained in culture at 20% O2 as triplicates from an early PD until senescence at late PDs. Each growth curve is measured in triplicate. Data points of all measurements are displayed (not the mean). (B & C) Percentage of SA-β gal positive cells at different time points of their growth in culture in the control WI-38 fibroblast cell line and in the cell lines where quiescence was induced for 100 or 150 days respectively. S3B and S3C Figs. are plotted with PDs and days in the y-axis respectively. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). (D) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2, Ki-67 and γH2A.X in WI-38 fibroblast cell lines (subjected to different culture conditions of 100 or 150 days quiescence by contact inhibition and no quiescence induction) maintained in culture at 20% O2 until they approached senescence at late PD. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (E, F, G, H, I, J, K) Comparison of mean fold change of protein expression levels of p16 (E), p21 (F), p27 (G), Cyclin D1 (H), Cyclin D2 (I), Ki-67 (J) and γH2A.X (K) in WI-38 cell lines where quiescence was induced for 100 or 150 days by contact inhibition respectively compared to controls at corresponding span of time in culture. Cell lines were maintained at 20% O2 as triplicates. The bars indicate the mean ± S.D. ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3 specifies the number of samples except for γH2A.X (S3K Fig. where n = 2).
(TIF)
Effect of long term quiescence induction (100 or 150 days) in WI-38 fibroblasts maintained at 3% O2. (A) Growth curve of 3 independent WI-38 fibroblast cell lines (control with no quiescence induction, and cell lines where quiescence was induced for 100 or 150 days respectively by contact inhibition and then maintained in culture till they approached senescence) maintained in culture at 3% O2 as triplicates from an early PD until senescence at late PDs. Each growth curve is measured in triplicate. Data points of all measurements are displayed (not the mean). (B & C) Percentage of SA-β gal positive cells at different time points of their growth in culture in the control WI-38 fibroblast cell line and in the cell lines where quiescence was induced for 100 or 150 days respectively. S4B and S4C Figs. are plotted with PDs and days in the y-axis respectively. Each curve is measured in triplicate, the mean value is displayed with error bar (± S.E). (D) The blots show the protein expression levels of p16, p21, p27, Cyclin D1, Cyclin D2, Ki-67 and γH2A.X in WI-38 fibroblast cell lines (subjected to different culture conditions of 100 or 150 days quiescence by contact inhibition and no quiescence induction) maintained in culture at 3% O2 until they approached senescence at late PD. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. (E, F, G, H, I, J, K) Comparison of mean fold change of protein expression levels of p16 (E), p21 (F), p27 (G), Cyclin D1 (H), Cyclin D2 (I), Ki-67 (J) and γH2A.X (K) in WI-38 cell lines where quiescence was induced for 100 or 150 days by contact inhibition respectively compared to controls at corresponding span of time in culture. Cell lines were maintained at 3% O2 as triplicates. The bars indicate the mean ± S.D. * p<0.05, ** p<0.01, *** p<0.001 - significantly different compared to fibroblasts with PD assigned 1. n = 3.
(TIF)
Impact of Staurosporine treatment in young and old PD MRC-5 fibroblasts. (A) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (young PD = 34) treated with different concentrations of Staurosporine for different time spans (B) Percentage of SA-β gal positive cells in MRC-5 fibroblast cell lines (young PD = 34) treated with different concentrations of Staurosporine for different time spans (C) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (old PD = 68) treated with different concentrations of Staurosporine for different time spans (D) Percentage of SA-β gal positive cells in MRC-5 fibroblast cell lines (old PD = 68) treated with different concentrations of Staurosporine for different time spans. In each instance, MRC-5 fibroblasts were maintained in culture at 20% O2 as triplicates. In A & C, the bars indicate the mean ± S.D. The mean values in B & D is displayed with error bar (± S.E). (E) The blots show the protein expression levels apoptotic protein Bax in MRC-5 fibroblast cell lines (young PD = 34 & old PD = 68) treated with 0.1 or 2.0 µM of Staurosporine for 96 hrs compared to controls (0 hrs). The MRC-5 fibroblasts were maintained in culture at 20% O2. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. n = 3.
(TIF)
Impact of Etoposide or Staurosporine treatment in MRC-5 fibroblasts subjected to short or long term quiescence. (A) Percentage of apoptotic cells in MRC-5 fibroblast cell lines (untreated and treated with 7.5 µM Etoposide or 2.0 µM Staurosporine for 96 hrs) maintained at different culture conditions - after 9 (MRC-5 PD 35) or 150 days (MRC-5 PD 32) of quiescence induction by contact inhibition, fibroblasts of young PD = 34 and in fibroblasts of old PD = 68. (B) The blots show the protein expression levels of apoptotic protein Bax in MRC-5 fibroblast cell lines (young PD = 34, MRC-5 cell lines subjected to 150 consecutive days of quiescence induction by contact inhibition) treated with 2.0 (Staurosporine) or 7.5 µM (Etoposide) of apoptotic agents compared to controls (0 hrs). The MRC-5 fibroblasts were maintained in culture at 20% O2. The up or down-regulation was signified by the presence or absence of the bands in Western Blots. Values statistically different from their controls (t-test) are indicated with an asterix: * p<0.05, ** p<0.01, *** p<0.001. n = 3.
(TIF)
Intersection of the most commonly differentially regulated genes with age (both up and down) in IMR-90 fibroblasts subjected to replicative senescence [97] compared to genes retrieved from MRC-5 fibroblasts subjected to long term quiescence induction compared to their controls. Venn plots of DEGs in young vs. senescent IMR-90 fibroblasts (red circles, data of [97]) and young (MRC-5, PD 32) vs. quiescent MRC-5 fibroblasts (PD 32+150 days quiescent; blue circles, data determined here), (A) up-, (B) down-regulated. Both numbers of DEGs in the intersection are significant with regards to the expected overlap of two independent groups.
(TIF)
Heatmap showing scaled gene-set level changes for KEGG pathways identified using gene set enrichment analysis. The left column represents the comparison of young and senescent IMR-90 cells [97] while the right column is based on expression changes between young and long-term quiescent MRC-5 fibroblasts. Each KEGG pathway was found to be significantly regulated in IMR-90, MRC-5 or both datasets. The histogram on the left explains the color coding (e.g. red boxes encode down-regulation in senescent IMR-90 and quiescent MRC-5 fibroblasts).
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All the data except sequencing data are available in the manuscript and the supplement section. Sequencing results can be found at: NCBI GEO under the accession number GSE60883.